Abstract
The eukaryotic genome is highly compacted into a protein-DNA complex called chromatin. The cell controls access of transcriptional regulators to chromosomal DNA via several mechanisms that act on chromatin-associated proteins and provide a rich spectrum of epigenetic regulation. Elucidating the mechanisms that fold chromatin fibers into higher-order structures is therefore key to understanding the epigenetic regulation of DNA accessibility. Here, using histone H4-V21C and histone H2A-E64C mutations, we employed single-molecule force spectroscopy to measure the unfolding of individual chromatin fibers that are reversibly cross-linked through the histone H4 tail. Fibers with covalently linked nucleosomes featured the same folding characteristics as fibers containing wild-type histones but exhibited increased stability against stretching forces. By stabilizing the secondary structure of chromatin, we confirmed a nucleosome repeat length (NRL)-dependent folding. Consistent with previous crystallographic and cryo-EM studies, the obtained force-extension curves on arrays with 167-bp NRLs best supported an underlying structure consisting of zig-zag, two-start fibers. For arrays with 197-bp NRLs, we previously inferred solenoidal folding, which was further corroborated by force-extension curves of the cross-linked fibers. The different unfolding pathways exhibited by these two types of arrays and reported here extend our understanding of chromatin structure and its potential roles in gene regulation. Importantly, these findings imply that chromatin compaction by nucleosome stacking protects nucleosomal DNA from external forces up to 4 piconewtons.
Keywords: chromatin, chromatin structure, cysteine-mediated cross-linking, nucleosome, single-molecule biophysics, force spectroscopy
Introduction
The eukaryotic genome is highly compacted into the protein-DNA complex called chromatin. The basic unit of chromatin organization comprises 147 base pairs of DNA wrapped in 1.7 turns around an octamer of proteins consisting of the core histones H2A, H2B, H3, and H4 (1). This nucleosome core particle has been shown to form a barrier (2) as well as a binding site (3) for proteins involved in DNA metabolism, such as those responsible for replication, transcription, and DNA repair. The cell modulates access to chromosomal DNA by remodeling it (4) via active nucleosome translocation (5), post-translational modifications of histones (6), and/or the binding of other proteins like the linker histone or HP1 (7, 8), providing a rich spectrum of regulation.
The accessibility of chromosomal DNA can further be modulated by folding strings of nucleosomes into higher-order chromatin structures, driven by nucleosome-nucleosome interactions. This secondary structure remains controversial (9) despite extensive efforts in exploring it using electron microscopy (EM) (10, 11), crystallography (12), analytical ultracentrifugation (13, 14), and structural modeling (15–17). These studies generally support a preferential stacking of nucleosomes in a face-to-face fashion, mediated by the H4 tail (12, 18). However, the geometrical constraints of short linker DNA between the consecutive nucleosomes may inhibit such interactions, and therefore variations in the DNA linker length may influence the resulting higher-order folding (19). This is especially relevant in the context of epigenetics because the expression of particular genes can be correlated with the occupancy and spacing of nucleosomes on the DNA (20, 21). For example, the expulsion of nucleosomes in the promotor regions facilitates the accessibility of the underlying DNA sequence to polymerases (22). However, a fundamental understanding of the mechanisms involved in this epigenetic regulation requires a significant advance in our knowledge of chromatin folding.
Although nucleosomal arrays readily fold into 30-nm fibers in vitro, their existence in vivo has been questioned (23). Nevertheless, it seems likely that interactions between nucleosomes will occur in a crowded eukaryotic nucleus (24), and given the heterogeneity in nucleosome spacing (25), it is relevant to uncover how the length of the linker DNA affects nucleosome-nucleosome contacts and the chromatin structure that is induced by those interactions. Tandem arrays of the Widom 601 sequence, which force the nucleosomes into well defined positions, have been instrumental in structural studies of chromatin (26–28). The most detailed structures show chromatin fibers that form two stacks of nucleosomes connected by straight linker DNA as shown in the crystal structure of a tetranucleosome (12). The same tetranucleosomal geometry was recently visualized by cryo-EM (10), implying that a two-start configuration is the predominant structure of chromatin fibers.
Other studies, however, indicate that arrays of nucleosomes may form alternative structures (29). In particular, the one-start or solenoid structure has been proposed early on (30). Robinson et al. (11) demonstrated that the diameter of folded chromatin fibers, as measured with EM, is independent of the DNA linker length, which is difficult to reconcile with a two-start helix with straight linker DNA. The relatively irregular folding of these fibers impeded averaging of structures, leaving the linker DNA unresolved.
Probing the mechanical response of chromatin to tension constrains some of the proposed models of the underlying structure. In single-molecule force spectroscopy, the extension of individual chromatin fibers is measured as a function of the applied force (31, 32). Previously, we introduced a detailed interpretation of the force-extension relation with a statistical mechanics model that quantifies extensions and transitions between all chromatin conformations upon stretching (33). From our analysis, we inferred a two-start folding of 167-bp nucleosome repeat length (NRL)3 fibers and a one-start folding of 197-bp NRLs. The two-start organization was recently confirmed using single-molecule force spectroscopy on 177- and 187-bp NRL fibers (34). The interpretation that 197-bp NRL fibers fold as a one-start solenoid, which was also inferred from EM studies (11), has been challenged (17, 35), arguing that the obtained linear extension of the chromatin fibers can be achieved by gradual unwrapping of DNA from the nucleosome cores. To clarify this issue, it is important to resolve at what force and extension the nucleosome-nucleosome contacts are broken.
Both of the proposed folding geometries imply stacking of nucleosomes, which has been proposed to be mediated by electrostatic interactions between the H4 tail and the acidic patch on the H2A-H2B dimer (36). An elegant way to study such tail-mediated nucleosome interactions was developed by Dorigo et al. (18), who cross-linked neighboring nucleosomes by introduction of two cysteines, one in the acidic patch and one at the end of the H4 tail. The reversible cross-linking process is controlled by the redox potential of the measurement buffer. Under oxidizing conditions, a covalent disulfide bond between two cysteines in adjacent nucleosomes mimics the suggested native structure of the H4 tails in stacked nucleosomes. Both gel electrophoresis and EM imaging confirmed the two-start folding of cross-linked fibers by showing that odd and even nucleosomes form two separate stacks (18).
Here, we combine cysteine-mediated nucleosome cross-linking with single-molecule dynamic force spectroscopy. This approach brings the advantage of being able to distinguish the signatures of rupturing nucleosome-nucleosome contacts from those of gradually unwrapping nucleosomal DNA. As long as the nucleosome-nucleosome stacking is not disrupted, i.e. H4-mediated interactions between nucleosomes are not broken, the introduction of disulfide links between nucleosomes should not change the force-extension relation. The obtained force-extension curves of cross-linked nucleosomes should in this case overlap with those of either arrays containing wild-type (WT) nucleosomes or reduced Cys mutant (MT) nucleosomes. Both the stretching stiffness and the extension at the rupture force would be unaffected by the disulfide bonds. Alternatively, if the linear stretching of folded chromatin fibers mainly involves DNA unwrapping, cross-linking the nucleosomes with the H4 tails should inhibit large extension of the fiber. Thus, single-molecule force spectroscopy of cross-linked chromatin fibers should be able to discriminate different unfolding pathways of a chromatin fiber. The results presented in this work clearly reveal H4 tail-mediated nucleosome stacking and one- and two-start structures for 167- and 197-bp NRLs, respectively.
Results
Chromatin with short NRL folds into a stiffer structure than chromatin with long NRL
We first briefly recapitulate our previous force spectroscopy results on chromatin fibers and how the force-extension data can be interpreted. We use a statistical physics model that quantifies the changes in extension of each intermediate nucleosome conformation upon stretching the fiber and explicitly takes variations in the fiber composition into account. The force-dependent extension of the tether is obtained by the sum of the extensions of each nucleosome and the extension of the bare DNA handles at the flanks of the chromatin fiber, which are modeled by a wormlike chain. The features of individual chromatin fibers are captured by fitting this model to the experimental force-extension curves (Fig. 1A).
Figure 1.
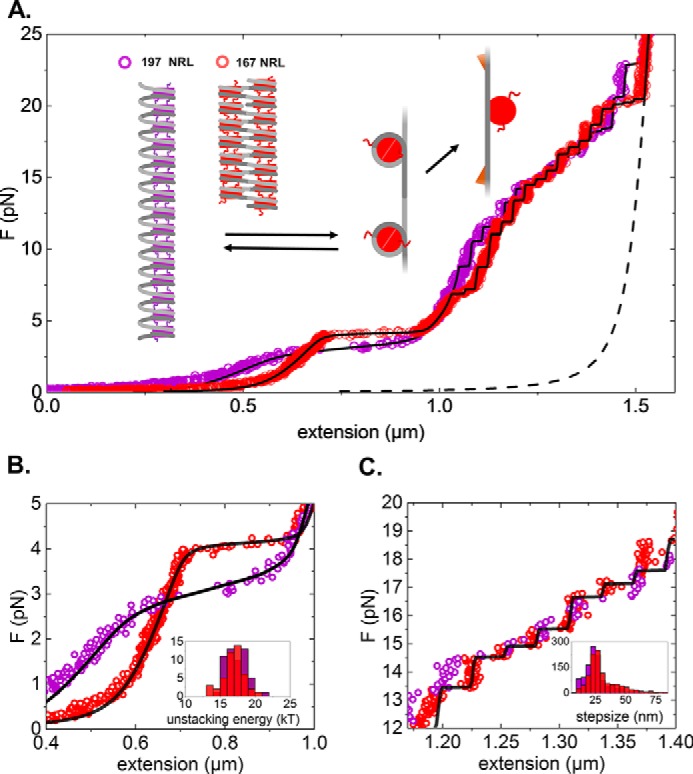
Wild-type histone chromatin fibers with 167- and 197-bp NRLs respond differently to applied force. A, force (F)-extension curve of a chromatin fiber reconstituted on 15 × 167-bp DNA (20-bp linker DNA; red) and 15 × 197-bp DNA (50-bp linker DNA; violet). As a reference, the WLC extension of a 4.4-kbp DNA is plotted (black dotted line). Fitted black curves correspond to the statistical mechanics model for chromatin unfolding described in Meng et al. (33). The graphs represent conformational changes upon stretching. A folded fiber initially stretches linearly until nucleosome-nucleosome interactions rupture and simultaneous release of 56 bp of nucleosomal DNA. The remaining nucleosomal DNA releases in two steps, a small 5-nm step around 6 pN and an ∼25-nm step at higher forces. Note that tetrasomes only feature the last step. B, zoom of the low-force part of the force-extension curve, representing the stretching of the folded fiber. Compared with 197-bp NRL fibers, 167-bp NRL fibers are more condensed (although extra tetrasomes or hexasomes in this fiber shift the curve to higher condensation), are stiffer, have a higher rupture force, and rupture cooperatively. The unstacking transition is in equilibrium, and the fiber refolds as the force is released (data not shown). Inset, histogram of the fitted unstacking energy per nucleosome for both types of fibers. C, zoom of the high-force regime of the force-extension diagram. The remaining wrapped DNA unfolds in a non-equilibrium fashion, resulting in a staircase-like extension curve. Inset, histogram of the step sizes.
A characteristic feature of the force-extension curves of folded chromatin fibers is the force plateau at 3–4 pN. We will refer to this region as the unstacking transition of the chromatin fiber. At forces below this transition, the fiber is folded into a condensed structure that is stabilized by nucleosome-nucleosome interactions. In this regime, the fiber extension increases linearly with applied force, like a Hookean spring. When the force exceeds the rupture force, the extension of the total fiber rapidly increases to about 1 μm (Fig. 1, A and B). This extension corresponds to about 30 nm per nucleosome and is consistent with a single wrap of DNA remaining around each histone core. The unstacking transition is modeled with a force-dependent equilibrium in which the stacking energy is balanced with the work done by the bead. For a more detailed description, we refer to Meng et al. (33).
Only at forces exceeding 7 pN does the second wrap of DNA start to be released. As opposed to the low-force ruptures, this second transition is not in equilibrium, resulting in clear, well defined steps of 25 nm for each unfolding event (Fig. 1C). We note that the release of DNA from singly wrapped octameric nucleosomes, as present after the unstacking transition, cannot be distinguished from the unfolding of tetrasomes, which consist of two copies of the histones H3 and H4 wrapped by a single turn of DNA; both would result in the appearance of 25-nm steps. The force-dependent extension of the entire fiber is defined by the sum of the force-dependent extensions of each nucleosome, tetrasome, hexasome, and the flanking DNA. The number of nucleosomes and tetrasomes are fitted to the force-extension curve of each individual fiber, yielding the black lines in Fig. 1. All force-extension curves feature a similar unfolding pathway, although the detailed composition and interaction parameters may differ between individual fibers.
Several features of the force-extension curves depend on the NRL. Compared with 197-bp NRL fibers, 167-bp NRL fibers exhibit an increased rupture force of 3.5 and 4 pN, respectively, and an almost 4 times larger slope in the low-force regime that precedes the unstacking transition (Fig. 1B). For the 167-bp NRL fibers, the latter feature results in a maximal extension of the fiber before the unstacking transition of about 5 nm per nucleosome, compared with roughly 10 nm per nucleosome for 197-bp NRL fibers. Furthermore, 167-bp NRL fibers feature a sharp force plateau, implicating cooperative unfolding, whereas the more gradual unstacking of 197-bp NRL fibers indicates independent ruptures of stacked nucleosomes. Interestingly, the stacking energy, which can be approximated by the product of the rupture force and the length of the force plateau divided by the number of stacked nucleosomes, is the same for both fibers (Fig. 1B, inset). This suggests that similar nucleosome-nucleosome interactions govern the folding of the two types of nucleosomal arrays into higher-order structures.
These results and those of our previous studies imply a two-start zig-zag structure for the 167-bp NRL fibers and a one-start solenoidal folding for the 197-bp NRL fibers. However, this interpretation has been questioned (9), arguing that the excessive bending of the linker DNA does not allow for stacking of neighboring nucleosomes. Moreover, it was suggested that stacking of nucleosomes involves more than just histone tail-mediated interactions and would require a closely packed stack of nucleosomes, which is inconsistent with an extension of up to 10 nm per nucleosome. Clearly, identifying the exact structural nature of the unstacking transition will help to address these issues.
The H4 tail mediates the nucleosome-nucleosome interactions in folded chromatin fibers
In the absence of DNA, H4-V21C and H2A-E64C mutant histones readily dimerize under oxidizing conditions but remain largely separate in the presence of a reducing agent such as DTT (Fig. 2A). When these histones are folded into a chromatin fiber, H2A-H4 cross-linking stabilizes the MT fiber against a stretching force (Fig. 2B). Indeed, cross-linked chromatin fibers remain condensed up to much larger forces than fibers in reduced conditions, which exhibit an unstacking plateau similar to WT fibers. The difference in extension is more than 500 nm at 5 pN for a fiber containing 15 nucleosomes, or about 30 nm per nucleosome. Moreover, at forces above 10 pN, we observed large, discrete, and irregular rupture events on cross-linked MT fibers (Fig. 2B) as opposed to clear 25-nm steps (Fig. 1B) for WT fibers. Similar to WT fibers, this transition is irreversible, and the refolding curve does not overlap with a stretching curve (supplemental Fig. 2). Not all histones dissociate from the DNA, however, as the refolding curve significantly deviates from that of bare DNA as described by a WLC at forces below 25 pN. Compared with WT fibers, this deviation appears to be slightly bigger for the cross-linked fibers, but large variations were observed.
Figure 2.
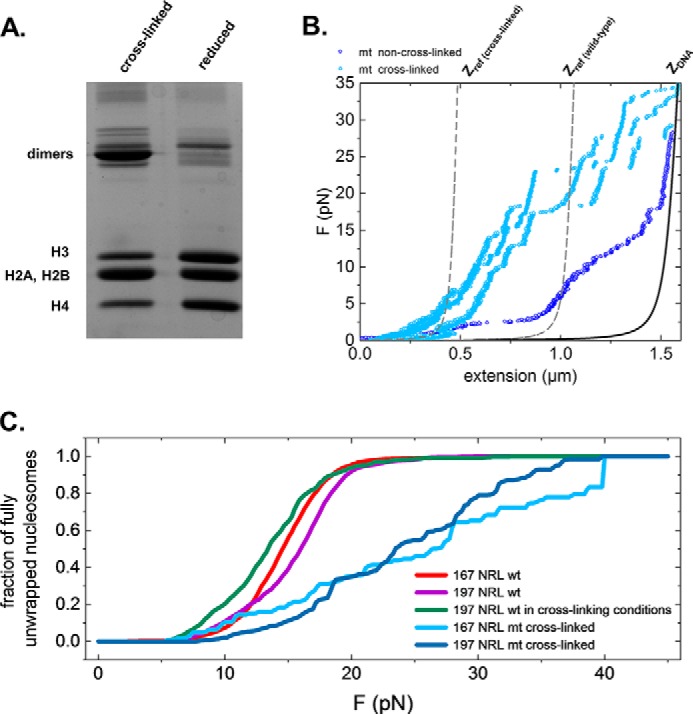
Cross-linking mutant histones introduces covalent bonds between nucleosomes in folded chromatin fibers. A, 18% SDS-PAGE of H4-V21C/H2A-E64C double mutant X. laevis histone octamers in reducing and oxidizing conditions. Apart from the three bands corresponding to H4, H2A-H2B, and H3 histones, additional bands are visible due to the formation of histone dimers through the disulfide bridge. Multiple dimers can be formed because of endogenous cysteines in H3 and newly introduced cysteines in both H4 and H2A. B, force (F)-extension curves of cross-linked 197-bp NRL fibers (light blue) and a WT 197-bp NRL fiber (dark blue). Dashed lines represent WLC curves of DNA with the same extension as chromatin fibers at 7 pN. These curves were used to calculate the fraction of fully unwrapped nucleosomes. C, fraction of unfolded nucleosomes as a function of force for the wild-type and cross-linked fibers. Wild-type and non-cross-linked fibers of both NRLs feature full unwrapping of nucleosomes in a force range between 6 and 22 pN. In cross-linked fibers, only 30% of the nucleosomes are fully unwrapped at 22 pN. Full unwrapping is only achieved at 40 pN.
To quantify the unfolding more precisely, we computed the fraction of fully unwrapped nucleosomes as a function of force (Fig. 2C) for all types of fibers. Whereas forces of 20 pN are sufficient to unwrap more than 90% of the nucleosomal DNA in WT fibers and MT fibers under reduced conditions, only about a third of the nucleosomal DNA is released in cross-linked MT fibers (Fig. 2C, 167- and 197-bp NRLs in light and dark blue curves, respectively). To unwrap more than 90% of the nucleosomal DNA in cross-linked MT fibers, forces in the range of 35–40 pN are required. Thus, efficient cross-linking of Cys residues is achieved in folded fibers when the redox potential is sufficiently low.
In contrast to the observed differences in the high-force regime, inspection of the low-force regime reveals that here the mechanical properties of the cross-linked MT fibers are very similar to those of folded WT fibers. For 167-bp NRL fibers (Fig. 3A), close overlap is observed for the force-extension curves below the 4-pN unstacking plateau. Both fibers yield the same stiffness (p = 4 × 10−12). At 4.5 pN, the WT fiber undergoes the unstacking transition, whereas the cross-linked MT fiber continues to stretch in almost linear fashion. The force-extension curves of the 197-bp NRL fibers also feature substantial overlap up to the unstacking plateau (Fig. 3B), whereupon the curves start to deviate. Again, both fibers yield the same stiffness (p = 2 × 10−12). As in the case of the WT fibers, the cross-linked 197-bp NRL MT fibers exhibit reduced stiffness relative to the cross-linked 167-bp NRL MT fibers. We therefore conclude that the mechanical properties of the folded fibers at forces below the unstacking transition are unaffected by cross-linking. This is noteworthy as it implies that contacts between the H4 tail and the H2A acidic patch remain intact throughout the linear stretching regime.
Figure 3.
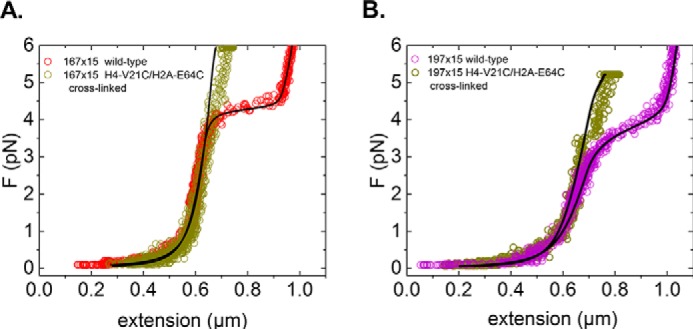
Cross-linked fibers do not feature the characteristic chromatin-unstacking transition at 4 pN. A, the typical force plateau of 167-bp NRL fibers at 4.5 pN is not observed in cross-linked fibers. Until unstacking, the force (F)-extension curves overlap. B, comparison between a cross-linked MT fiber and a WT 197-bp NRL fiber shows a similar stabilization of the fiber. Note that these fibers have a different number of tetrasomes and hexasomes, resulting in a small offset between the curves. The small transition at 3.5 pN in the cross-linked 197-bp NRL MT fiber may indicate that one of the nucleosome pairs is not properly cross-linked. Fitting the curves yields the same values for the fiber stiffness of cross-linked and non-cross-linked fibers.
H4-V21C/H2A-E64C mutations affect chromatin stacking
One would expect that MT fibers under reducing conditions and WT fibers exhibit similar force-extension behavior, but instead we found that MT fibers are generally more extended at low force (Fig. 4, A and B). This difference can be understood by explicitly accounting for the numbers of nucleosomes, hexasomes, and tetrasomes present in the fibers. Fitting the data to our physical model of chromatin stretching shows that MT fibers generally contain fewer nucleosomes, but more tetrasomes, than their WT equivalents. Consider, for example, the 167-bp NRL WT fiber and the corresponding MT fiber shown in Fig. 4A. The WT fiber is found to contain 14 nucleosomes and five tetrasomes, whereas the MT fiber contains nine nucleosomes and 10 tetrasomes (fit results with residuals are listed in supplemental Figs. 3 and 4). As both fibers contain 19 nucleosomal particles in total, their extensions at forces exceeding 5 pN are very similar. In a similar fashion, we fitted 15 nucleosomes and eight tetrasomes for the 197-bp NRL WT fiber shown in Fig. 4B, whereas the 197-bp NRL MT fiber contained nine nucleosomes and 15 tetrasomes. These fit results are typical for our reconstitutions and may reflect an inherently lower stability of nucleosomes in MT fibers. If so, H2A-H2B dimers may more easily dissociate. H2A-H2B dissociation will result in less nucleosome-wrapped DNA (as tetrasomes only wrap a single turn of DNA) and fewer nucleosomes that stack on each other (for which the acidic patch on H2A-H2B is required). Both effects will increase the extension of the fiber at forces below the unstacking transition. These differences in fiber composition, however, do not impact the stiffness of WT and cross-linked MT fibers, which continue to reflect the stiffness characteristic for their NRL (supplemental Figs. 3 and 4).
Figure 4.
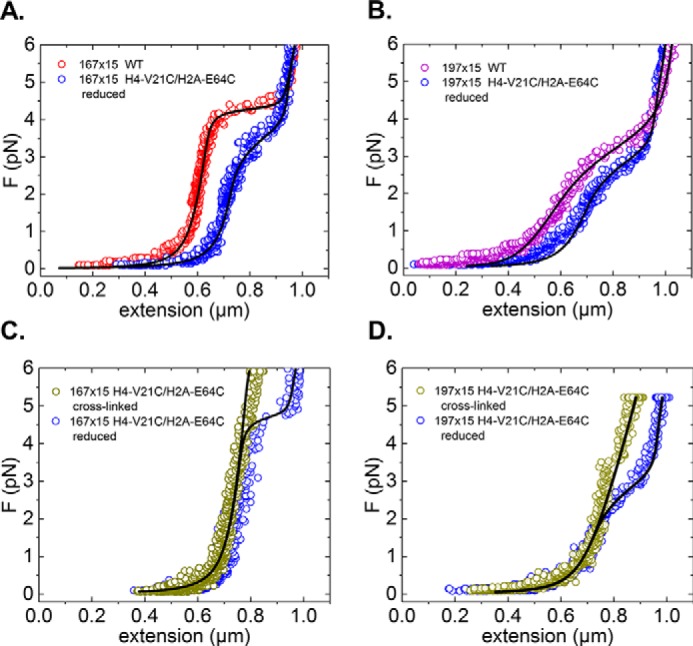
Cross-linking of individual fibers does not affect their mechanical properties. A, low-force regime of the force (F)-extension curve of a chromatin fiber assembled on 15 × 167-bp DNA with WT octamers (red curve) and H4-V21C/H2A-E64C octamers (dark blue) under reducing conditions. B, low force regime of the force-extension curve of a chromatin fiber assembled on 15 × 197-bp DNA with WT octamers (purple curve) and H4-V21C/H2A-E64C octamers (dark blue) under reducing conditions. The difference in the plateau height between WT and MT fibers in both graphs indicates a reduced interaction strength. The smaller width of force plateaus of the MT fibers should be attributed to a smaller number of H2A-H2B dimers than in the WT fibers. C and D, low-force regime of the force-extension curves of the same MT chromatin fibers as shown in A and B under reduced (blue) and oxidized conditions (green). The tether was first pulled when cross-linked in oxidizing buffer and then in reducing buffer (light blue). Removal of cross-links recovers the force plateau. Importantly, before fiber rupture, the force-extension behaviors of the MT fibers overlap with the curves obtained when cross-linked.
We can further exclude the effects of differences in the composition of individual fibers by exposing individual fibers sequentially to cross-linking and reducing conditions. We first applied forces up to 6 pN on MT fibers under cross-linking conditions (Fig. 4, C and D, green curves). Next, we flushed in DTT to disrupt cross-linking and repeated the pulling experiment (Fig. 4, C and D, blue curves). As before, the force-extension curves of cross-linked and reduced MT fibers largely overlap at forces below the unstacking transition with the differences in stiffness between 167- and 197-bp NRL fibers being independent of the presence of cross-linking. Because cross-linking of H4-V21C to H2A-E64C does not affect the mechanical properties of the folded fiber but does inhibit the unstacking transition (Fig. 3), we can unequivocally attribute this unstacking transition in both 167- and 197-bp NRL fibers to the rupture of H4 tail-mediated nucleosome stacking rather than the gradual unwrapping of the nucleosomal DNA.
Discussion
The folding of chromatin fibers is a highly debated topic that has been addressed mainly by studying regularly spaced 601 arrays reconstituted in vitro. To elucidate how the nucleosome repeat length influences the secondary structure and dynamics of chromatin, we performed a single-molecule force spectroscopy study on 167- and 197-bp NRL nucleosomal arrays reconstituted with wild-type histone octamers and compared their response with that of fibers composed of double mutant (H4-V21C/H2A-E64C) histone octamers. Using these two point mutations, we could attribute the nature of nucleosome stacking in folded chromatin fibers to these specific residues. Importantly, up to the unstacking transition, we observed the same mechanical properties in cross-linked and WT fibers, which underscores the role of the H4 tail in nucleosome-nucleosome interactions that drive higher-order folding.
Successful cross-linking in the flow cell was confirmed by the increased rupture forces of the individual fibers (Fig. 4) and could be reversed by flushing a reducing agent into the buffer. We speculate that the large changes in extension at forces exceeding 10 pN (Fig. 2B) represent the disassembly of clusters of covalently linked nucleosomes from the DNA.
All force-extension curves showed a linear extension of the folded fibers in the low-force regime. The overlap between WT fibers and MT fibers below 3 pN (Fig. 3) implies that cross-linked MT fibers share the same structure as equivalent fibers assembled with WT histone octamers. We can therefore pinpoint the predominant stacking interaction to the two mutated residues, although additional interactions between other tails and neighboring nucleosomes cannot be excluded. Linking the nucleosomes by the disordered, flexible tails explains the very low stiffness of the fibers and the large linear extension that we report here and in previous work.
Our MT nucleosomes still carried the native H3 Cys-110 residue, which could form disulfide bonds with the cysteines in H4-V21C and H2A-E64C. We cannot exclude the formation of such cross-links in our chromatin fibers. However, the H3 Cys-110 residues are more deeply buried in the nucleosome structure and therefore less accessible. In the low-force range, we observed the same mechanical properties for the fibers under oxidizing and reducing conditions, suggesting that only the anticipated H4-V21C–H2A-E64C disulfide bonds are formed, mimicking native H4 tail-H2A acidic patch interactions.
The cross-linking approach has a significant advantage in the single-molecule force spectroscopy assay. Covalently fixing the state of the fiber in the absence of force allowed us to decouple the mechanical properties of 197-bp NRL fibers from spontaneous fiber unfolding. The results obtained with the WT fibers show that the maximum extension of the folded 197-bp NRL fiber is ∼10 nm per nucleosome. Considering a height of a nucleosome of ∼6 nm, a gap of ∼4 nm remains between two stacked nucleosomes in the stretched fiber. The H4 tails have just enough length to span this gap. The cross-linked MT fibers can be stretched several nm per nucleosome more, prior to the occurrence of rupture transitions at ∼8 pN. This additional length may result from elastic deformation of the nucleosome, and it cannot be excluded that part of the H4 tail is pulled out of the nucleosome core at such high forces. The observed extension shows, however, that nucleosome-nucleosome stacking does not require close packing of the stacked nucleosomes.
We detected a higher abundance of tetrasomes in MT fibers, which could result from weaker stacking interactions between nucleosomes. We observed that the unstacking plateau occurs at lower forces, 3.5 versus 4 pN for 167-bp NRL and 2.5 versus 3 pN for 197-bp NRL for non-cross-linked MT fibers. Replacing the glutamic acid residue in H2A by a polar cysteine reduces the charge of the acidic patch and may consequently decrease electrostatic nucleosome-nucleosome interactions and therefore the stability of nucleosome stacking. The small extension of the 197-bp NRL fiber at 3.5 pN (Fig. 3B) may represent the unstacking of a few non-cross-linked nucleosomes, indicating that cross-linking may not have been achieved fully in these fibers. Interestingly, the rupture force for single nucleosomes (which cannot stack and can only unwrap) is 2.5 pN (33). Thus, unwrapping of nucleosomal DNA in the absence of stacking could account for the appearance of the unstacking transition at 2.5 pN; consistent with this interpretation, we observed a more gradual unstacking transition for 167-bp NRL MT fibers under reducing conditions (Fig. 4A). Likewise, sedimentation velocity analysis of 167- and 177-bp NRL MT fibers under reducing conditions indicated a reduced degree of condensation for MT fibers relative to WT fibers (18). As a secondary effect, destabilization of the nucleosome-nucleosome interactions may enhance H2A-H2B dimer dissociation, leading to fibers with a larger tetrasome/nucleosome ratio. This may be particularly relevant for the single-molecule force spectroscopy assay presented here, in which the fibers are highly diluted (37).
We can exclude, however, that the increased number of tetrasomes in MT fibers inhibited H2A-H4–mediated nucleosome stacking in the case that all nucleosomes would be interspersed by tetrasomes. The same fibers under cross-linking conditions continue to stretch linearly beyond 3.5 pN (Fig. 4, C and D), demonstrating the establishment of covalent H2A-H4 cross-linking. Thus, both comparisons between ensembles of WT and cross-linked MT fibers and between individual cross-linked and reduced MT fibers indicate that the unstacking plateau should be attributed to the unstacking of nucleosomes rather than to the unwrapping of nucleosomal DNA.
The most detailed structural studies on folded chromatin fibers used short NRLs (10, 12) and clearly resolved a two-start helix. Our experiments on WT chromatin arrays with a short NRL (167 bp) showed that these fibers fold into rather stiff tethers that unstack cooperatively, in line with such a two-start helix. Longer repeat lengths, however, are more common in higher eukaryotes, which makes it highly relevant to resolve their topology. Robinson et al. (11) reported that the dimensions of folded fibers with larger NRLs are incompatible with two-start folding. Compared with 167-bp NRL fibers, 197-bp NRL fibers appear less stiff and rupture at smaller forces. The distribution of the unstacking energy in 197-bp NRL fibers is slightly broader than that of 167-bp NRL fibers (Fig. 1B, inset), which may indicate more heterogeneity. Consequently, thermal fluctuations will induce larger variations in extension and enhanced spontaneous unstacking of nucleosomes in the folded fibers. Such a more disordered and dynamic structure of chromatin fibers with a long NRL renders studying their organization using structural techniques more challenging.
It is relevant to point out that our measurements were performed at physiological salt concentrations (i.e. 100 mm NaCl; see “Experimental Procedures”). The recent results obtained with force spectroscopy and cryo-EM on 167- and 177-bp NRL fibers by Li et al. (34) were acquired at much lower salt concentrations (10 mm HEPES, pH 8.0, 0.1 mm EDTA, and optionally 1.5 mm MgCl2), which could strengthen interactions between histones and DNA, driving the fibers in more compacted, closely packed structures. Whereas Li et al. (34) report steplike transitions for unstacking, a sign of non-equilibrium transitions, our force-extension curves on WT fibers feature a gradual unstacking plateau that can be reversibly refolded. Such behavior indicates more frequent nucleosome unstacking and restacking events, which may be relevant in vivo where nucleosomal DNA seems readily accessible.
The current results place strong constraints on structural models of 30-nm fibers. Such models should be compatible, upon the application of force, with an extension of up to 10 nm per nucleosome while maintaining the base of the H4 tail (residue H4 Asn-24) and the acidic patch (residue H2A Glu-64) in the adjoining nucleosome within ∼3.5 nm (the maximal extension of the H4 tail (16)). For 197-bp NRL fibers, this spatial constraint excludes any higher-order nucleosome-nucleosome stacking other than a one-start model. For shorter fibers such as the 167-bp NRL, the constraint, set experimentally by the maximal extension per nucleosome, is partially relieved as these fibers are found to have a smaller maximal extension before rupture; in accordance with all other experimental evidence, this points to a two-start structure. The 10-nm linear extension per nucleosome for 197-bp NRL fibers clearly points to a one-start helix as such extension in two-start models would require distances between stacked nucleosomes that exceed the length of the H4 tail.
An alternative interpretation of the extension of the fiber before the unstacking transition would be the gradual unwrapping of nucleosomal DNA (9). Such a model would be incompatible with our data as DNA unwrapping increases the distance between the nucleosomes to lengths that cannot be bridged by the H4 tail. The covalent bond that we introduced by cross-linking furthermore prohibits any conformational changes that require changes of interacting nucleosomes upon stretching by unstacking and restacking with other nucleosomes. We therefore conclude that stacking between neighboring nucleosomes in a one-start helix is the most likely structure for the 197-bp NRL fibers studied here.
Additional measurements can be envisioned that further test the folding pathway reported here. Clipping of the histone tails, acetylation of H4 Lys-16, and the depletion of Mg2+ should all affect fiber folding by reducing the histone tail-mediated stacking interaction between the nucleosomes. We did observe a lower interaction energy for these cases (data not shown), but as for the MT fibers under reducing conditions, we predominantly observed accompanying degradation of fiber composition, leading to fibers containing mostly tetrasomes. We speculate that the lack of higher-order structure makes the fiber more fragile when exposed to the drag forces that are unavoidable when flushing the flow cell. Therefore, it appears that nucleosome stacking and nucleosome stability in fibers may be correlated in our experimental setup. The current approach of strengthening nucleosome stacking, rather than weakening, allows for better control of the fiber's composition.
Although our work focuses on the physical properties of regular arrays of nucleosomes in vitro, the linker length dependence of nucleosome stacking may have important consequences for chromatin structure in vivo. It is conceivable that the variable linker length that has been reported for native chromatin can result in irregular chromatin structures, which would match recent reports on the absence of higher-order structure (38) or the observation of pebbles of locally condensed structures (24). Alternating stacking between neighbors, next neighbors, or more distant nucleosomes could locally modulate the accessibility of the chromatin and hence play an important role in epigenetic regulation.
For linker lengths of 20 and 50 bp, we established that nucleosome stacking protects nucleosomal DNA from external forces up to about 4 pN. This may be the most important ramification of the current work. Despite extensive extension of the chromatin fibers, nucleosome stacking remains intact in this force regime, which will reduce accessibility of the DNA and will therefore affect the activity of all nuclear processes that require access to DNA.
Experimental procedures
Chromatin reconstitution and cross-linking
A plasmid based on pUC18 (Novagen) containing 15 tandem repeats of the Widom 601 sequence spaced by either 20 or 50 bp of a linker DNA (NRL = 167 and 197 bp, respectively) was digested with BsaI and BseYI, yielding linear fragments of 4535 bp for 167-bp NRL and 4985 bp for 197-bp NRL substrates. A Klenow reaction was used to fill in the single-stranded ends with dUTP-digoxigenin (at the BsaI site) or dUTP-biotin (at the BseYI site). Wild-type and H4-V21C/H2A-E64C double mutant Xenopus laevis histone octamers were purified and reconstituted as described previously (33). Prior to reconstitution, MT histone octamers were diluted in high-salt buffer (2 m NaCl, 1× Tris-EDTA, pH 7.5) containing 100 mm DTT to reduce the cysteine residues as described previously (18). For both WT and MT histone octamers, reconstitution was performed using salt gradient dialysis under reducing conditions (20 mm β-mercaptoethanol). Optimal reconstitution was selected from a titration of histone/DNA ratios evaluated by agarose gel electrophoresis (supplemental Fig. 1). To induce the disulfide bridging between neighboring nucleosomes in arrays reconstituted with H4-V21C/H2A-E64C, a second dialysis was performed with buffer without any oxidizing agents but supplemented with 2 mm MgCl2.
Sample preparation
A clean coverslip was coated with 0.1% nitrocellulose (Ladd Research) in amyl acetate solution and then mounted onto a polydimethylsiloxane (Dow Corning) flow cell containing a 10 × 40 × 0.4-mm3 flow channel. The flow cell was incubated with 3 μg/ml anti-digoxigenin (Roche Applied Science) for 2 h and then passivated with 4% bovine serum albumin (w/v) and 0.1% Tween 20 overnight. 20 ng/ml reconstituted fibers in the measurement buffer (MB; 100 mm NaCl, 2 mm MgCl2, 10 mm HEPES, pH 7.6, 10 mm NaN3, 0.2% BSA, 0.1% Tween 20) were flushed into the flow cell and incubated at room temperature for 10 min followed by flushing in 1- or 2.8-μm-diameter streptavidin-coated superparamagnetic beads (MyOne and M270, respectively, Invitrogen) diluted in MB. Untethered beads were flushed out with MB 10 min later.
Magnetic tweezers
We used home-built multiplexed magnetic tweezers.4 The extensions of the molecules were measured in real time at a frame rate of 30 Hz with a complementary metal-oxide-semiconductor camera (CMOS Vision Condor). We applied a continuously increasing force by moving the magnet toward the flow cell at a constant velocity. Force ramps typically lasted 30 s. Subsequently, we reversed the magnet trajectory to decrease the force. We used a double-exponential function that was calibrated prior to the experiment to calculate the exerted force (32). The resulting force-extension plots were aligned at high force with a wormlike chain with a contour length equal to that of the DNA used in reconstitution (Fig. 1A).
Data analysis
We used LabVIEW software to analyze the obtained time traces of tether extension as a function of magnet position. The resulting force-extension curves were fitted to a statistical mechanics-based model described previously (33).
The cumulative distribution of rupture forces was calculated using the following equation,
| (Eq. 1) |
where zmeas.(F) is the extension of the tether at force F, zDNA(F) is the extension of the DNA fully unwrapped from all nucleosomes, and zref(F) is the extension of the tether above a force threshold of 7 pN. Punwrapped(F) is set to 0 for negative values, which would correspond to more than one turn of wrapped DNA per nucleosome. The cumulative distribution is normalized using the total number of nucleosomal steps that were found in multiple pulling curves.
To determine the significance of the overlap between two force-extension curves, we performed a Student's t test,
| (Eq. 2) |
where kWT and kMT are the fitted slopes of the force-extension curves between 0.5 and 3 pN for WT and MT fibers, and S.E.WT and S.E.MT are the standard error of the fit for WT and MT fibers. The two-tailed p value was calculated for n degrees of freedom, equal to the number of data points minus the number of variables in the fit.
Author contributions
A. K., L. N., N. H. D., and J. v. N. conceived and coordinated the study and wrote the paper. A. A. prepared mutant histones. T. B. B. set up the multiplexed magnetic tweezers. A. K. designed, performed, and analyzed the experiments shown in Figs. 1–4. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Nikolay Korolev, Nicolaas Hermans, Orkide Ordu, and Josko de Boer for valuable discussions and comments and Chi Pham and Ineke de Boer for DNA preparation.
This work was supported by the Netherlands Organisation for Scientific Research (NWO/OCW), as part of the Frontiers of Nanoscience program, by NWO-VICI Research Program Project 680-47-616; the Singapore Ministry of Education Academic Research Fund through Tier 3 Grant MOE2012-T3-1-001; and the Singapore Agency for Science Technology and Research through Biomedical Research Council Grant 10/1/22/19/666. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. 1–4.
T. B. Brouwer, A. Kaczmarczyk, N. Hermans, and J. van Noort, manuscript in preparation.
- NRL
- nucleosome repeat length
- pN
- piconewton(s)
- MT
- mutant
- WLC
- wormlike chain
- MB
- measurement buffer.
References
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome resolution core particle at 2.8 Å. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Bednar J., Studitsky V. M., Grigoryev S. A., Felsenfeld G., and Woodcock C. L. (1999) The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol. Cell 4, 377–386 [DOI] [PubMed] [Google Scholar]
- 3. Park Y. J., and Luger K. (2008) Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 18, 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lorch Y., Maier-Davis B., and Kornberg R. D. (2010) Mechanism of chromatin remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Workman J. L. (2006) Nucleosome displacement in transcription. Genes Dev. 20, 2009–2017 [DOI] [PubMed] [Google Scholar]
- 6. Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M. J., Davie J. R., and Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 7. Routh A., Sandin S., and Rhodes D. (2008) Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eissenberg J. C., and Elgin S. C. (2000) The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204–210 [DOI] [PubMed] [Google Scholar]
- 9. Victor J. M., Zlatanova J., Barbi M., and Mozziconacci J. (2012) Pulling chromatin apart: unstacking or unwrapping? BMC Biophys. 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R. M., Zhu P., and Li G. (2014) Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 [DOI] [PubMed] [Google Scholar]
- 11. Robinson P. J., Fairall L., Huynh V. A., and Rhodes D. (2006) EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. U.S.A. 103, 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schalch T., Duda S., Sargent D. F., and Richmond T. J. (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 13. Allahverdi A., Chen Q., Korolev N., and Nordenskiöld L. (2015) Chromatin compaction under mixed salt conditions: opposite effects of sodium and potassium ions on nucleosome array folding. Sci. Rep. 5, 8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Correll S. J., Schubert M. H., and Grigoryev S. A. (2012) Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 31, 2416–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Langowski J., and Schiessel H. (2006) Computational Studies of RNA and DNA, pp. 605–634, Springer, Dordrecht, The Netherlands [Google Scholar]
- 16. Norouzi D., Katebi A., Cui F., Zhurkin V. B. (2015) Topological diversity of chromatin fibers: interplay between nucleosome repeat length, DNA linking number and the level of transcription. AIMS Biophys. 2, 613–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nikitina T., Norouzi D., Grigoryev S. A., and Zhurkin V. B. (2017) DNA topology in chromatin is defined by nucleosome spacing. bioRxiv 10.1101/104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., and Richmond T. J. (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 19. Grigoryev S. A. (2012) Nucleosome spacing and chromatin higher-order folding. Nucleus 3, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang C., and Pugh B. F. (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilbert N., and Ramsahoye B. (2005) The relationship between chromatin structure and transcriptional activity in mammalian genomes. Brief. Funct. Genomic. Proteomic. 4, 129–142 [DOI] [PubMed] [Google Scholar]
- 22. Tompitak M., Vaillant C., and Schiessel H. (2017) Genomes of multicellular organisms have evolved to attract nucleosomes to promoter regions. Biophys. J. 112, 505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maeshima K., Hihara S., and Eltsov M. (2010) Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 22, 291–297 [DOI] [PubMed] [Google Scholar]
- 24. Ricci M. A., Manzo C., García-Parajo M. F., Lakadamyali M., and Cosma M. P. (2015) Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158 [DOI] [PubMed] [Google Scholar]
- 25. Clark D. J. (2010) Nucleosome positioning, nucleosome spacing and the nucleosome code. J. Biomol. Struct. Dyn. 27, 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowary P. T., and Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 27. Widom J. (2001) Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 34, 269–324 [DOI] [PubMed] [Google Scholar]
- 28. Flaus A. (2011) Principles and practice of nucleosome positioning in vitro. Front. Life Sci. 5, 5–27 [Google Scholar]
- 29. Koslover E. F., Fuller C. J., Straight A. F., and Spakowitz A. J. (2010) Local geometry and elasticity in compact chromatin structure. Biophys. J. 99, 3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finch J. T., and Klug A. (1976) Solenoidal model for superstructure in chromatin. Proc. Natl. Acad. Sci. U.S.A. 73, 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chien F.-T., and van Noort J. (2009) 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr. Pharm. Biotechnol. 10, 474–485 [DOI] [PubMed] [Google Scholar]
- 32. Kruithof M., Chien F., de Jager M., and van Noort J. (2008) Subpiconewton dynamic force spectroscopy using magnetic tweezers. Biophys. J. 94, 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meng H., Andresen K., and van Noort J. (2015) Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res. 43, 3578–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li W., Chen P., Yu J., Dong L., Liang D., Feng J., Yan J., Wang P. Y., Li Q., Zhang Z., Li M., and Li G. (2016) FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol. Cell 64, 120–133 [DOI] [PubMed] [Google Scholar]
- 35. Schlick T., Hayes J., and Grigoryev S. (2012) Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 287, 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Q., Yang R., Korolev N., Liu C. F., and Nordenskiöld L. (2017) Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail-H2A acidic patch interaction. J. Mol. Biol. 429, 2075–2092 [DOI] [PubMed] [Google Scholar]
- 37. Claudet C., Angelov D., Bouvet P., Dimitrov S., and Bednar J. (2005) Histone octamer instability under single molecule experiment conditions. J. Biol. Chem. 280, 19958–19965 [DOI] [PubMed] [Google Scholar]
- 38. Maeshima K., Ide S., Hibino K., and Sasai M. (2016) Liquid-like behavior of chromatin. Curr. Opin. Genet. Dev. 37, 36–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


