Summary
Antimicrobial resistance is known to be an emerging problem, but the extent of the issue remains incomplete. The aim of this study was to determine the presence or absence of nine resistance genes (bla TEM, catI, mecA, qnrS, sulI, sul II, tet(A), tet(Q), vanA) in the faeces of 141 pigeons from four urban parks in Alajuela, Guadalupe, Tres Ríos and San José in Costa Rica. The genes were identified by real‐time PCR directly from enema samples. About 30% of the samples were positive for genes catI and sulI; between 13% and 17% were positive for qnrS, sul II, tet(A) and tet(Q); and 4% were positive for bla TEM. The mecA and vanA genes were not detected. The average of antimicrobial resistance genes detected per pigeon was 2. Eight different patterns of resistance were identified, without differences in the sampling areas, being the most common pattern 2 (sul II positive samples). During rainy season, the genes more frequently found were sulI and tet(A). In conclusion, the urban inhabiting pigeons tested are currently carrying antimicrobial resistance genes, potentially acting as reservoirs of resistant bacteria and vectors to humans. To the authors’ knowledge, this is the first study carried out on direct detection of resistance genes in the digestive metagenomes of pigeons.
Keywords: Antimicrobial resistances, antimicrobial resistance genes, Costa Rica, Columba livia, urban parks, ecosystem health
Impacts.
This work provides information about the presence of antimicrobial resistance genes in four urban areas in Costa Rica, taking pigeons as biomarkers, considering that they share their habitat with people.
Pigeons can carry antimicrobial resistance genes, potentially acting as reservoirs of resistant bacteria and vectors to humans.
This work provides information on potential routes of environmental exposure of pigeons and humans through them.
Introduction
Since the discovery of the first antibiotics, the extent of antimicrobial resistances (AMR) has increased in a variety of habitats, including animal feeding systems (Durso et al., 2011), municipal waste streams (Nagulapally et al., 2009) and environments with little or no human impact (Durso et al., 2012). Because of its impact on public health, this has been considered as an emerging global problem (WHO, 2014). The Pan American Health Organization (PAHO) has pointed out that there is an urgent need to strengthen surveillance and monitoring the use of antimicrobial agents in different activities (Acar and Moulin, 2013).
One of the most important mechanisms for the acquisition of AMR is the incorporation of mobile genetic elements, mainly antibiotic resistance genes (ARGs), which facilitate horizontal gene transfer between bacteria (Mazodier and Davies, 1991). This can be caused by different environmental factors, given that they are not necessarily linked to exposure to antibiotics. The rate of development of microbial resistance is more rapid in environments where the use of antibiotics is greater (van de Sande‐Bruinsma et al., 2008), as in human medicine, veterinary medicine, production systems and agriculture (Kümmerer, 2009).
Generally used diagnostic methods that involve bacterial culture and subsequent detection of ARGs in the isolates could affect the results of analysis because non‐culturable fractions include most of the microbial community (only 0.1–1% of soil bacteria can be grown in laboratory conditions) (Torsvik and Øvreås, 2002). This hinders the understanding of potential negative effects of AMR on the ecosystem and on human and animal health, and is the reason for which some authors recommend genetic studies rather than those based only on cultures (Jiang et al., 2013; Marti et al., 2013).
Urbanization is a continuous process that leads to different densities and patterns of human settlements, reducing and fragmenting the native vegetation, and modifying communities of resident fauna (Marzluff and Ewing, 2001). In Costa Rica, about 60% of the population lives in the so‐called greater metropolitan area (GMA), where four conurbations are located (San José, Alajuela, Cartago and Heredia), and represents just over 4% of the area of the country. It is the most urbanized, populated and economically active region of Costa Rica and it is characterized by urban sprawl, overcrowding and low quality of urban space (INEC, 2012). Thirty‐one per cent of national and specialized hospitals of the country are located in the GMA. However, in the country 72% of the population uses septic tanks and only 3.6% of sewage is treated in centralized plants. This contamination could influence the finding of ARGs in other scenarios in the country (Rodriguez et al., 2006), including urban ecosystems.
Birds are a good choice for monitoring urban ecosystems because they can be surveyed on a large scale, their occurrence and abundance is influenced by habitat characteristics, and they are easy to see and attractive to the population (Carignan and Villard, 2002). The Columbia livia pigeon is common in cities in many countries and can transmit more than 30 diseases to humans through the air or their excreta (Weber et al., 1995), although, until now, it is unknown whether ARGs can be spread by their faeces.
Numerous studies about AMR in humans and domestic animals are currently underway, but little is known about its presence in animals and birds living in an environment not controlled by man. The objective of this research is to identify ARGs directly in faeces from pigeons that inhabit urban parks, which could serve as indicators of pollution.
Materials and Methods
The study was conducted in the towns of Alajuela, Guadalupe, Tres Ríos and San José, located in the central region of Costa Rica (GMA), belonging to three of the provinces with the largest urban populations (coordinates: 476 538.952 to 1 107 612.622 m, 491 550.327 to 1 098 323.532 m, 494 116.374 to 1 099 811.839 m, 501 609.231 to 1 095 552.201 m). The capture of 141 birds was carried out in two different terms between March 2013 and May 2014 in the parks of Guadalupe (55), San José (44), Alajuela (29) and Tres Ríos (16), using a net and corn or peanuts as bait (Table 1). They were immediately transferred to the Veterinary School of the Universidad Nacional de Costa Rica, where enema samples were taken approximately 1.5 h after the capture introducing 0.5 ml of sterile DNAse‐ and Rnase‐free phosphate‐buffered saline (PBS) into the cloaca, and then retrieving the PBS together with the cloacal contents. The recovered suspension was diluted with PBS to yield a final volume of 2 ml. All samples were stored at −80°C until they were analysed. All procedures were performed according to national standards of environmental well‐being, permission #EMV‐CBA‐06‐2012. Little is known about ARG movement in the environment through wildlife species. For this reason, studies based on culture‐independent methods are strongly recommended (Allen et al., 2010).
Table 1.
Results obtained about the presence of ARGs in enemas from pigeons
| Location | N | β‐lactamases† | Chloramphenicol† | Methicillin (MRSA gene)‡ | Fluoroquinolones§ | Sulphonamides† | Tetracyclines† | Vancomycin† | ANG*/ sample | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % bla TEM | % catI | % mecA | % qnrS | % sulI | % sulII | % tet(A) | % tet(Q) | % vanA | |||
| San José | 41 | 9.8 | 31.7 | 0.0 | 9.8 | 39.0 | 19.5 | 22.0 | 14.6 | 0.0 | 2.46 |
| Alajuela | 29 | 0.0 | 24.1 | 0.0 | 13.8 | 48.3 | 17.2 | 3.4 | 10.3 | 0.0 | 2.17 |
| Tres Ríos | 16 | 0.0 | 31.3 | 0.0 | 18.8 | 12.5 | 31.3 | 43.8 | 12.5 | 0.0 | 2.50 |
| Guadalupe | 55 | 3.6 | 29.1 | 0.0 | 14.5 | 20.0 | 12.7 | 5.5 | 14.5 | 0.0 | 2.00 |
| Total | 141 | 4.3 | 29.1 | 0.0 | 13.5 | 30.5 | 17.7 | 14.2 | 13.5 | 0.0 | 2.23 |
DNA was extracted directly from all enema samples with the QuickGene DNA Tissue Kit S (Fujifilm, Japan) following the manufacturer's instructions. The 16S rRNA gene was identified to detect the presence of bacterial genetic material in each sample, following the methodology proposed by Doi and Arakawa (2007). A sample was considered validated when a 10‐fold dilution showed a cycle threshold (ct) less than 30. ARGs were detected by real‐time PCRs, using SYBR Green in all of them, except for the gene mecA, where a TaqMan probe was used (Francois et al., 2003; Jiang et al., 2013; Marti and Balcazar, 2013). The ARGs analysed (bla TEM, catI, mecA, qnrS, sulI, sulII, tet(A), tet(Q), vanA) were selected as representatives of some of the leading groups of antimicrobial agents (Table 1).
In order to investigate the patterns of resistance, a k‐means clustering method was applied (genesis software vs 1.7.7, Graz University of Technology, Graz, Austria). Each sample was assigned to one cluster. Since multiresistant bacteria are defined by the resistance to more than two different classes of antimicrobials (Karczmarczyk et al., 2011; Sacristán et al., 2014), we apply the term ‘multiresistant microbiome’ when a faecal sample present at least three ARG encoding resistance to different groups of antimicrobials. Thus, samples were also classified into ‘multiresistant microbiome’ and ‘non‐multiresistant microbiome’. Besides, a nonparametric test (Mann–Whitney U‐test) was applied (spss vs. 14.0) to establish differences among sampling areas for the following parameters: presence of each ARG, average number of ARGs per sample, resistance pattern and multiresistance. Sampling dates were classified into dry (December to April) and wet (May to November) seasons, and the same nonparametric test was performed. Additionally, all the hospitals, clinics, contaminated rivers and vegetable markets whose crop farming has been associated with the use of antibiotics (Rodriguez et al., 2006) were identified in a radius of 3 km from every park (Table 2), in order to identify possible routes of infection to the pigeons.
Table 2.
Characterization of the area within a radius of 3 km from the parks
| Park | Population living in the canton where the park is located* | Human hospitals or clinics | Rivers contaminated with faecal matter |
|---|---|---|---|
| San José | 336 792 | 8 | 2 |
| Guadalupe | 133 557 | 5 | 1 |
| Alajuela | 297 879 | 2 | 2 |
| Tres Rios | 107 755 | 1 | 2 |
*National Institute of Statistics and Censuses of Costa Rica, INEC (2012).
Results and Discussion
Enema samples from 141 pigeons were obtained, and in all of them the 16S rRNA gene was detected. Of the nine ARGs searched for, seven were found in at least one sample; catI and sulI were the most common (29.1% and 30.5%, respectively), followed by sulII (17.7%), tet(A) (14.2%), qnrS (13.5%), tet(Q) (13.5%) and bla TEM (4.3%). vanA and mecA genes were not detected. The average of antimicrobial resistance genes detected per sample was two, which is low compared with findings reported in aquatic resources, where nine of 11 ARGs were identified in all samples (Table 1) (Marti et al., 2013).
The k‐means clustering identified eight different patterns (Table 3). No differences were found between sample locations and the distribution on different clusters. Cluster 1 was the most frequent (23.4%) and represents all the samples negative to all ARGs. Cluster 2 includes all the sulII positive samples, being the third most frequent (17.7%), and represents the 80% of the total of ‘multiresistant microbiomes’. Multiresistance was associated with sul genes as well as tet(A) and qnrS, also with season. All the multiresistant microbiomes were detected during wet season (12.7%), when a higher percentage of positive samples to sulI (40.5%) and tet(A) (21.5%) was observed. In addition, a higher average number of ARGs per sample (1.4) was found during rainy season than in dry season (17.7%, 4.8% and 0.9, respectively). These findings match with a peak in respiratory and digestive bacterial infections that normally occur in this season in the country.
Table 3.
Patterns of resistance obtained in the enema samples by k‐means clustering
| Cluster | Resistance pattern | Location | No. samples included in the pattern / No. total samples (%) | Multiresistant microbiomes (%)* |
|---|---|---|---|---|
| 1 | No resistance | Alajuela | 9/29 (31.0) | 0 (0) |
| Guadalupe | 13/55 (23.6) | 0 (0) | ||
| San José | 9/41 (22.0) | 0 (0) | ||
| Tres Ríos | 2/16 (12.5) | 0 (0) | ||
| Total | 33/141 (23.4) | 0 (0) | ||
| 2 | sulII + | Alajuela | 5/29 (17.2) | 2 (20) |
| Guadalupe | 7/55 (12.7) | 0 (0) | ||
| San José | 8/41 (19.5) | 6 (60) | ||
| Tres Ríos | 5/16 (31.3) | 0 (0) | ||
| Total | 25/141 (17.7) | 8 (80) | ||
| 3 | catI +, qnrS/sulII/tet(A)/tet(Q) − | Alajuela | 5/29 (17.2) | 0 (0) |
| Guadalupe | 12/55 (21.8) | 0 (0) | ||
| San José | 7/41 (17.1) | 0 (0) | ||
| Tres Ríos | 4/16 (25.0) | 0 (0) | ||
| Total | 28/141 (19.9) | 0 (0) | ||
| 4 | qnrS +, sulI/sulII − | Alajuela | 1/29 (3.4) | 0 (0) |
| Guadalupe | 5/55 (9.1) | 0 (0) | ||
| San José | 2/41 (4.9) | 1 (10) | ||
| Tres Ríos | 2/16 (12.5) | 0 (0) | ||
| Total | 10/141 (7.1) | 1 (10) | ||
| 5 | tet(Q) +, qnrS/sulII − | Alajuela | 2/29 (6.9) | 0 (0) |
| Guadalupe | 8/55 (14.5) | 0 (0) | ||
| San José | 4/41 (9.8) | 0 (0) | ||
| Tres Ríos | 1/16 (6.3) | 0 (0) | ||
| Total | 15/141 (10.6) | 0 (0) | ||
| 6 | tet(A) +, qnrS/sulII/tet(Q) − | Alajuela | 1/29 (3.4) | 0 (0) |
| Guadalupe | 2/55 (3.6) | 0 (0) | ||
| San José | 2/41 (4.9) | 0 (0) | ||
| Tres Ríos | 1/16 (6.3) | 0 (0) | ||
| Total | 6/141 (4.3) | 0 (0) | ||
| 7 | sulI +, catI/sulII/tet(Q) − | Alajuela | 6/29 (20.7) | 0 (0) |
| Guadalupe | 7/55 (12.7) | 0 (0) | ||
| San José | 8/41 (19.5) | 0 (0) | ||
| Tres Ríos | 1/16 (6.3) | 1 (10) | ||
| Total | 22/141 (15.6) | 1 (10) | ||
| 8 | bla TEM +, rest − | Alajuela | 0/29 (0) | 0 (0) |
| Guadalupe | 1/55 (1.8) | 0 (0) | ||
| San José | 1/41 (2.4) | 0 (0) | ||
| Tres Ríos | 0/16 (0) | 0 (0) | ||
| Total | 2/141 (1.4) | 0 (0) |
*Number of ‘multiresistant microbiomes’. The percentage was calculated from the total number of ‘multiresistant microbiomes’ (n = 10).
To date, few studies have analysed the genetic level of AMR in wildlife and most of them differ in the genes monitored and in the diagnostic methodology used, normally based on culture, which constitutes a limiting factor in the interpretation of results (Francois et al., 2003; Jiang et al., 2013; Marti et al., 2013). This is the case of previous research with domestic pigeons (Poeta et al., 2005; Sacristán et al., 2014), in which, as in the present study, the resistance to tetracycline highlights the widespread distribution among different ecological niches. A higher percentage of animals with resistance to sulphonamides, chloramphenicol and fluoroquinolones was found in this study compared to data reported by Sacristán et al. (2014) (18.9% sulphamethoxazole, 14% trimethoprim‐sulphamethoxazole, 4.9% chloramphenicol and 0.6% ciprofloxacin) and Poeta et al. (2005) (0% chloramphenicol and 8.6% ciprofloxacin). Sulphonamide resistance has been associated with human activities (Radimersky et al., 2010), which may explain the findings in all city parks. The case of the phenicols is interesting, because in Costa Rica its use is limited to human medicines, and chloramphenicol is banned in all species producing food for human consumption (Rojas Martínez, 2009). So, it is necessary to identify the pathways of contamination with this antibiotic in future studies.
Results for the representatives of the β‐lactam group were very different from those of the studies cited above: 19.5% for ampicillin (Sacristán et al., 2014), 0% for ampicillin (Poeta et al., 2005) and in the present research (4.3%). Other authors found that the close contact with antimicrobial products or by‐products derivate of human activities (i.e. human and veterinary medicine) increases the number of antibiotic resistance in wild and domestic animals (Guardabassi et al., 2004; Dolejska et al., 2007). No methicillin‐resistant genes were found (0% mecA) in the sampled areas, which is highly important for public health in these spaces (Grundmann et al., 2006); this is different from the amount reported for birds of prey in Portugal (Sousa et al., 2014).
Although fluoroquinolone resistance can be regarded as higher than that detected in previous studies, it is less than that reported in E. coli isolates among diseased pigeons farmed in China (21.9% qnrS gene) (Yang et al., 2015), where there is a possibility that the birds had been treated previously with this antibiotic for a period of time. Our result is worrisome because fluoroquinolones are used for treating a wide variety of infections (Hopkins et al., 2005). All the samples were negative for vanA, a plasmid‐mediated vancomycin resistance gene. This result contrasts with those found in wildlife from natural parks of Portugal (4.3%) (Poeta et al., 2005), feral pigeons from Czech Republic (3%) (Radimersky et al., 2010) and birds of the Azores Archipelago (0.72%) (Santos et al., 2013). Resistant bacterial strains have been reported in Costa Rica since the early 1990s (Olivo Meza et al., 1998).
Domestic pigeons do not travel long distances (an approximate maximum distance of 5.29 km) (Rose et al., 2006), and they have to meet their needs with what they find within the above‐mentioned distance. In this study, each park was located in environments with high human population, closed to national hospitals or clinics and rivers contaminated with faecal matter (Table 2), where they can get water and food contaminated with pharmaceutical products among them antibiotics, faecal matter or ARGs, or even be in contact with contaminated airdrops (Fig. 1) (Rodriguez et al., 2006; Aali et al., 2014; McEachran et al., 2015).
Figure 1.
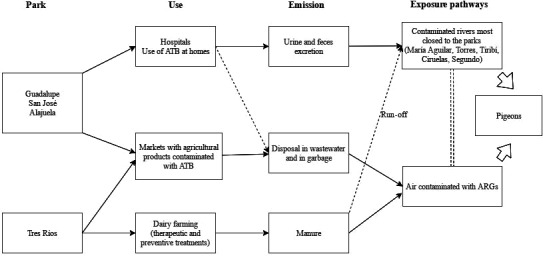
Possible routes of exposure to ARGs by the doves.
No differences in resistance patterns were found in the four locations, a fact related to the homogeneous presence of ARGs. However, some variances were observed in the percentage of appearance of some ARGs, such as sul genes, tet(A) and qnrS, which could be attributed to differences in the exposure sources. San José, Guadalupe and Alajuela correspond to locations with a high density of human population (e.g. San Jose is the area most crowded in the country with a population density of 5000–10 000 inhabitants/km2), and in a radius of 3 km from the parks, it is possible to find human clinics, hospitals and rivers (influenced by these clinics and hospitals) with high levels of contamination with faecal matter (Table 2) (Calvo Brenes et al., 2012). Conventional and hospital discharges have been described as a potential route for dissemination of ARGs into the natural environment and pose a hazard to environmental and public health, because ARGs can be transferred among the bacteria from the different areas through plasmids, integrons and transposons (Rahube and Yost, 2015). Besides, transduction can occur even without the presence of the ‘donor’ and ‘recipient’ cells at the same place or at the same time, between bacteria belonging to different genera (Mazaheri Nezhad Fard et al., 2011; Muniesa et al., 2013). On the other hand, Tres Ríos is an urban centre characterized by its agricultural production, mainly coffee fields, and dairy farming. This park showed higher percentage of tet(A) positive samples than the others from San José, Alajuela and Guadalupe, which could be due to the water or air transportation of particles contaminated with tetracyclines used in the livestock activities performed in the area (Fig. 1) (Calvo Brenes et al., 2012; de la Cruz et al., 2014).
The importance of rainfall is not well recognized in urban ecosystems. It is known that the rainy season can affect the metabolism of birds (Wilson et al., 2004), taking into account that it is possible that they be forced to seek or find shelter during heavy rainfall (Elkins, 1988), but there is no evidence about its influence in the contamination with ARGs. In Costa Rica, the rainy season is characterized by high precipitation during several months and Calvo Brenes et al. (2012) proposed that rivers showed a higher level of faecal contamination due to the leaching of contaminants to them, which can come from human activities and excreta of animals, both domestic and wild. So, it is possible that the transmission of ARGs to the birds is higher at this season, which could explain the detection of multiresistant microbiomes and the higher percentage of sulI and tet(A). However, the areas with higher resistance were all sampled during the rainy season, so the localization or the season could act as confounding factors.
Pigeons have been identified as sources that disseminate multiple infectious agents to human populations (Weber et al., 1995), so it is possible that they can spread ARGs by their faeces. Other authors have suggested that they can interact with other birds, facilitating the acquisition and propagation of ARGs to other species (Santos et al., 2013). This could produce an ecological impact given Costa Rica's rich avifauna. However, it is most important to determinate the mechanisms by which pigeons could have acquired the ARGs, especially considering that the direction of gene transmission could be from humans, livestock or other activities to pigeons. This information can help to generate management measures to diminish the risk of contamination with ARGs by other animals and humans.
Although the percentages of detected ARGs may be considered low, some of them are important for public and animal health. A pending task for the future is to determinate whether there is a link between our results and the presence of resistant infectious agents in hospitals.
This work clearly illustrates that pigeons in close proximity to people are potential indicators that can facilitate rapid dissemination of AMR in urban areas, and opens the door for further investigations in which the presence of ARGs will be compared in pigeons from different areas, as well as to birds and animals of different ecological niches.
Acknowledgements
The authors would like to acknowledge the technical support of Elena Neves and Veronica Nogal, as well as the Publications Fund of the Universidad Nacional and Dr. David Lean for the linguistic review of this paper. This work was supported by the Fondo de Educación Superior Estatal del Consejo Nacional de Rectores de Costa Rica (CONARE) [SIA 0536‐12], by the Instituto Nacional de Tecnología Agraria y Alimentaria (INIA) [RTA2014‐00012‐C03‐02] and by the Community of Madrid [S2013/ABI‐2747TAVS].
References
- Aali, R. , Nikaeen M., Khanahmad H., and Hassanzadeh A., 2014: Monitoring and comparison of antibiotic resistant bacteria and their resistance genes in municipal and hospital wastewaters. Int. J. Prev. Med. 5, 887–894. [PMC free article] [PubMed] [Google Scholar]
- Acar, J. F. , and Moulin G., 2013: Integrating animal health surveillance and food safety: the issue of antimicrobial resistance. Rev. Sci. Tech. Int. Off. Epizoot. 32, 383–392. [DOI] [PubMed] [Google Scholar]
- Allen, H. K. , Donato J., Wang H. H., Cloud‐Hansen K. A., Davies J., and Handelsman J., 2010: Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. [DOI] [PubMed] [Google Scholar]
- Calvo Brenes, G. , Brenes G. C., and Molina J. M., 2012: Contaminación fecal en varios ríos de la Gran Área Metropolitana y la Península de Osa. Rev. Tecnol. En Marcha 25, 33–39. [Google Scholar]
- Carignan, V. , and Villard M.‐A., 2002: Selecting indicator species to monitor ecological integrity: a review. Environ. Monit. Assess. 78, 45–61. [DOI] [PubMed] [Google Scholar]
- de la Cruz, E. , Fournier M. L., García F., Molina A., Chavarría G., Alfaro M., Ramírez F., and Rodríguez C., 2014: Hazard prioritization and risk characterization of antibiotics in an irrigated Costa Rican region used for intensive crop, livestock and aquaculture farming. J. Environ. Biol. Acad. Environ. Biol. India 35, 85–98. [PubMed] [Google Scholar]
- Doi, Y. , and Arakawa Y., 2007: 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 45, 88–94. [DOI] [PubMed] [Google Scholar]
- Dolejska, M. , Cizek A., and Literak I., 2007: High prevalence of antimicrobial‐resistant genes and integrons in Escherichia coli isolates from black‐headed gulls in the Czech Republic. J. Appl. Microbiol. 103, 11–19. [DOI] [PubMed] [Google Scholar]
- Durso, L. M. , Harhay G. P., Bono J. L., and Smith T. P. L., 2011: Virulence‐associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J. Microbiol. Methods 84, 278–282. [DOI] [PubMed] [Google Scholar]
- Durso, L. M. , Miller D. N., and Wienhold B. J., 2012: Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non‐agricultural metagenomes. PLoS ONE 7, e48325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins, N. , 1988: Weather and Bird Behaviour. 2nd edn Bath Press, Avon. [Google Scholar]
- Francois, P. , Pittet D., Bento M., Pepey B., Vaudaux P., Lew D., and Schrenzel J., 2003: Rapid Detection of methicillin‐resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41, 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann, H. , Aires‐de‐Sousa M., Boyce J., and Tiemersma E., 2006: Emergence and resurgence of meticillin‐resistant Staphylococcus aureus as a public‐health threat. Lancet Lond. Engl. 368, 874–885. [DOI] [PubMed] [Google Scholar]
- Guardabassi, L. , Schwarz S., and Lloyd D. H., 2004: Pet animals as reservoirs of antimicrobial‐resistant bacteria. J. Antimicrob. Chemother. 54, 321–332. [DOI] [PubMed] [Google Scholar]
- Hopkins, K. L. , Davies R. H., and Threlfall E. J., 2005: Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25, 358–373. [DOI] [PubMed] [Google Scholar]
- INEC , 2012. 2011‐2025.Proyecciones distritales. Población total por grupos de edades, según provincia y cantón [WWW Document]. Inst. Nac. Estad. Censos. URL http://www.inec.go.cr (accessed 7.9.16).
- Jiang, L. , Hu X., Xu T., Zhang H., Sheng D., and Yin D., 2013: Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai. China. Sci. Total Environ. 458–460, 267–272. [DOI] [PubMed] [Google Scholar]
- Karczmarczyk, M. , Abbott Y., Walsh C., Leonard N., and Fanning S., 2011: Characterization of multidrug‐resistant escherichia coli isolates from animals presenting at a University Veterinary Hospital. Appl. Environ. Microbiol. 77, 7104–7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer, K. , 2009: Antibiotics in the aquatic environment–a review–part II. Chemosphere 75, 435–441. [DOI] [PubMed] [Google Scholar]
- Marti, E. , and Balcazar J. L., 2013: Real‐Time PCR Assays for Quantification of qnr genes in environmental water samples and chicken feces. Appl. Environ. Microbiol. 79, 1743–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, E. , Jofre J., and Balcazar J. L., 2013: Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 8, e78906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, J. M. , and Ewing K., 2001: Restoration of fragmented landscapes for the conservation of birds: a general framework and specific recommendations for urbanizing landscapes. Restor. Ecol. 9, 280–292. [Google Scholar]
- Mazaheri Nezhad Fard, R. , Barton M. D., and Heuzenroeder M. W., 2011: Bacteriophage‐mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 52, 559–564. [DOI] [PubMed] [Google Scholar]
- Mazodier, P. , and Davies J., 1991: Gene transfer between distantly related bacteria. Annu. Rev. Genet. 25, 147–171. [DOI] [PubMed] [Google Scholar]
- McEachran, A. D. , Blackwell B. R., Hanson J. D., Wooten K. J., Mayer G. D., Cox S. B., and Smith P. N., 2015: Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ. Health Perspect. 123, 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa, M. , Colomer‐Lluch M., and Jofre J., 2013: Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human‐body associated bacterial populations? Mob. Genet. Elem. 3, e25847‐1–e‐25847‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagulapally, S. R. , Ahmad A., Henry A., Marchin G. L., Zurek L., and Bhandari A., 2009: Occurrence of ciprofloxacin‐, trimethoprim‐sulfamethoxazole‐, and vancomycin‐resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. Res. Publ. Water Environ. Fed. 81, 82–90. [DOI] [PubMed] [Google Scholar]
- Olivo Meza, M. , Avila‐Agüero M. L., Odio Pérez C., Falngezlcht‐Gutman I., Herrera M. L., and Rivera‐Brenes R., 1998: Colonización por enterococos resistentes a la vancomicina en una Unidad de cuidados intensivos pediátricos (Colonization of vancomycin‐resistant enterococci in a children's intensive care unit). Acta Pediátrica Costarric. 12, 66–69. [Google Scholar]
- Poeta, P. , Costa D., Sáenz Y., Klibi N., Ruiz‐Larrea F., Rodrigues J., and Torres C., 2005: Characterization of antibiotic resistance genes and virulence factors in faecal enterococci of wild animals in Portugal. J. Vet. Med. B Infect. Dis. Vet. Public Health 52, 396–402. [DOI] [PubMed] [Google Scholar]
- Radimersky, T. , Frolkova P., Janoszowska D., Dolejska M., Svec P., Roubalova E., Cikova P., Cizek A., and Literak I., 2010: Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. J. Appl. Microbiol. 109, 1687–1695. [DOI] [PubMed] [Google Scholar]
- Rahube, T. O. , and Yost C. K., 2015: Antibiotic resistance plasmids in wastewater treatment plants and their possible dissemination into the environment. Afr. J. Biotechnol. 9, 9183–9190. [Google Scholar]
- Rodriguez, C. , Lang L., Wang A., Altendorf K., Garcia F., and Lipski A., 2006: Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline‐ and gentamicin‐resistant bacteria. Appl. Environ. Microbiol. 72, 5870–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Martínez, J. L. , 2009: Resultados Plan Nacional de Residuos (Results of theNational Waste Plan). Ministry of Agriculture and Cattle Raising, National Animal Health Service, Heredia. [Google Scholar]
- Rose, E. , Nagel P., and Haag‐Wackernagel D., 2006: Spatio‐temporal use of the urban habitat by feral pigeons (Columba livia). Behav. Ecol. Sociobiol. 60, 242–254. [Google Scholar]
- Sacristán, C. , Esperón F., Herrera‐León S., Iglesias I., Neves E., Nogal V., Muñoz M. J., and de la Torre A., 2014: Virulence genes, antibiotic resistance and integrons in Escherichia coli strains isolated from synanthropic birds from Spain. Avian Pathol. 43, 172–175. [DOI] [PubMed] [Google Scholar]
- van de Sande‐Bruinsma, N. , Grundmann H., Verloo D., Tiemersma E., Monen J., Goossens H., Ferech M. And European Antimicrobial Resistance Surveillance System Group, European Surveillance of Antimicrobial Consumption Project Group , 2008: Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 14, 1722–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, T. , Silva N., Igrejas G., Rodrigues P., Micael J., Rodrigues T., Resendes R., Gonçalves A., Marinho C., Gonçalves D., Cunha R., and Poeta P., 2013: Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 24, 25–31. [DOI] [PubMed] [Google Scholar]
- Sousa, M. , Silva N., Igrejas G., Silva F., Sargo R., Alegria N., Benito D., Gómez P., Lozano C., Gómez‐Sanz E., Torres C., Caniça M., and Poeta P., 2014: Antimicrobial resistance determinants in Staphylococcus spp. recovered from birds of prey in Portugal. Vet. Microbiol. 171, 436–440. [DOI] [PubMed] [Google Scholar]
- Torsvik, V. , and Øvreås L., 2002: Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5, 240–245. [DOI] [PubMed] [Google Scholar]
- Weber, A. , Popel J., and Schafer‐Schmidt R., 1995: Untersuchungen zumVorkommen von Listeria monocytogenes in Kotproben von Tauben. Berl. Münch. Tierärztl. Wochenschr. 108, 26–27. [PubMed] [Google Scholar]
- WHO , 2014: Antimicrobial Resistance: Global Report on Surveillance. WHO Press, Geneva. [Google Scholar]
- Wilson, G. R. , Cooper S. J., and Gessaman J. A., 2004: The effects of temperature and artificial rain on the metabolism of American kestrels (Falco sparverius). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 139, 389–394. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Yang L., Lü D.‐H., Zhang W.‐H., Ren S.‐Q., Liu Y.‐H., Zeng Z.‐L., and Jiang H.‐X., 2015: Co‐prevalance of PMQR and 16S rRNA methylase genes in clinical Escherichia coli isolates with high diversity of CTX‐M from diseased farmed pigeons. Vet. Microbiol. 178, 238–245. [DOI] [PubMed] [Google Scholar]


