Abstract
RAD52 motif containing 1 (RDM1) encodes the RAD52 protein involved in DNA double-strand break repair and recombination events. However, the importance of RDM1 in papillary thyroid carcinoma (PTC) is largely unknown. In the present study, we examined the role of RDM1 in thyroid cancer. The RDM1 expression in PTC patients was examined using immunohistochemistry. The expression levels of RDM1 mRNA in thyroid cancer cells were measured by quantitative real-time PCR (qRT-PCR). Lentivirus-mediated small interfering RNAs (siRNAs) were used to knock down the RDM1 expression in the K1 and TPC1 cells. Then, changes in the RDM1 target gene expression were determined by qRT-PCR and Western blot. Cell proliferation was examined by a high content screening assay. Cell cycle distribution and apoptosis were detected by flow cytometric analysis and MTT analysis. We showed that the RDM1 expression was higher in PTC tissue compared to pericarcinous tissue. RDM1 mRNA was found to be expressed by qRT-PCR. Using a lentivirus-based RNA interference (RNAi) approach, the RDM1 expression was significantly inhibited. The inhibition of RDM1 expression by RNAi significantly impaired cell proliferation, increased apoptosis and arrested cells in the G2/M phase. These data showed that RDM1 was highly expressed in PTC tissue and thyroid cancer cell lines. Moreover, RDM1 may play an important role in cell proliferation, cell cycle distribution and apoptosis of human PTC cells.
Keywords: RDM1, thyroid cancer, papillary thyroid carcinoma, siRNA, lentivirus
Introduction
Papillary thyroid carcinoma (PTC) is the most common subtype of thyroid cancer. In 2008, the incidence of thyroid cancer was 1.4/100,000, and it accounts for 0.8% of all malignancies in China (0.4% of those in men, 1.3% in women) (1). Through surgery and radioactive iodine ablation, most PTC patients have a good prognosis; however, a significant proportion of patients have persistent or recurrent disease and suffer from frequent recurrences or even distant metastases, leading to an unfavourable outcome. A minority of these patients will die from thyroid cancer (2).
The RAD52 motif containing 1 (RDM1) gene showed upregulation in PTC compared with normal thyroid tissues through an analysis of the gene expression dataset of Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TCGA). RDM1 is located at 17q11.2 and belongs to the gene-binding motif containing family. RDM1 contains a small amino acid motif, named the RD motif, which it shares with the recombination and repair protein RAD52 (3). RDM1 can bind to RNA as well as single- and double-stranded DNA and is involved with DNA double-strand break (DSB) repair and recombination events. When the repair of DNA damage occurs, many genes are involved in DNA repair, including excision repair, post-replication repair and recombination/DSB repair. The repair of DSBs is mediated extensively by RAD52-dependent recombination (4).
To explore the role of RDM1 in PTC, an analysis of RDM1 expression was conducted in thyroid cancer cell lines. Lentivirus-mediated small interfering RNA (siRNA) targeting of RDM1 was used to downregulate gene expression. Then, experiments were performed to examine the effect of RDM1 silencing on the cell cycle, apoptosis and proliferation.
Materials and methods
Patient clinical data
Before this study began, written informed consent was obtained from all patients who participated in the study, which was approved by the Ethics Committee of Tianjin Medical University General Hospital. A group of 23 PTC patients who underwent total thyroidectomy and neck lymph dissection in Tianjin Medical University General Hospital, China, was studied. Paraffin-embedded tissue material/tissue blocks were chosen for histological studies.
Analysis of gene expression database
To analyse comparative of RDM1 expression, several web-based bioinformatics tools were used. GEO statistical tool (http://www.ncbi.nlm.nih.gov/geo/) and TCGA website (https://cancergenome.nih.gov/) were reciprocally employed (5). The microarray data were downloaded from website, relative gene expression and log2 fold change values of gene expression, and survival (Kaplan–Meier) curves were calculated, compared and statistically analysed through Excel and GEO2R, cBioPortal online tool (http://www.cbioportal.org/).
Cell lines
The human thyroid cancer cell lines K1 and TPC1 were used in our laboratory (6). All cell lines were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS and 1% penicillin/streptomycin. All culture reagents were purchased from Gibco. The cell cultures were stored in a humidified atmosphere containing 5% CO2 buffered with ambient air at 37°C.
Immunohistochemical staining for RDM1
The specimens were immunostained using the PV-6000 immunohistochemistry method to evaluate the expression of the RDM1 protein. The primary antibody used was RDM1 rabbit polyclonal antibody (HPA024708, Sigma). The samples stained with RDM1 antibodies were examined using a microscope (BX51, Olympus). The immunoreactivity of the samples was evaluated using a scoring scale that measured both the intensity and the percentage of cells positively stained. The staining intensity was scored as follows: 0 (none), 1 (weak), 2 (moderate) or 3 (intense). The percentage of positively stained cells was scored as follows: 0 (<1%), 1 (1–25%), 2 (25–50%), 3 (50–75%) or 4 (>75%). Immunoreactivity scores were then assigned according to the product of the two parameters evaluated (intensity and positivity) as follows: ≤6 was low expression and >6 was high expression (7).
Quantitative real-time PCR (qRT-PCR) for gene expression
Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer’s instructions. One-step RT-PCR was carried out in triplicate with the following primers: RDM1 (forward: 5′-GCCCATCCTGGTTTCTATGCCC-3′; reverse: 5′-AGACGAACCTTGACTGGAGAT-3′); GAPDH (forward: 5′-TGACTTCAACAGCGACACCCA-3; reverse: 5′-CACCCTGTTGCTGTAGCCAAA-3′). The RDM1 gene expression was calculated using ΔCT and normalised to GAPDH. The relative gene expression after lentivirus infection was calculated using the 2−ΔΔCT method and normalised to GAPDH (8).
Lentivirus vectors for RDM1 siRNA construction and transduction
Lentiviral vectors for RDM1 siRNA were used to silence the RDM1 gene expression. Lentivirus was used to express siRNAs targeting the RDM1 sequence (Genbank No. NM_145654). The siRNA sequence was 5′-GGCCCATCCTGGTTTCTAT-3′. The sequences were cloned into pGCSIL (GeneChem, Shanghai, China) to generate the lentiviral vectors. RDM1-siRNA lentivirus is referred to as shRDM1; the negative control lentivirus that inserts a meaningless sequence (5′-TTCTCCGAACGTGTCACGT-3′) is referred to as shCtrl. shCtrl has no homology to any known human genes. K1 and TPC1 cells were infected with shRDM1 and shCtrl, respectively. The interference efficiency of the template was detected by the Western blot analysis.
For lentivirus transduction, K1 and TPC1 cells were incubated with a 1:1 combination of culture medium and lentivirus medium. After 8 h of incubation, the medium was replaced with a complete culture medium, and the cells were allowed to grow for 3 days. The lentivirus-transduced cells were treated with 1 mg/mL puromycin for 48 h for stable clone selection (9).
Western blot analysis
The expression of the RDM1 protein was studied in K1 and TPC1 cells by Western blotting as described earlier (10). Cells were harvested with a scraper after being washed with ice-cold phosphate-buffered saline and lysed in a buffer containing 10 mM Tris·HCl (pH 7.5), 1 mM DTT, 20% (v/v) glycerol, 1 mM ethylenediaminetetraacetic acid and a protease inhibitor mixture. Proteins were separated by SDS-PAGE, transferred onto PVDF membranes, stained with the primary antibodies RDM1 rabbit-antibody (HPA024708, Sigma, 1:200) and GAPDH mouse-antibody (sc-32233, Santa-Cruz, 1:2000) at 4°C overnight and then incubated with secondary antibody at room temperature for 30 min. ECL Western blotting reagents were used for detection. Exposures were made at room temperature for 1–3 min using Fujifilm films. Prestained protein marker (26620, ThermoFisher) was run in the same gels to compare molecular weights and estimate transfer efficiency.
Flow cytometric analysis of cell cycle distribution and apoptosis
Flow cytometric analysis was performed to define the cell cycle distribution and induction of apoptosis in K1 and TPC1 cells. Values are an average of at least 3 independent experiments. Cells infected with RDM1-siRNA lentivirus were plated onto 6-well plates in triplicate and incubated at 37°C for 4 days. Cells were then collected, washed twice with ice-cold D-Hanks, fixed with 75% ice-cold ethanol, washed twice with ice-cold D-Hanks and stained with propidium iodide (PI, 2 mg/mL) in the presence of RNase (10 mg/mL). The cell cycle phase of K1 and TPC1 cells was analysed by flow cytometry. For induction of apoptosis, an Annexin V Apoptosis Detection Kit APC (eBioscience, USA) was used. Cells were plated and infected with RDM1-siRNA lentivirus as described earlier for the cell cycle assay. Following incubation at 37°C for 3 days, cells were collected and stained with 10 μL of Annexin V-APC away from light. The percentage of apoptotic cells was identified.
Plate analysis with the adherent cell cytometry system Celigo
K1 and TPC1 cells were analysed with fluorescence staining after lentivirus transduction to quantify the cellular siRNA gene knockdown rate. The adherent cell cytometry system allowed rapid quantification of cellular fluorescence as previously described (11). 96-Well plates were analysed using an adherent cell cytometer equipped with bright field and green filter fluorescent channels. Gating parameters were adjusted for the fluorescence channel to exclude background and other non-specific signals. The Celigo system provided a gross quantitative analysis for each fluorescence channel and individual well, including the total count and the average integrated red fluorescence intensity of gated events.
MTT cell viability assay
The inhibition of cell proliferation following treatment with the extract was measured using an MTT assay. Briefly, K1 and TPC1 cells infected with shRDM1 or shCtrl were separately plated in 96-well plates (2000 cells/well). Following 1–5 days of incubation at 37°C, 20 μL of MTT solution (5 mg/mL) was added to the cells, and then the cells were incubated for 4 h. Then, the supernatant was removed, 150 μL of DMSO was added, and then the samples were shaken for 5 min until the crystal dissolved. The absorbance was measured using a microplate reader (Infinite 200 PRO series; TECAN) at a wavelength of 490 nm. The optical density (OD) value at 490 nm was measured by an enzyme-linked immunosorbent assay reader. The negative control well had no cells and was used as the zero point of absorbance. All the experiments in each group were performed in triplicate (12).
Statistical analysis
Data were expressed as the mean ± s.d. Comparison between two groups and two or more independent groups was done with independent sample T-test, and one-way ANOVA. P < 0.05 indicates significant difference.
Results
RDM1 is upregulated in PTC
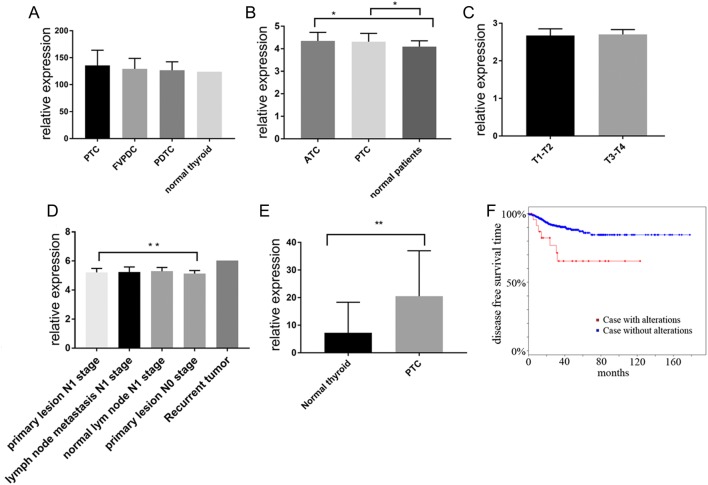
Using the GEO data mining platform, we performed a comprehensive analysis of RDM1 expression profiles in various types of thyroid cancer vs normal thyroid tissues (a total of 21 clinical samples including PTC, the follicular variant of papillary thyroid carcinoma (FVPTC), poorly differentiated thyroid carcinoma (PDTC) and normal thyroid tissue) that was reported in a previous study (13). The data showed no statistical significance, because normal thyroid tissue had only one patient (Fig. 1A). Then, we performed a comprehensive analysis of RDM1 expression profiles in various types of thyroid cancer vs normal thyroid tissues (a total of 105 clinical samples including papillary, anaplastic thyroid cancer and patients with no-tumour control) that was reported in a previous study (14). This analysis described that PTC showed a consistent upregulation of RDM1 compared to normal thyroid tissues, and the relative gene expression of RDM1 in PTC was about 1.2-fold higher than normal thyroid tissue (Fig. 1B). Moreover, we also performed a comprehensive analysis of RDM1 expression in different PTC tissues (compared with PTC primary lesion, lymph node metastasis, normal lymph node and recurrent tumour at N0 and N1 stages); the result described an upregulation RDM1 expression of primary lesion and lymph node at N1 stage patients (Fig. 1D) than primary lesion of N0 stage. But compared with different tumour T stages (T1 or T2 vs T2 or T3), the data showed no statistical significance (Fig. 1C).
Figure 1.
RDM1 expression in thyroid cancer from the GEO and TCGA datasets. (A) RDM1 expression in PTC, FVPTC, PDTC and normal thyroid tissue (PTC, n = 7; FVPTC, n = 8; PDTC, n = 5; normal thyroid tissue, n = 1). Data source: GSE53157. PTC=135.8 ± 28.18, FVPTC = 129.2 ± 6.91, PDTC = 126.8 ± 6.99, normal thyroid tissue = 123.9. P = 0.88. (B) RDM1 expression in ATC, PTC and normal patients’ thyroid (ATC, n = 11; PTC, n = 49; normal patients’ thyroid, n = 45). Data source: GSE33630. ATC = 4.35 ± 0.38, PTC = 4.31 ± 0.37, normal thyroid = 4.09 ± 0.26. *P < 0.05. (C) RDM1 expression in T1–T2 and T3–T4 PTC patients (T1–T2, n = 21; T3–T4, n = 15). Data source: GSE65074. T1–T2 = 2.68 ± 0.04; T3–T4 = 2.71 ± 0.03. P = 0.57. (D) RDM1 expression in PTC primary lesion, lymph node metastasis, normal lymph node and recurrent tumour at N0 and N1 stages. Primary lesion N1 stage, n = 20; primary lesion N0 stage, n = 14; lymph node metastasis N1 stage, n = 21; normal lymph node N1 stage, n = 4; Recurrent tumour, n = 1. Data source: GSE60542. Primary lesion N1 stage = 5.21 ± 0.28; primary lesion N0 stage = 5.14 ± 0.21; lymph node metastasis N1 stage = 5.24 ± 0.35; normal lymph node N1 stage = 5.29 ± 0.26; Recurrent tumour = 6.02. **P < 0.01. (E) RDM1 expression in 568 pairs of PTCs and normal thyroid tissue from TCGA RNA-seq data (normal thyroid, n = 58; PTC, n = 510). PTC = 20.51 ± 0.73; normal thyroid = 7.26 ± 1.45. **P < 0.01. (F) Disease-free survival (Kaplan–Meier) curves. Cases with alteration (mRNA expression z-scores choose >2 or <−2) were 24 (relapsed n = 7, 29.17%), without alteration were 469 (relapsed n = 41, 8.74%).
The RDM1 expression of thyroid tumour and normal thyroid tissue (a total of 568 clinical samples, include 58 normal thyroid tissue and 508 PTC) was analysed using the RNAseqV2 datasets for thyroid cancer deposited on TCGA (https://cancergenome.nih.gov/) website (5). The data indicated that the RDM1 expression was increased by more than 2-fold (Fig. 1E). Based on TCGA data, we used cBioPortal (http://www.cbioportal.org) to draw disease-free survival (Kaplan–Meier) curves and genomic profiles condition: mRNA expression z-scores choose >2 or <−2. Kaplan–Meier curves for the survival of patients with the RDM1 expression being changed more than 2-fold were significantly lower than control (logrank test P-value: 0.00321). Survival probabilities (95% confidence intervals) at 40, 80 and 120 months are as follows: for 89.67% vs 65.45%, 84.61% vs 65.45% and 84.61% vs 65.45%, respectively (Fig. 1F).
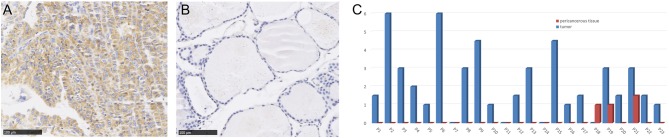
To further validate the expression of RDM1 in thyroid cancer, we used immunoreactivity scores to examine the RDM1 expression in the tumours and pericarcinous tissue (the same patient’s normal thyroid tissues) of 23 pairs PTC patients. The immunoreactivity scores of RDM1 positive percentage were significantly higher in the tumours than in the pericarcinous tissue (immunoreactivity scores: 2.2 ± 0.4, 0.2 ± 0.1, P < 0.01) (Fig. 2C). In agreement with the TCGA data, the RDM1 expression was higher in most thyroid tumour tissues in comparison with their pericarcinous tissue. Examples of typical immunostaining of positive and negative RDM1 expressions are presented in Fig. 2A and B.
Figure 2.
RDM1 expression in the immunohistochemical analysis. (A) Typical immunohistochemistry staining of RDM1 positive in PTC tissue (×40). (B) Typical immunohistochemistry staining of RDM1 negative in pericarcinous tissue/normal thyroid tissue (×40). (C) Expression of RDM1 in 23 paired PTCs and pericarcinous tissue by the immunoreactivity score analysis.
The clinical characteristics of 23 PTC patients were as follows. The patients consisted of 2 males and 21 females. The ages of the patients were from 47 to 76 years old, with an average age of 55.5 ± 7.6 years. One patient had a bilateral tumour, and 22 patients had unilateral tumours. Extrathyroidal extension occurred in 17 patients. Twenty-two patients presented with lymph node metastases and none reported distant metastasis. Three patients were diagnosed with Hashimoto thyroiditis by pathological examination.
The RDM1 expression is downregulated by shRDM1 in K1 and TPC1 cells
After 8 h of infection with shRDM1 or shCtrl, the thyroid cancer cells were incubated for 72 h and then observed with a fluorescence microscope. The infection efficiency was more than 80% and the cells remained healthy.
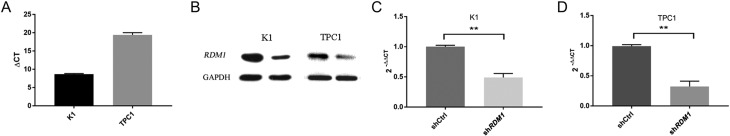
The RDM1 mRNA expression level in K1 and TPC1 cells was high when normalised to the GAPDH expression, as analysed by qRT-PCR (Fig. 3A). K1 and TPC1 cells were infected with either shRDM1 or shCtrl lentivirus. The RDM1 protein level detected by Western blot was reduced in RDM1-siRNA infected cells, indicating effective knockdown of the target sequence (Fig. 3B). Compared with cells infected with control lentivirus shCtrl, the RDM1 mRNA expression in cells infected with shRDM1 was lower (P < 0.01). The knockdown efficiency of shRDM1 was about 51% and 70% in K1 and TPC1 cells, respectively, as determined by qRT-PCR (Fig. 3C and D). The results indicated that the interference efficiency was valid in K1 and TPC1 cells.
Figure 3.
RDM1 expression examined by qRT-PCR and Western blot. CT is the threshold cycle for target amplification in K1 cells. ΔCT is equal to the difference in threshold cycles for the target and reference; ΔΔCT calculates the relative quantification of target. (A) RDM1 mRNA expression level in K1 and TPC1 cells. ΔCT = CTRDM1 − CTGAPDH. (B) RDM1 protein in K1and TPC1 cells. Compared with shCtrl, the level of RDM1 protein in K1 and TPC cells decreased after the RDM1 expression was silenced by RNAi. GAPDH is a control. The RDM1 and GAPDH proteins were observed as major bands corresponding to molecular weights of 26 and 34 kDa, respectively. (C) and (D) Knock down efficiency was determined by qRT-PCR. The RDM1 mRNA expression level in K1 cells after infection with different lentiviruses was measured using the 2−ΔΔCT method. ΔΔCT = mean (CTRDM1/CTGAPDH) − CTRDM1. 2−ΔΔCT of shRDM1 and shCtrl in K1 and TPC1 cells were 0.5 ± 0.1 vs 1.0 ± 0.0, 0.3 ± 0.1 vs 0.9 ± 0.0, respectively (**P < 0.01).
Knockdown o RDM1 expression by RNA interference (RNAi) induces apoptosis and G2/M arrest
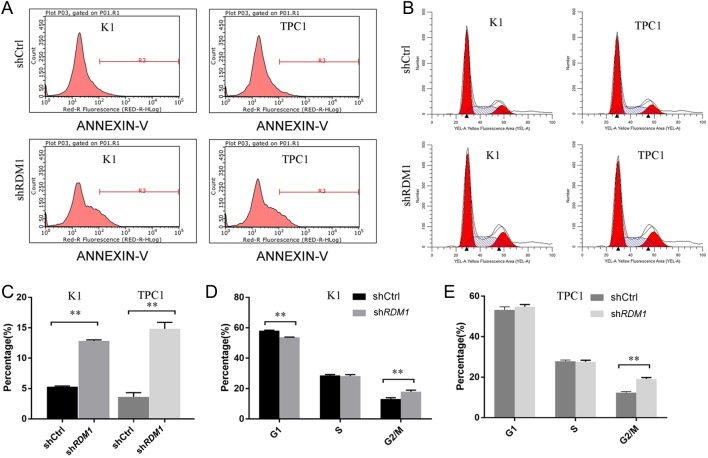
In addition, to determine whether RDM1-siRNA enhanced cell apoptosis by the induction of cell cycle arrest, K1 and TPC1 cells were infected with lentivirus and incubated for 3–4 days, and then the cell cycle distribution and apoptosis were evaluated by the flow cytometric analysis. As shown in Fig. 4A and C, the inhibited expression of RDM1 resulted in an increase in the apoptosis of K1 and TPC1 cells. The cell cycle analysis showed an increased percentage of cells in the G2/M phase (P < 0.01) in K1 and TPC1 cells and a decreased population in G1 phase (P < 0.01) in K1 cells (Fig. 4B, C and D). The population showed no significant difference in the S phase.
Figure 4.
Effect of the downregulation of RDM1 on K1 cells. (A) and (C) RDM1-siRNA enhanced cell apoptosis (n = 3). Flow cytometry analyses of apoptosis were shown in shRDM1 and control vector in K1 and TPC1 cells. The apoptosis rate was observed to be 5.3 ± 0.1% (shCtrl) and 12.8 ± 0.2% (shRDM1) in K1 cells. The apoptosis rate was observed to be 3.6 ± 0.7% (shCtrl) and 14.8 ± 1.1% (shRDM1) in TPC1 cells. **P < 0.01, compared with shCtrl. (B), (C) and (E) RDM1 expression affects cell cycle distribution (n = 3). The cell cycle analyses of shRDM1 and control vector in K1 cells were 18.0 ± 1.0% and 13.2 ± 0.8% in the G2/M phase, and 58.2 ± 0.3% and 53.8 ± 0.2% in the G1/S phase. The cell cycle analyses of shRDM1 and control vector in TPC1 cells were 19.1 ± 0.7% and 12.3 ± 0.5% in the G2/M phase, and 54.8 ± 1.1% and 53.2 ± 1.6% in the G1/S phase. **P < 0.01, compared with shCtrl.
The ability of cell proliferation was significantly inhibited by cell count analysis and MTT
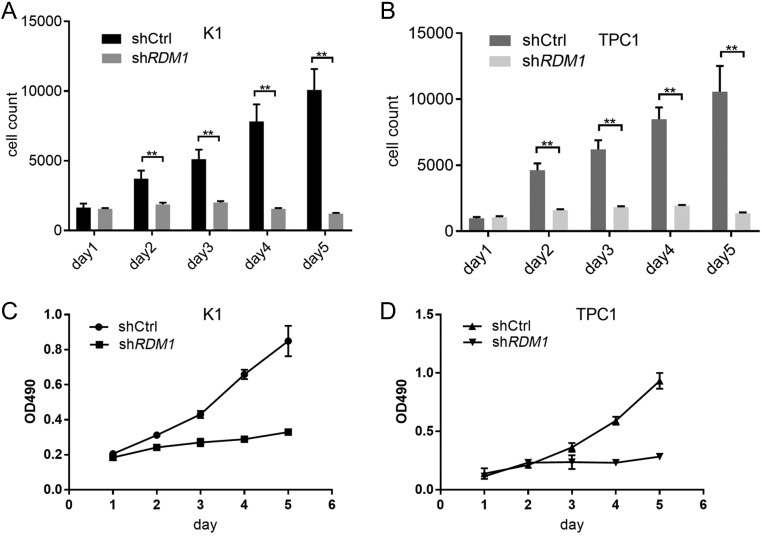
Celigo software was used to generate representative scatter plots that depict the green fluorescence area (in μm2) and integrated fluorescence intensity (sum of the pixel intensities within a gated event) of the gated events in the individual treatment groups (11, 15). Strong differences in total count were observed between cells infected with shRDM1 and shCtrl. From 1 day to 5 days, the fluorescence pixel counts/cell in K1 and TPC1 cells infected with shRDM1 were significantly lower than those in cells transfected with shCtrl (Fig. 5A and B). The K1 and TPC1 cell growths were significantly inhibited by RDM1-siRNA.
Figure 5.
Growth analysis of K1 and TPC1 cells after infected with lentivirus (n = 3). The proliferation of K1 and TPC1 cells was inhibited slightly by RDM1-siRNA, and analysed by Celigo cytometer and MTT. (A) and (B) Cell growth rate using Celigo cytometer. Cell growth rate was defined as the cell count of Nth day/cell count of 1st day. K1 and TPC1 cells infected shCtrl grew quickly; however, those cells infected shRDM1 which showed growth retardation. **P < 0.01. (C) and (D) Growth curves by MTT. The K1 cell density OD 490 in the presence of RDM1-siRNA and in the control group were 0.24 ± 0.01 and 0.31 ± 0.00, respectively, on day 2; 0.33 ± 0.01 and 0.85 ± 0.09, respectively, on day 5. The TPC1 cell density compared with shRDM1 and shCtrl were 0.21 ± 0.00 and 0.23 ± 0.01, respectively, on day 2; 0.29 ± 0.03 and 0.93 ± 0.07, respectively, on day 5.
The cell density OD 490 was measured to calculate cell growth by the MTT analysis. As shown in Fig. 5C and D, the proliferation inhibition of RDM1-siRNA in K1 and TPC1 cells was time-dependent: it was noticeable on day 5.
Discussion
RDM1 encodes a 284-amino-acid-long protein and binds RNA, ssDNA and dsDNA. RDM1 can increase sensitivity of chemotherapy drug cisplatin and may have a possible role during spermatogenesis and in the maintenance of stemness (3, 16). The mechanism of RDM1 increases sensitivity of cisplatin such that the RD motif of RDM1 plays a role in the interaction of the protein with nucleic acids, possibly as a modulator of the RRM, and/or that it is involved in the ability of RDM1 to self-interact, as illustrated in this study by the filament-like structures observed on dsDNA (16). In 2007, a study showed that RDM1 has a function in the heat-shock response (8).
Currently, there are few studies of RDM1. RDM1 expression and function in thyroid cancer had not been studied. Thus, this study first explored the expression levels of RDM1 in thyroid cancer tissue and thyroid cancer cell lines and found that it was more expressed in thyroid cancer tissue compared with pericancerous tissue. The expression of RDM1 was also analysed at both the gene and miRNA levels in a large dataset of tumour and normal samples obtained from TCGA and GEO. As discussed in detail in the Results section, the high expression of RDM1 was also confirmed at our immunohistochemistry samples. To assess the RDM1 effect in thyroid cancer cell lines, these cells were infected with RDM1-siRNA lentivirus or control lentivirus. Compared to the controls, RDM1-siRNA-treated cells showed decreased proliferation, increased apoptosis and an increase in the proportion of cells in the G2/M phases. Therefore, RDM1 may promote thyroid cancer cell growth. To further clarify the mechanism of RDM1-siRNA growth inhibition, its ability to arrest cell cycle has been studied. The data suggest that RDM1-siRNA has the ability to arrest the cell cycle in the G2/M phase. Cell cycle progression and cell division are driven by the sequential activation of a group of serine–threonine kinases called cyclin-dependent kinases (CDKs). The activity of CDKs is positively regulated by cyclins and is negatively regulated by CDK inhibitors. The G2/M transition is regulated by the sequential activation and deactivation of CDK-regulatory proteins and cyclin complexes Moreover, DNA DSB repair is known to proceed via two major mechanisms: homologous recombination (HR) and non-homologous end joining. HR includes the exchange of genetic information between homologous or homoeologous DNA sequences and is thus generally active in the S and G2 phases of the cell cycle (17, 18). RD motif is the RRM domain of RDM1 conserved in the DNA recombination and repair protein Radiation sensitive 52 (RAD52). RAD52 is an important HR protein (19). Experiments have shown that when recombinational repair is absent, hRAD52 expresses very low levels during the G1 phase, but through homologous repair rising, its level rises steadily through the S phase, reaching a maximum in the G2 phase (20). However, why RDM1-siRNA influences the G2/M phase and how RDM1 plays a role in thyroid cancer are currently unknown and warrant further study.
Conclusion
This study highlights critical roles for RDM1 in the PTC cell line. The data may provide the basis for further exploration of the role of RDM1 in the occurrence and development of thyroid cancer. A follow-up study on the mechanism by which RDM1 affects PTC cell biology is needed. RDM1 may become a new target of PTC molecular therapy.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
Author contribution statement
L W and T J conceived the study and participated in its design and coordination. Z G Z helped to draft the manuscript. L W and S D Y carried out the molecular genetic studies. H Q participated in statistical analysis, GEO and TCGA data analysis. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Genechem for providing them with lentiviral particles and technical assistance.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians 2016. 66 115–132. ( 10.3322/caac.21338) [DOI] [PubMed] [Google Scholar]
- 2.Duman BB, Kara OI, Uguz A, Ates BT. Evaluation of PTEN, PI3K, MTOR, and KRAS expression and their clinical and prognostic relevance to differentiated thyroid carcinoma. Contemporary Oncology 2014. 18 234–240. ( 10.5114/wo.2014.43803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamimes S, Bourgeon D, Stasiak AZ, Stasiak A, Van Dyck E. Nucleic acid-binding properties of the RRM-containing protein RDM1. Biochemical and Biophysical Research Communications 2006. 344 87–94. ( 10.1016/j.bbrc.2006.03.154) [DOI] [PubMed] [Google Scholar]
- 4.Milne GT, Ho T, Weaver DT. Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics 1995. 139 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang W, Guan H, He X, Ke W, Xu L, Liu L, Xiao H, Li Y. Down-regulation of SOSTDC1 promotes thyroid cancer cell proliferation via regulating cyclin A2 and cyclin E2. Oncotarget 2015. 6 31780–31791. ( 10.18632/oncotarget.5566) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka J, Ogura T, Sato H, Hatano M. Establishment and biological characterization of an in vitro human cytomegalovirus latency model. Virology 1987. 161 62–72. ( 10.1016/0042-6822(87)90171-1) [DOI] [PubMed] [Google Scholar]
- 7.Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL, Zeng ZL, Wang RY, Huang YX, Jin Y, Wang F, et al. Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer. World Journal of Gastroenterology 2014. 20 18404–18412. ( 10.3748/wjg.v20.i48.18404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messaoudi L, Yang YG, Kinomura A, Stavreva DA, Yan G, Bortolin-Cavaille ML, Arakawa H, Buerstedde JM, Hainaut P, Cavaille J, et al. Subcellular distribution of human RDM1 protein isoforms and their nucleolar accumulation in response to heat shock and proteotoxic stress. Nucleic Acids Research 2007. 35 6571–6587. ( 10.1093/nar/gkm753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Xie Q, Yu Z, Zhou H, Huang Y, Bi X, Wang Y, Shi W, Sun H, Gu P, et al. A regulatory loop containing miR-26a, GSK3beta and C/EBPalpha regulates the osteogenesis of human adipose-derived mesenchymal stem cells. Scientific Reports 2015. 5 15280 ( 10.1038/srep15280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki T, Lorkovic ZJ, Liang SC, Matzke AJ, Matzke M. The ability to form homodimers is essential for RDM1 to function in RNA-directed DNA methylation. PLoS ONE 2014. 9 e88190 ( 10.1371/journal.pone.0088190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabzdyk CS, Chun M, Pradhan L, Logerfo FW. High throughput RNAi assay optimization using adherent cell cytometry. Journal of Translational Medicine 2011. 9 48 ( 10.1186/1479-5876-9-48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu GS, Zou SQ, Liu ZR, Tang ZH, Wang JH. Celecoxib inhibits proliferation and induces apoptosis via prostaglandin E2 pathway in human cholangiocarcinoma cell lines. World Journal of Gastroenterology 2003. 9 1302–1306. ( 10.3748/wjg.v9.i6.1302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pita JM, Banito A, Cavaco BM, Leite V. Gene expression profiling associated with the progression to poorly differentiated thyroid carcinomas. British Journal of Cancer 2009. 101 1782–1791. ( 10.1038/sj.bjc.6605340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomás G, Tarabichi M, Gacquer D, Hébrant A, Dom G, Dumont JE, Keutgen X, Fahey TJ, 3rd, Maenhaut C, Detours V. A general method to derive robust organ-specific gene expression-based. Oncogene 2012. 31 4490–4498. ( 10.1038/onc.2011.626) [DOI] [PubMed] [Google Scholar]
- 15.Nabzdyk CS, Chun M, Pradhan Nabzdyk L, Yoshida S, LoGerfo FW. Differential susceptibility of human primary aortic and coronary artery vascular cells to RNA interference. Biochemical and Biophysical Research Communications 2012. 425 261–265. ( 10.1016/j.bbrc.2012.07.078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamimes S, Arakawa H, Stasiak AZ, Kierzek AM, Hirano S, Yang YG, Takata M, Stasiak A, Buerstedde JM, Van Dyck E. RDM1, a novel RNA recognition motif (RRM)-containing protein involved in the cell response to cisplatin in vertebrates. Journal of Biological Chemistry 2005. 280 9225–9235. ( 10.1074/jbc.M412874200) [DOI] [PubMed] [Google Scholar]
- 17.Barlow JH, Rothstein R. Timing is everything: cell cycle control of Rad52. Cell Division 2010. 5 7 ( 10.1186/1747-1028-5-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Research 2008. 18 99–113. ( 10.1038/cr.2008.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Game JC, Mortimer RK. A genetic study of X-ray sensitive mutants in yeast. Mutation Research 1974. 24 281–292. ( 10.1016/0027-5107(74)90176-6) [DOI] [PubMed] [Google Scholar]
- 20.Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutation Research 1997. 384 205–211. ( 10.1016/S0921-8777(97)00020-7) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a