Abstract
Burning incense to worship deities is a popular religious ritual in large parts of Asia, and is a popular custom affecting more than 1.5 billion adherents. Due to incomplete combustion, burning incense has been well recognized to generate airborne hazards to human health. However, the correlation between burning incense and lung cancer in epidemiological studies remains controversy. Therefore, we speculated that some unknown materials in incense smoke are involved in the initiation or progression of lung cancer. Based on this hypothesis, we identified a major compound auramine O (AuO) from the water‐soluble fraction of incense burned condensate using mass spectrometry. AuO is commonly used in incense manufacture as a colorant. Due to thermostable, AuO released from burned incenses becomes an unexpected air pollutant. AuO is classified as a Group 2B chemical by the International Agency of Research on Cancer (IARC), however, the damage of AuO to the respiratory system remains elusive. Our study revealed that AuO has no apparent effect on malignant transformation; but, it dramatically promotes lung cancer malignancy. AuO accumulates in the nucleus and induces the autophagy activity in lung tumor cells. AuO significantly enhances migration and invasive abilities and the in vitro and in vivo stemness features of lung tumor cells through activating the expression of aldehyde dehydrogenase family 1 member A1 (ALDH1A1), and ALDH1A1 knockdown attenuates AuO‐induced autophagy activity and blocks AuO‐induced lung tumor malignancy. In conclusion, we found that AuO, an ingredient of incense smoke, significantly increases the metastatic abilities and stemness characters of lung tumor cells through the activation of ALDH1A1, which is known to be associated with poor outcome and progression of lung cancer. For public health, reducing or avoiding the use of AuO in incense is recommended.
Keywords: ALDH1A1, auramine O, incense, lung cancer, mass spectrometry
1. INTRODUCTION
Burning incense is a popular religious ritual and culture of worshipping activity throughout countries of Asia‐Pacific region, and this custom influences 1.5 billion adherents of major such as Hinduism, Buddhism, Taoism, and Chinese folk religions in this area.1 In Taiwan, there are more than 1.2 million temple goers who frequently visit temples and burn incense inside the temples.2 Incense stick is the most common form of incense used in Taiwan. Agarwood and sandalwood are the two major ingredients in incense, other materials such as spices, herbs, and essential oils are usually incorporated into incense components to increase the fragrance of incense smoke and make incense smolder effectively.3 Due to incomplete combustion, incense burning has been recognized to generate airborne hazards to human health, especially to the respiratory system. Particulate matters (PMs),4, 5, 6 polycyclic aromatic hydrocarbons (PAHs),7, 8, 9 and volatile organic compounds (VOCs)10, 11, 12 are hazardous materials commonly detected in incense smoke. However, the correlation between burning incense and lung tumor progression/malignancy remains controversy.13 Therefore, we speculated that the unidentified components in incense smoke could result in inconsistency on this issue.
Water‐soluble fraction in incense burned condensate (IBC) has long been ignored because organic matters are decomposed by combustion into small chemical compounds, most of which are organic‐soluble or volatile such as PMs, PAHs, and VOCs. Whether water‐soluble materials exist in incense smoke or they are correlated with cancer progression/malignancy is fully unknown. In this study, we utilized mass spectrometry (MS) to identify the water‐soluble materials of IBC and found auramine O (AuO) is the major compound in them. AuO (bis[4‐(dimethylamino)phenyl] methaniminium chloride), a yellow dye, is commonly used in incense manufacture as a colorant. AuO has a high melting point of 267°C and thermostability, which results in the existence of AuO in incense smoke as an unexpected air pollutant.
Oral administration of high‐dose AuO has been shown to induce liver tumors in both mice and rats14 and cause genotoxicity in primary rat and human hepatocytes,15 and the International Agency of Research on Cancer (IARC) classified AuO into Group 2B as a possible carcinogen to humans.16 To date, the exposure and damage of AuO to the respiratory system has never been recognized. Therefore, we investigated the carcinogenic potential of AuO for the initiation and progression of lung tumor. In this study, we found that AuO undergoes nuclear translocation and induces autophagy activity in lung tumor cells. AuO has no apparent effect on inducing lung tumor initiation. However, AuO significantly increases cell migration and invasive abilities through activating the stemness of tumor cells, which finally leads to tumor malignancy.
2. MATERIAL AND METHODS
2.1. Chemicals, antibodies, and plasmids
Acridine orange (#A9231) and auramine O (#861030) were purchased from Sigma‐Aldrich. The antibodies used in this study include ALDH1A1 (#12035; Cell Signaling), CD133 (#PAB12663; Abnova), CD44 (#LS‐C44932; LSBio), LC3B (#ab51520; abcam), and β‐actin (#3700; Cell Signaling). The shRNAs against ALDH1A1 gene were purchased from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). The target sequences of shALDH1A1#1 and shALDH1A1#2 are: CACCGATTTGAAGATTCAATA and GCTGATTTAATCGAAAGAGAT, respectively.
2.2. Incense and incense smoke collection
Incense sticks including Chen‐Shiang (CS; agarwood) and Tan‐Shiang (TS; sandalwood) were purchased from vendors surrounding Dajia Jenn Lann Temple (Taichung, Taiwan) and local incense manufacture (Taichung, Taiwan). Incense sticks (three CS and three TS) collected from vendors were used for the estimation of AuO content, whereas those (two CS and two TS) collected from local incense manufacture were used as reference because their AuO content was no detection. The sampling device and sampling method have been described in an earlier publication.17 Personal environmental monitor (SKC 761–203) with a size cut of 2.5 μm mounted with 37 mm Teflon filter (SKC 225–1709) was used for smoke particle sampling. After 5 min collection, the filters were cut into small pieces and extracted by 20 mL ddH2O. The extracted solution was lyophilized and resolved with assigned volume of ddH2O.
2.3. Mass spectrometry
Materials extracted from IBC were assayed by Apex Qe FT‐ICR MS equipped with a 9.4 T actively shielded magnet (Bruker Daltonics). The analysis was set in positive ion mode using an Apollo II electrospray source. The MS scan was set m/z 100 to 1000 with a mass resolution of 66,000 at m/z 400. The spectra were processed using Data Analysis 4.0 (Bruker Daltonics). For comparative proteomic analysis, the trypsinized peptides were identified using the linear ion trap‐Fourier transform ion cyclotron resonance mass spectrometer (LTQ‐FTICR MS, Thermo Fisher). The survey scan of MS analysis (m/z 320–2000) was performed on LTQ‐FTICR MS with a mass resolution of 100,000 at m/z 400. Top 10 most abundant multiply charged ions were sequentially isolated for MS/MS by LTQ. The MaxQuant18 and MaxLFQ19 software were used for protein identification and label‐free quantification. The significance threshold for protein identification was set to P < .01.
2.4. Molecular modeling
The molecular modeling was performed to predict the binding affinity of AuO to DNA using Autodock software, and the similar structures of AuO were analyzed using Autodock 4.2 software with Lamarckian Genetic Algorithm.20 The crystal structure of DNA was obtained from the protein data bank (PDB ID: 1Z3F) (http://www.rcsb.org/pdb/home/home.do).21 The substrates including ligands, water, and small molecules were removed from co‐crystallized DNA. Polar hydrogens and Kallman united atom charges were added to the DNA for docking calculation using Autodock Tool 1.5.6 interfaces (ADT).22, 23 The optimization of AuO and its similar compounds were performed by using MMFF94 force field by ChemBio3D software (version 11.0; Cambridge Soft Corp.). Hydrogens and Gasteiger charges were added to the AuO for docking.22 The Grid box calculated by AutoGrid program was centered at the binding site of co‐crystal ligand of DNA with dimensions 68 × 64 × 44 Å grid points at spacing of 0.375Å. All docking parameters were set to default except for the following parameter: maximum number of energy evaluation increase to 25,000,000 per run. The docking results were analyzed by using cluster analysis and were shown by ADT. The models of docking results were shown by Discovery Studio Visualizer 4.5 (Accelrys).
2.5. Isothermal titration calorimetry
The energetic process of AuO‐dsDNA association was measured by the isothermal titration calorimetry (ITC, TA Instrument). The titration of dsDNA (five GC pairs of DNA) with AuO was performed by adding AuO from a rotating syringe in 20 injections with an interval of 310 s. All solutions were appropriately degassed to avoid the interference of bubble formation during the process of titration. The measurement in phosphate buffer was used as the background control to correct for the heat of dilution and deconvoluted on NanoAnalyze software (version 2.4.1) according to a single site binding model.
2.6. Cell culture
The human nontumorigenic bronchial epithelial cell line BEAS‐2B was cultured in DMEM/F‐12 medium [with 1 μM hydrocortisone, 5 μg/mL insulin, 10 μM HEPES, 10% fetal bovine serum (Gibco), and 1% penicillin/streptomycin (Gibco)]. The human lung adenocarcinoma cell line CL1–0 was gifts from Dr. Pan‐Chyr Yang (National Taiwan University, Taipei, Taiwan). Both CL1–0 and A549 cells were maintained in RPMI 1640 media (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotics (Gibco). All cells were grown in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
2.7. Protein extraction and tryptic peptide preparation
Total proteins were extracted using RIPA lysis and extraction buffer (Thermo Fisher) and protein quantification was determined using the Bio‐Rad Protein Assay by the measurement of absorbance at 595 nm. Total 20 μg of protein was separated using 10% SDS‐PAGE and divided into 10 gel fractions, which were then cut into small gel pieces (<1 mm3) individually. The in‐gel digestion procedure includes the following steps in order: (1) destaining using 50% acetonitrile (ACN) and 25 mM ammonium bicarbonate (ABC); (2) reduction using 10 mM dithiothreitol at 58°C for 45 min; (3) alkylation using 55 mM iodoacetamide at RT for 45 min in the dark; (4) enzyme digestion using 4 ng/μL sequencing grade trypsin in 25 mM ABC solution at 37°C for 16 h; (5) extraction of tryptic peptides using a 60% ACN/1% trifluoroacetic acid solution. After drying to remove the solvent, the tryptic peptides were dissolved and used for MS.
2.8. Western blot analysis
After SDS‐PAGE separation, proteins were transferred onto a PVDF membrane using electroblot at 400V at 4°C for 3 h in 25 mmol/L Tris‐HCl, 197 mmol/L glycine, and 13.3% (v/v) methanol. Membranes were blocked with 5% (w/v) skim milk in TBST for 1 h, and then incubated with primary antibodies by a gentle shaking at room temperature for overnight. After gently agitating in three TBST washes and one TBS wash for 15 min each, horseradish peroxidase (HRP)‐conjugated secondary antibody was added to further incubate at RT for 1 h. Immunoreactive signals were revealed using an enhanced chemiluminescence substrate kit (NEN Life Science), which includes luminescent substance luminol that is oxidized to luminesce by HRP using H2O2 as an oxidizing agent and enhancers that increases the light intensity, and the signals were recorded by developing photographic film under optimum exposure.
2.9. Wound healing and matrigel invasion assay
The wound healing assay was used to study the migration ability of tumor cell in vitro. Tumor cells (70 µL; concentration: 3 × 105 cells/mL) were added to Culture‐Insert well (ibidi) and cultured for 24 h. A “wound gap” was created by removal of Culture‐Insert, and the “healing” of this gap by cell migration was recorded per 4 h until 24 h. The migration area of tumor cells was measured using Image J software. The matrigel invasion assay was used to determine the invasion ability of tumor cell in vitro. Tumor cells (2.5 × 105 cells in 200 µL) were suspended in DMEM medium and added to the upper half of a PET membrane transwell insert chamber (BD Biosciences), which was coated with Matrigel (1 mg/mL; BD Biosciences) on a 24‐well plate. DMEM medium supplemented with 10% FBS was added as a chemoattractant to the lower half. After incubation at 37°C for 24 h, tumor cells that passed through the insert were fixed with 3% formalin (Sigma‐Aldrich) and stained with 0.2% crystal violet (Sigma‐Aldrich).
2.10. Spheroid formation assay
Spheroid formation assay was performed in 6 cm culture dish coated with 1% agarose. Tumor cells suspended in serum free medium were seeded at a density 5000 cells/dish and incubated in a humidified atmosphere of 5% CO2 and 95% air at 37°C for 7 days. The numbers of spheroid were counted manually at 7th day under microscope.
2.11. Mouse model of subcutaneous inoculation
The animal procedure (2016–047) was approved by the Institutional Animal Care and Use Committee (IACUC) at CMUH. Lung cancer cell A549 was pretreated with 1 μM AuO for 7 days. Both AuO‐treated and control A549 cells (1 × 106) were mixed with matrigel and injected into 5‐week‐old male BALB/c nude mice (purchased from BioLASCO, Taiwan). Left flank was injected with control, whereas right flank was injected with AuO‐treated A549 cell. Tumor size was measured by caliper weekly for 6 weeks once tumors became visible. Tumor volume was calculated with the formula: (length × width2)/2. After animal sacrifice, tissue samples were fixed in formalin and embedded with paraffin, and then analyzed using immunohistochemical (IHC) staining. The IHC procedure followed our previously described method.24 Briefly, rabbit anti‐human ALDH1A1 antibody (#12035; Cell Signaling) was used to perform IHC staining by horseradish peroxidase‐conjugated avidin‐biotin complex from the Vectastain Elite ABC Kit (Vector Laboratories) and AEC chromogen (Vector Laboratories).
2.12. Statistical analysis
Data are expressed as means ± SD. The significance of difference was examined by Student's t‐test (two‐tailed). P < .05 was considered to be significant.
3. RESULTS
3.1. AuO is the major water‐soluble component of IBC
The Personal Environmental Monitor (SKC Inc., PA) was used to collect the soot materials emitted from commercial Chen‐Shiang (CS, agarwood) and Tan‐Shiang (TS, sandalwood) incenses.17 The water‐soluble components were extracted from Teflon membranes (PM10 was collected) and analyzed using an Apex‐Qe Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics). A major compound with m/z 268.18 is detected in both incense samples (Figure 1A), and it has two daughter fragments with m/z 122 and m/z 147 in the tandem mass spectrum (Figure 1B). The chemical formula of target compound was deduced from the precise molecular weight of parent ion and the tandem mass signals of daughter ions, the search result showed that the potential candidate is AuO. Using commercial standard AuO, we confirmed that it exhibits the consistent mass signal (Figure 1A, upper panel) and tandem mass signal with the compound extracted form IBC.
Figure 1.
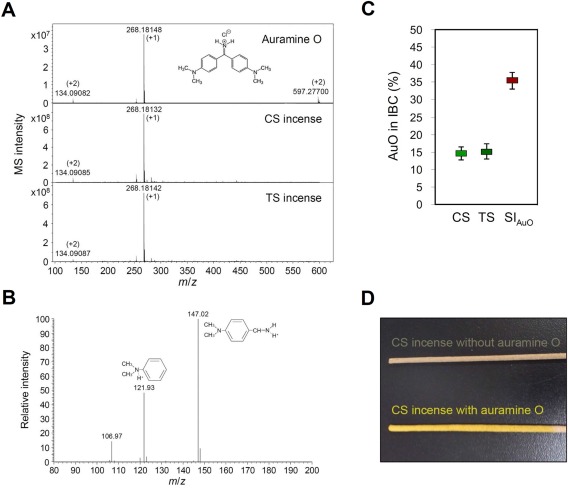
AuO is the major water‐soluble component of IBC. A, The compositions of commercial standard AuO and water‐soluble IBC compounds extracted form CS and TS incenses were analyzed by an Apex Qe FT‐ICR MS. B, Tandem mass profile of parental ion m/z 268.18 was generated by a Velos Pro MS. C, The weight percentage of AuO in commercial and AuO‐soaked incenses was determined by MS signal intensity derived quantitative calibration curve. D, The different appearances between incenses with and without AuO [Color figure can be viewed at wileyonlinelibrary.com]
To quantify the content of AuO in IBC and incense, we used AuO standard compound to construct a calibration curve using a Velos Pro MS (Thermo Fisher) coupled with a nano‐HPLC system. The linear relationship for AuO quantity and peak‐area intensity is calculated as y = 8.95x + 3044 and an R 2 of 0.9965, with AuO quantity ≧10 femtomole. Based on this linear equation, the content of AuO in CS and TS incenses is calculated as 5.25 mg/g and 4.85 mg/g (average concentration), respectively. The weight percentage of AuO in IBC of CS and TS incenses is 14.4% and 14.9%, respectively (Figure 1C). In addition, we soaked AuO‐free incenses with the saturated AuO solution (Figure 1D), which results in an up to 35.4% of weight percentage of AuO in IBC (Figure 1C).
According to our previous study,25 the average exposure concentration of PM10 in temples was around 600 μg/m3, approximately equal to 88 μg/m3 of AuO (assuming AuO accounts for 14.7% of PM10). Consequently, we estimate that a temple worker (8 h working time per day) inhales 338 μg (1.27 μmol) of AuO into the lung, and the maximum accumulation concentration in blood is 289 nM in 1 day (Supporting Information Table 1).
3.2. AuO accumulates in the nucleus and binds DNA of lung tumor cells
To understand the mode of action of AuO in the lung, we first observed the cellular location of AuO based on its fluorescent feature using confocal microscopy. After AuO treatment, AuO enters tumor cell and accumulates in the nucleus of A549 within 2 h (Figure 2), suggesting the DNA binding potential of AuO. However, the nuclear translocation is not observed in bronchial epithelial cell BEAS‐2B up to 4 h, implying normal lung cells might be more resistant to AuO (Figure 2). Further, we conducted the molecular modeling to understand the motif of AuO is essential for DNA interaction. As shown in Figure 3A, the methylamino group is important for DNA binding ability of AuO (A4 compound Ki: 8.2 μM vs. A2 compound 164.9 μM). However, the dimethylamino group is not essential for enhancing the binding affinity (A4 compound 8.2 μM vs. A1 compound 13.4 μM). Interestingly, the iminum group of AuO does not affect the interaction of AuO‐dsDNA (A1 compound 13.4 μM vs. A3 compound 15.9 μM). To analyze the binding affinity of AuO to DNA, isothermal titration calorimetry (ITC) was used for assessing the change in Gibbs free energy during the interaction of AuO and double‐strand DNA through calculating the thermodynamic parameters of interactions in solution. The dissociation constant for AuO and double strand DNA is 7.9 μM (Figure 3B). In summary, these results point out the nuclear translocation capability and DNA binding ability of AuO, especially in lung tumor cells.
Figure 2.
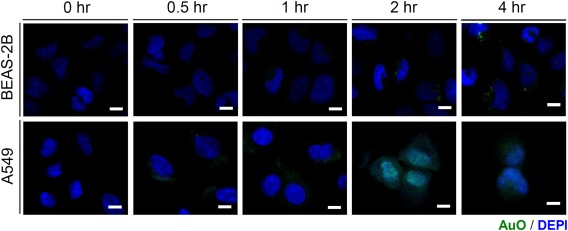
AuO accumulates in the nucleus of A549 cells. Human bronchial epithelial cell BEAS‐2B and lung tumor cell A549 were treated with 10 μM AuO for different times. The accumulation of AuO in cells was observed by confocal microscopy. Scale bars, 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
Figure 3.
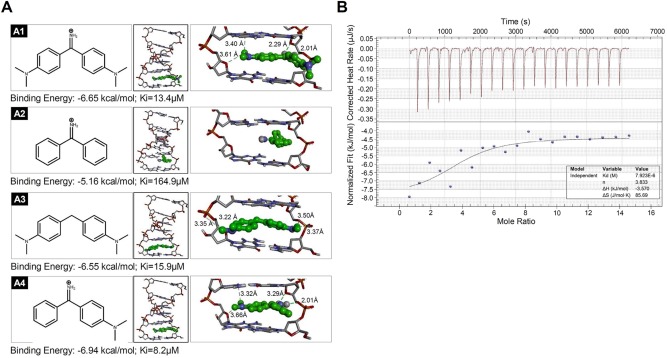
AuO binds the genomic DNA. A, The binding model of four simulated AuO derivatives in DNA. Compound A1–A4 (green ball‐stacked form) gets the docked binding energy values −6.65, −5.16, −6.55, and −6.94 kcal/mol, and Ki values 13.4, 164.9, 15.9, and 8.2 μM, respectively. The binding affinity of compound to DNA is dependent on the hydrogen bond (green dash line) interacted with DNA double helix. Compound A1 forms four hydrogen bonds with bond length 2.01 Å, 2.29 Å, 3.40 Å, and 3.61 Å. Compound A2 does not form any hydrogen bond. Compound A3 forms four hydrogen bonds with bond length 3.22 Å, 3.35 Å, 3.37 Å, and 3.50 Å. Compound A4 forms four hydrogen bonds with bond length 2.01 Å, 3.29 Å, 3.32 Å, and 3.66 Å. B, Isothermal titration calorimetry (ITC) was used to determine the binding of AuO to five pairs of dsDNA (GCs) at pH 7.0. The upper figure shows raw data curve and the down figure shows the fitted integrated data curve [Color figure can be viewed at wileyonlinelibrary.com]
3.3. AuO induces lung tumor cell autophagy
In comparison to normal culture condition, BEAS‐2B cells do not have remarkably morphological change under AuO‐containing culture condition (Figure 4A). However, AuO treatment leads to the granular structures at the surface of A549 cells (Figure 4A). We speculated that the granular structures might be AuO‐induced autophagosomes. To verify this hypothesis, acridine orange was used to stain the autophagosomal vacuoles based on their acidic feature. The AuO‐treated A549 cells exhibit stronger orange to red fluorescent signals compared with untreated control cells, but BEAS‐2B cells do not show significant fluorescent change (Figure 4A). The microtubule‐associated proteins 1 light chain 3B (LC3B) is an essential autophagosomal protein and LC3B puncta represent a convenient marker of autophagy.26 To confirm AuO induces autophagy activity, we compared the protein expression of LC3B in both BEAS‐2B and A549 cells with or without AuO treatment. Confocal images showed that an increase of LC3 puncta is observed in AuO‐treated A549 cells in comparison to the control cells (Figure 4B). Chloroquine (CQ) treatment is used as positive control because CQ inhibits both fusion of autophagosome with lysosome and lysosomal protein degradation, which leads to LC3B accumulation.27 In addition, significantly higher levels of LC3B II/I ratio (a definitive autophagy biomarker) were observed in A549 cells within 5‐day AuO treatment (Figure 4D). However, AuO does not have an obvious effect on enhancing autophagy activity in bronchial epithelial cell BEAS‐2B (Figure 4B,C). These results are consistent with the confocal observations. Collectively, our data demonstrate that AuO exposure increases autophagy activity of lung cancer cell.
Figure 4.
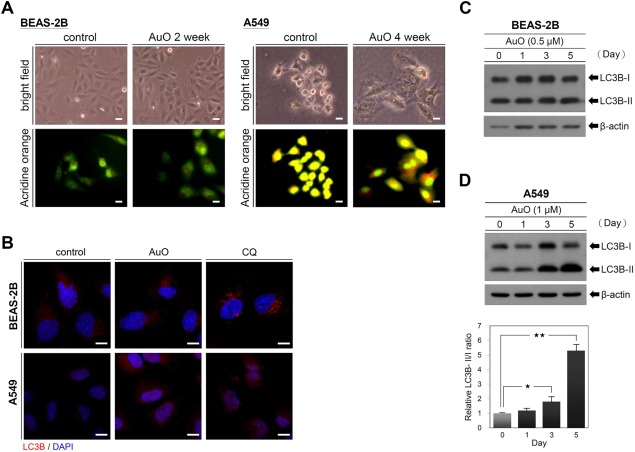
AuO induces autophagy in A549 cells. A, The morphologies of BEAS‐2B and A549 without and with AuO treatment (0.5 μM AuO for BEAS‐2B and 1 μM AuO for A549) were observed by a bright‐field microscopy. The staining of acridine orange was observed by a fluorescent microscopy. B, Confocal images of BEAS‐2B and A549 with antibody to LC3B. Both cells were treated with AuO for 3 days or with 150 μM chloroquine for 2 h. C, Western blot analysis of LC3B in BEAS‐2B. D, Western blot analysis of LC3B in A549. The treated times of AuO as indicated. Relative LC3B‐II/I ratio was analyzed by software image J. β‐actin, loading control [Color figure can be viewed at wileyonlinelibrary.com]
3.4. AuO has no apparent effect on lung cancer initiation
Since AuO is known as a possible carcinogen to humans, we wonder whether AuO promotes lung cancer initiation. To determine the effect of AuO on lung cell transformation, BEAS‐2B cells were treated with various doses of AuO, and the result showed that AuO at 0.5 µM (the maximal concentration) does not dramatically affect cell viability (Figure 5A). After 7‐day AuO treatment, BEAS‐2B cells do not significantly change cell migration and invasive abilities (Figure 5B,C), nor the ability of anchorage‐independent growth in soft agar (Figure 5D). The cell cycle of BEAS‐2B is also unchanged under AuO treatment (Figure 5E). Combined with the previous observations, all our findings suggest that normal lung cells exhibit insensitive to AuO stimulation and AuO could be an ineffective carcinogen to directly induce normal lung cell transformation, at least at the current experimental condition.
Figure 5.
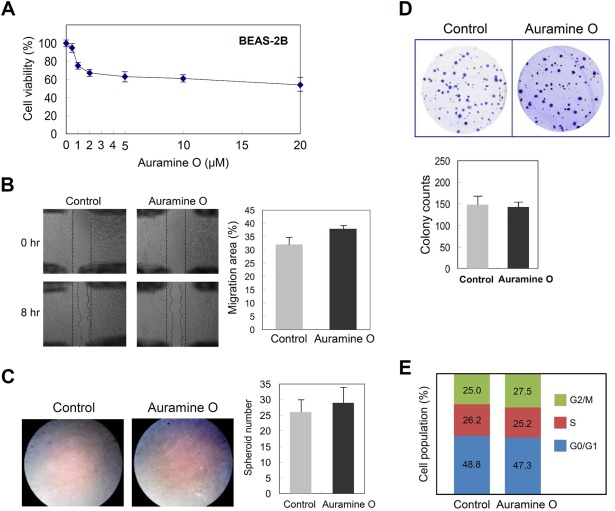
AuO has no apparent effect on promoting neoplastic transformation of BEAS‐2B. A, BEAS‐2B cells were treated with various doses of AuO for 1 day and the cell viability was determined by MTT assay. Before performing the following analyses B to E, BEAS‐2B was pretreated with 0.5 μM AuO for 7 days. B, Wound healing assay to determine the migration ability of cells in vitro. C, Transwell assay to determine the invasion ability of cells in vitro. D, Soft agar assay. BEAS‐2B cells (2 × 104/well) were cultured in 0.3% agarose gel (based agarose gel 0.6%), and cell colonies were stained with crystal violet. E, The cell cycle was analyzed by flow cytometry. The illustration showed cell population in distinct stages of cell cycle. Experiments were performed in triplicate (mean ± SD) [Color figure can be viewed at wileyonlinelibrary.com]
3.5. AuO promotes lung cancer malignancy
Next, we determined whether AuO promotes lung cancer malignancy. Two human lung cancer cell lines A549 and CL1–0 were used to examine their metastatic abilities after AuO treatment. Various doses of AuO were tested and 1 µM AuO was used for the following treatment because this dose shows a limited inhibition to cancer viability (Figure 6A). After 7 culture days, both cancer cells increase their migration and invasive abilities (Figure 6B,C). Moreover, AuO enhances the spheroid formation of lung cancer cells in number and size (Figure 6D), suggesting that AuO could increase the stemness of lung tumor cells. These data indicate that AuO significantly promotes lung cancer malignancy.
Figure 6.
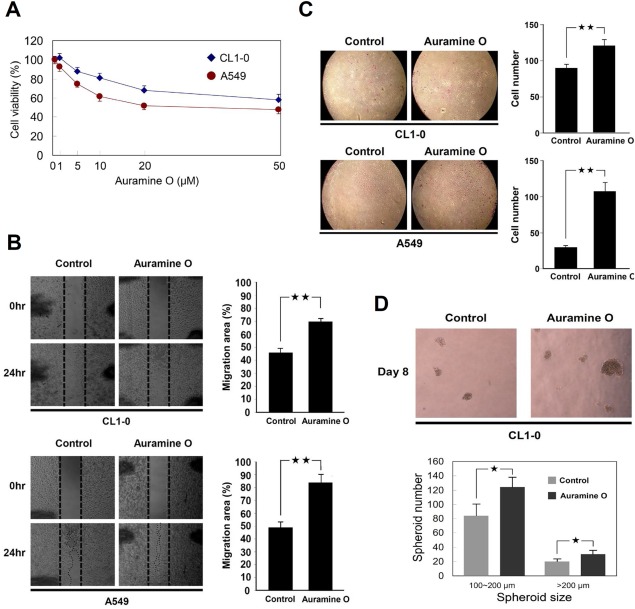
AuO promotes lung cancer malignancy. A, A549 and CL1–0 cells were treated with various doses of AuO for 1 day and the cell viability was determined by MTT assay. After pretreated with 1 μM AuO for 7 days, AuO‐treated and the control A549 and CL1–0 cells were used to perform the following analyses. B, Wound healing assay. C, Transwell assay. D, Spheroid formation assay. Experiments were performed in triplicate (mean ± SD). *P < .05; **P < .01 [Color figure can be viewed at wileyonlinelibrary.com]
3.6. AuO increases the metastatic abilities of lung tumor cells through activating ALDH1A1 expression
To investigate the molecular effect of AuO on lung cancer malignancy, a comparative proteomic analysis was performed to determine the protein changes between AuO‐treated A549 cells and their control cells. Total 2712 proteins are identified, and the overexpressed proteins in AuO‐treated cells with a fold increase of more than five are listed in Table 1. Of these, we noticed that aldehyde dehydrogenase family 1 member A1 (ALDH1A1), which has been characterized as a potential cancer stem cell (CSC) marker of lung cancer and its expression is associated with poor outcome and progression of lung cancer.28, 29 Western blot validation showed that AuO induces ALDH1A1 expression in a dose‐dependent manner (Figure 7A). Although other CSC‐related markers such as CD133 and CD44 were not included in the list, they also showed an increase of protein levels under AuO treatment in western blot assay (Figure 7A). To evaluate the efficiency of AuO in ALDH1A1 induction, the shortest time and minimum dose of AuO treatment were determined in A549 cells. As presented in Figure 7B,C, AuO is able to induce ALDH1A1 at the minimum dose of 5 nM within 1 day, which is an achievable dose for people under daily exposure based on our previous estimation in temple workers.
Table 1.
The overexpressed proteins in AuO‐treated A549 cells in comparison with the control cells
| Uniprot | Protein name | Gene name | Unique no. | Coverage (%) | MW (kDa) | PEPa |
|---|---|---|---|---|---|---|
| P00352 | Aldehyde dehydrogenase family 1 member A1 | ALDH1A1 | 25 | 62.9 | 54.86 | <1.0E‐307 |
| B9EGG0 | Anoctamin‐6 | ANO6 | 3 | 4.4 | 108.43 | 6.19E‐06 |
| P10398 | Serine/threonine‐protein kinase A‐Raf | ARAF | 1 | 4.1 | 67.93 | 3.78E‐03 |
| O75787 | Renin receptor | ATP6AP2 | 5 | 19.7 | 39.01 | 1.93E‐12 |
| O60826 | Coiled‐coil domain‐containing protein 22 | CCDC22 | 4 | 10.4 | 70.76 | 6.81E‐56 |
| Q9UFE4 | Coiled‐coil domain‐containing protein 39 | CCDC39 | 2 | 2.7 | 109.90 | 9.11E‐04 |
| Q8WVB6 | Chromosome transmission fidelity protein 18 homolog | CHTF18 | 3 | 3.6 | 129.40 | 1.99E‐13 |
| Q8N668 | COMM domain‐containing protein 1 | COMMD1 | 3 | 27.4 | 21.18 | 3.23E‐11 |
| Q9H9Q2 | COP9 signalosome complex subunit 7b | COPS7B | 3 | 19.7 | 29.62 | 5.90E‐13 |
| Q13618 | Cullin‐3 | CUL3 | 4 | 9.4 | 88.93 | 1.10E‐40 |
| P63172 | Dynein light chain Tctex‐type 1 | DYNLT1 | 2 | 30.1 | 12.45 | 2.60E‐11 |
| O43432 | Eukaryotic translation initiation factor 4 gamma 3 | EIF4G3 | 5 | 8.5 | 180.50 | 6.38E‐17 |
| Q9NYY8 | FAST kinase domain‐containing protein 2 | FASTKD2 | 3 | 7.0 | 81.46 | 6.04E‐65 |
| Q86VS8 | Protein Hook homolog 3 | HOOK3 | 3 | 5.0 | 83.13 | 6.13E‐09 |
| P56937 | 3‐keto‐steroid reductase | HSD17B7 | 3 | 12.3 | 38.21 | 3.35E‐11 |
| P42858 | Huntingtin | HTT | 7 | 3.6 | 347.86 | 2.18E‐174 |
| Q86VI3 | Ras GTPase‐activating‐like protein IQGAP3 | IQGAP3 | 5 | 5.6 | 184.70 | 2.16E‐13 |
| Q12768 | WASH complex subunit strumpellin | KIAA0196 | 6 | 7.4 | 134.28 | 4.20E‐14 |
| Q92845 | Kinesin‐associated protein 3 | KIFAP3 | 2 | 3.9 | 91.20 | 1.52E‐03 |
| P45985 | Dual specificity mitogen‐activated protein kinase kinase 4 | MAP2K4 | 3 | 17.0 | 44.29 | 1.70E‐16 |
| O95983 | Methyl‐CpG‐binding domain protein 3 | MBD3 | 3 | 14.8 | 32.84 | 8.06E‐11 |
| O15091 | Mitochondrial ribonuclease P protein 3 | MRPP3 | 4 | 8.9 | 67.32 | 1.19E‐04 |
| Q8NC60 | Nitric oxide‐associated protein 1 | NOA1 | 3 | 7.3 | 78.46 | 9.84E‐121 |
| P37198 | Nuclear pore glycoprotein p62 | NUP62 | 3 | 10.0 | 53.25 | 5.08E‐14 |
| P19174 | 1‐Phosphatidylinositol 4,5‐bisphosphate phosphodiesterase gamma‐1 | PLCG1 | 4 | 4.0 | 148.66 | 1.18E‐07 |
| Q8TF05 | Serine/threonine‐protein phosphatase 4 regulatory subunit 1 | PPP4R1 | 4 | 6.7 | 107.00 | 8.23E‐24 |
| P49643 | DNA primase large subunit | PRIM2 | 3 | 7.3 | 58.81 | 1.94E‐04 |
| P34896 | Serine hydroxymethyltransferase, cytosolic | SHMT1 | 3 | 12.4 | 53.08 | 6.27E‐10 |
| Q05519 | Serine/arginine‐rich splicing factor 11 | SRSF11 | 3 | 8.1 | 53.54 | 1.32E‐03 |
| Q8TC07 | TBC1 domain family member 15 | TBC1D15 | 5 | 8.5 | 79.49 | 1.78E‐08 |
| Q00059 | Transcription factor A, mitochondrial | TFAM | 3 | 9.8 | 29.10 | 3.00E‐03 |
| Q9UPU5 | Ubiquitin carboxyl‐terminal hydrolase 24 | USP24 | 14 | 7.7 | 294.36 | 9.70E‐25 |
Posterior error probability (PEP) was obtained from statistical analysis of total peptide identification for a protein in one sample. The value essentially operates as a statistical value, and low PEP indicates high statistical significance.
Figure 7.
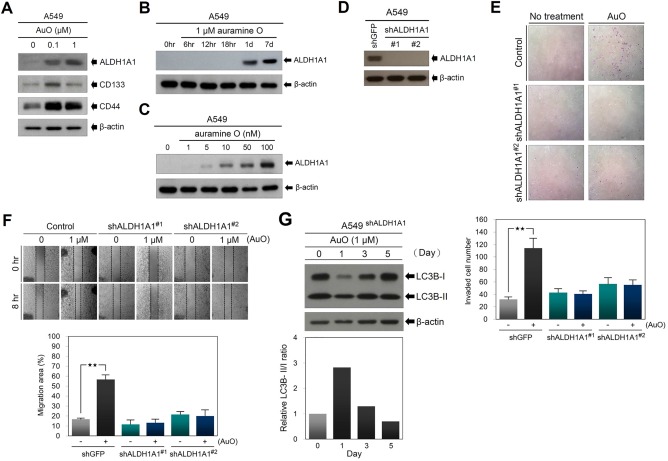
AuO enhances the metastatic abilities of A549 cells through increasing ALDH1A1 expression. A, A549 cells were treated with indicated dose of AuO and the expression of ALDH1A1, CD133, and CD44 was determined by western blot. B, Western blot assay of ALDH1A1 in A549 cells treated with 1 μM AuO at different times. C, Western blot assay of ALDH1A1 in A549 cells treated with various doses of AuO for 1 day. D, Western blot assay of ALDH1A1 in A549 cells was used to validate the shRNA knockdown efficiency. E, Transwell assay was used to determine the effect of ALDH1A1 knockdown on cell invasion of A549. F, Wound healing assay was used to determine the effect of ALDH1A1 knockdown on cell migration of A549. G, Western blot analysis of LC3B in AuO‐treated A549 cells with ALDH1A1 knockdown. β‐actin, loading control. **P < .01 [Color figure can be viewed at wileyonlinelibrary.com]
To address whether AuO increases lung cancer metastatic abilities through ALDH1A1 activation, small hairpin RNAs (shRNAs) were used to knock down ALDH1A1 expression in A549 cells. Western blot confirmed that two shRNAs inhibit AuO‐induced ALDH1A1 expression (Figure 7D) and significantly abolish AuO‐induced migration and invasive abilities in A549 cells (Figure 7E,F). Moreover, because autophagy has been shown to be involved in modulating tumor cell motility and invasion,30 we checked the effect of ALDH1A1 knockdown on AuO‐induced autophagy activity. Compared with the data in Figure 4D, ALDH1A1 knockdown attenuates AuO‐induced autophagy activity (Figure 7G). These results suggest that ALDH1A1 plays an important role in mediating AuO‐induced lung cancer metastasis.
3.7. AuO enhances the stemness of lung tumor cells in vitro and in vivo through the induction of ALDH1A1 expression
To determine whether ALDH1A1 is essential for AuO‐induced stemness features, we examined the effect of ALDH1A1 knockdown on in vitro stemness character by spheroid formation assay. The result showed that the spheroid number and size of A549 cells are significantly attenuated by ALDH1A1 knockdown (Figure 8A). To evaluate whether AuO increases in vivo tumorigenesis, we subcutaneously injected control (shGFP) and AuO‐pretreated A549 cells into both flanks of immunodeficient mice and monitored tumor growth. The data showed that AuO treatment promotes tumorigenesis of A549 cells and increases the speed of tumor growth (Figure 8B,C). Additionally, ALDH1A1 has higher expression levels in tissues of AuO‐treated mice in comparison with the control in the IHC analysis (Figure 8D). Taken together, these results demonstrate that AuO enhances the stemness characters of lung tumor cell by increasing ALDH1A1 expression.
Figure 8.
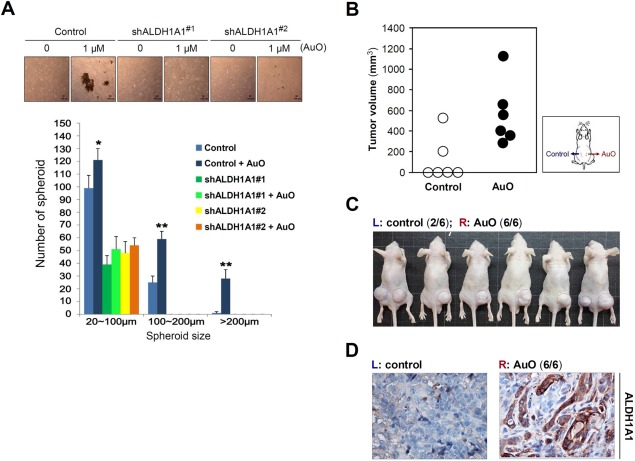
AuO enhances the stemness feature of A549 cells. A, spheroid formation assay was used to determine the effect of ALDH1A1 knockdown on in vitro stemness feature of A549 cells. Various sizes of spheroids in A549 cells with and without AuO treatment were counted at day 8. *P < .05; **P < .01. B, AuO‐treated and the control A549 cells (1 × 106) were subcutaneously injected into both flanks of 5‐week‐old male BALB/c nude mice (N = 6) as indicated. Tumor volume was calculated with the formula: (length × width2)/2. The tumor volumes show the measurement after 6‐week injection. C, The formed tumors in experimental mice after 6‐week injection. D, Representative IHC images of ALDH1A1 expression in AuO‐treated and the control tumor tissues. [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
AuO is a multifunctional dye used in the coloring of paper, textile, leather, incense as well as acid‐fast bacteria such as Mycobacterium tuberculosis.31 Due to low price, high intensive color, and excellent stability, AuO has been found to extensively add in foodstuff and pharmaceuticals as an illegal additive. The abuse of AuO is regarded as an important public health issue because of its carcinogenic potential.15 Therefore, AuO is forbidden in food matrices by many international food regulation acts, including European Union,32 China,33 Japan,34 and the United States.35 Meanwhile, many sensitive and rapid methods are developed for the detection and removal of AuO to ensure a safe living environment.36, 37, 38 However, whether AuO damages the respiratory system remains fully unknown. In this study, we identified AuO from water‐soluble fraction of IBC as unexpected air pollution. AuO is able to induce the stemness of lung tumor cells at the minimum concentration of 5 nM within 1 day culture. Based on our measurement and estimation of AuO exposure from temple workers, their maximum accumulation concentration of AuO is 289 nM in blood in 1 day, arguing that the environmental exposure of AuO could dramatically cause damage to the respiratory system.
Previous studies indicated that AuO causes DNA damage after in vitro treatment in culture cells and oral administration in rats and mice.39, 40 Different from previously experimental designs, low dose of AuO is used for mimicking an achievable exposure dosage in this study, consequently cell viability is not affected nor DNA damage is observed. Whether the damage effect is caused by direct AuO‐DNA interaction remains unclear. Recently, AuO is demonstrated to selectively bind the G‐quadruplex structure of DNA with higher affinity than single or double strand DNA.41 G‐quadruplex is a square planar structure formed in guanine rich nucleic acids such as telomeres through Hoogsteen hydrogen bond.42 Recent studies implicated that G‐quadruplex involves in the protection of telomere ends from nuclease attack,43 the programmed recombination of immunoglobin genes,44 and both positive and negative transcriptional regulation.45 This information suggests a potential mechanism that AuO induces ALDH1A1 expression through regulating the G‐quadruplex of ALDH1A1 gene. Using QGRS Mapper46 to predict the potential G‐quadruplex sequence, we indeed find G‐quadruplex sequences located at the promoter of ALDH1A1. However, the precise mechanism remains to be investigated.
Autophagy is an important mechanism used to clear pathogenic organism and deal with environmental stress such as starvation for maintaining cellular homeostasis. During the transformation of normal cells, autophagy protects genomic stability and inhibits the formation of chronic inflammatory microenvironment, thus preventing tumor generation.47 Autophagy has a complex and context‐dependent role in tumor cells, that is, autophagy suppresses primary tumor growth but is required for tumor maintenance and progression to advanced disease.48 Additionally, recent investigations have suggested that autophagy promotes multiple steps in the metastatic cascade.30 In this study, we find that AuO induces autophagy activity (Figure 3) as well as increasing the metastatic abilities of lung tumor cells to promote lung cancer malignancy, suggesting a positive correlation between autophagy and cancer metastasis, which is consistent with the current knowledge. Autophagy activity is not significantly induced in the bronchial epithelial cell line BEAS‐2B under low dose of AuO treatment (final 0.5 μM). Thus, we cannot evaluate whether the protective effect of autophagy involves in preventing AuO‐induced transformation or normal lung cell is intrinsically resistant to AuO. Does autophagy keep its protective ability or DNA damage cause malignant transformation in normal lung cell under heavy AuO exposure? These questions need to be further explored.
CSC defined as a subset of tumor cells with self‐renewability involves in cancer initiation and progression. CSC is also associated with the resistance to radiation and chemotherapy as well as the metastasis of carcinoma,49, 50 thus increased CSC population has been viewed as a poor prognosis indicator of cancer patients.51 ALDH1A1 overexpression has been known to associate with lung cancer malignancy.29 Our study reveals that ALDH1A1 plays an important role in mediating AuO‐induced stemness features and metastatic abilities of lung tumor cells, whereas ALDH1A1 knockdown significantly blocks these effects. In clinic, ALDH1A1 overexpression might be relevant to AuO exposure, especially in Asian countries with the custom of burning incense. Their correlation could be addressed clearly through epidemiological study in the future. In recent years, accumulating efforts have been put to develop drugs for targeting CSC.52 Since ALDH1A1 is crucial for AuO‐induced lung malignancy, ALDH1A1 targeting therapy could be benefit to the patients suffering from AuO exposure.53, 54
In conclusion, we identify AuO released from incense smoke as an unexpected air pollutant due to its thermostability. The damage of AuO to the respiratory system has never been awakened. In this study, we find that AuO promotes lung cancer malignancy through increasing autophagy activity and the stemness of lung cancer cells. AuO is currently used as an additive colorant in incense manufacture, to some degree meaning the control of its usage could be accessible. For public health, reducing or avoiding the use of AuO in incense is recommended.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Supporting Information
ACKNOWLEDGMENTS
We thank the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) for providing the shRNAs. We are grateful to the National Center for High‐Performance Computing for computer time and facilities. We also thank the Center for Resources, Research, and Development of Kaohsiung Medical University for the ChemBioOffice technical support.
Tung J‐C, Huang W‐C, Yang J‐C, et al. Auramine O, an incense smoke ingredient, promotes lung cancer malignancy. Environmental Toxicology. 2017;32:2379–2391. https://doi.org/10.1002/tox.22451
Contributor Information
Shih‐Chun Candice Lung, Email: sclung@rcec.sinica.edu.tw.
Wei‐Chao Chang, Email: T21443@mail.cmuh.org.tw.
REFERENCES
- 1.Pew Research Center. The Future of World Religions: Population Growth Projections, 2010‐2050. 2015; http://www.pewforum.org/2015/04/02/religious-projections-2010-2050/.
- 2. Chiang KC, Liao CM. Heavy incense burning in temples promotes exposure risk from airborne PMs and carcinogenic PAHs. Sci Total Environ. 2006;372:64–75. [DOI] [PubMed] [Google Scholar]
- 3. Chuang HC, BéruBé K, Lung SC, Bai KJ, Jones T. Investigation into the oxidative potential generated by the formation of particulate matter from incense combustion. J Hazard Mater. 2013;244:142–150. [DOI] [PubMed] [Google Scholar]
- 4. Lung SC, Guo KJ, Chen PY, Tsai PF, Chen PC. Participants' exposure to PM2.5 and gaseous/particulate polycyclic aromatic hydrocarbons during the Ma‐tsu Goddess parade. J Expo Anal Environ Epidemiol. 2004;14:536–543. [DOI] [PubMed] [Google Scholar]
- 5. Wang B, Lee SC, Ho KF, Kang YM. Characteristics of emissions of air pollutants from burning of incense in temples, Hong Kong. Sci Total Environ. 2007;377:52–60. [DOI] [PubMed] [Google Scholar]
- 6. Lui KH, Bandowe BA, Ho SS, et al. Characterization of chemical components and bioreactivity of fine particulate matter (PM2.5) during incense burning. Environ Pollut. 2016;213:524–532. [DOI] [PubMed] [Google Scholar]
- 7. Lin TC, Chang FH, Hsieh JH, Chao HR, Chao MR. Characteristics of polycyclic aromatic hydrocarbons and total suspended particulate in indoor and outdoor atmosphere of a Taiwanese temple. J Hazard Mater. 2002;95:1–12. [DOI] [PubMed] [Google Scholar]
- 8. Yang CR, Lin TC, Chang FH. Particle size distribution and PAH concentrations of incense smoke in a combustion chamber. Environ Pollut. 2007;145:606–615. [DOI] [PubMed] [Google Scholar]
- 9. Chiang KC, Chio CP, Chiang YH, Liao CM. Assessing hazardous risks of human exposure to temple airborne polycyclic aromatic hydrocarbons. J Hazard Mater. 2009;166:676–685. [DOI] [PubMed] [Google Scholar]
- 10. Dewangan S, Chakrabarty R, Zielinska B, Pervez S. Emission of volatile organic compounds from religious and ritual activities in India. Environ Monit Assess. 2013;185:9279–9286. [DOI] [PubMed] [Google Scholar]
- 11. Manoukian A, Quivet E, Temime‐Roussel B, Nicolas M, Maupetit F, Wortham H. Emission characteristics of air pollutants from incense and candle burning in indoor atmospheres. Environ Sci Pollut Res Int. 2013;20:4659–4670. [DOI] [PubMed] [Google Scholar]
- 12. Manoukian A, Buiron D, Temime‐Roussel B, Wortham H, Quivet E. Measurements of VOC/SVOC emission factors from burning incenses in an environmental test chamber: influence of temperature, relative humidity, and air exchange rate. Environ Sci Pollut Res Int. 2016;23:6300–6311. [DOI] [PubMed] [Google Scholar]
- 13. Seow WJ, Lan Q. Domestic incense use and lung cancer in Asia: a review. Rev Environ Health. 2016;31:155–158. [DOI] [PubMed] [Google Scholar]
- 14. Williams MH, Bonser GM. Induction of hepatomas in rats and mice following the administration of auramine. Br J Cancer 1962;16:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martelli A, Campart GB, Canonero R, et al. Evaluation of auramine genotoxicity in primary rat and human hepatocytes and in the intact rat. Mutat Res. 1998;414:37–47. [DOI] [PubMed] [Google Scholar]
- 16. International Agency for Research on Cancer . Auramine Auramine Prod. 2012;110:101–104. [Google Scholar]
- 17. Lung SC, Hu SC. Generation rates and emission factors of particulate matter and particle‐bound polycyclic aromatic hydrocarbons of incense sticks. Chemosphere 2003;50:673–679. [DOI] [PubMed] [Google Scholar]
- 18. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 19. Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 2014;13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Canals A, Purciolas M, Aymamí J, Coll M. The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs. Acta Crystallogr D Biol Crystallogr. 2005;61:1009–1012. [DOI] [PubMed] [Google Scholar]
- 22. Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model 1999;17:57–61. [PubMed] [Google Scholar]
- 23. Freitas RF, Prokopczyk IM, Zottis A, et al. Discovery of novel Trypanosoma cruzi glyceraldehyde‐3‐phosphate dehydrogenase inhibitors. Bioorg Med Chem. 2009;17:2476–2482. [DOI] [PubMed] [Google Scholar]
- 24. Chou CK, Fan CC, Lin PS, et al. Sciellin mediates mesenchymal‐to‐epithelial transition in colorectal cancer hepatic metastasis. Oncotarget 2016;7:25742–25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lung SC, Kao MC. Worshippers' exposure to particulate matter in two temples in Taiwan. J Air Waste Manage Assoc. 2003;53:130–135. [DOI] [PubMed] [Google Scholar]
- 26. Chauhan S, Ahmed Z, Bradfute SB, et al. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat Commun. 2015;6:8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shintani T, Klionsky DJ. Autophagy in health and disease: a double‐edged sword. Science 2004;306:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alamgeer M, Ganju V, Szczepny A, et al. The prognostic significance of aldehyde dehydrogenase 1A1 (ALDH1A1) and CD133 expression in early stage non‐small cell lung cancer. Thorax 2013;68:1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hashida S, Yamamoto H, Shien K, et al. Acquisition of cancer stem cell‐like properties in non‐small cell lung cancer with acquired resistance to afatinib. Cancer Sci. 2015;106:1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene 2016;36:1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng AG, Chang A, Farwell DG, Agoff SN. Auramine orange stain with fluorescence microscopy is a rapid and sensitive technique for the detection of cervical lymphadenitis due to mycobacterial infection using fine needle aspiration cytology: a case series. Otolaryngol Head Neck Surg. 2005;133:381–385. [DOI] [PubMed] [Google Scholar]
- 32. European Food Safety Authority . Opinion of the scientific panel on food additives, flavourings. Processing aids and materials in contact with food on a request from the commission to review the toxicology of a number of dyes illegally present in food in the EU. EFSA J. 2005;263:1–71., [Google Scholar]
- 33.Chinese Standards for Food Additives. GB 2760–2011. People's Republic of China: Chinese Standards for Food Additives; 2011.
- 34. Japan Society for Occupational Health . Recommendation of occupational exposure limits (2007–2008). J Occup Health 2007;49:328–344. [PubMed] [Google Scholar]
- 35.U.S. FDA. Summary of Color Additives Listed for Use in the United States in Food, Drugs, Cosmetics and Medical Devices. 2004.
- 36. Tatebe C, Zhong X, Ohtsuki T, Kubota H, Sato K, Akiyama H. A simple and rapid chromatographic method to determine unauthorized basic colorants (rhodamine B, auramine O, and pararosaniline) in processed foods. Food Sci Nutr. 2014;2:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghaedi M, Khodadoust S, Sadeghi H, Khodadoust MA, Armand R, Fatehi A. Application of ultrasonic radiation for simultaneous removal of auramine O and safranine O by copper sulfide nanoparticles: experimental design. Spectrochim Acta A Mol Biomol Spectrosc. 2015;136(PtB):1069–1075. [DOI] [PubMed] [Google Scholar]
- 38. Yan J, Huang X, Liu S, et al. A simple and sensitive method for auramine O detection based on the binding interaction with bovin serum albumin. Anal Sci. 2016;32:819–824. [DOI] [PubMed] [Google Scholar]
- 39. Parodi S, Santi L, Russo P, et al. DNA damage induced by auramine O in liver, kidney, and bone marrow of rats and mice, and in a human cell line (alkaline elution assay and SCE induction). J Toxicol Environ Health 1982;9:941–952. [DOI] [PubMed] [Google Scholar]
- 40. Kitchin KT, Brown JL. Dose‐response relationship for rat liver DNA damage caused by 49 rodent carcinogens. Toxicology 1994;88:31–49. [DOI] [PubMed] [Google Scholar]
- 41. Xu H, Geng F, Wang Y, et al. A label‐free fluorescent molecular switch for a DNA hybridization assay utilizing a G‐quadruplex‐selective auramine O. Chem Commun (Camb). 2015;51:8622–9625. [DOI] [PubMed] [Google Scholar]
- 42. Smith FW, Feigon J. Quadruplex structure of oxytricha telomeric DNA oligonucleotides. Nature 1992;356:164–168. [DOI] [PubMed] [Google Scholar]
- 43. Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G‐quadruplex structures. Nat Rev Genet. 2012;13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siddiqui‐Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G‐quadruplex in a promoter region and its targeting with a small molecule to repress c‐MYC transcription. Proc Natl Acad Sci. USA 2002;99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kikin O, D'antonio L, Bagga PS. QGRS Mapper: a web‐based server for predicting G‐quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun K, Deng W, Zhang S, et al. Paradoxical roles of autophagy in different stages of tumorigenesis: protector for normal or cancer cells. Cell Biosci. 2013;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rao S, Tortola L, Perlot T, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056–3070. [DOI] [PubMed] [Google Scholar]
- 49. Charafe‐Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1‐positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007;1:313–323. [DOI] [PubMed] [Google Scholar]
- 51. Mertins SD. Cancer stem cells: a systems biology view of their role in prognosis and therapy. Anticancer Drugs 2014;25:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khan IN, Al‐Karim S, Bora RS, Chaudhary AG, Saini KS. Cancer stem cells: a challenging paradigm for designing targeted drug therapies. Drug Discov Today 2015;20:1205–1216. [DOI] [PubMed] [Google Scholar]
- 53. Liu X, Wang L, Cui W, et al. Targeting ALDH1A1 by disulfiram/copper complex inhibits non‐small cell lung cancer recurrence driven by ALDH‐positive cancer stem cells. Oncotarget 2016;7:58516–58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Supporting Information


