Abstract
Background
Nuclear receptors (NRs) can regulate gene expression; therefore, they are classified as transcription factors. Despite the extensive research carried out on NRs, still several issues including (1) the expression profile of NRs in human tissues, (2) how the NR expression is modulated during atherosclerosis and metabolic diseases, and (3) the overview of the role of NRs in inflammatory conditions are not fully understood.
Methods
To determine whether and how the expression of NRs are regulated in physiological/pathological conditions, we took an experimental database analysis to determine expression of all 48 known NRs in 21 human and 17 murine tissues as well as in pathological conditions.
Results
We made the following significant findings: (1) NRs are differentially expressed in tissues, which may be under regulation by oxygen sensors, angiogenesis pathway, stem cell master regulators, inflammasomes, and tissue hypo-/hypermethylation indexes; (2) NR sequence mutations are associated with increased risks for development of cancers and metabolic, cardiovascular, and autoimmune diseases; (3) NRs have less tendency to be upregulated than downregulated in cancers, and autoimmune and metabolic diseases, which may be regulated by inflammation pathways and mitochondrial energy enzymes; and (4) the innate immune sensor inflammasome/caspase-1 pathway regulates the expression of most NRs.
Conclusions
Based on our findings, we propose a new paradigm that most nuclear receptors are anti-inflammatory homeostasis-associated molecular pattern receptors (HAMPRs). Our results have provided a novel insight on NRs as therapeutic targets in metabolic diseases, inflammations, and malignancies.
Electronic supplementary material
The online version of this article (10.1186/s13045-017-0526-8) contains supplementary material, which is available to authorized users.
Keywords: Nuclear receptors (NRs), Homeostasis-associated molecular pattern receptors, Atherosclerosis, Metabolic disease, Cardiovascular disease
Background
Pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) generated during microbial invasion or tissue injury act as stimuli and activate the innate immune system to respond to infection or injury [1]. The key cellular receptors that recognize the “threat” signals initiated by PAMPs and DAMPs are referred to as PRRs (pattern recognition receptors). One of the receptor families that are highly characterized as PRRs is the Toll-like receptor (TLR) family. Most of the TLRs are mainly located on the plasma membrane and activate inflammatory genes to counteract tissue injury and mediate repair. Moreover, TLRs work in synergy with cytosolic PRR families like NLRs (NOD (nucleotide-binding oligomerization domain)-like receptors) to recognize DAMPs, particularly in what we proposed—inflammation-privileged tissues where inflammasome component genes that initiate inflammation are not constitutively expressed [2, 3]. Additionally, four other PRR families including C-type lectin receptors, retinoid acid-inducible gene 1 (RIG-1), absent in melanoma-2 (AIM-2), and receptor for advanced glycation end products (RAGE, also a receptor for high-mobility group box 1 (HMGB1)) have also been characterized [4].
Previously, using endogenous metabolite lysophospholipids (LPLs) as a prototype, we proposed a new paradigm for the first time that certain metabolites that play cellular functions during normal physiological status can adapt as pro-inflammatory mediators at elevated concentrations. We named such metabolites as “conditional DAMPs” and their endogenous receptors as “conditional DAMP receptors.” We further pointed out significant loopholes in the current danger model which identify only the six receptors mentioned above as PRRs, which we named as “classical DAMP receptors” [5]. Along the line, we recently reported a series of significant findings on the expression and roles of caspase-1 in the NLR pathway in vascular inflammation [2, 6–15]. In the same publication mentioned above, we concluded that activation of inflammation by conditional DAMPs may be realized via binding to their own intrinsic receptors and may not necessarily always involve or “converge to” TLRs, NLRs, and other classical DAMP receptors [5].
Another significant problem associated with the current danger theory is that it fails to recognize the roles played by potential endogenous metabolites in anti-inflammatory responses, inflammation resolution, and maintenance of homeostasis. Therefore, we further advanced the current paradigm by proposing endogenous metabolites such as lysophosphatidylserine and lysophosphatidylethanolamine that not only maintain homeostasis at physiological levels, but also act as anti-inflammatory mediators to inhibit inflammation and promote inflammation resolution at pathologically elevated levels as homeostasis-associated molecular patterns (HAMPs). Furthermore, we proposed that these HAMPs bind to their receptors (HAMP receptors) to initiate anti-inflammatory/homeostatic signaling and promote inflammation resolution [5]. However, an outstanding issue of whether endogenous lipophilic metabolites that bind to nuclear receptors can serve as HAMPs remains unknown.
The nuclear hormone receptor superfamily has 48 lipophilic ligand-activated receptors including 32 nuclear hormone receptors (NHRs) for thyroid and steroid hormones, retinoids, and vitamin D, as well as 16 orphan nuclear receptors where the ligands are yet unknown [16–18]. Nuclear receptors (NRs), as transcription factors, have the ability to directly bind to DNA and regulate the expression of adjacent genes [19, 20]. Ligands for some of these NRs have been recently identified, including lipid metabolites such as fatty acids, prostaglandins, or cholesterol derivatives. These ligands can regulate gene expression by binding to NRs [21]. Ligand binding to a NR results in a conformational change and activation of the receptor, leading to up- or downregulation of the target gene expression. Thus, NRs are involved in the regulation of various physiological processes including development, homeostasis, and metabolism of the organism [22] and pathogenesis of metabolic disease in response to metabolic/environmental changes [23].
However, despite the recent progress, there are many aspects of NRs that have not yet been explored: first, the expression profile of NRs under physiological conditions in various human tissues have not been studied; second, whether the expression of certain NRs are either upregulated or downregulated in atherogenic and metabolic disease-related pathological conditions are not clear; third, mechanistically, whether pro-/anti-inflammatory signaling is negatively/positively associated with the expression of NRs is not known; and fourth, whether NRs have the capacity to function as our newly proposed HAMP receptors, which suppress inflammatory responses and maintain tissue homeostasis in response to the stimulation of exogenous and endogenous PAMPs/DAMPs. To address these questions, we took a “panoramic view” at the tissue expression pattern of all 48 identified human and mouse NRs. Our results demonstrated that NRs are differentially expressed among tissues at physiological conditions, which may be regulated by oxygen sensors, vascular endothelial growth factor pathways, stem cell master regulators, innate immune sensors, and DNA hypo-/hypermethylation status. We also found that the expressions of certain NRs have less tendency to be upregulated than to be downregulated in atherogenic conditions, metabolic diseases, which may be contributed by significant regulation of innate immune sensor caspase-1/inflammasome pathway. Our findings provide novel insights into the upstream regulation of nuclear receptors in physiological, autoimmune arthritis, and cardiovascular and metabolic disease conditions.
Methods
Tissue expression profiles of genes encoding nuclear receptors
An experimental data mining strategy (Fig. 1) was used to analyze the expression profiles of mRNA transcripts of NR genes in 21 different human and 17 mouse tissues including the heart and vasculature. We utilized an experimentally verified mRNA expression in the expressed sequence tag (EST) databases of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) UniGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene) to determine the transcription profile of nuclear receptors in tissues of interest. Transcripts per million of genes of interest were normalized to that of housekeeping gene β-actin in each given tissue to calculate the arbitrary units of gene expression. A confidence interval of the expression variation of housekeeping genes was generated by calculating the mean plus two times that of the standard deviation of the arbitrary units of three randomly selected housekeeping genes (PRS27A, GADPH, and ARHGDIA in human; Ldha, Nono, and Rpl32 in mouse) normalized by β-actin in the given tissues. If the expression variation of a given gene in the tissues was larger than the upper limit of the confidence interval, the high expression levels of genes in the tissues were considered statistically significant. Gene transcripts where the expression level was lower than one per million were technically considered as no expression.
Fig. 1.
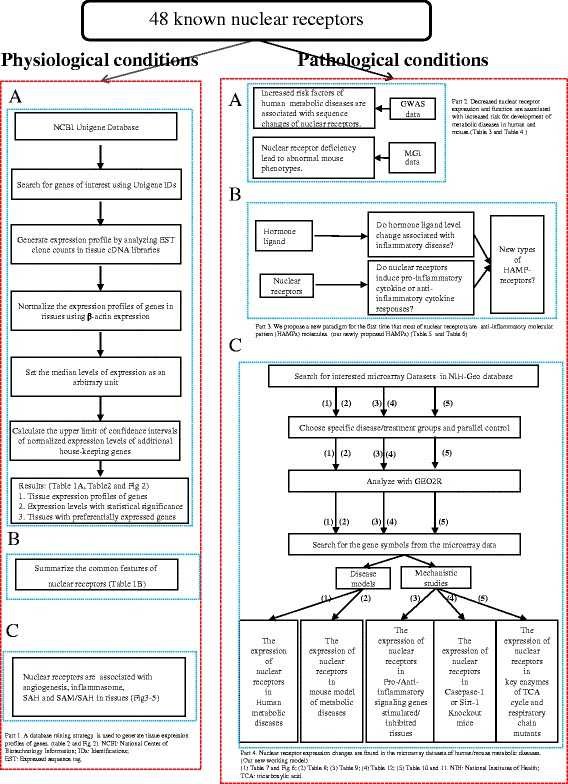
Flow chart of database mining strategy and two parts of data organization. Part1: shows the database mining strategy utilized to generate tissue nuclear receptor expression profile. Part 2: the strategy utilized to measure nuclear receptor expression in human and mouse metabolic diseases. Parts 3 and 4: shows the strategy that is used to analyze the microarray data sets and identifying nuclear receptors as homeostasis-associated molecular pattern receptors (HAMPRs)
Expression profiles of nuclear receptors in disease models and cell activity
Microarray datasets were collected from the Array Express of European Bioinformatics Institute, which stores data from high-throughput functional genomics experiments (https://www.ebi.ac.uk/arrayexpress). These data include the information of the expression of nuclear receptors through experiments submitted directly to Array Express or imported from the NCBI Gene Expression Omnibus database. We used data from the following databases: (1) Metabolic disease: (a) adipose tissue and liver in high fat diet-induced obese mouse model versus normal diet controls, (b) aortic arch segment of the atherogenic apolipoprotein E gene knockout (apolipoprotein E (ApoE−/−)) mice versus wild-type mouse aorta controls, (c) pancreatic islets and white fat of leptin receptor mutant db/db type II diabetic mice versus control mice, (d) oxidized low-density lipoprotein (Ox-LDL)-stimulated mouse endothelial cells versus control endothelial cells, and (e) high-concentration homocysteine (Hcy)-treated human aortic smooth muscle cells (HASMCs) versus low-concentration homocysteine (Hcy)-treated vascular smooth muscle cells (VSMCs); (2) CD4+Foxp3+ regulatory T cell (Treg) polarization/differentiation—we examined the expression changes of the nuclear receptors in Tregs versus effector T cells in mice, as well as in vitro, with cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) ligation; (3) mRNA expression of NR changes due to the stimulus with pro-/anti-inflammation conditions; and (4) we screened the datasets among energy metabolic nuclear receptors of tricarboxylic acid (TCA) cycle and respiratory chain. The modulation of nuclear receptor expression in cancers were determined by analyzing the Cancer Genome Atlas database.
Application of big GWAS data to clarify the relationship between nuclear receptors and metabolic disease
Genome-wide association studies (GWASs) continue to be a widely used approach to detect genetic association with a phenotype of interest in well-defined populations. Various anthropometric measures serve as surrogates for obesity, with body mass index (BMI) (HGVPM 1111 and 564) and waist-hip ratio (HGVPM 1114) as the most frequently used markers in epidemiologic studies aimed at assessing obese disease risks. Anti-cyclic citrullinated peptide-positive rheumatoid arthritis status (HGVPM 38) and rheumatoid arthritis (HGVPM 235) are the most frequently used markers in epidemiologic studies aimed at rheumatoid arthritis risk. Fasting plasma glucose (HGVPM 825), homeostatic model assessment of β-cell function (HGVPM 827), fasting insulin (HGVPM 822), homeostatic model assessment of insulin resistance (HGVPM 826), glycated hemoglobin levels (HGVPM 1081), glycosylated hemoglobin (HGVPM 569), 2-h glucose challenge (HGVPM 769), type II diabetes status (HGVPM 4 and 5), early onset type II diabetes mellitus (HGVPM 74), proinsulin levels (HGVPM 1538), and a-glucose (HGVPM 3639) are the most frequently used markers in epidemiologic studies aimed at diabetes risks. Serum cholesterol (HGVPM 568), lipids (CH3) (HGVPM 3602), lipids (CH2) (HGVPM 3611), lipids (CH2CO) (HGVPM 3616), lipids (CH=CH*CH2CH2) (HGVPM 3640), and systolic blood pressure (HGVPM 563) are the most frequently used markers in epidemiologic studies aimed at vascular atherosclerosis risks. With the advent of large GWASs, we now have the ability to identify NRs associated with dangerous risks for specific disease.
Application of MGI data to clarify the abnormal mouse phenotypes in nuclear receptor knockout mouse adipose, cardiovascular, metabolism, and endocrine systems
MouseMine (www.informatics.jax.org) is a new data warehouse for accessing mouse data from Mouse Genome Informatics (MGI). The main source of MouseMine data is MGI, which includes a wealth of information about the structure and function of the mouse genome, developmental gene expression patterns, phenotypic effects caused by mutations, and annotations of human disease models. The “Human-mouse: disease connection” tool (www.informatics.jax.org/humanDisease.shtml) supports uploading a list of nuclear receptor gene IDs/symbols and getting back certain information about those nuclear receptors, such as those associated human diseases and abnormal mouse phenotypes reported in adipose, cardiovascular, metabolic, and endocrine systems.
Tissue SAH and SAM measurements in mice
The concentrations of S-adenosyl methionine (SAM) and S-adenosyl homocysteine (SAH) were measured in six tissues (heart, liver, lung, kidney, spleen, and brain) in C57BL/6J (n = 4) mice from 13.4 to 18 weeks of age. Mouse tissues were collected and homogenized in 0.4 mol/L perchloric acid (PCA) solution. The homogenized tissues were centrifuged for 10 min at 2000 rpm. Supernatant was collected and stored at −80 °C. SAM and SAH levels were analyzed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS; Institute of Metabolic Disease, Baylor Research Institute, Dallas, TX). The unit of SAH level in tissues is nanomole per gram [24].
Results
Nuclear receptors are differentially expressed in tissues. Nuclear receptor expression is associated with angiogenesis pathway, stem cell master genes, PRRs, and tissue hypomethylation/hypermethylation indices
As summarized in Table 1, the NR superfamily includes 48 NRs classified into seven families, such as class I-thyroid hormone receptor-like family (19 members), class II-retinoid X receptor-like family (12 members), class III-estrogen receptor-like family (9 members), class IV-nerve growth factor IB-like family (3 members), class V-steroidogenic factor-like receptor family (2 members), class VI-germ cell nuclear receptor-like family (1 member), and class O-miscellaneous family (2 members). In addition, we summarized seven common features of the NR superfamily in Table 2. One of the most striking features of NRs is that in addition to transduce steroid, thyroid, retinoid, and other hormone signals, NRs can also serve as metabolic sensors and xenobiotic sensors for high-affinity ligands and low-affinity molecular patterns [25]. Several reports showed that NRs not only bind to specific ligands but also recognize structural patterns (Table 3), which raises a possibility for NRs to recognize many endogenous metabolites that can act as HAMPs that are yet to be identified/characterized [5].
Table 1.
The UniGene ID of 48 human nuclear receptors and mouse homologs
| Gene name (full name) | NRNC symbol | Receptor | Ligand(s) | ID | ||
|---|---|---|---|---|---|---|
| Human | Mouse (Mm.) | |||||
| (Hs.) | ||||||
| Class I—thyroid hormone receptor-like | ||||||
| THRA | Thyroid hormone receptor, alpha | NR1A1 | Thyroid hormone receptor | Thyroid hormone | 724 | 265917 |
| THRB | Thyroid hormone receptor, beta | NR1A2 | 187861 | 32563 | ||
| RARA | Etinoic acid receptor, alpha | NR1B1 | Retinoic acid receptor | Vitamin A and related compounds | 654583 | 439744 |
| RARB | Etinoic acid receptor, beta | NR1B2 | 654490 | 259318 | ||
| RARG | Etinoic acid receptor, gamma | NR1B3 | 1497 | 1273 | ||
| PPARA | Peroxisome proliferator-activated receptor alpha | NR1C1 | Peroxisome proliferator-activated receptor | Fatty acids, prostaglandins | 103110 | 212789 |
| PPARD | Peroxisome proliferator-activated receptor delta | NR1C2 | 696032 | 328914 | ||
| PPARG | Peroxisome proliferator-activated receptor gamma | NR1C3 | 162646 | 3020 | ||
| NR1D1 | Nuclear receptor subfamily 1 group D member 1 | NR1D1 | Rev-ErbA | Heme | 592130 | 390397 |
| NR1D2 | Nuclear receptor subfamily 1 group D member 2 | NR1D2 | 37288 | 26587 | ||
| RORA | RAR-related orphan receptor A | NR1F1 | Cholesterol | 560343 | 427266 | |
| RORB | RAR-related orphan receptor B | NR1F2 | 494178 | 234641 | ||
| RORC | RAR-related orphan receptor C | NR1F3 | 256022 | 4372 | ||
| NR1H3 | Nuclear receptor subfamily 1 group H member 3 | NR1H3 | Liver X receptor-like receptor | Oxysterols | 438863 | 22690 |
| NR1H2 | Nuclear receptor subfamily 1 group H member 2 | NR1H2 | 432976 | 968 | ||
| NR1H4 | Nuclear receptor subfamily 1 group H member 4 | NR1H4 | 282735 | 3095 | ||
| VDR | Vitamin D (1,25-dihydroxyvitamin D3) receptor | NR1I1 | Vitamin D receptor-like receptor | Vitamin D | 524368 | 245084 |
| NR1I2 | Nuclear receptor subfamily 1group I member 2 | NR1I2 | Xenobiotics | 7303 | 8509 | |
| NR1I3 | Nuclear receptor subfamily 1 group I member 3 | NR1I3 | Androstane | 349642 | 486506 | |
| Class II—retinoid X receptor-like | ||||||
| HNF4A | Hepatocyte nuclear factor 4, alpha | NR2A1 | Hepatocyte nuclear factor-4 receptor | Fatty acids | 116462 | 202383 |
| HNF4G | Hepatocyte nuclear factor 4, gamma | NR2A2 | 241529 | 330897 | ||
| RXRA | Retinoid X receptor alpha | NR2B1 | Retinoid X receptor | Retinoids | 590886 | 24624 |
| RXRB | Retinoid X receptor beta | NR2B2 | 388034 | 1243 | ||
| RXRG | Retinoid X receptor gamma | NR2B3 | 26550 | 3475 | ||
| NR2C1 | Nuclear receptor subfamily 2 group C member 1 | NR2C1 | Testicular receptor | UD | 108301 | 107483 |
| NR2C2 | Nuclear receptor subfamily 2 group C member 1 | NR2C2 | 555973 | 87062 | ||
| NR2E1 | Nuclear receptor subfamily 2 group E member 1 | NR2E1 | Tailless-like receptors | UD | 157688 | 287100 |
| NR2E3 | Nuclear receptor subfamily 2 group E member 3 | NR2E3 | 187354 | 103641 | ||
| NR2F1 | Nuclear receptor subfamily 2 group F member 1 | NR2F1 | COUP-TF-like receptors | UD | 347991 | 439653 |
| NR2F2 | Nuclear receptor subfamily 2 group F member 2 | NR2F2 | 519445 | 158143 | ||
| NR2F6 | Nuclear receptor subfamily 2 group F member 6 | NR2F6 | 466148 | 28989 | ||
| Class III—estrogen receptor-like | ||||||
| ESR1 | Estrogen receptor 1 | NR3A1 | Estrogen receptor | Estrogens | 208124 | 9213 |
| ESR2 | Estrogen receptor 2 | NR3A2 | 660607 | 2561 | ||
| ESRRA | Estrogen-related receptor alpha | NR3B1 | Estrogen-related receptor | UD | 110849 | 386776 |
| ESRRB | Estrogen-related receptor beta | NR3B2 | 435845 | 235550 | ||
| ESRRG | Estrogen-related receptor gamma | NR3B3 | 444225 | 89989 | ||
| NR3C1 | Nuclear receptor subfamily 3 group C member 1 | NR3C1 | 3-Ketosteroid receptors | Cortisol | 122926 | 129481 |
| NR3C2 | Nuclear receptor subfamily 3 group C member 2 | NR3C2 | Aldosterone | 163924 | 324393 | |
| PGR | Progesterone receptor | NR3C3 | Progesterone | 32405 | 12798 | |
| AR | Androgen receptor | NR3C4 | Testosterone | 76704 | 439657 | |
| Class IV—nerve growth factor IB-like | ||||||
| NR4A1 | Nuclear receptor subfamily 4 group A member 1 | NR4A1 | Nerve growth factor IB-like receptors | UD | 524430 | 119 |
| NR4A2 | Nuclear receptor subfamily 4 group A member 2 | NR4A2 | 563344 | 3507 | ||
| NR4A3 | Nuclear receptor subfamily 4 group A member 3 | NR4A3 | 279522 | 247261 | ||
| Class V—steroidogenic factor-like | ||||||
| NR5A1 | Nuclear receptor subfamily 5 group A member 1 | NR5A1 | Fushi tarazu F1-like receptors | Phosphatidylinositols | 495108 | 31387 |
| NR5A2 | Nuclear receptor subfamily 5 group A member 2 | NR5A2 | 33446 | 16794 | ||
| Class VI—germ cell nuclear factor-like | ||||||
| NR6A1 | Nuclear receptor subfamily 6 group A member 1 | NR6A1 | Germ cell nuclear factor receptors | UD | 586460 | 439703 |
| Class O—miscellaneous | ||||||
| NR0B1 | Nuclear receptor subfamily 0 group B member 1 | NR0B1 | DAX-like Receptors | UD | 268490 | 5180 |
| NR0B2 | Nuclear receptor subfamily 0 group B member 2 | NR0B2 | 427055 | 346759 | ||
UD undetermined
Table 2.
The common features of nuclear receptors
| Common features of nuclear receptors | PMID |
|---|---|
| 1. Five domain structures including N-terminal regulatory domain, DNA binding domain, hinge region, ligand-binding domain, and C-terminal domain | 10406480/10751636/12893880 |
| 2. Lipophilic ligand-activated transcription factors including orphan receptors for unknown endogenous ligands | 8,807,884/10671476 |
| 3. 48 known super human family members including seven groups, mice (49), rats (47), C. elephant (270) | 10219237/9460643/15059999 |
| 4. 350 co-regulators to facilitate their functions | 22733267 |
| 5. Transduce steroid, thyroid, retinoid, and other hormonal signals | 11729302/8521507 |
| 6. Metabolic sensors and xenobiotic sensors for high-affinity ligands and low-affinity molecular patterns | 20615454 |
| 7. Serve as the targets for 13% FDA-approved drugs | 17139284 |
Table 3.
Nuclear receptors can recognize and bind many ligands which have similar structures/patterns via its ligand-binding domain
| Features of nuclear receptors’ ligand-binding domain | PMID |
|---|---|
| 1. Ligand-binding domains have the capacity to bind coactivator segments with LXXLL sequences, and corepressor segments with LXXXLXXX[I/L] sequences (where L = leucine, I = isoleucine, and X = any amino acid) | 9808622 |
| 2. A single nuclear receptor controls the multitude of gene expressions | 20148675 |
| 3. The ligand-binding domain consists of a hydrophobic pocket that can bind a hydrophobic ligand | 20615454 |
| 4. Flexible ligands can contort to fit in the ligand-binding pocket | 9501913 |
| 5. Pharmacological antagonists and have been shown to bind to the receptor in the ligand-binding site and to inhibit hormone-activated receptor function | |
| (1). NR1A1 ligand-binding domain can bind 3,5-dimethyl-3-isopropylthyronine except thyroid hormone | 8523397 |
| (2). NR1B3 ligand-binding domain can bind to all-trans retinoic acid except vitamin A and related compounds | 7501014 |
| (3). NR3A1 ligand-binding domain can bind to estradiol and raloxifene | 9338790 |
To determine whether tissues have functional differences in sensing metabolic stressors and xenobiotic stressors via NRs, we hypothesized that various tissues express differential levels and certain types of NRs under physiological conditions. To examine this hypothesis, the expression of 48 NR genes in 21 human tissues and 17 mouse tissues were examined (fewer mouse tissues were examined due to unavailability of gene expression data for four types of mouse tissues, i.e., nerve, trachea, stomach, and vascular tissues in the NIH UniGene database) (Additional file 1: Figure S1). The results showed that some human tissues such as muscle (17), trachea (14), and nerve (10) express a large variety of NRs at high expression levels (Tables 4 and 5). This data suggests that the gene expression, differentiation, and function of these tissues may largely be regulated by NRs under normal physiological levels. Comparatively, eyes (7), adrenal gland (6), kidney (5), and adipose tissue (5) express more variety of NRs than the heart, liver, and pancreas (Table 4). Similarly, when comparing the human NR expression profile to that of the mouse, human tissues express much more types of NRs at high expression levels than mice. For example, although human and mouse muscles contain more variety of NRs at high levels relative to other tissues studied, human muscle expresses 17 NRs whereas mouse muscle expresses only 7 NRs. Among the 17 human muscle-expressed NRs, the higher expression of THRB, RORA, ESR1, ESRRA, NR3C2, and NR4A3 in human muscle is not seen in mouse muscle (Tables 4 and 5). Therefore, this indicates that these receptors were evolutionally gained, and addition of these NRs in humans may be responsible for the development of new muscle functions in response to environmental changes/nutritional changes that humans face. Furthermore, nearly half of the tissues examined (including the heart, liver, pancreas, brain, and lymph node) did not contain a large variety of NRs at high expression levels. These results suggested that the gene expression, differentiation, and function of these tissues may be largely dependent on those expressed NRs rather than the non-expressed NRs. Similarly, the human skin, spleen, stomach, vascular, blood, and lung tissue had minimal varieties of nuclear receptors in physiological conditions, since less than 4 out of 48 nuclear receptors are highly expressed (Additional file 2: Figure S2).
Table 4.
28 out of 43 nuclear receptors in classes I–IV are highly expressed in the human muscle, trachea, nerve, and other tissues
| Gene | Human tissues | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adipose tissue | Adrenal gland | Brain | Eye | Heart | Intestine | Kidney | Liver | Lymph node | Muscle | Nerve | Pancreas | Skin | Spleen | Stomach | Trachea | |
| Class I—thyroid hormone receptor-like (15 out of 19) | ||||||||||||||||
| THRA | * | * | * | * | * | |||||||||||
| THRB | * | * | ||||||||||||||
| RARA | * | |||||||||||||||
| RARB | * | * | * | * | ||||||||||||
| RARG | * | |||||||||||||||
| PPARA | * | * | * | * | * | |||||||||||
| PPARD | * | * | ||||||||||||||
| PPARG | * | * | * | |||||||||||||
| NR1D1 | * | * | * | * | * | |||||||||||
| NR1D2 | * | * | ||||||||||||||
| RORA | * | * | * | |||||||||||||
| RORC | * | * | * | * | * | * | * | |||||||||
| NR1H3 | * | * | * | * | ||||||||||||
| NR1H2 | * | |||||||||||||||
| VDR | * | |||||||||||||||
| Class II—retinoid X receptor-like (5 out of 12) | ||||||||||||||||
| RXRA | * | |||||||||||||||
| RXRB | * | |||||||||||||||
| NR2C2 | * | * | * | |||||||||||||
| NR2F2 | * | * | * | * | * | |||||||||||
| NR2F6 | * | |||||||||||||||
| Class III—estrogen receptor-like (5 out of 9) | ||||||||||||||||
| ESR1 | * | * | * | |||||||||||||
| ESRRA | * | * | * | * | ||||||||||||
| NR3C2 | * | * | * | * | * | |||||||||||
| PGR | * | * | * | * | * | |||||||||||
| AR | * | * | * | |||||||||||||
| Class IV—nerve growth factor IB-like (3 out of 3) | ||||||||||||||||
| NR4A1 | * | |||||||||||||||
| NR4A2 | * | * | * | * | ||||||||||||
| NR4A3 | * | * | * | * | ||||||||||||
*High expression
Table 5.
15 out of 41 nuclear receptors in classes I–VI are highly expressed in the mouse muscle, skin, and other tissues
| Gene | Mouse tissues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenal gland | Blood | Brain | Eye | Heart | Intestine | Kidney | Liver | Lung | Lymph node | Muscle | Pancreas | Skin | Spleen | |
| Class I—thyroid hormone receptor-like (7 out of 19) | ||||||||||||||
| Thra | * | * | * | * | * | |||||||||
| Rara | * | * | * | * | ||||||||||
| Ppara | * | * | * | * | * | * | * | |||||||
| Nr1d1 | * | * | ||||||||||||
| Nr1d2 | * | * | ||||||||||||
| Nr1h2 | * | * | ||||||||||||
| Vdr | * | * | ||||||||||||
| Class II—retinoid X receptor-like (5 out of 12) | ||||||||||||||
| Rxra | * | * | ||||||||||||
| Nr2c1 | * | * | ||||||||||||
| Nr2c2 | * | |||||||||||||
| Nr2f2 | * | * | * | |||||||||||
| Nr2f6 | * | * | * | |||||||||||
| Class III—estrogen receptor-like (2 out of 9) | ||||||||||||||
| Nr3c1 | * | * | * | |||||||||||
| Ar | * | |||||||||||||
| Class VI—germ cell nuclear factor-like (1 out of 1) | ||||||||||||||
| Nr6a1 | * | * | * | * | ||||||||||
*High expression
Based on the distribution pattern of highly expressed NRs among the tissues, we classified NRs into following four groups: very highly distributed, highly distributed, moderately distributed, and scarcely distributed (Table 6). In order to determine whether very highly distributed and highly distributed groups of NRs have any functional differences from that of moderately distributed and scarcely distributed group of NRs, we analyzed the potential signaling pathways with the Ingenuity Pathway Analyzer for these two major groups of NRs. The results in Table 6 show that among the top 10 pathways examined for each group, the two major NR groups share four signaling pathways such as FXR/retinoid X receptor (RXR) activation, hepatic cholestasis, aryl hydrocarbon receptor signaling, and RAR activation. The very highly distributed and highly distributed group of NRs have six specific top pathways including peroxisome proliferator-activated receptor (PPAR) signaling, glucocorticoid receptor signaling, melatonin signaling, estrogen receptor signaling, adipogenesis pathway, and PPARα/RARα activation. In contrast, the moderately distributed and scarcely distributed groups of NRs have another six specific top pathways including Oct4 stem cell pluripotency, pregnane X receptor (PXR)/RXR activation, LPS/IL-1-mediated inhibition of RXR function, retinoic acid-mediated apoptosis signaling, 25-dihydroxyvitamin D3 (vitamin D3) receptor (VDR)/RXR activation, and liver X receptor (LXR)/RXR activation. Of note, the NRs that have vitamin A, vitamin D, and retinoids as ligands are all included in the scarcely distributed group. Therefore, these data suggest that the tissue expression levels and distribution pattern of NRs can be used as an indicator of functional differences in tissues.
Table 6.
Nuclear receptors can be classified into four groups including very highly, highly, moderately, and scarcely distributed based on their distribution in tissues. Very highly/highly distributed nuclear receptors and moderately/scarcely expressed nuclear receptors regulate different signal pathways
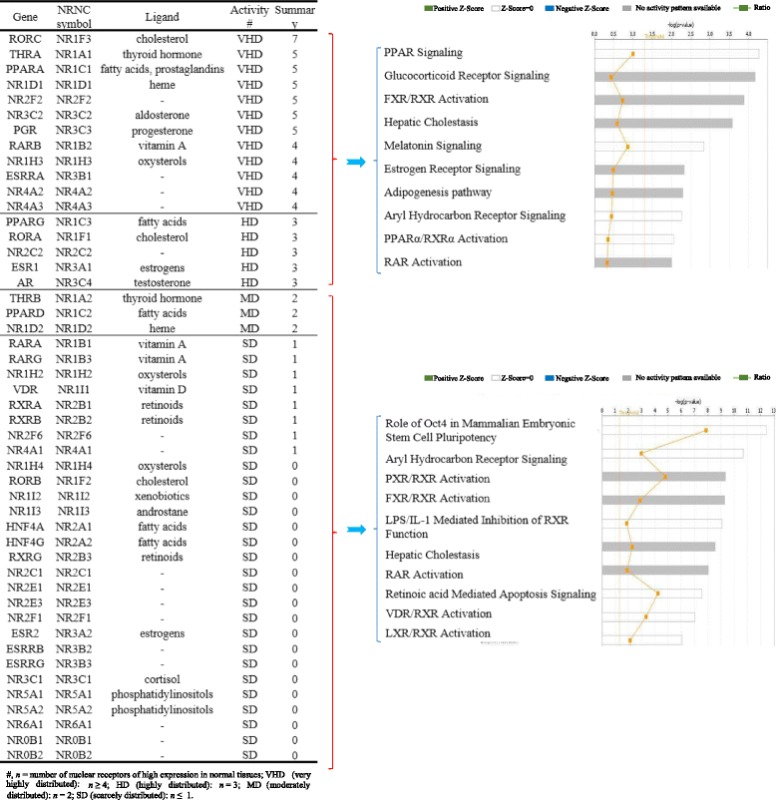
A previous paper reported mouse nuclear receptor tissue expression profile using nucleic acid-binding-based RT-PCR technique [26, 27]. However, NR superfamily expression using more accurate DNA sequencing-based technique has not been profiled in human tissues. Comparing with that reported for mouse NR expression by Bookout et al. [26], our results on highly expressed NRs have the following features (Table 7): (1) our expression sequence tag (EST)-based data were more precise; (2) our data included 21 human tissues, but the previous report only examined mouse tissues; and (3) our data implicated that there are significant differences between human and mouse NR expressions, which had never been investigated before. Our data shows that humans have more NRs expressed in the central nerve system (CNS, 19 versus 11), metabolic system (40 versus 13), and cardiovascular system (19 versus 8). Therefore, our data of NR tissue expression profiles have provided valuable insight over potential NR functions in human tissues.
Table 7.
Several findings in this study are significantly novel in comparing to what is published
| Items | Expression profile of Nuclear receptors | |
|---|---|---|
| Our findings | Cell paper (PMID: 16923397) | |
| The number of nuclear receptors | 48 known human NR | 49 known mouse NR |
| Species | Human and mouse | Mouse |
| The number of tissues | 21 human tissues and 17 mouse tissues | Only 39 mouse tissues |
| Analysis method | cDNA cloning and DNA sequencing experiments (EST database) | RT-PCR (high-throughput capacity) |
| Advantage of the method | More precise | – |
| NR groups based on their tissue distribution | – | Restricted (11), widespread (17), all tissues (21) |
| NR groups based on the expression level of nuclear receptors | Super high (12), high (5), low (3), super low activation (28) | – |
| Tissue groups based on number of highly expressed nuclear receptors | Super high (3/2 in human/mouse), high (4/5 in human/mouse), low (3/5 in human/mouse), supper low varieties (6/2 in human/mouse) | – |
| CNS (# human/mouse) | Brain, eye, nerve (19/1) | Eye, brainstem, cerebellum, cerebrum, corpus striatum, olfactory bulb, spinal cord, hypothalamus, and pituitary (11) |
| Gastroenteric system (# human/mouse) | Stomach, pancreas (5/5) | Tongue, stomach, duodenum, jejunum, ileum, colon, and gall bladder (13) |
| Metabolic system (# human/mouse) | Liver, kidney, adrenal gland, adipose, intestine, and muscle (40/14) | Liver, kidney, brown and white adipose, and muscle (13) |
| Immune system (# human/mouse) | Spleen and lymph node (4/6) | Spleen and thymus (2) |
| Cardiovascular system (# human/mouse) | Heart, lung, blood and trachea (19/6) | Aorta, heart, and lung (8) |
| Structural system (# human/mouse) | Skin (1/7) | Bone and skin (5) |
Based on the variety of NRs expressed in tissues, we classified tissues examined into three categories (Fig. 2), high variety (expressed NRs n ≥ 10; n = number of different types of highly expressed NRs), moderate variety (expressed NRs 5 ≤ n < 10), and low variety (expressed NRs n ≤ 4) in a new nuclear receptor pyramid model shown in Fig. 2 in humans. Similarly, we classified mouse NR pyramid model as high variety (expressed NRs n ≥ 7; n = numbers of the highly expressed NRs), moderate variety (expressed NRs 3 ≤ n < 7), and low variety (expressed NRs n < 3) (Fig. 2). These results suggested that the super high variety and moderate variety of NRs are found in tissues such as the muscle, trachea, and nerves in humans and in the muscle and skin in mice. Therefore, it can be concluded that these tissues may use NR pathways the most to regulate gene expression in response to developmental, physiological, and environmental stimulation. However, a high variety expression of NRs in the trachea has not been extensively reported [28]. It has been reported that NRs regulate skeletal muscle mitochondrial function [29] and the nervous system [30]. In addition, those tissues that have low variety of NRs may need fewer variety of NR pathways to regulate genes in response to developmental, physiological, and environmental stimuli; thus, they may also have other redundant pathways to carry out similar functions to that of NRs.
Fig. 2.
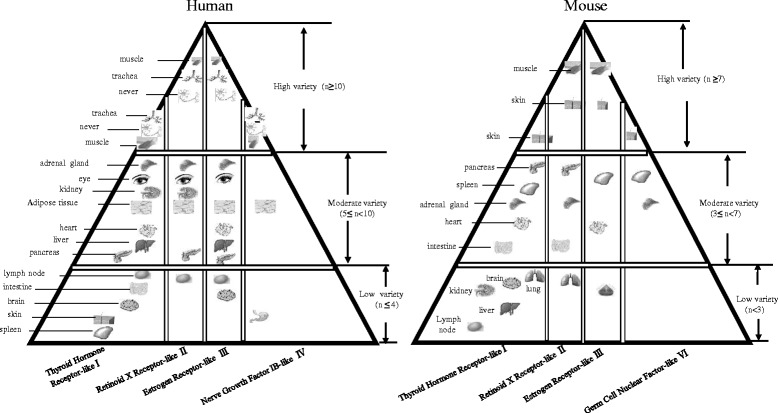
Our newly proposed “nuclear receptor pyramid” model in humans and mice constructed based on the number of variety of nuclear receptors expressed in tissues. n: the number of highly expressed nuclear receptors. Based on the numbers of NRs expressed in tissues, we classified tissues examined into three categories: high variety (expressed NRs n ≥ 10; n = numbers of the highly expressed NRs), moderate variety (expressed NRs 5 ≤ n < 10), and low variety (expressed 4 ≤ n) in a new nuclear receptor pyramid model in humans and high variety (expressed NRs n ≥ 7; n = numbers of the highly expressed NRs), moderate variety (expressed NRs 3 ≤ n < 7), and low variety (expressed NRs n < 3) in a new nuclear receptor pyramid model in mice
Correlation with oxygen sensors, angiogenic genes, and stem cell master regulators in human tissues
As shown in Table 6, NR functions in tissues may be involved in metabolism and stem cell-mediated tissue regeneration. However, it has been poorly characterized whether oxygen sensor genes such as prolyl hydroxylase domain-containing protein 2 (PHD2), hypoxia-inducible factor 1B (HIF1B), HIF1A, and HIF2A regulate NR expressions in tissues [31]. To determine the extents to which factors and NRs are related, we conducted correlation studies, with the hypothesis that if there is a positive functional correlation, the expression of the given factor (such as oxygen sensors, genes that regulate angiogenesis pathway, stem cell master genes, PRR, and inflammasome components) and the NR will increase or decrease together [1]. Similarly, we analyzed the correlation between NR expression and tissue methylation indices determined by the ratios between S-adenosyl methionine (SAM—the universal methyl donor)/S-adenosyl homocysteine (SAH—a methyltransferase inhibitor) and SAH levels [32].
As shown in Fig. 3a, b, we examined whether highly expressed NR potential (highly expressed NRs/total NRs × 100%) in tissues are correlated with tissue expression of four oxygen-sensing genes including PHD2, HIF1B, HIF1A, and HIF2A and seven vascular endothelial growth factor (VEGF) pathway genes including VEGFA, VEGFB, VEGFC, FIGF, FLT1, KDR, and FLT4, as well as six stem cell master genes including CD34, KIT, and four Yamanaka’s inducible pluripotent stem cell (IPSC) genes such as Myc, Kruppel-like factor 4 (KLF4), POU5F1 (octamer-binding transcription factor 4 (Oct4)), and sex determining region Y (SRY)-box 2 (Sox2) [33]. As shown in Fig. 3b, c, among 17 genes examined, the correlation of seven genes achieved statistical significance (p < 0.05). The highly expressed NR potentials were highly correlated with oxygen-sensing genes PHD2, HIF1B, and stem cell master regulator gene Sox2 (high correlation r 2 > 0.7). A moderate correlation was observed between highly expressed NRs and HIF1A, VEGFB, and KIT genes (0.5 ≤ r 2 ≤ 0.7). Low level correlation was observed between FLT1 and highly expressed NRs (r 2 < 0.5). These results suggested that the expression of oxygen-sensing genes PHD2, HIF1B, and HIF1A, VEGF pathway gene VEGFB and stem cell master gene SOX2, and KIT have a positive correlation with NR expression, and these genes may be either upstream regulators or downstream targets of NR signaling pathways.
Fig. 3.
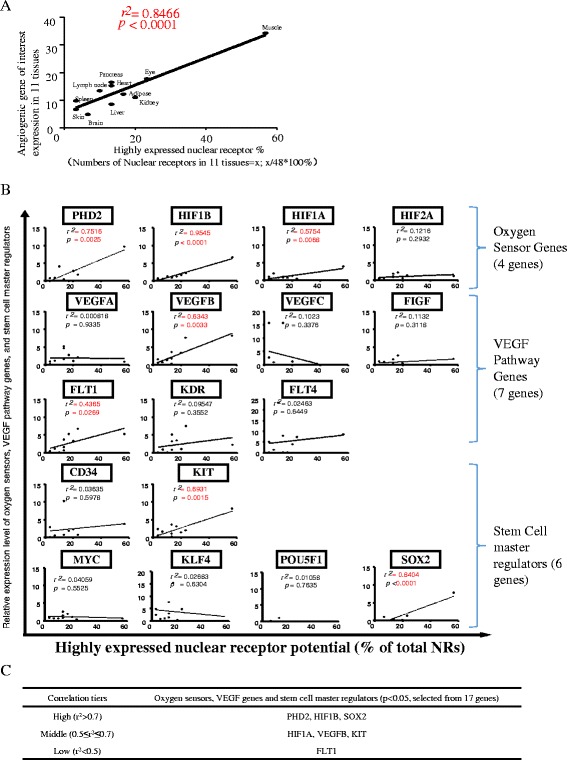
Oxygen sensors, VEGF pathway regulators, and stem cell master regulators may regulate nuclear receptor expression in human tissues (tissues: adipose, brain, eye, heart, kidney, liver, lymph node, muscle, pancreas, skin, spleen). a Highly expressed nuclear receptors in 11 tissues were strongly associated with angiogenic gene expression. b Correlation between highly expressed nuclear receptors and gene that regulate oxygen sensing, angiogenesis, and stem cells. c Correlation tiers between genes of interests and nuclear receptor expression in tissues. Abbreviations: PHD2: prolyl hydroxylase domain-containing protein 2; HIF1B: hypoxia-inducible factor-1 beta; HIF1/2A: hypoxia-inducible factor 1/2-alpha; VEGFA/B/C: vascular endothelial growth factor A/B/C; FIGF: C-fos-induced growth factor; FLT1/4: Fms related tyrosine kinase ¼; KDR: kinase insert domain receptor; MYC: MYC proto-oncogene; KIT: KIT proto-oncogene receptor tyrosine kinase; KLF4: Kruppel-like factor 4; POU5F1: POU class 5 homeobox 1; SOX2: SRY-box 2
Correlation with PRRs in human tissues
Additionally, we addressed the question whether the highly expressed NRs have a positive correlation with the expression of PRR genes such as NLRs, AIM-2 (absent in melanoma-2), and IFI16 (interferon gamma-inducible protein 16) or genes of inflammasome components such as ASC (apoptosis speck-like CARD-containing protein) and CARD8 (caspase recruitment domain family member 8) [2, 8, 34]. As shown in Fig. 4, among 14 inflammasome-related genes examined, four PRR genes achieved statistically significant correlations (p < 0.05). The highly expressed NR potentials were highly correlated with microbial infection-sensing NOD1 [35] (high correlation r 2 > 0.7), moderately correlated with NOD2 and NOD4 (0.5 ≤ r 2 ≤ 0.7), and a weak correlation with nuclear DNA damage-sensing PRR IFI16 (r 2 < 0.5) [36].
Fig. 4.
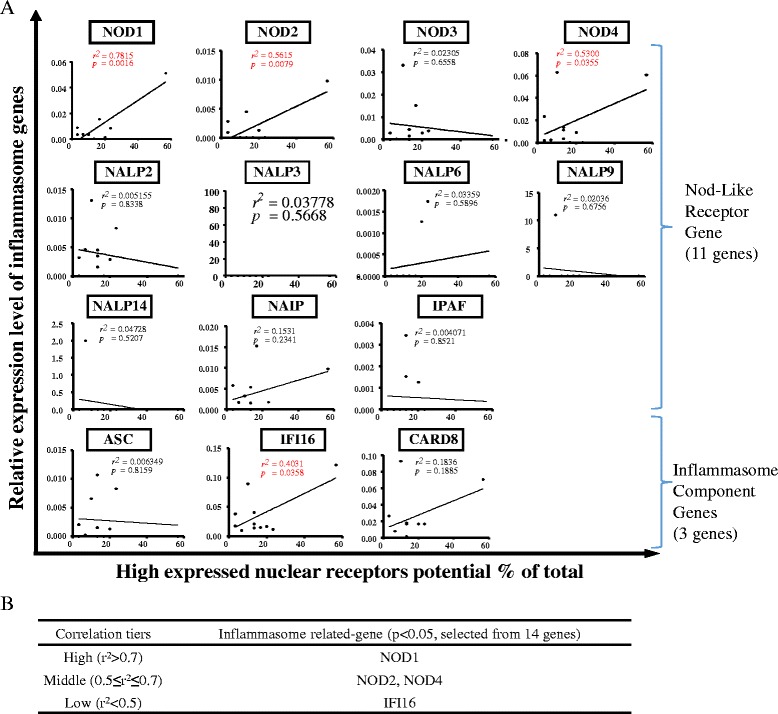
NLRs/inflammasome sensors may be either upstream regulators or downstream targets of nuclear receptors in human tissues (tissues: adipose, brain, eye, heart, kidney, liver, lymph node, muscle, pancreas, skin, spleen). a Correlation between inflammasome genes and highly expressed nuclear receptors. b Different correlation tiers show the level of statistically significant correlation between inflammasome genes and highly expressed nuclear receptors. Abbreviations: NOD1/2/3/4: nucleotide-binding oligomerization domain-like receptors 1/2/3/4; NALP2/3/6/9/14: NLR family pyrin domain containing 2/3/6/9/14; NAIP: NLR family apoptosis inhibitory protein; NLRC4: NLR family CARD domain containing 4; ASC: PYD and CARD domain containing; IFI16: interferon gamma-inducible protein 16; CARD8: caspase recruitment domain family member 8
It has been reported that nucleotide-binding oligomerization domain (NOD) proteins such as NOD1 and NOD2 are founding members of the NLR family, sense conserved motifs in bacterial peptidoglycan, and induce pro-inflammatory and anti-microbial responses [35]. It should be noted that three out of four PRRs, which include NOD1, NOD2, and NOD4 that are positively correlated with highly expressed NRs, activate inflammatory cascade independent of caspase-1 inflammasome complex. Nevertheless, IFI16, which is a PRR dominantly localized in the nucleus is a constituent of caspase-1 inflammasome complex, but it has a weak correlation with highly expressed NRs [37, 38]. Furthermore, NLRP3 [34], a PRR that is well identified as a component of caspase-1 inflammasome complex also failed to achieve a statistically significant correlation with highly expressed NRs. Therefore, this suggests that PRRs such as NOD1, NOD2, and NOD4 that function independently of caspase-1 are either upstream regulators or downstream targets of highly expressed NRs.
Correlation with methylation index in mouse tissues
DNA methylation has been recognized as one of the regulatory mechanisms underlying the expression of some NRs [39]. However, the question remains whether tissue methylation status regulates NR expression. There are two main intermediate compounds that determine the potential for methylation/demethylation in biological systems. S-adenosyl methionine (SAM) acts as a major methyl donor for many cellular methylation reactions of DNA, RNA, proteins, and lipids. In contrast, S-adenosyl homocysteine (SAH) is a potent inhibitor of biological transmethylation [40].
To determine whether tissue methylation level determines NR expression, first we measured the tissue levels of SAH and SAM in six mouse tissues including the liver, brain, heart, kidney, lung, and spleen using liquid chromatography-electrospray ionization-tandem mass spectrometry [41]. We then analyzed the potential correlation between highly expressed NRs and the tissue hypomethylation determined by SAH (methyltransferase inhibitor) levels. Similarly, we examined whether a positive correlation exists between highly expressed NRs and tissue hypermethylation status determined by SAM/SAH (Fig. 5). Our data implicated that the NRs that undergo expression changes based on tissue methylation and demethylation status are mutually exclusive as we reported before [32].
Fig. 5.
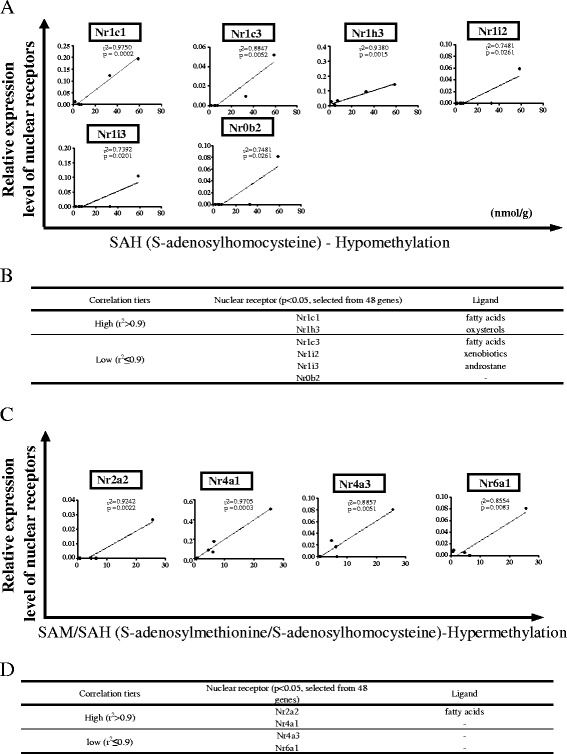
Tissue methylation status may determine the expression level of nuclear receptors in mouse tissues (tissues: mouse tissues: liver, brain, heart, kidney, lung, and spleen). a Correlation between nuclear receptors and hypomethylation status of the mouse tissues. b Different correlation tiers that depict the degree of association between hypomethylation status and nuclear receptor expression in mouse tissues. c Correlation between nuclear receptor expression and hypermethylation status of the mouse tissues. d Correlation tiers of hypermethylation status of the mouse tissues and nuclear receptor expression
As shown in Fig. 5a, among 48 NRs examined, 6 NRs showed a statistically significant positive correlation between NR expression and tissue hypomethylation status (p < 0.05). Two NRs including Nr1c1 (Pparα) and Nr1h3 were highly correlated with SAH levels in six tissues including the liver, brain, heart, kidney, lung, and spleen (high correlation r 2 > 0.9); four NRs including Nr1c3 (Pparγ), Nr1i2, Nr1i3, and Nr0b2 were moderately correlated with SAH levels (0.7 ≤ r 2 ≤ 0.9). Notably, most of the receptors that had increased expression levels in the presence of hypomethylation fall to class I NRs. Previously, it was shown that nutritional status can alter the methylation status of the PPARα gene and subsequently regulate its expression level both in rodent models and in humans [42]. It is highly likely that function of these receptors may also be increased during hypomethylation status as it provides easy access to these NRs to reach their response elements. Despite the observation that expression of certain class I NRs are increased in hypomethyation status, further experiments are needed to validate whether the function of these receptors are also enhanced.
In addition, as shown in Fig. 5c, d, among 48 NRs examined, 4 NRs achieved statistically significant correlation (p < 0.05) with tissue hypermethylation status. Two NRs including Nr2a2 and Nr4a1 were highly correlated with the SAM/SAH ratio in six tissues including the liver, brain, heart, kidney, lung, and spleen (high correlation r 2 > 0.9); two NRs including Nr4a3 and Nr6a1 show moderate correlation with the SAM/SAH ratio (0.8 ≤ r 2 ≤ 0.9). These results suggested that tissue hypermethylation status differentially regulates the tissue expression of NRs, and the tissue expression of five NRs may be significantly upregulated by hypermethylation. These results have demonstrated for the first time that tissue hypomethylation and hypermethylation status may have an impact on expression levels of two mutually exclusive groups of NRs [43].
We acknowledge that our data is not adequate to conclude that the NR expression changes that we observed are due to direct hypermethylation/hypomethylation of the particular NR gene. Tissue methylation status may regulate NR expression indirectly via other mediators. Though we did not observe expression changes on estrogen receptor-alpha (ER-α) in our mouse dataset, previously it was shown that methylation status of the ER-α gene determines its expression in the colon, blood, lung, heart, prostrate, and ovary in humans [44–48]. This was extensively studied in human breast cancer cell lines, where loss of ER expression and acquired hormone resistance was attributed to hypermethylation of the ER gene [44, 49]. Therefore, to conclude whether the NR expression changes we observed are due to direct methylation of the particular genes requires further experimental validation. Furthermore, pathophysiological relevance of the methylation status of the tissues and NR expression need to be tested in the future. Also, it is possible that upregulation of certain NRs may regulate the tissue methylation status via unknown pathways.
It should be noted that the values that determined the correlation tiers in Fig. 5 are different from those of Figs. 3 and 4. This is because the basal levels were different as Fig. 5 demonstrates the data obtained from mice and Figs. 3 and 4 depict the data obtained from human tissues. Also, when analyzing the correlation, NR potential was taken into account in Figs. 3 and 4, while the correlation was calculated for each and individual NR in Fig. 5.
Nuclear receptor sequence changes and mutations are associated with increased risk for development of metabolic, cardiovascular, and autoimmune diseases, hormone insensitivity/resistance, and cancers
Genome-wide association studies (GWASs) have investigated potential genetic factors that explain inter-individual variations in response to NR ligand stimulations in various pathologies [50]. Given that susceptibility to complex human metabolic diseases is likely a result of genes operating as part of functional modules rather than individual effects, association analysis methods hold promise in discovering additional associations from existing GWAS data [51]. Previous GWAS studies have been reported for NRs in some diseases such as liver injury [50], osteoporosis, sarcopenia, and obesity. However, it is unclear whether the GWAS data on NRs are associated with globally increased genetic risks for metabolic diseases and autoimmune disease, such as rheumatoid arthritis, obesity, diabetes, and vascular atherosclerosis in human populations.
To address this issue, we examined the GWAS database (http://www.gwascentral.org/) for all the NRs. As shown in Tables 8 and 9, 45 out of 48 NRs with sequence changes or mutations were associated with rheumatoid arthritis, obesity, diabetes, and vascular disease and atherosclerosis. In addition, two NRs such as PPARA and NR3C2 variations were associated with certain lipid metabolite traits (Table 8). Despite the fact that AR exerts pro-inflammatory effects like PPARD and RXRA, it was much less associated with development of obesity and diabetes unlike PPARD and RXRA (Table 8). Finally, NR2F2 variations were not associated with the diseases examined except in one diabetes study.
Table 8.
45 out of 48 nuclear receptors with sequence changes or mutations are associated with increased risks of human rheumatoid arthritis, obese, diabetes, and metabolic vascular diseases
| Gene | Diseases | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rheumatoid arthritis | Obese | Diabetes | Vascular dis. and atherosclerosis | ||||||||||||||||||||
| Phenotype ID (HGVPM) | Phenotype ID (HGVPM) | Phenotype ID (HGVPM) | Phenotype ID (HGVPM) | ||||||||||||||||||||
| 38 | 235 | 564 | 1111 | 1114 | 4 | 5 | 74 | 822 | 825 | 826 | 827 | 569 | 769 | 1081 | 1538 | 3639 | 563 | 568 | 3602 | 3611 | 3640 | 3616 | |
| THRA | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| THRB | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||
| RARA | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||||
| RARB | * | * | * | * | * | * | * | * | * | * | |||||||||||||
| RARG | * | * | * | * | * | * | * | * | * | ||||||||||||||
| PPARA | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||
| PPARD | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| PPARG | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||||
| NR1D1 | * | * | * | * | * | * | * | * | * | * | * | * | |||||||||||
| NR1D2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| RORA | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||
| RORB | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| RORC | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| NR1H3 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR1H2 | * | * | * | * | * | * | * | * | |||||||||||||||
| NR1H4 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| VDR | * | * | * | * | * | * | * | * | * | * | * | ||||||||||||
| NR1I2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| NR1I3 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| HNF4A | * | * | * | * | * | * | * | * | * | * | * | * | |||||||||||
| HNF4G | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| RXRA | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| RXRG | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR2C1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR2C2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR2E1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR2E3 | * | * | * | * | * | * | * | * | * | ||||||||||||||
| NR2F1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| NR2F2 | * | ||||||||||||||||||||||
| NR2F6 | * | * | * | * | * | * | * | * | * | ||||||||||||||
| ESR1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| ESR2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| ESRRA | * | * | * | * | * | * | * | ||||||||||||||||
| ESRRB | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| ESRRG | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR3C1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR3C2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||
| PGR | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| AR | * | * | * | * | * | * | * | ||||||||||||||||
| NR4A1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR4A2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR4A3 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR5A1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||
| NR5A2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| NR6A1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
Nuclear receptors marked with bold have a pro-inflammatory role
Table 9.
Study ID and phenotype ID from Table 8
| Disease | Study ID (HGVST) | Study name (GWAS) | Phenotype ID (HGVPM) | Phenotype property | Title (phenotype HGVPM) |
|---|---|---|---|---|---|
| Rheumatoid arthritis | 27 | Rheumatoid arthritis | 38 | Anti-cyclic citrullinated peptide-positive rheumatoid arthritis | 38: Stage 1 anti-CCP-positive rheumatoid arthritis status |
| 185 | Rheumatoid arthritis in the Spanish population | 235 | Rheumatoid arthritis | 235: Rheumatoid arthritis | |
| Obese | 640 | Body mass index | 1111 | Body mass index | 1111: Phenotype method for body mass index |
| 308 | Adult body mass index in a British population | 564 | Body mass index | 564: Adult body mass index measurement | |
| 641 | Meta-analysis of 32 genome-wide association studies for waist-hip ratio adjusted for body mass index | 1114 | Waist-hip ratio | 1114: Phenotype method for waist-hip ratio | |
| Diabetes | 463 | Glycemic traits | 825 | Fasting glucose-related: fasting plasma glucose | 825: Phenotype method for fasting glucose-related: fasting plasma glucose |
| 463 | Glycemic traits | 827 | Fasting glucose-related: homeostatic model assessment of beta-cell function | 827: Phenotype method for fasting glucose-related: homeostatic model assessment of beta-cell function | |
| 463 | Glycemic traits | 822 | Fasting insulin-related: fasting insulin | 822: Phenotype method for fasting insulin-related: fasting insulin | |
| 463 | Glycemic traits | 826 | Fasting insulin-related: homeostatic model assessment of insulin resistance | 826: Phenotype method for fasting insulin-related: homeostatic model assessment of insulin resistance | |
| 618 | Glycated hemoglobin levels | 1081 | Glycated hemoglobin levels | 1081: Phenotype method for glycated hemoglobin levels | |
| 313 | Log10 glycosylated hemoglobin in a British population | 569 | Log10 glycosylated hemoglobin | 569: Log10 glycosylated hemoglobin measurement | |
| 433 | Glucose levels 2 h after an oral glucose challenge | 769 | 2-h glucose challenge | 769: Phenotype method for 2-h glucose challenge | |
| 5 | Type II diabetes mellitus | 4 | Type II diabetes | 4: T2D status | |
| 3 | Type II diabetes mellitus | 5 | Type II diabetes | 5: T2D status | |
| 52 | Type II diabetes mellitus in American Indians | 74 | Early onset type II diabetes mellitus | 74: Phenotype method forearly onset type II diabetes mellitus | |
| 907 | Proinsulin levels | 1538 | Proinsulin levels | 1538: Phenotype method for proinsulin levels | |
| 1827 | Metabolite quantitative traits | 3639 | a-Glucose | 3639: Phenotype method for a-glucose | |
| Metabolic vascular disease | 312 | Serum cholesterol levels in a British population | 568 | Serum cholesterol | 568: Serum cholesterol measurement |
| 1827 | Metabolite quantitative traits | 3602 | Lipids (CH3) | 3602: Phenotype method for Lipids (CH3) | |
| 1827 | Metabolite quantitative traits | 3611 | Lipids (CH2) | 3611: Phenotype method for Lipids (CH2) | |
| 1827 | Metabolite quantitative traits | 3616 | Lipids (CH2CO) | 3616: Phenotype method for Lipids (CH2CO) | |
| 1827 | Metabolite quantitative traits | 3640 | Lipids (CH=CH*CH2CH2) | 3640: Phenotype method for Lipids (CH=CH*CH2CH2) | |
| 307 | Systolic blood pressure in a British population | 563 | Systolic blood pressure | 563: Systolic blood pressure measurement |
These results suggest that NRs may be very important factors in determining the susceptibility and progression of metabolic disorders including obesity, diabetes, and atherosclerosis. Also, our GWAS analysis suggests that NRs may play an important role in the progression of autoimmune disorders such as rheumatoid arthritis. Most interestingly, the NR mutations associated with various metabolic disorders and autoimmune diseases are different. This observation can be supported by multiple publications that had demonstrated NRs play an important role in immune cells. Especially, the PPARs are highly expressed in human CD4+ T cells [52], and the role of PPAR agonists in the treatment of autoimmune disorders had been extensively discussed [53, 54]. It was shown that activation of T cells was dramatically decreased in the presence of PPARA and PPARG agonists and suppressed pro-inflammatory cytokine secretion [52]. In addition to PPARs, other NRs such as ROR-γt were found to regulate differentiation of CD4+ T helper 17 (Th17) subset [55]. However, RAR/RXR dimerization exerts contrasting effects to that of ROR-γt by enhancing Foxp3 transcription factor positive inducible T-regulatory cells (Tregs) while inhibiting Th17 differentiation [56]. Therefore, it is evident that the cross talk between NRs play a critical role in the immunity and development of autoimmune disorders.
In addition to the GWAS analysis, we further performed the cause-effect analysis using the mouse genome informatics (MGI) database (www.mousemine.org) that contains a comprehensive compilation of genomic and phenotypic data from NR transgenic and gene knockout mouse models. In Table 10, 26 NR deficiencies lead to four groups of abnormalities, including (1) hormone insufficiency/insensitivity/resistance, (2) cancers, (3) autoimmune/immunodeficiency diseases, and (4) metabolic cardiovascular diseases. The NRs in the hormone insufficiency/insensitivity/resistance group include NRs THRA, THRB, RARB, NR1H3, VDR, NR2E3, NR2F1, NR2F2, ESR1, ESRRB, NR3C1, NR3C2, PGR, AR, NR4A2, NR4A3, NR5A1, and NR0B1. The NRs in the cancer group include RARA (acute promyelocytic leukemia), NR1H4 (hepatocellular carcinoma), ESR1 (breast cancer), and AR (prostate cancer). The NRs in the autoimmune/immunodeficiency disease group include PPARG (systemic lupus erythematosus), RORC (immunodeficiency), and RXRA (systemic lupus erythematosus). The NRs in the metabolic cardiovascular diseases group include RARA (cardiomyopathy), PPARD (diabetes mellitus), PPARG (diabetes mellitus, lipodystrophy, obesity, pulmonary hypertension), HNF4A (diabetes mellitus), NR2F2 (congenital heart defects), ESR1 (myocardial infarction), NR3C2 (hypertension), AR (obesity), and NR0B2 (obesity). Taken together, the results of the GWAS association studies and MGI casual analysis suggest that NRs play significant roles in maintaining the homeostasis and suppressing various hormonal diseases, cancers, autoimmune diseases/immunodeficiency, and metabolic cardiovascular diseases.
Table 10.
The Mouse Genome Informatics (MGI) database shows that 26 nuclear receptor deficiencies lead to abnormal metabolism and endocrine and cardiovascular phenotypes in mice
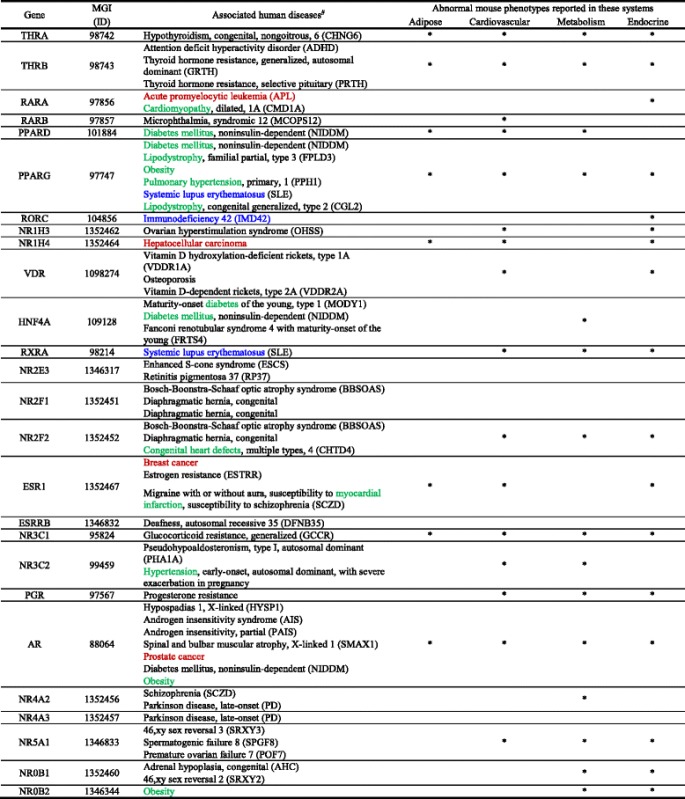
#Disease association of human genes are from the NCBI mim2gene_medgen file and include annotations from OMIM, NCBI curation, Gene. *Abnormal mouse phenotypes. Of note, red fonts indicate cancers, blue fonts indicate autoimmune diseases, and green fonts indicate metabolic disorders such as obesity
To further determine whether many NRs serve as homeostasis-associated molecular pattern receptors (HAMPRs) by inhibiting inflammation, we conducted an extensive literature survey to find out experimentally validated data to prove our hypothesis. As shown in Table 11, the level of 10 hormone ligands of NRs was changed with inflammatory diseases. The ligands of class-I thyroid hormone receptor-like group including vitamin A, fatty acids, and prostaglandins levels were reduced in the presence of inflammatory disorders, suggesting that they have the potential to exert anti-inflammatory effects. In addition, retinoids and estrogen inhibited inflammatory intestinal disease and atherosclerosis respectively. Moreover, testosterone suppressed Crohn’s disease.
Table 11.
Hormone ligand level changes are associated with inflammatory diseases
| NRNC symbol | Ligand(s) | Inflammatory disease | Ligand level change | PMID |
|---|---|---|---|---|
| Class I—thyroid hormone receptor-like | ||||
| NR1A1 | Thyroid hormone | Inflammatory bowel diseases | ↑ | 8562993 |
| NR1A2 | ||||
| NR1B1 | Vitamin A | Chronic obstructive pulmonary disease | ↓ | 26339144 |
| NR1B2 | ||||
| NR1B3 | ||||
| NR1C1 | Fatty acids, prostaglandins | Inflammatory bowel disease | ↓ | 27631140 |
| NR1C2 | ||||
| NR1C3 | ||||
| NR1F1 | Cholesterol | Atherosclerotic cardiovascular disease | ↑ | 21686232 |
| NR1F2 | ||||
| NR1F3 | ||||
| NR1H3 | Oxysterols | Inflammatory bowel diseases | ↑ | 24024145 |
| NR1H2 | ||||
| NR1H4 | ||||
| Class II—retinoid X receptor-like | ||||
| NR2B1 | Retinoids | Inflammatory intestinal disease | ↓ | 23690441 |
| NR2B2 | ||||
| NR2B3 | ||||
| Class III—estrogen receptor-like | ||||
| NR3A1 | Estrogens | Atherosclerosis | ↓ | 12816884 |
| NR3C1 | Cortisol | Obesity | ↑ | 12466357 |
| NR3C2 | Aldosterone | Renal fibrosis | ↑ | 26730742 |
| NR3C4 | Testosterone | Crohn’s disease | ↓ | 26020563 |
Finally, we also searched for the evidence in the literature where gene knockout and activation approaches of NRs were used to determine the pathological phenotypes. Twelve out of 15 NRs including NR1A1, NR1C3, NR1D1, NR1H3, NR1H2, NR1H4, NR2F2, NR3A1, NR3B2, NR4A1, NR4A3, and NR0B2 have anti-inflammatory roles as shown in Table 12. The three NRs NR1C2, NR2B1, and NR3C4 did not show any anti-inflammatory properties. Taken together, these results suggested that most human and mouse NRs have anti-inflammatory functions in various tissues and cell types.
Table 12.
12 out of 15 nuclear receptors have anti-inflammatory roles reported in the literature
| Gene name (full name) | NRNC symbol | Tissue/cell type | Purpose | Treat | Suppressed | Induced | PMID | Inflam. |
|---|---|---|---|---|---|---|---|---|
| Cytokines/signaling | ||||||||
| Class I—thyroid hormone receptor-like | ||||||||
| THRA | NR1A1 | Aorta macrophages | Atherosclerosis | KO | – | IL-1β, NFκB, TNF-α | 24797634 | Anti |
| PPARG | NR1C3 | Mouse cancer model | Tumor growth and angiogenesis | Act | IL-17 | – | 23619236 | Anti |
| NR1D1 | NR1D1 | Peritoneal macrophages | Aging- or obesity-associated impairment of clockwork and inflammation | Act | Ccl2, ERK, p38 | – | 24307731 | Anti |
| Mice macrophages | Circadian clockwork and inflammatory disease | KO | – | IL-6 | 22184247 | |||
| NR1H3 | NR1H3 | Mice plasma and kidney | Normal and diabetic kidney | KO | – | Nox2, Ncf1, MDA, TLR2, ICAM1, IL-1β, CD68 | 24201575 | Anti |
| NR1H2 | NR1H2 | Mice plasma and kidney | Normal and diabetic kidney | KO | – | Nox2, Ncf1, TLR2, ICAM1, IL-1β, CD69, MDA(urinary) | 24201575 | Anti |
| Macrophage cell line | LPS treat | Act | TNF-α, IL-1β, IL-6, IL-12p40 | – | 23099324 | |||
| ob/ob mouse liver | Cellular lipid metabolism | Block | – | Cox-2, MCP-1, MIP-2 | 24206663 | |||
| NR1H4 | NR1H4 | Obese mice liver | Obesity-related metabolite disorder | Ace | – | Mmp13, Cxcl2, Cxcl8, Cxcl14, IL-1β, IL-6, TNF-α | 25425577 | Anti |
| PPARD | NR1C2 | Epithelial cells | Act | – | COX-2 | 24763687 | Pro | |
| Class II—retinoid X receptor-like | ||||||||
| RXRA | NR2B1 | Spleen macrophages | Age-related disease | Act | – | COX-2, NF-kB, IL-6 | 24051096 | Pro |
| NR2F2 | NR2F2 | Prostate cancer | Prostate cancer | Act | TGF-β | – | 23201680 | Anti |
| Class III—estrogen receptor-like | ||||||||
| ESR1 | NR3A1 | Male mice | Obesity | KO | IL-10 | IL-1β, TNF-α, IL-6 | 25373903 | Anti |
| Astrocytes | Neuroprotective | Act | CCL2, CCL7 | – | 23804112 | |||
| ESR3 | NR3B2 | Mice | Intestine tumor | KO | – | TGF-β | 24104551 | Anti |
| AR | NR3C4 | Prostate cancer cells | Prostate tumorigenesis | KD | – | AKT | 25527506 | Pro |
| Hepatocellular carcinoma cells | Cell adhesion and migration | KO | – | PI3K/AKT | 24944078 | |||
| Class IV—nerve growth factor IB-like | ||||||||
| NR4A1 | NR4A1 | Macrophages | Atherosclerotic lesions | KO | – | IL-4 | 23288947 | Anti |
| Bone marrow-derived macrophages (BMM) | Atherosclerotic lesions | KO | – | IL-12, IFN-δ, SDF-1α | 22194623 | |||
| Macrophage | Atherosclerotic lesions | KO | – | TNF-α, TLR-4, NFκB | 22194622 | |||
| NR4A3 | NR4A3 | Mast cells | Vascular biology and inflammation | KO | – | IL-13, MCP-1, TNF-α | 24586680 | Anti |
| Hematopoietic stem cells | Atherosclerotic lesions | KO | – | Ly6C(+) monocytes | 24806827 | |||
| Endothelial cells | Atherosclerotic lesions | KO | – | VCAM-1, ICAM-1 | 20558821 | |||
| Class O—miscellaneous | ||||||||
| NR0B2 | NR0B2 | Mice kidney | Inflammasome | KO | – | IL-1β, IL-18, NLRP3, ASC | 25655831 | Anti |
Abbreviations: KO knockout, Act activation, Ace acetylation, Cxcl Cxc ligand, IL interleukin, MCP monocyte chemotactic protein, Mmp matrix metallopeptidase, TLR Toll-like receptor, VCAM vascular cell adhesion molecule, ICAM intercellular adhesion molecule, Inflam inflammation, Anti anti-inflammatory, Pro pro-inflammatory
Nuclear receptors have the tendency to be downregulated than being upregulated in autoimmune and metabolic diseases and cancers
In order to determine the overall roles of NRs in modulating the pathogenesis of human autoimmune diseases, metabolic diseases, and cancer, we examined the expression changes of 48 NRs in eight human diseases using the microarray datasets (https://www.ncbi.nlm.nih.gov/gds/) deposited by other investigators in the NIH-GEO dataset database. The microarray datasets we analyzed were conducted on various pathological settings including autoimmune disease rheumatoid arthritis, and five metabolic diseases such as familial hypercholesterolemia, type 2 diabetes, type 1 diabetes, obesity, hyperhomocysteinemia, and also hypertension. We analyzed The Cancer Genome Atlas (TCGA) database to determine NR expression changes in human cancers.
As shown in Table 13 (A), three NRs were upregulated but nine NRs were downregulated in the synovial tissue of patients with rheumatoid arthritis. Similarly, in Table 13 (B), 7 NRs were upregulated and 11 NRs were downregulated in T cells from patients with familial hypercholesterolemia. Also, we analyzed the monocytes isolated from patients with familial hypercholesterolemia, peripheral blood from patients with metabolic syndrome, arterial tissue from patients with type 2 diabetes, peripheral blood mononuclear cells from patients with type 1 diabetes, adipose stem cells and omental adipose tissue from morbidly obese patients, aortic smooth muscle cells from patients with hyperhomocysteinemia, and carotid artery atheromatous plaques from patients with hypertension. The results showed that NRs have the tendency to be downregulated during metabolic disorders and autoimmune disorders rather than being upregulated. However, this trend was not observed in morbidly obese patients where equal numbers of NRs were upregulated and downregulated (Table 13 (B)). To further consolidate the finding, we analyzed the NR expression changes in the presence of proatherogenic stimulus oxidized low-density lipoprotein (Ox-LDL) in human aortic endothelial cells (HAECs). This analysis also showed that NRs tend to be downregulated than upregulated with prolonged Ox-LDL treatment (Table 13 (C)). Taken together, these results suggested that NRs have the tendency to be downregulated than upregulated during human autoimmune rheumatoid arthritis and metabolic diseases, and this tendency of NRs was more obvious in autoimmune arthritis than in metabolic diseases.
Table 13.
Nuclear receptors are more downregulated than upregulated in human diseases
| Disease | Tissue/cell type | Number | Upregulated gene | Downregulated gene | PMID/GEO ID | |||
|---|---|---|---|---|---|---|---|---|
| Up | Down | |||||||
| A. Nuclear receptors have the tendency to be downregulated than being upregulated in rheumatoid arthritis | ||||||||
| Fold* | Fold* | |||||||
| ∆ | ∆ | |||||||
| Rheumatoid arthritis | Synovial tissue | 3 | 9 | NR1H3 | 2.08 | NR1A1 | −20 | 24690414/GSE55235 |
| NR1I1 | 2.12 | NR1C3 | −2.56 | |||||
| NR3A1 | 2.59 | NR1D1 | −20 | |||||
| NR2F1 | −2.94 | |||||||
| NR3C3 | −2 | |||||||
| NR3C4 | −2.22 | |||||||
| NR4A1 | −4.76 | |||||||
| NR4A2 | −10 | |||||||
| NR4A3 | −3.85 | |||||||
| B. Nuclear receptors are more downregulated than upregulated in metabolic diseases in humans | ||||||||
| Fold** | Fold** | |||||||
| ∆ | ∆ | |||||||
| Family hypercholesterolemia | T cells | 7 | 11 | NR1B3 | 1.41 | NR1A1 | −1.3 | –/GSE6088 |
| NR1C3 | 1.94 | NR1B1 | −1.22 | |||||
| NR1I1 | 1.75 | NR1B2 | −1.72 | |||||
| NR3C4 | 1.97 | NR1C1 | −1.72 | |||||
| NR4A2 | 2.03 | NR1F1 | −2 | |||||
| NR4A3 | 1.5 | NR1F2 | −1.79 | |||||
| NR0B1 | 2.36 | NR1H3 | −1.28 | |||||
| NR3B3 | −2.08 | |||||||
| NR3C1 | −1.22 | |||||||
| Family hypercholesterolemia | Monocytes | 10 | 12 | NR1B1 | 1.23 | NR1A1 | −2.27 | 19040724/GSE6054 |
| NR1F3 | 2.38 | NR1B2 | −1.72 | |||||
| NR1I1 | 1.31 | NR1C1 | −1.59 | |||||
| NR2F6 | 2.61 | NR1F1 | −1.52 | |||||
| NR3B2 | 1.33 | NR1H3 | −1.28 | |||||
| NR4A1 | 1.94 | NR2A2 | −2.13 | |||||
| NR4A2 | 2.06 | NR2C1 | −1.47 | |||||
| NR5A1 | 2.1 | NR2C2 | −1.19 | |||||
| NR6A1 | 1.62 | NR2E3 | −2.63 | |||||
| NR0B1 | 2.02 | NR3A1 | −2 | |||||
| NR3A2 | −1.79 | |||||||
| NR3C1 | −1.28 | |||||||
| Metabolic syndrome | Peripheral blood | 0 | 1 | NR4A3 | −1.54 | 21368773/GSE23561 | ||
| Type 2 diabetes | Arterial tissue | 0 | 2 | NR1B2 | −1.28 | 22340758/GSE13760 | ||
| NR3C3 | −1.12 | |||||||
| Type 1 diabetes | Peripheral blood mononuclear cell | 1 | 2 | NR3C4 | 1.2 | NR1F1 | −1.89 | –/GSE55100 |
| NR4A3 | −1.2 | |||||||
| Morbidly obese | Adipose stem cells | 3 | 3 | NR4A1 | 8.06 | NR1A1 | −1.2 | 24040759/GSE48964 |
| NR4A2 | 12.64 | NR1D1 | −1.2 | |||||
| NR4A3 | 4.44 | NR2C2 | −1.25 | |||||
| Morbidly obese | Human omental adipose tissue | 1 | 1 | NR4A2 | 3.5 | NR2B3 | −2.44 | 20678967/GSE15773 |
| Homocysteine (100 μM) | Human aortic smooth muscle cells | 3 | 3 | NR2B3 | 1.2 | NR1H3 | −1.33 | 18602108/GSE9490 |
| NR3A2 | 1.61 | NR2F2 | −1.7 | |||||
| NR4A3 | 1.49 | NR4A2 | −1.24 | |||||
| Hypertension | Carotid artery atheromatous plaques | 0 | 2 | NR1A2 | −2.04 | 23660665/GSE43292 | ||
| NR3C3 | −2 | |||||||
| C. Nuclear receptors are significantly downregulated than upregulated in human aortic endothelial cells (HAECs) treated with oxidized low-density lipoproteins (Ox-LDLs) in a time-dependent manner | ||||||||
| Fold* | Fold* | |||||||
| ∆ | ∆ | |||||||
| Treated with Ox-LDL for 6 h | HAEC | 4 | 3 | NR1I2 | 2.18 | NR1B2 | −3.57 | 19279231/GSE13139 |
| NR2A2 | 3.8 | NR1F1 | −5.26 | |||||
| NR3A2 | 3.04 | NR4A1 | −2.56 | |||||
| NR5A2 | 4 | |||||||
| Treated with Ox-LDL for 12 h | 0 | 7 | NR1B1 | −2.86 | ||||
| NR1B2 | −5.56 | |||||||
| NR1F1 | −2.86 | |||||||
| NR1H4 | −5.88 | |||||||
| NR2A2 | −2.04 | |||||||
| NR3A1 | −2.94 | |||||||
| NR3C3 | −2.33 | |||||||
| Treated with Ox-LDL for 24 h | 5 | 7 | NR1C1 | 2.24 | NR1B1 | −2.56 | ||
| NR1I1 | 2.01 | NR1B2 | −2.17 | |||||
| NR3A2 | 5.09 | NR1B3 | −2.04 | |||||
| NR3B2 | 2.34 | NR1H4 | −4.55 | |||||
| NR5A2 | 5.81 | NR1I2 | −2.5 | |||||
| NR2A2 | −2.13 | |||||||
| NR2F6 | −4.55 | |||||||
Abbreviations: HAECs human aortic endothelial cells, Ox-LDL oxidized low-density lipoprotein
*Fold change > 2
**Fold change > 1.2
Specifically, our data shows that NR1C1 (PPARα) is among the downregulated genes in familial hypercholesterolemia. NR1C1 is one of the primary modulators in fatty acid oxidation and apolipoprotein synthesis [23]. This receptor was also found abundantly in the vascular wall and in human macrophages and was shown to exert anti-inflammatory and anti-atherogenic effects [57]. Therefore, downregulation of this gene may contribute to hypercholesterolemia and also to progression of atherosclerotic events. PPARα agonists are widely used to correct hyperlipidemia and were shown to reduce mortality and morbidity due to cardiovascular events [58]. Furthermore, we observed that NR1C3 (PPARγ) is downregulated in patients with rheumatoid arthritis. Previously, PPARγ was reported to have a negative effect on oxidative stress, and therefore, it was suggested that concomitant use of PPARγ agonists with other treatments will give additional therapeutic benefits against rheumatoid arthritis [59].
NRs play an important role in the development and progression of cancers. For an example, the roles of androgen receptors in breast and prostate cancers are well documented [60–62]. We analyzed the NR expression in 17 different types of cancers in TCGA database. Similar to the observation we elaborated above, our data revealed that the tendency of NRs to be downregulated is more than being upregulated. NR1H2 receptor was downregulated in as many as seven types of cancers, NR2B2 in six types, and NR1B1 and NR1A1 in five types of cancers (Table 14). However, specifically NR1A1 (RAR α) and also NR1B1 (RAR β) are associated with progression of estrogen-dependent breast cancers [63]. This is contrasting to our observation of the expression of these two receptors in other types of cancers. Nevertheless, activation of NR1H2 which falls in to liver X receptors was shown to inhibit proliferation of HT29 colorectal cancer cells [64]. Therefore, this suggests that NR1H2 can be a potential therapeutic target for the treatment of many types of cancers.
Table 14.
More nuclear receptors are downregulated in 17 different types of human cancers
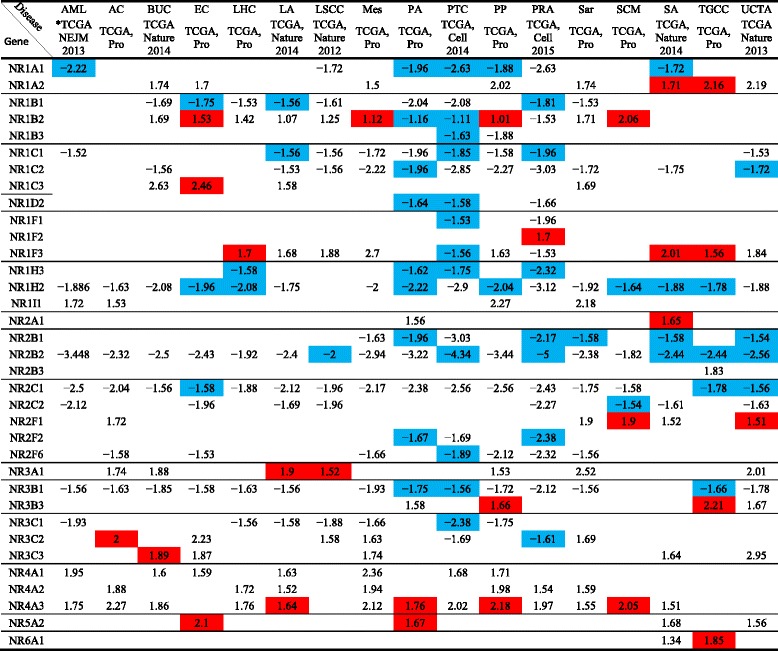
The numbers in the cells represent fold change (≥ 1.5); positive symbol means upregulation while negative symbol means downregulation. Red color means significant upregulation while blue color means significant downregulation (p < 0.05). No color means no significance (p ≥ 0.05), and blank means no data is available
Abbreviations: TCGA The Cancer Genome Atlas, Pro provisional, AML acute myeloid leukemia, AC adrenocortical carcinoma, BUC bladder urothelial carcinoma, EC esophageal carcinoma, LHC liver hepatocellular carcinoma, LA lung adenocarcinoma, LSCC lung squamous cell carcinoma, Mes mesothelioma, PA pancreatic adenocarcinoma, PTC papillary thyroid carcinoma, PP pheochromocytoma and paraganglioma, PRA prostate adenocarcinoma, Sar sarcoma, SCM skin cutaneous melanoma, SA stomach adenocarcinoma, TGCC testicular germ cell cancer, UCEC Uterine Corpus Endometrial Carcinoma
*Reference
To determine the features of those human diseases-modulated NRs, we performed Venn analysis as we previously reported [15]. The Venn analysis/diagram is a very useful analytical tool as it helps to clearly visualize the NRs that are shared between the different diseases analyzed. In Fig. 6a, b, the results show that NR expression changes in human diseases are not shared. In four human diseases analyzed by the Venn analysis, 13 NRs were upregulated, 20 NRs were downregulated, and 15 NRs were not changed in their expression levels (Fig. 6c). Of note, 10 out of 13 upregulated NRs in human diseases were from the scarcely distributed group shown in Table 6, 11 out of 20 NRs downregulated in human diseases were from the very highly distributed and highly distributed groups in Table 6, and 13 out of 15 NRs whose expressions were not changed in human diseases were from the moderately distributed and scarcely distributed groups shown in Table 6. Notably, the tissue expression of 3 NRs out of 20 disease-mediated downregulated NRs including NR1C1, NR1H3, and NR1C3 were correlated with tissue hypomethylated index SAH levels (Fig. 5b), 4 out of 15 NRs whose expressions were not changed were correlated with hypomethylated index SAH levels, and none of the NRs in the disease-upregulated group were correlated with hypomethylated index SAH levels (Fig. 6c). These findings are in a good correlation with tissue hypomethylation function in promoting inflammation as we reported [32, 65], suggesting that hypomethylation-promoting hyperhomocysteinemia may facilitate inflammation via inhibiting the expression of those human disease-downregulated NRs and also keep the stable expression of those non-disease-changed NRs.
Fig. 6.
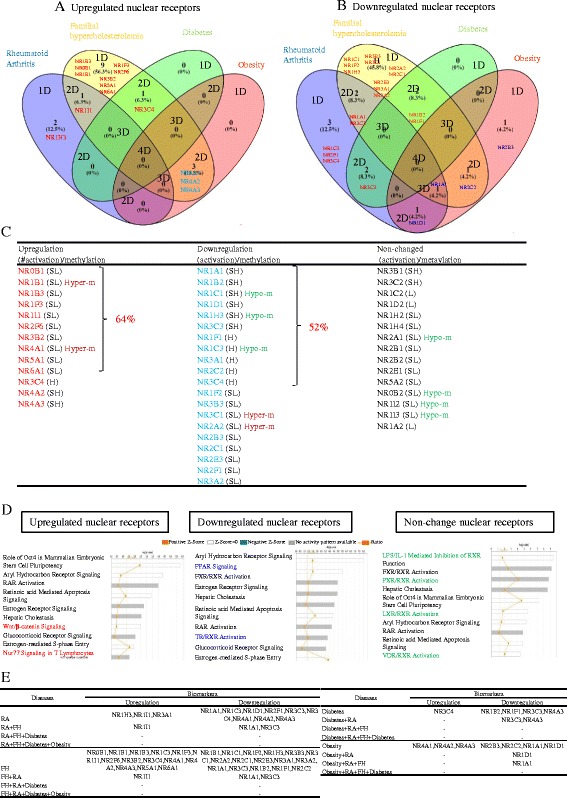
Venn analysis of significantly changed nuclear receptor expression among four different tissues. a, b Venn diagram shows the number of significantly upregulated and downregulated nuclear receptors in four different pathologies respectively (blue, yellow, green and red represent rheumatoid arthritis/ familial hypercholesterolemia/diabetes/obesity). c The nuclear receptor genes that are upregulated, downregulated, and without any expression changes in four pathologies of interest. d The signaling pathways that are regulated by nuclear receptor genes that are upregulated, downregulated, and have no expression changes in four pathologies of interest. e A list of nuclear receptors that can be used as biomarkers to detect indicated pathologies
In order to determine functional significances of disease-modulated NR expression, we analyzed the top 10 signaling pathways with the disease-upregulated NRs, disease-downregulated NRs, and non-disease-modulated NRs using the Ingenuity Pathway Analyzer (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). As shown in Fig. 6d; in addition to the shared pathways among three groups of NRs, two pathways, Wnt/β-catenin signaling, and Nur77 signaling were specifically associated with the disease-upregulated NRs; two other pathways such as peroxisome proliferator-activated receptor (PPAR) signaling and thyroid hormone receptor (TR)/retinoid X receptor (RXR) activation were specifically associated with the downregulated NRs; and four pathways including LPS/IL-1 inhibition, pregnane X receptor (PXR)/RXR activation, liver X receptor (LXR)/RXR activation and 1α, and 25-dihydroxyvitamin D3 (vitamin D3) receptor (VDR)/RXR activation were specifically associated with the non-disease-modulated NRs. These results have provided novel insight on the potential functions of various NRs in modulating the pathogenesis of human autoimmune arthritis and metabolic diseases.
Since we found that upregulation and downregulation of certain NRs can be shared in several human diseases (Fig. 6a), we examined whether the NRs shared in human diseases can be used as biomarkers for the diseases and complications. To test this issue, we organized the analysis results in Fig. 6e. The results showed that upregulation of three NRs such as NR1H3, NR1I1, and NR3A1 can be used as biomarkers for rheumatoid arthritis and that upregulation of NR1I1 alone and downregulation of only two NRs NR1A1 and NR3C3 can be used as potential biomarkers for rheumatoid arthritis with familial hypercholesterolemia. In addition, upregulation of NR3C4 can be used as potential biomarker for type 2 diabetes whereas downregulation of two NRs including NR3C3 and NR4A3 can only be used as biomarkers for diabetes with rheumatoid arthritis as a complication. Moreover, upregulation of three NRs such as NR4A1, NR4A2, and NR4A3 can be used for the potential biomarkers for obesity while downregulation of NR1D1 can be used as a potential biomarker for obesity with rheumatoid arthritis as a complication. Finally, downregulation of NR1A1 alone can be used as the biomarker for obesity complicated with rheumatoid arthritis and familial hypercholesterolemia. The results suggest that the NRs shared in human diseases may be highly valuable in serving as potential biomarkers for detection of autoimmune arthritis, metabolic diseases, and their complications.
The expression of nuclear receptors are regulated by numerous inflammation-modulating pathways and mitochondrial energy metabolic enzymes
We then hypothesized that the expression of NRs is regulated by numerous inflammation-modulating pathways. To test this hypothesis, we examined the expression of NRs in various gene-deficient mouse models and cells with overexpression of genes of interests. First, two NRs such as Nr1h3 and Nr1i1 were found to be upregulated, and four other NRs were downregulated in the aortic arch in apolipoprotein E (ApoE)−/− mice fed with 24 weeks of high fat diet (Table 15). However, only one NR, Nr2f1, was found to be downregulated in the aortic arch of ApoE−/− mice fed with 8 weeks of high fat diet. These findings suggest that NR modulation in ApoE−/− in the aortic arch requires prolonged high fat diet feeding.
Table 15.
A large nuclear receptor is downregulated in proatherogenic mouse models ApoE−/−, LDL-R−/−, and type 2 diabetes mouse model db/db
| Disease | Tissue/cell type | Number | Upregulated gene | Fold* | Downregulated gene | Fold* | PMID/GEO ID | |
|---|---|---|---|---|---|---|---|---|
| Up | Down | ∆ | ∆ | |||||
| ApoE−/− 8-week HFD | Aortic arch | 0 | 1 | Nr2f1 | −1.2 | 20577049/GSE18443 | ||
| ApoE−/− 24-week HFD | 2 | 4 | Nr1h3 | 1.26 | Nr1a2 | −1.25 | ||
| Nr1i1 | 1.27 | Nr1b2 | −1.2 | |||||
| Nr1d1 | −1.28 | |||||||
| Nr3a1 | −1.22 | |||||||
| LDL-R−/− VS. WT | Macrophages of aorta | 2 | 9 | Nr1c3 | 1.29 | Nr1a2 | −1.3 | 21868699/GSE24342 |
| Nr3a1 | 1.3 | Nr1b2 | −1.2 | |||||
| Nr1d2 | −1.28 | |||||||
| Nr1i1 | −1.3 | |||||||
| Nr2b3 | −1.25 | |||||||
| Nr3c1 | −1.2 | |||||||
| Nr3c4 | −1.27 | |||||||
| Nr4a1 | −1.32 | |||||||
| Nr4a2 | −1.28 | |||||||
| db/db VS. WT | Glomerular endothelial cell | 1 | 3 | Nr1b2 | 1.74 | Nr1i1 | −1.47 | 20706631/GSE21324 |
| Pgr | −1.52 | |||||||
| Ar | −1.67 | |||||||
Abbreviations: WT wild type, ApoE−/− apolipoprotein E-deficient mice; LDL-R−/− low-density lipoprotein receptor deficient mice, db/db mice leptin receptor gene mutant mice, HFD high fat diet, VS. versus
*Fold change > 1.2
In another study, it was shown that expression of NRs were upregulated and nine NRs were downregulated in aortic macrophages of low-density lipoprotein receptor (LDL-R)-deficient mouse aortic macrophages relative to wild type (Table 15). In a separate study, one NR Nr1b2 was found to be upregulated but three NRs were downregulated in diabetic db/db glomerular endothelial cells (Table 15). These results suggest that once again in proatherogenic models and type 2 diabetes model, there is a less tendency for NRs to be upregulated than downregulated.
Second, we examined whether inflammatory cytokine signaling pathways can downregulate NR expression. In Table 16, in interferon-γ (IFN-γ)-stimulated endothelial cells, interleukin-1β (IL-1β)-stimulated endothelial cells, IL-1β-stimulated human peripheral mononuclear cells (PBMCs) and tumor necrosis factor-α (TNF-α)-stimulated PBMCs, the NR expressions were either upregulated and downregulated in similar numbers or less upregulated than downregulated.
Table 16.
Pro-inflammatory cytokine signaling negatively regulates the expression of nuclear receptors
| Disease | Tissue/cell type | Number | Upregulated gene | Fold* | Downregulated gene | Fold* | PMID/GEO ID | ||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | ∆ | ∆ | ||||||
| IFN-γ stimulation | EC | 2 | 2 | NR2F1 | 1.39 | NR2B1 | −1.26 | 19553003/GSE3920 | |
| NR3C1 | 1.53 | NR2F6 | −1.36 | ||||||
| IL-1β stimulation | EC | 3 | 7 | NR1H4 | 5.32 | NR1B2 | −3.84 | 21469100/GSE19240 | |
| NR3A1 | 2.21 | NR1B3 | −4.93 | ||||||
| NR5A2 | 6.57 | NR1F1 | −4.72 | ||||||
| NR1F2 | −2.58 | ||||||||
| NR3A2 | −4.94 | ||||||||
| NR3B3 | −11.03 | ||||||||
| NR4A1 | −3.43 | ||||||||
| Human PBMCs-IL-1β and TNF-α stimulations | IL-1β 2 h | PBMC | 0 | 3 | NR1A1 | −14.93 | 23104095/GSE40838 | ||
| NR1I1 | −5.1 | ||||||||
| NR3A2 | −6.54 | ||||||||
| IL-1β 6 h | 0 | 1 | NR5A2 | −23.26 | |||||
| TNF-α 2 h | 1 | 1 | NR1B2 | 38.32 | NR2A1 | −21.11 | |||
| TNF-α 6 h | 2 | 3 | NR1B2 | 27.28 | NR1A2 | −23.92 | |||
| NR3B3 | 4.92 | NR1B3 | −16.22 | ||||||
| NR4A1 | −3.86 | ||||||||
Abbreviations: IFN-γ interferon gamma, IL-1β interleukin-1β, TNF-α tumor necrosis factor-α-like, PBMCs peripheral blood mononuclear cells
Third, in Table 17, we examined whether anti-inflammatory cytokine pathways and inhibition of the pro-inflammatory transcription factor regulate NR expressions. We observed that NR expressions were modulated in hepatocellular carcinoma cells stimulated with transforming growth factor-β (TGF-β), palatal mesenchyme cells from TGF-β knockout (KO) mice, in conventional T cells stimulated with anti-CTLA-4 (cytotoxic T-lymphocyte-associated protein 4, also known as CD152, a T cell co-suppressor) antibody, in regulatory T cells (Tregs) stimulated with anti-CTLA-4 antibody and in hearts extracted from cardiac-specific transgenic PPARα mice. In a study with NF-kB inhibitor-treated cells, six NRs were upregulated and five NRs were downregulated. These results demonstrated that immune suppressor pathways CTLA-4, NF-kB inhibitor, and Treg suppress inflammation by significantly upregulating the expression of nine NRs including NR4A1 (5.6–8.3 folds), NR4A2 (6.7–15.8 folds), NR4A3 (4.6–8.1 folds), NR1B2 (6.7 folds), NR1D1 (3.4 folds), NR2A2 (3.2 folds), NR1H4 (2.7 folds), NR2C1 (5.4 folds), and NR3A2 (4.2 folds).
Table 17.
Anti-inflammatory cytokine signaling and Tregs positively regulate the expression of nuclear receptors
| Disease | Tissue/cell type | Number | Upregulated gene | Fold* | Downregulated gene | Fold* | PMID/GEO ID | ||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | ∆ | ∆ | ||||||
| TGF-β stimulation | HCC Huh-7 cells | 8 | 7 | NR1B1 | 1.19 | NR1B2 | −1.21 | 19723656/GSE10393 | |
| NR1H2 | 1.37 | NR1C3 | −1.23 | ||||||
| NR2B1 | 1.75 | NR1D2 | −1.35 | ||||||
| NR2B2 | 1.23 | NR1F1 | −1.42 | ||||||
| NR2F1 | 1.27 | NR1H4 | −1.45 | ||||||
| NR2F2 | 1.36 | NR3C1 | −1.21 | ||||||
| NR2F6 | 1.37 | NR5A2 | −1.29 | ||||||
| NR0B2 | 1.59 | ||||||||
| TGF-β KO | PM cells | 2 | 3 | NR2C2 | 1.21 | NR2C1 | −1.26 | 23975680/GSE46150 | |
| NR3C1 | 1.23 | NR2F6 | −1.3 | ||||||
| NR3C4 | −1.67 | ||||||||
| Tconv stimulate by anti-CTLA-4 | Spleen and lymph node | 7 | 3 | NR1D2 | 1.69 | NR1B3 | −1.14 | 23277554/GSE42267 | |
| NR1H3 | 1.2 | NR2B1 | −1.18 | ||||||
| NR2C2 | 1.23 | NR2C1 | −1.28 | ||||||
| NR3C4 | 1.64 | ||||||||
| NR4A1 | 5.61 | ||||||||
| NR4A2 | 15.78 | ||||||||
| NR4A3 | 8.12 | ||||||||
| Treg stimulated with anti-CTLA-4 | Spleen and lymph node | 5 | 4 | NR1D2 | 1.53 | NR1B1 | −1.25 | 23277554/GSE42267 | |
| NR1F1 | 2.4 | NR1B3 | −1.32 | ||||||
| NR3C4 | 2.36 | NR1F1 | −1.23 | ||||||
| NR4A1 | 3.3 | NR3A1 | −1.23 | ||||||
| NR4A2 | 6.67 | ||||||||
| Cardiac-specific transgenic (Tg-PPARα) mice | Heart | 5 | 13 | NR1F2 | 1.22 | NR1A1 | −1.27 | 22055503/GSE33101 | |
| NR1H3 | 1.46 | NR1C1 | −1.67 | ||||||
| NR1I2 | 0.83 | NR1F1 | −3.84 | ||||||
| NR2A1 | 1.24 | NR1F3 | −2 | ||||||
| NR0B2 | NR2B1 | −1.22 | |||||||
| NR2B2 | −1.22 | ||||||||
| NR2B3 | −1.59 | ||||||||
| NR2F2 | −1.22 | ||||||||
| NR2F6 | −1.27 | ||||||||
| NR3B1 | −1.22 | ||||||||
| NR3B2 | −1.23 | ||||||||
| NR3C1 | −1.25 | ||||||||
| NR3C3 | −1.27 | ||||||||
| NFκB inhibitor | 4 h | EKC | 6 | 5 | NR1B1 | 2.33 | NR1C2 | −2.81 | 15722350/GSE2489 |
| NR1B2 | 6.73 | NR1F1 | −6.54 | ||||||
| NR1D1 | 3.36 | NR1I3 | −2.93 | ||||||
| NR2A2 | 3.16 | NR3A2 | −2.91 | ||||||
| NR2C1 | 2.01 | NR5A2 | −5.17 | ||||||
| NR4A1 | 8.34 | ||||||||
| 48 h | 4 | 4 | NR1H4 | 2.71 | NR1C3 | −5.46 | |||
| NR2C1 | 5.35 | NR2A1 | −3.63 | ||||||
| NR3A2 | 4.2 | NR2F1 | −14.03 | ||||||
| NR4A3 | 4.56 | NR3B3 | −4.82 | ||||||
Abbreviations: HCC hepatocellular carcinoma cells, TGF-β transforming growth factor-β, PM palatal mesenchyme, Tconv conventional T cells, Treg regulatory T cell, EKC epidermal keratinocytes
Fourth, in Table 18, we examined whether a key enzyme of tricarboxylic acid (TCA) cycle, isocitrate dehydrogenase (IDH), regulates the expression of nuclear receptors. The results showed that IDH mutation in isogenic epithelial cells results in significant upregulation of six NRs and downregulation of eight NRs. Fifth, in Table 19, we examined whether four key enzymes of the mitochondrial respiratory chain including nicotinamide adenine dinucleotide (quinone) (NADH) dehydrogenase (Nd2), succinate dehydrogenase (SDH), cytochrome c oxidase-4 (COX4), and mitochondrial respiratory chain complex IV regulate the expression of nuclear receptors. The results showed that the mutations of these enzymes result in significant changes in the expression of NR2F2 (Nd2 mutation induced sevenfold upregulation), NR5A2 (Nd2 mutation induced 27-fold downregulation), NR1A2 (COX4 mutation induced 46.5-fold upregulation), and NR2F6 (Cox4 mutation induced 3.3-fold downregulation). Taken together, these results suggest that the expressions of nuclear receptors are regulated by numerous inflammation-modulating pathways and mitochondrial energy metabolic enzymes.
Table 18.
A key enzyme of tricarboxylic acid (TCA) cycle, isocitrate dehydrogenase, regulates the expression of nuclear receptors
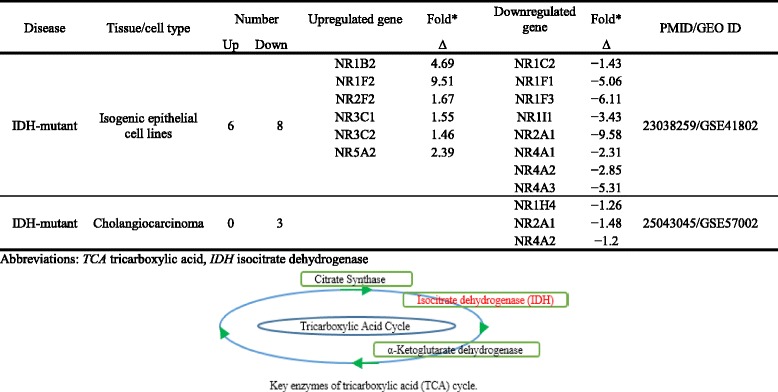
Table 19.
Key enzymes of the mitochondrial respiratory chain regulate the expression of nuclear receptors
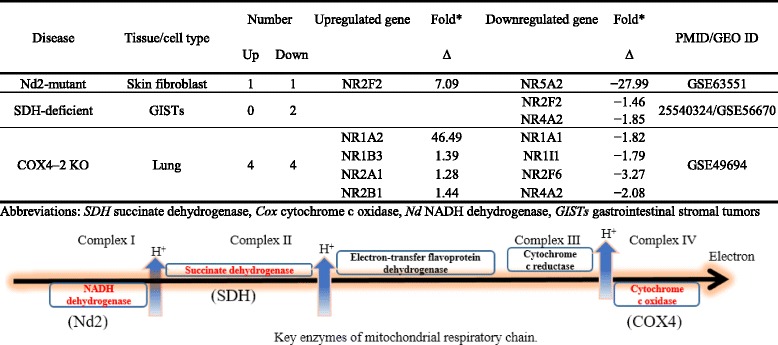
Innate immune sensor inflammasome/caspase-1 pathway plays a critical role in regulating the expression of most nuclear receptors
In Fig. 4, we found that innate immune sensor PRRs such as NOD1, NOD2, NOD4, and IFI16 may either act as upstream regulators or downstream targets of NRs in tissues and that the NLR-mediated regulation on NR expression in tissues are evolutionally conserved and mainly act toward suppression of NRs during microbial infection-triggered inflammations. To consolidate this finding, we determined whether gene deficiencies of caspase-1 and other inflammasome components affect the expression of NRs.
As shown in Table 20, deficiency of caspase-1 in ApoE−/− mouse aorta, adipose tissue, deficiency of caspase-1 in associated speck-like protein containing a CARD (ASC)−/− background, deficiency of histone deacetylase and caspase-1 substrate sirtuin 1 (Sirt1) in Tregs, and deficiency of NLRP3 in adult and children PBMCs led to mostly upregulation of NRs instead of downregulation of NRs. For example, the deficiency of NLRP3 led to upregulation of 26 NRs (54%) but downregulation of 9–11 NRs (19–23%). These results suggest that caspase-1/NLRP3 inflammasome pathways play a critical role in regulating the expression of NRs. In addition, in Table 21, we also noticed that the inflammasome/caspase-1 deficiencies upregulated 29 NRs (60%), downregulated 10 NRs (21%), but did not change the expression of 9 NRs (19%).
Table 20.
Nuclear receptors are significantly changed in caspase-1 and Sirt1 knockout mice, indicating that caspase-1-Sirt1 pathway negatively regulates nuclear receptor expression
| Disease | Tissue/cell type | Number | Upregulated gene | Fold* | Downregulated gene | Fold* | PMID/GEO ID | ||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | ∆ | ∆ | ||||||
| AopE−/−/Casp1−/− vs. ApoE−/− | Aorta | 3 | 0 | NR1A2 | 1.22 | GSE72448 | |||
| NR1D2 | 1.17 | ||||||||
| NR2C2 | 1.19 | ||||||||
| Adipose | 10 | 1 | NR1A1 | 1.35 | NR6A1 | −1.32 | |||
| NR1C3 | 1.84 | ||||||||
| NR1F1 | 1.78 | ||||||||
| NR1I3 | 1.22 | ||||||||
| NR2A1 | 2.24 | ||||||||
| NR2B1 | 1.52 | ||||||||
| NR3A1 | 1.39 | ||||||||
| NR3C1 | 1.41 | ||||||||
| NR3C4 | 1.66 | ||||||||
| NR4A2 | 1.16 | ||||||||
| Casp1−/−/ASC−/− vs. ASC−/− | White adipose tissue | 7 | 0 | NR1A1 | 1.35 | 21876127/GSE25205 | |||
| NR1C3 | 1.84 | ||||||||
| NR1I3 | 1.22 | ||||||||
| NR2A1 | 1.17 | ||||||||
| NR2B1 | 1.52 | ||||||||
| NR3C4 | 1.65 | ||||||||
| NR4A2 | 1.16 | ||||||||
| Sirt1−/− vs. WT | Treg | 4 | 0 | NR1B1 | 1.2 | 21199917/GSE26425 | |||
| NR1F1 | 1.29 | ||||||||
| NR2F6 | 1.22 | ||||||||
| NR4A3 | 1.29 | ||||||||
| Cardiac-specific transgenic (Tg-Sirt1) mice | Heart | 2 | 6 | NR3C4 | 1.52 | NR1B1 | −3.57 | 22055503/GSE33101 | |
| NR0B2 | 1.35 | NR1B3 | −1.25 | ||||||
| NR1F3 | −2 | ||||||||
| NR1I1 | −1.27 | ||||||||
| NR2B3 | −1.22 | ||||||||
| NR2C1 | −1.22 | ||||||||
| NLRP3 mutation | Adult control | PBMC | 26 | 9 | NR1B1 | 1.22 | NR1F1 | −2.93 | –/GSE43553 |
| NR1B2 | 1.47 | NR1F2 | −1.4 | ||||||
| NR1B3 | 1.43 | NR2C1 | −1.93 | ||||||
| NR1C2 | 1.18 | NR3C1 | −1.38 | ||||||
| NR1C3 | 1.2 | NR3C2 | −1.68 | ||||||
| NR1I1 | 1.73 | NR4A1 | −1.32 | ||||||
| NR1I2 | 1.2 | ||||||||
| NR2A1 | 1.53 | ||||||||
| NR2B3 | 1.25 | ||||||||
| NR2E1 | 1.31 | ||||||||
| NR2E3 | 1.57 | ||||||||
| NR2F1 | 1.2 | ||||||||
| NR3A1 | 1.61 | ||||||||
| NR3A2 | 1.28 | ||||||||
| NR3B1 | 1.34 | ||||||||
| NR4A3 | 1.27 | ||||||||
| NR5A1 | 1.42 | ||||||||
| NR6A1 | 1.3 | ||||||||
| NR0B2 | 1.34 | ||||||||
| Children control | 26 | 11 | NR1A1 | 1.4 | NR1D1 | −1.82 | |||
| NR1A2 | 1.24 | NR1D2 | −1.56 | ||||||
| NR1B1 | 1.31 | NR1F1 | −2.53 | ||||||
| NR1B2 | 1.51 | NR2B2 | −1.44 | ||||||
| NR1B3 | 1.49 | NR2C1 | −2.25 | ||||||
| NR1C3 | 1.32 | NR2C2 | −1.29 | ||||||
| NR1I1 | 1.69 | NR3C1 | −1.3 | ||||||
| NR1I2 | 1.27 | NR3C2 | −1.53 | ||||||
| NR2A1 | 1.66 | NR4A1 | −1.37 | ||||||
| NR2A2 | 1.22 | NR4A2 | −4.23 | ||||||
| NR2E1 | 1.21 | ||||||||
| NR2E3 | 1.55 | ||||||||
| NR2F1 | 1.24 | ||||||||
| NR2F6 | 1.26 | ||||||||
| NR3A1 | 1.23 | ||||||||
| NR3A2 | 1.41 | ||||||||
| NR3B1 | 1.65 | ||||||||
| NR3C3 | 1.21 | ||||||||
| NR4A3 | 1.23 | ||||||||
| NR5A1 | 1.41 | ||||||||
| NR6A1 | 1.29 | ||||||||
| NR0B2 | 1.34 | ||||||||
Abbreviations: ApoE−/− apolipoprotein E-deficient mice, Casp1−/− caspase-1-deficient mice, HFD high fat diet, ASC−/− PYD and CARD domain-containing deficient mice, Sirt 1−/− sirtuin 1-deficient mice; WT wild-type mice, NLRP3 NLR family pyrin domain containing 3 deficient mice; vs. versus
Table 21.
The expression changes of NRs in the presence of inflammasome/caspase-1 deficiencies
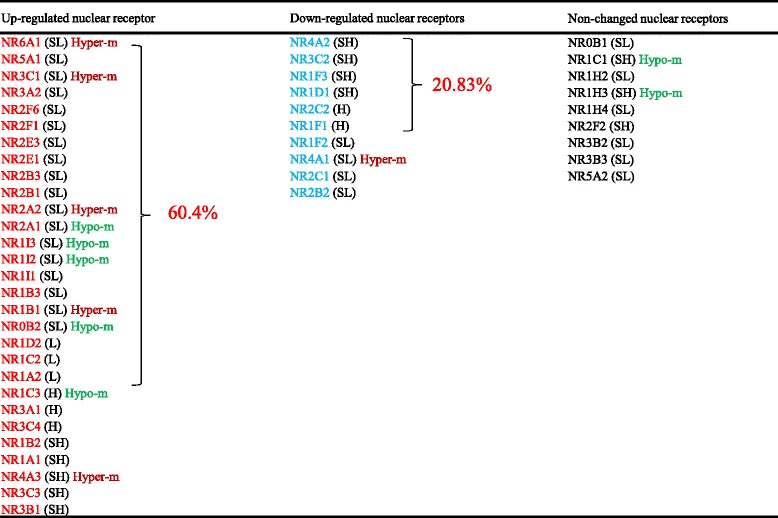
Moreover, we noticed that among top 10 pathways identified with the Ingenuity Pathway Analysis for the inflammasome/caspase-1 deficiency-upregulated NRs, three pathways including aryl hydrocarbon receptor signaling, RAR activation, and estrogen receptor signaling were unique. Similarly, among the top 10 pathways identified with the Ingenuity Pathway Analysis for the inflammasome/caspase-1 deficiency-downregulated NRs, seven pathways including circadian rhythm signaling, thyroid cancer signaling, Nur77 signaling in T lymphocytes, calcium-induced T-lymphocyte apoptosis, melatonin signaling, T helper cell differentiation, and non-small cell lung cancer signaling were specific. Furthermore, among the top 10 pathways identified with the Ingenuity Pathway Analysis for the inflammasome/caspase-1 deficiency-non-changed NRs, three pathways including LPS/IL-1-mediated inhibition of RXR function, LXR/RXR activation, and Toll-like receptor signaling were unique. Therefore, these pathways may not play a significant role in progression of inflammatory pathologies. Taken together, these results suggest that inflammasome/caspase-1 pathway deficiencies regulate the expressions of most NRs (81%) and that inflammasome/caspase-1 innate immune sensors control the expression of most NRs.
We propose a new paradigm that most nuclear receptors are anti-inflammatory HAMPs for regulating the balance of inflammation, inhibition of inflammation, and resolution of inflammation
HAMPs (homeostasis-associated molecular pattern molecules), the new concept we proposed, are designated for mitigating the progression of inflammation or inhibition of inflammation under sterile inflammation. These HAMP receptors initiate anti-inflammatory/homeostatic signaling and promote inflammation resolution [5]. Since most endogenously metabolite-nuclear receptor signals inhibit inflammation and maintain the tissue homeostasis, we propose that most NRs act as HAMP receptors. To consolidate this new hypothesis, we conducted an extensive literature search (Parts 2, 3, and 4 in Fig. 1). We found the following supporting evidences.
The first supporting evidence for classifying most of nuclear receptors as HAMP receptors is presented in Tables 8, 9, and 10 (Part 2 in Fig. 1): (1) Mutations in NR significantly increase the risk for development of human metabolic diseases (Tables 4, 5, 6, and 7), suggesting that NR sequence changes may weaken the NR functions in suppressing human metabolic diseases and inflammation; (2) NR deficiencies lead to abnormal mouse phenotypes and inflammation from the MGI database (Tables 8 and 9), suggesting that NRs’ expression and functions are essential for maintaining the homeostasis and inhibition of inflammation and that NR deficiencies increase the likelihood of developing metabolic diseases in mice and potentially in humans.
The second supporting evidence for classifying most of nuclear receptors as HAMP receptors is presented in Table 12 (Part 3 in Fig. 1). NRs inhibit inflammation signaling gene functions and inflammation readouts. Of note, 12 out of 15 NRs have anti-inflammatory roles verified by published papers (Table 12).
The third supporting evidence for classifying most NRs as HAMP receptors is demonstrated in Tables 13, 14, and 15 (Part 4 (1) and (2) in Fig. 1). NRs were less upregulated than downregulated during the progression of metabolic, cardiovascular, and autoimmune diseases and cancers, suggesting that NRs’ physiological expression and functions may block the pathogenesis and progression of those diseases.
The fourth supporting evidence for classifying most of nuclear receptors as HAMPs is demonstrated in Tables 16 and 17 (Part 4 (3) in Fig. 1). Inflammation signaling genes regulated nuclear receptor expression levels as judged by the following results: (1) Most NRs were downregulated when stimulated with pro-inflammatory agents, suggesting that the pro-inflammatory signals suppress the NRs expression, and (2) some NRs were downregulated when anti-inflammatory signaling genes were deficient. In contrast, those NRs were upregulated when anti-inflammatory signals were activated.
The fifth supporting evidence for classifying most of nuclear receptors as HAMP receptors is demonstrated in Tables 20, 21, and 22 (Part 4 (4) in Fig. 1). Most NRs were more upregulated than downregulated when innate immune sensor inflammasome/caspase-1 genes were deficient. In contrast, caspase-1-degrading gene histone deacetylase Sirt1 [8] transgene may have anti-inflammatory functions by increasing the expression of certain NRs.
Table 22.
Signal pathways that are upregulated by genes listed in Table 21
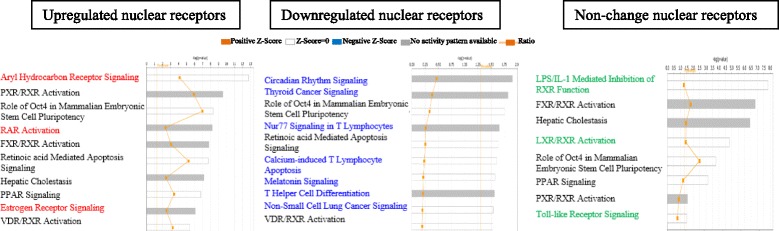
Discussion
NRs are a class of 48 lipophilic ligand-activated transcription factors identified as key players of metabolic and developmental processes. Upon activation by the ligand messenger, NRs typically function as transcription factors where they bind to recognition elements on the genomic DNA and regulate the expression of target genes via type I, II, and III signaling formats [66]. Regardless of the significant progress that has been made in characterizing NR functions and expression, the global profiling of NR expression in human immune and cardiovascular tissues and potential mechanisms underlying the physiological expression of NRs remained poorly defined. In addition, the important issue of how innate immune sensor inflammasome/caspase-1 and other inflammatory signaling globally regulate NR expression in tissues and cells also remained unknown. To examine these issues, we took panoramic profiling database analysis approaches and made the following important findings: (1) NRs are differentially expressed in human and mouse tissues and NR expression may be under regulation by oxygen sensors, angiogenesis pathway, stem cell master genes, PRRs, and tissue hypomethylation/hypermethylation indices; (2) NR sequence changes and mutations are associated with increased risks for development of metabolic diseases, cardiovascular diseases, hormone insensitivity/resistance, cancers, and autoimmune diseases; (3) NRs have less tendency to be upregulated than downregulated in human autoimmune diseases, metabolic diseases, and cancers, which may be regulated by numerous inflammation-modulating pathways and mitochondrial energy metabolic enzymes; (4) The innate immune sensor inflammasome/caspase-1 pathway plays a critical role in regulating the expression of most NRs (Table 23); and (5) We propose a new paradigm that most NRs are anti-inflammatory HAMPs for regulating the balance of inflammation, inhibition of inflammation, and resolution of inflammation.
Table 23.
Nuclear receptor expression was regulated by ApoE and LDL-R, pro/anti-inflammatory cytokines, and inflammasomes in pathology
| Human metabolic disease | NRNC symbol | ApoE KO | LDL-R KO | IFN-γ stimulation | IL-1β stimulation | TNF-α stimulation | NFκB inhibitor | TGF-β KO | Tg-PPARα | CAS1 KO | ASC KO | Tg-Sirt1 | NLRP3 mutant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upregulation (from Fig 6c) | NR0B1 | ||||||||||||
| NR1B1 | ↑ | ↓ | ↑ | ||||||||||
| NR1B3 | ↓ | ↓ | ↓ | ↑ | |||||||||
| NR1F3 | ↓ | ↓ | |||||||||||
| NR1I1 | ↑ | ↓ | ↓ | ↑ | |||||||||
| NR2F6 | ↓ | ↓ | ↓ | ↑ | |||||||||
| NR3B2 | ↓ | ||||||||||||
| NR4A1 | ↓ | ↓ | ↓ | ↑ | ↓ | ||||||||
| NR5A1 | ↑ | ||||||||||||
| NR4A2 | ↓ | ↑ | ↓ | ||||||||||
| NR6A1 | ↓ | ↑ | |||||||||||
| NR4A3 | ↑ | ↑ | |||||||||||
| Downregulation (from Fig 6c) | NR1A1 | ↓ | ↑ | ↑ | ↑ | ||||||||
| NR1B2 | ↓ | ↓ | ↓ | ↑ | ↑ | ↑ | |||||||
| NR1C1 | ↓ | ||||||||||||
| NR1D1 | ↑ | ↑ | |||||||||||
| NR1H3 | ↑ | ↑ | |||||||||||
| NR3C3 | ↓ | ↑ | |||||||||||
| NR1F1 | ↓ | ↓ | ↓ | ↑ | ↓ | ||||||||
| NR1C3 | ↑ | ↓ | ↑ | ↑ | ↑ | ||||||||
| NR3A1 | ↓ | ↑ | ↑ | ↑ | ↑ | ||||||||
| NR2C2 | ↑ | ↑ | ↓ | ||||||||||
| NR3C4 | ↓ | ↓ | ↑ | ↑ | ↑ | ||||||||
| NR1F2 | ↓ | ↑ | ↓ | ||||||||||
| NR3B3 | ↓ | ↑ | ↓ | ||||||||||
| NR3C1 | ↓ | ↑ | ↑ | ↓ | ↑ | ↓ | |||||||
| NR2A2 | ↑ | ↑ | |||||||||||
| NR2B3 | ↑ | ↓ | ↓ | ↑ | |||||||||
| NR2C1 | ↑ | ↑ | ↓ | ↓ | |||||||||
| NR2E3 | ↑ | ||||||||||||
| NR2F1 | ↓ | ↑ | ↓ | ↑ | |||||||||
| NR3A2 | ↓ | ↑ | ↑ |
Abbreviations: KO knockout, ApoE apolipoprotein E, LDL-R−/− low-density lipoprotein receptor, IFN-γ interferon gamma, IL-1β interleukin 1 beta, TNF-α tumor necrosis factor-α-like; TGF-β transforming growth factor-β, Casp1−/− caspase-1-deficient mice; ASC−/− PYD and CARD domain-containing deficient mice, Tg-Sirt1 transgenic sirtuin 1 mice, NLRP3 NLR family pyrin domain containing 3 deficient mice, PBMC peripheral blood mononuclear cells
We utilized an experimental database mining approach that was pioneered and developed in our laboratory throughout the years [2, 67–69]. By analyzing DNA sequencing data from tissue cDNA libraries, we were able to study expression profiles of NRs in various tissues. Since the gene expression sequencing tag (EST) data deposited in the NIH-NCBI-UniGene database have been established based on DNA sequencing data, the data extracted from EST database mining are more precise in providing the tissue expression profiles of genes than traditional hybridization- and primer annealing-based approaches like Northern blots and RT-PCRs [2]. Of note, since the UniGene database does not have many non-tumor cell line-related gene expression data in the presence of various gene deficiencies and stimulation conditions, we analyzed microarray-based gene expression data deposited in NIH-GEO datasets to determine NR expression changes under pathological conditions. Also, as all the data we provided in this manuscript were collected from cDNA cloning, DNA sequencing experiments, and microarray datasets rather than theoretical data derived from computer modeling, we believe that our findings are relevant for many biological and pathological scenarios. Nevertheless, herein we acknowledge that further well-designed experiments are needed to consolidate our findings.
As we pointed out in Table 7, a previous paper reported a mouse NR tissue expression profile using nucleic acid binding based RT-PCR technique [26, 27]. However, NR superfamily expressions using a more accurate DNA sequencing-based technique have not been profiled for human tissues. Other reports have confirmed the tissue distribution of few NRs. For example, as previously mentioned, the rat tissue distribution and/or the relative level of NR3A1 and NR3A2 expression seems to be quite different, i.e., moderate to high expression in the uterus, testis, pituitary, ovary, kidney, epididymis, and adrenal for NR3A1 and the prostate, ovary, lung, bladder, brain, uterus, and testis for NR3A2 [70]. Another study showed that NR2E3 mRNA was detected in the adrenal gland, thyroid gland, prostate, testis, uterus, trachea, and salivary gland [71]. A study assessed the expression patterns of NRs in peripheral blood mononuclear cells and found that 33/48 NRs were expressed in peripheral blood mononuclear cells [72]. In order to clearly summarize our findings, study the expression profile of NRs, and offer a simple, powerful way to obtain highly relational information about their physiologic functions as individual proteins and as a superfamily, we proposed a novel pyramid model to highlight several categories of NR activities in many important tissues. This pyramid model is significant as it improves our understanding of the tissue differences of NR machinery. This model is also significant for understanding the potential pharmacological side effects of new drugs targeting NRs in those tissues. Based on the different distributions and relative levels of the NRs in different target tissues, ligands could be used to elicit beneficial hormone-like activities and reduce adverse side effects of NR-targeted drugs.
The current DAMP receptor model emphasizes only the danger signals generated from endogenous metabolic processes. It fails to recognize the roles of potential endogenous metabolites in anti-inflammatory responses, inflammation resolution, and maintenance of homeostasis. As we pointed out in our previous report [5], it is significant for us to address these limitations and shift the paradigm to form a new model [73] to recognize novel anti-inflammatory and homeostatic signals derived from endogenous metabolites. Recent advances in immunology have clearly demonstrated the well-published “two arms model.” This model states that in addition to the pro-inflammatory immunoeffector and T cell co-stimulatory mechanisms, there are several immunotolerance and anti-inflammatory mechanisms mediated by the immune system. These anti-inflammatory mechanisms include T cell co-inhibition/co-suppression pathways, T cell anergy, regulatory T cells [74], and secretion of anti-inflammatory/immunosuppressive cytokines such as transforming growth factor-β (TGF-β), interleukin-10 (IL-10), IL-35 [69, 75], and IL-37 as we and others reported, etc. We have reported two types of lysophospholipids such as lysophosphatidylserine (LysoPS) and lysophosphatidylethanolamine (LPE) [5] and a few uremic toxins as anti-inflammatory homeostasis-associated molecular patterns [76]. In addition, along the same line, endogenous specialized pro-resolving mediators have been identified as regulators of infection and inflammation [77].
Our new classification of most NRs as homeostasis-associated molecular pattern (HAMP) receptors was that some NRs have been experimentally proved to bind promiscuously to certain types of “patterns” but not exclusively stick to highly specific ligands (Table 3). For example, activation of NRs by a variety of endo- and exogenous chemicals are elemental to induction and repression of drug-metabolism pathways. The master xenobiotic-sensing NRs, the promiscuous pregnane X receptor (PXR), and less-promiscuous constitutive androstane receptor (CAR) are crucial to initial ligand recognition, jump-starting the metabolic process [78]. In addition, phytoestrogens are natural endocrine disruptors that interfere with estrogenic pathways. They insert directly within the hormone-binding domain of estrogen receptor-α (ER-α) and β, with a preference for the β isoform of which the concentration predominates in the normal mammary epithelium [79]. Moreover, bisphenol A (BPA) is widely used as a component in polycarbonate plastics for food and beverage packaging, epoxy linings for canned foods, and dental sealants, among other applications. Experimental literature demonstrates BPA’s affinity for estrogen receptors and downstream effects on estrogen-responsive genes [80]. Those examples have clearly demonstrated that some NRs can promiscuously bind to certain types of “patterns” but not exclusively stick to highly specific ligands.
However, little is known how and why some receptors such as PXR and CAR develop promiscuity. The most widely accepted speculation is that both narrow and broad specificity seen for receptors or proteins are a result of natural selection process [81]. Less specificity of receptors provides evolutionary advantage to organisms that had to conduct a broad set of biological activities with limited protein repertoire and also allowed the organisms to evolve new responses to many endogenous and external ligands [82–84]. Promiscuity of such receptors complicate identification of the physiological ligands that activate them in vivo [85]. One way to identify candidate ligands for orphan NRs is to identify their three dimensional structure [86, 87]. However, receptor affinity for the ligand and the physiological concentrations of the ligand in the tissues have to be taken into account when determining the potential relevance of the specific ligand for the receptor function [85, 88]. Additionally, if it is known that the promiscuous NRs require intracellular lipid binding proteins to shuttle the ligand toward it (like PPAR utilizing certain FABPs—fatty acid binding proteins), the nuclear translocation of the particular protein in response to a compound can be used to determine potential ligands for the NR [85]. Nevertheless, identifying the potential endogenous ligands bound to the NR of interest in vivo by using mass spectroscopy, high-performance liquid chromatography (HPLC) or gas chromatography are the most relevant methods than the ones mentioned above [85, 89, 90].
We acknowledge that some of the fold changes shown in our tables are less than twofold. However, complex diseases such as diabetes, obesity, cancer, and autoimmune disorders are regulated by myriad of genes similar to quantitative traits [91, 92]. Previously, for most of the continuous traits, the strongest genetic association could explain only a small fraction of the genetic variance [93, 94]. However, later analyses revealed that casual loci with small effect size are also important in determining continuous traits and complex diseases such as schizophrenia [94]. Moreover, recent publications demonstrated that complex and chronic diseases are driven by accumulation of weak effects on the key genes and regulatory pathways [95, 96]. It is evident that polygenic effects are important across a wide variety of traits and diseases such as diabetes [97]. Therefore, it is our understanding that even a low fold change in potent transcription factors such as NRs can significantly impact progression of complex diseases.
Conclusions
To improve our understanding on most NRs as anti-inflammatory sensors and regulators, we propose a new working model and classified most NRs as homeostasis-associated molecular pattern receptors (HAMPRs) as shown in Fig. 7. First, NRs can sense lipophilic metabolites, hormones, and xenobiotics in a ligand-receptor-specific manner and “pattern” recognition manner; second, most NRs inhibit inflammation; third, a list of tissue homeostasis regulator pathways, tissue regeneration, and angiogenesis pathways including hypoxia sensors, VEGFR pathways, stem cell master regulators, and hypomethylation/hypermethylation may regulate the tissue expression of NRs (as shown in Fig. 7a); fourth, metabolic diseases [98, 99] and autoimmune arthritis have less tendency to upregulate than downregulate NR expression; fifth, comparing to conventional receptor-mediated pathways that signal via multiple steps for checking and relaying, NRs’ signals are much faster and require much less signal relay (as shown in Fig. 7b); and sixth, innate immune sensor inflammasome/caspase-1 pathways suppress the majority of NR expressions, suggesting that most NRs play critical roles in counteracting the role of DAMPs during sterile inflammatory pathologies and maintaining the homeostasis of tissues and cells in addition to their functions in metabolic, developmental [100], and growth processes [101–107]. Our new findings have significantly improved our understanding on NRs in the regulation of inflammation and tissue homeostasis (as shown in Fig. 7c).
Fig. 7.
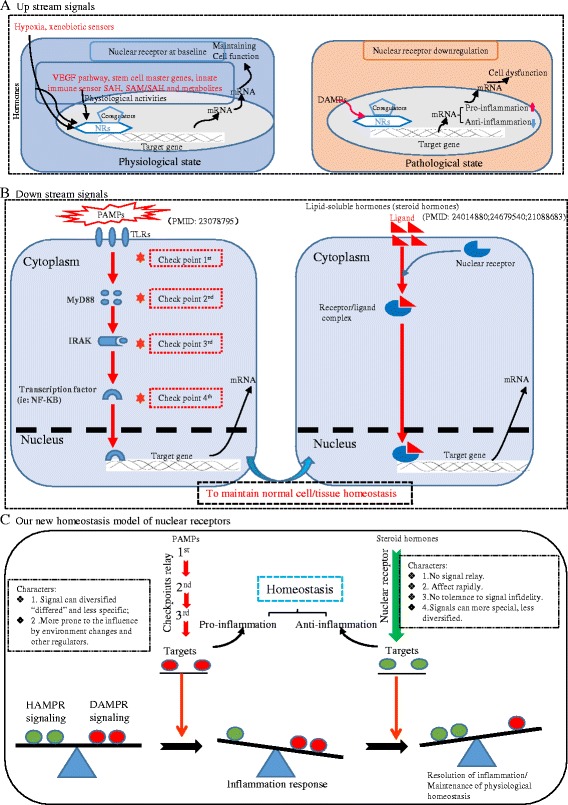
a–c Newly proposed working model which describes that most of the nuclear receptors can be classified as a family of homeostasis-associated molecular pattern receptors
Additional files
Nuclear receptors are differently expressed in human and mouse tissues. A: representative tissue mRNA distribution profile of housekeeping gene ARHGDIA in humans and Ldha in mice. See “Experimental Procedures” for details. B: mRNA distribution profiles of 26 nuclear receptors in 21 human tissues. C: MRNA distribution profiles of 15 nuclear receptors in 17 mouse tissues. The statistical significance was defined as when gene expression was larger than the upper limit of the confidence interval. Gene symbols are listed in Tables 1, 2, and 3. (PDF 296 kb)
Forty-eight nuclear receptors are associated with SAH and SAM/SAH ratio in mouse tissues. A: shows the correlation of 48 nuclear receptors associated with SAH levels (hypomethylation status) in mouse tissues. B. shows the correlation of 48 nuclear receptors associated with SAM/SAH ratio in mouse tissues. (PDF 648 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by an NIH grant to Drs. XF Yang, H. Wang, and ET Choi (Grant No. HL131460-01); National Natural Science Foundation of China (Grant No. 81560051) and the National Key R&D Program in the Twelfth Five-year Plan (Grant No. 2014ZX09303305) to XS Cheng; and Natural Science Foundation of China (Grant Nos. 81370371, 81570394) and Ministry of Education of the People’s Republic of China (Grant No. B13037) to HM Tan.
Availability of data and materials
Experimentally verified mRNA expressions of NRs in various tissues were obtained from databases of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) UniGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene). The microarray datasets that were used in this study were retrieved from NIH-GEO database (https://www.ncbi.nlm.nih.gov/gds), and the numbers of datasets that were used are as follows: GSE55235, GSE6088, GSE6054, GSE23561, GSE13670, GSE55100, GSE48964, GSE15773, GSE9490, GSE43292, GSE13139, GSE18443, GSE24342, GSE21324, GSE3920, GSE19240, GSE40838, GSE10939, GSE46150, GSE42267, GSE33101, GSE2489, GSE41802, GSE57002, GSE56670, and GSE49694. MouseMine database (http://www.mousemine.org/mousemine/begin.do) was used to analyze how the deficiency of NRs lead to development of metabolic and cardiovascular pathologies.
Abbreviations
- AIM-2
Absent in melanoma-2
- ASC
Apoptosis speck-like CARD-containing protein
- BPA
Bisphenol A
- CARD8
Caspase recruitment domain family member 8
- COX4
Cytochrome c oxidase-4
- CTLA-4
Cytotoxic T-lymphocyte-associated protein 4
- DAMP
Danger-associated molecular patterns
- ER
Estrogen receptor
- GWASs
Genome-wide association studies
- HAMPs
Homeostasis-associated molecular patterns
- HAMPRs
Homeostasis-associated molecular pattern receptors
- HIF
Hypoxia-inducible factor
- HMGB1
High-mobility group box 1
- HPLC
High-performance liquid chromatography
- IDH
Isocitrate dehydrogenase
- IFI16
Interferon gamma-inducible protein 16
- IL-1β
Interleukin-1β
- KLF4
Kruppel-like factor 4
- LPLs
Lysophospholipids
- NOD
Nucleotide-binding oligomerization domain
- NR
Nuclear receptor
- Ox-LDL
Oxidized low-density lipoprotein
- PAMPs
Pathogen-associated molecular patterns
- PBMCs
Peripheral mononuclear cells
- PHD2
Prolyl hydroxylase domain-containing protein 2
- PRRs
Pattern recognition receptors
- PXR
Pregnane X receptor
- RAGE
Receptor for advanced glycation end products
- RIG-1
Retinoid acid-inducible gene 1
- SAH
S-adenosyl homocysteine
- SAM
S-adenosyl methionine
- SDH
Succinate dehydrogenase
- TCA
Tricarboxylic acid
- TCGA
The Cancer Genome Atlas
- TGF-β
Transforming growth factor-β
- TLR
Toll-like receptor
- TNF-α
Tumor necrosis factor-α
- Tregs
Regulatory T cells
- VEGF
Vascular endothelial growth factor
Authors’ contributions
LW carried out the data gathering, data analysis, figure/table preparation, and manuscript writing. The other authors provided material input and helped revising the manuscript. XY supervised the experimental design and data analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to disclose.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13045-017-0526-8) contains supplementary material, which is available to authorized users.
Contributor Information
Luqiao Wang, Email: wlq8360@163.com.
Gayani Nanayakkara, Email: gayani@temple.edu.
Qian Yang, Email: 1091251425@qq.com.
Hongmei Tan, Email: tanhm@mail.sysu.edu.cn.
Charles Drummer, Email: tug65470@temple.edu.
Yu Sun, Email: tug36537@temple.edu.
Ying Shao, Email: tuf43098@temple.edu.
Hangfei Fu, Email: tuf07694@temple.edu.
Ramon Cueto, Email: tue54600@temple.edu.
Huimin Shan, Email: tue44606@temple.edu.
Teodoro Bottiglieri, Email: Teodoro.Bottiglieri@BSWHealth.org.
Ya-feng Li, Email: tuf22569@temple.edu.
Candice Johnson, Email: tuf65151@temple.edu.
William Y. Yang, Email: yingyang1612@gmail.com
Fan Yang, Email: gayanikn@gmail.com.
Yanjie Xu, Email: 745950710@qq.com.
Hang Xi, Email: hangxi@temple.edu.
Weiqing Liu, Email: 43362230@qq.com.
Jun Yu, Email: junyu@temple.edu.
Eric T. Choi, Email: Eric.Choi@tuhs.temple.edu
Xiaoshu Cheng, Email: xiaoshumenfan@126.com.
Hong Wang, Email: hongw@temple.edu.
Xiaofeng Yang, Email: xfyang@temple.edu.
References
- 1.Yang XF, Yin Y, Wang H. Vascular inflammation and atherogenesis are activated via receptors for PAMPs and suppressed by regulatory t cells. Drug Discov Today Ther Strateg. 2008;5(2):125–142. doi: 10.1016/j.ddstr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin Y, Yan Y, Jiang X, Mai J, Chen NC, Wang H, Yang XF. Inflammasomes are differentially expressed in cardiovascular and other tissues. Int J Immunopathol Pharmacol. 2009;22(2):311–322. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin Y, Pastrana JL, Li X, Huang X, Mallilankaraman K, Choi ET, Madesh M, Wang H, Yang XF. Inflammasomes: sensors of metabolic stresses for vascular inflammation. Front Biosci. 2013;18:638–649. doi: 10.2741/4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venereau E, Ceriotti C, Bianchi ME. DAMPs from cell death to new life. Front Immunol. 2015;6:422. doi: 10.3389/fimmu.2015.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Li YF, Nanayakkara G, Shao Y, Liang B, Cole L, Yang WY, Li X, Cueto R, Yu J, et al. Lysophospholipid receptors, as novel conditional danger receptors and homeostatic receptors modulate inflammation—novel paradigm and therapeutic potential. J Cardiovasc Transl Res. 2016;9(4):343–359. doi: 10.1007/s12265-016-9700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen J, Yin Y, Mai J, Xiong X, Pansuria M, Liu J, Maley E, Saqib NU, Wang H, Yang XF. Caspase-1 recognizes extended cleavage sites in its natural substrates. Atherosclerosis. 2010;210(2):422–429. doi: 10.1016/j.atherosclerosis.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y, Cheng Z, Li X, Chernaya V, Wang H, Yang XF. Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction- a novel mechanism for maintaining vascular function. J Hematol Oncol. 2014;7(1):80. doi: 10.1186/s13045-014-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin Y, Li X, Sha X, Xi H, Li YF, Shao Y, Mai J, Virtue A, Lopez-Pastrana J, Meng S, et al. Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arterioscler Thromb Vasc Biol. 2015;35(4):804–816. doi: 10.1161/ATVBAHA.115.305282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Pastrana J, Ferrer L, Li YF, Xiong X, Xi H, Cueto R, Nelson JZ, Sha X, Li X, Cannella AL, et al. Inhibition of caspase-1 activation in endothelial cells improves angiogenesis—a novel therapeutic potential for ischemia. J Biol Chem. 2015;290(28):17485–94. doi: 10.1074/jbc.M115.641191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li YF, Ren LN, Guo G, Cannella LA, Chernaya V, Samuel S, Liu SX, Wang H, Yang XF. Endothelial progenitor cells in ischemic stroke: an exploration from hypothesis to therapy. J Hematol Oncol. 2015;8(1):33. doi: 10.1186/s13045-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y-F, Huang X, Li X, Gong R, Yin Y, Nelson J, Gao E, Zhang H, Hoffman NE, Houser SR, Madesh M, Tilley DG, Choi ET, Jiang X, Huang C-X, Wang H, Yang X-F. Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Front Biosci (Landmark Ed) 2016;21:178–191. doi: 10.2741/4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer LM, Monroy AM, Lopez-Pastrana J, Nanayakkara G, Cueto R, Li YF, Li X, Wang H, Yang XF, Choi ET. Caspase-1 plays a critical role in accelerating chronic kidney disease-promoted neointimal hyperplasia in the carotid artery. J Cardiovasc Transl Res. 2016;9(2):135–144. doi: 10.1007/s12265-016-9683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi H, Zhang Y, Xu Y, Yang WY, Jiang X, Sha X, Cheng X, Wang J, Qin X, Yu J, et al. Caspase-1 inflammasome activation mediates homocysteine-induced pyrop-apoptosis in endothelial cells. Circ Res. 2016;118(10):1525–39. doi: 10.1161/CIRCRESAHA.116.308501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Fu H, Nanayakkara G, Li Y, Shao Y, Johnson C, Cheng J, Yang WY, Yang F, Lavallee M, et al. Novel extracellular and nuclear caspase-1 and inflammasomes propagate inflammation and regulate gene expression: a comprehensive database mining study. J Hematol Oncol. 2016;9(1):122. doi: 10.1186/s13045-016-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YF, Nanayakkara G, Sun Y, Li X, Wang L, Cueto R, Shao Y, Fu H, Johnson C, Cheng J, et al. Analyses of caspase-1-regulated transcriptomes in various tissues lead to identification of novel IL-1beta-, IL-18- and sirtuin-1-independent pathways. J Hematol Oncol. 2017;10(1):40. doi: 10.1186/s13045-017-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14(4):580–590. doi: 10.1101/gr.2160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97(2):161–3. [DOI] [PubMed]
- 18.Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19(3):207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 19.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olefsky JM. Nuclear receptor minireview series. J Biol Chem. 2001;276(40):36863–36864. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- 21.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 22.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog Horm Res. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 23.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582(1):2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang P, Zhang D, Cheng Z, Yan C, Jiang X, Kruger WD, Meng S, Arning E, Bottiglieri T, Choi ET, et al. Hyperhomocysteinemia potentiates hyperglycemia-induced inflammatory monocyte differentiation and atherosclerosis. Diabetes. 2014;63(12):4275–90. [DOI] [PMC free article] [PubMed]
- 25.Wang YM, Ong SS, Chai SC, Chen T. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol. 2012;8(7):803–817. doi: 10.1517/17425255.2012.685237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 28.Lee KS, Park SJ, Hwang PH, Yi HK, Song CH, Chai OH, Kim JS, Lee MK, Lee YC. PPAR-gamma modulates allergic inflammation through up-regulation of PTEN. FASEB J. 2005;19(8):1033–1035. doi: 10.1096/fj.04-3309fje. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Schindler J, Philp A. Regulation of skeletal muscle mitochondrial function by nuclear receptors: implications for health and disease. Clin Sci (Lond) 2015;129(7):589–599. doi: 10.1042/CS20150246. [DOI] [PubMed] [Google Scholar]
- 30.Forthmann B, Aletta JM, Lee YW, Terranova C, Birkaya B, Stachowiak EK, Stachowiak MK, Claus P. Coalition of nuclear receptors in the nervous system. J Cell Physiol. 2015;230(12):2875–2880. doi: 10.1002/jcp.25036. [DOI] [PubMed] [Google Scholar]
- 31.Abergel Z, Chatterjee AK, Zuckerman B, Gross E. Regulation of neuronal oxygen responses in C. elegans is mediated through interactions between globin 5 and the H-NOX domains of soluble guanylate cyclases. J Neurosci. 2016;36(3):963–978. doi: 10.1523/JNEUROSCI.3170-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, Yang XF, Wang H. Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB J. 2010;24(8):2804–2817. doi: 10.1096/fj.09-143651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Yin Y, Pastrana JL, Li X, Huang X, Mallilankaraman K, Choi ET, Madesh M, Wang H, Yang XF. Inflammasomes: sensors of metabolic stresses for vascular inflammation. Front Biosci (Landmark Ed) 2013;18:638–649. doi: 10.2741/4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41(6):898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choubey D, Panchanathan R. IFI16, an amplifier of DNA-damage response: role in cellular senescence and aging-associated inflammatory diseases. Ageing Res Rev. 2016;28:27–36. doi: 10.1016/j.arr.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Gurung P, Kanneganti TD. Immune responses against protozoan parasites: a focus on the emerging role of Nod-like receptors. Cell Mol Life Sci. 2016;73(16):3035–3051. doi: 10.1007/s00018-016-2212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Liu Z, Xiao TS. Post-translational regulation of inflammasomes. Cell Mol Immunol. 2017;14(1):65–79. doi: 10.1038/cmi.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koukoura O, Sifakis S, Spandidos DA. DNA methylation in endometriosis (review) Mol Med Rep. 2016;13(4):2939–2948. doi: 10.3892/mmr.2016.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Z, Yang X, Wang H. Hyperhomocysteinemia and endothelial dysfunction. Curr Hypertens Rev. 2009;5(2):158–165. doi: 10.2174/157340209788166940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Fang P, Yu D, Zhang L, Zhang D, Jiang X, Yang WY, Bottiglieri T, Kunapuli SP, Yu J, et al. Chronic kidney disease induces inflammatory CD40+ monocyte differentiation via homocysteine elevation and DNA hypomethylation. Circ Res. 2016;119(11):1226–1241. doi: 10.1161/CIRCRESAHA.116.308750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras AV, Torres N, Tovar AR. PPAR-alpha as a key nutritional and environmental sensor for metabolic adaptation. Adv Nutr. 2013;4(4):439–452. doi: 10.3945/an.113.003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medici V, Schroeder DI, Woods R, LaSalle JM, Geng Y, Shibata NM, Peerson J, Hodzic E, Dayal S, Tsukamoto H, et al. Methylation and gene expression responses to ethanol feeding and betaine supplementation in the cystathionine beta synthase-deficient mouse. Alcohol Clin Exp Res. 2014;38(6):1540–1549. doi: 10.1111/acer.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54(10):2552–2555. [PubMed] [Google Scholar]
- 45.Issa JP, Zehnbauer BA, Civin CI, Collector MI, Sharkis SJ, Davidson NE, Kaufmann SH, Baylin SB. The estrogen receptor CpG island is methylated in most hematopoietic neoplasms. Cancer Res. 1996;56(5):973–977. [PubMed] [Google Scholar]
- 46.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res. 1996;56(16):3655–3658. [PubMed] [Google Scholar]
- 47.Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43(4):985–991. doi: 10.1016/s0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 48.Sasaki M, Tanaka Y, Perinchery G, Dharia A, Kotcherguina I, Fujimoto S, Dahiya R. Methylation and inactivation of estrogen, progesterone, and androgen receptors in prostate cancer. J Natl Cancer Inst. 2002;94(5):384–390. doi: 10.1093/jnci/94.5.384. [DOI] [PubMed] [Google Scholar]
- 49.Lapidus RG, Nass SJ, Butash KA, Parl FF, Weitzman SA, Graff JG, Herman JG, Davidson NE. Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer Res. 1998;58(12):2515–2519. [PubMed] [Google Scholar]
- 50.Aithal GP, Grove JI. Genome-wide association studies in drug-induced liver injury: step change in understanding the pathogenesis. Semin Liver Dis. 2015;35(4):421–431. doi: 10.1055/s-0035-1567829. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Matsushita T, Madireddy L, Mousavi P, Baranzini SE. PINBPA: cytoscape app for network analysis of GWAS data. Bioinformatics. 2015;31(2):262–264. doi: 10.1093/bioinformatics/btu644. [DOI] [PubMed] [Google Scholar]
- 52.Racke MK, Gocke AR, Muir M, Diab A, Drew PD, Lovett-Racke AE. Nuclear receptors and autoimmune disease: the potential of PPAR agonists to treat multiple sclerosis. J Nutr. 2006;136(3):700–703. doi: 10.1093/jn/136.3.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shirinsky IV, Shirinsky VS. Targeting nuclear hormone receptors: PPARalpha agonists as potential disease-modifying drugs for rheumatoid arthritis. Int J Rheumatol. 2011;2011:937843. doi: 10.1155/2011/937843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burris TP, Busby SA, Griffin PR. Targeting orphan nuclear receptors for treatment of metabolic diseases and autoimmunity. Chem Biol. 2012;19(1):51–59. doi: 10.1016/j.chembiol.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park BV, Pan F. The role of nuclear receptors in regulation of Th17/Treg biology and its implications for diseases. Cell Mol Immunol. 2015;12(5):533–542. doi: 10.1038/cmi.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeuchi H, Yokota-Nakatsuma A, Ohoka Y, Kagechika H, Kato C, Song SY, Iwata M. Retinoid X receptor agonists modulate Foxp3(+) regulatory T cell and Th17 cell differentiation with differential dependence on retinoic acid receptor activation. J Immunol. 2013;191(7):3725–3733. doi: 10.4049/jimmunol.1300032. [DOI] [PubMed] [Google Scholar]
- 57.Li AC, Palinski W. Peroxisome proliferator-activated receptors: how their effects on macrophages can lead to the development of a new drug therapy against atherosclerosis. Annu Rev Pharmacol Toxicol. 2006;46:1–39. doi: 10.1146/annurev.pharmtox.46.120604.141247. [DOI] [PubMed] [Google Scholar]
- 58.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116(3):571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shahin D, Toraby EE, Abdel-Malek H, Boshra V, Elsamanoudy AZ, Shaheen D. Effect of peroxisome proliferator-activated receptor gamma agonist (pioglitazone) and methotrexate on disease activity in rheumatoid arthritis (experimental and clinical study) Clin Med Insights Arthritis Musculoskelet Disord. 2011;4:1–10. doi: 10.4137/CMAMD.S5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knutson TP, Truong TH, Ma S, Brady NJ, Sullivan ME, Raj G, Schwertfeger KL, Lange CA. Posttranslationally modified progesterone receptors direct ligand-specific expression of breast cancer stem cell-associated gene programs. J Hematol Oncol. 2017;10(1):89. doi: 10.1186/s13045-017-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schweizer MT, Yu EY. Persistent androgen receptor addiction in castration-resistant prostate cancer. J Hematol Oncol. 2015;8:128. doi: 10.1186/s13045-015-0225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sartor AO. Progression of metastatic castrate-resistant prostate cancer: impact of therapeutic intervention in the post-docetaxel space. J Hematol Oncol. 2011;4:18. doi: 10.1186/1756-8722-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24(2):171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Savic D, Ramaker RC, Roberts BS, Dean EC, Burwell TC, Meadows SK, Cooper SJ, Garabedian MJ, Gertz J, Myers RM. Distinct gene regulatory programs define the inhibitory effects of liver X receptors and PPARG on cancer cell proliferation. Genome Med. 2016;8(1):74. doi: 10.1186/s13073-016-0328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng S, Ciment S, Jan M, Tran T, Pham H, Cueto R, Yang XF, Wang H. Homocysteine induces inflammatory transcriptional signaling in monocytes. Front Biosci (Landmark Ed) 2013;18:685–695. doi: 10.2741/4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sever R, Glass CK. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol. 2013;5(3):a016709. doi: 10.1101/cshperspect.a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng Z, Elmes M, Kirkup SE, Abayasekara DR, Wathes DC. Alteration of prostaglandin production and agonist responsiveness by n-6 polyunsaturated fatty acids in endometrial cells from late-gestation ewes. J Endocrinol. 2004;182(2):249–256. doi: 10.1677/joe.0.1820249. [DOI] [PubMed] [Google Scholar]
- 68.Yang XF, Mirkovic D, Zhang S, Zhang QE, Yan Y, Xiong Z, Yang F, Chen IH, Li L, Wang H. Processing sites are different in the generation of HLA-A2.1-restricted, T cell reactive tumor antigen epitopes and viral epitopes. Int J Immunopathol Pharmacol. 2006;19(4):853–870. doi: 10.1177/039463200601900415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Mai J, Virtue A, Yin Y, Gong R, Sha X, Gutchigian S, Frisch A, Hodge I, Jiang X, et al. IL-35 is a novel responsive anti-inflammatory cytokine—a new system of categorizing anti-inflammatory cytokines. PLoS One. 2012;7(3):e33628. doi: 10.1371/journal.pone.0033628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138(3):863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 71.Nishimura M, Naito S, Yokoi T. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug Metab Pharmacokinet. 2004;19(2):135–149. doi: 10.2133/dmpk.19.135. [DOI] [PubMed] [Google Scholar]
- 72.D'Amore S, Vacca M, Graziano G, D'Orazio A, Cariello M, Martelli N, Di Tullio G, Salvia R, Grandaliano G, Belfiore A, et al. Nuclear receptors expression chart in peripheral blood mononuclear cells identifies patients with Metabolic Syndrome. Biochim Biophys Acta. 2013;1832(12):2289–2301. doi: 10.1016/j.bbadis.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Kuhn TS. The structure ofscientific revolutions(3rd ed.) Chicago: University of Chicago Press; 1996. [Google Scholar]
- 74.Yang WY, Shao Y, Lopez-Pastrana J, Mai J, Wang H, Yang XF. Pathological conditions re-shape physiological Tregs into pathological Tregs. Burns Trauma. 2015;3(1). [DOI] [PMC free article] [PubMed]
- 75.Sha X, Meng S, Li X, Xi H, Maddaloni M, Pascual DW, Shan H, Jiang X, Wang H, Yang XF. Interleukin-35 inhibits endothelial cell activation by suppressing MAPK-AP-1 pathway. J Biol Chem. 2015;290(31):19307–19318. doi: 10.1074/jbc.M115.663286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Y, Johnson C, Zhou J, Wang L, Li Y-F, Lu Y, Nanayakkara G, Fu H, Shao Y, Sanchez C, Yang WY, Wang X, Choi ET, Li R, Wang H, Yang X-F. Uremic toxins are conditional danger- or homeostasis-associated molecular patterns. Front Biosci. 2017;22(23):348–87. doi: 10.2741/4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Basil MC, Levy BD. Specialized pro-resolving mediators: endogenous regulators of infection and inflammation. Nat Rev Immunol. 2016;16(1):51–67. doi: 10.1038/nri.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallace BD, Redinbo MR. Xenobiotic-sensing nuclear receptors involved in drug metabolism: a structural perspective. Drug Metab Rev. 2013;45(1):79–100. doi: 10.3109/03602532.2012.740049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leclercq G, Jacquot Y. Interactions of isoflavones and other plant derived estrogens with estrogen receptors for prevention and treatment of breast cancer-considerations concerning related efficacy and safety. J Steroid Biochem Mol Biol. 2014;139:237–244. doi: 10.1016/j.jsbmb.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 80.Bloom MS, Mok-Lin E, Fujimoto VY. Bisphenol A and ovarian steroidogenesis. Fertil Steril. 2016;106(4):857–863. doi: 10.1016/j.fertnstert.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 81.Eick GN, Colucci JK, Harms MJ, Ortlund EA, Thornton JW. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 2012;8(11):e1003072. doi: 10.1371/journal.pgen.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440(7087):1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 83.Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 84.O'Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6(4):R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 85.Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46(47):13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- 86.Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, Tripathy A, Raetz CR, McDonnell DP, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12(4):357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 87.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5(2):289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 88.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296(5571):1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 89.Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature. 1992;355(6358):359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 90.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68(2):397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 91.Shi H, Kichaev G, Pasaniuc B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am J Hum Genet. 2016;99(1):139–153. doi: 10.1016/j.ajhg.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169(7):1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Furlong LI. Human diseases through the lens of network biology. Trends Genet. 2013;29(3):150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 96.Chakravarti A, Turner TN. Revealing rate-limiting steps in complex disease biology: the crucial importance of studying rare, extreme-phenotype families. Bioessays. 2016;38(6):578–586. doi: 10.1002/bies.201500203. [DOI] [PubMed] [Google Scholar]
- 97.Vassy JL, Hivert MF, Porneala B, Dauriz M, Florez JC, Dupuis J, Siscovick DS, Fornage M, Rasmussen-Torvik LJ, Bouchard C, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes. 2014;63(6):2172–2182. doi: 10.2337/db13-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–421. [PubMed]
- 99.Alberti KG, Zimmet P, Shaw J. The metabolic syndrome—a new worldwide definition. Lancet. 2005;366(9491):1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 100.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 101.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 102.Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, Iwamoto Y, Liao R, Zirlik A, Scherer-Crosbie M. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res. 2014;114(10):1611–1622. doi: 10.1161/CIRCRESAHA.114.303204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103(4):1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Giudici M, Goni S, Fan R, Treuter E. Nuclear receptor coregulators in metabolism and disease. 2015. [DOI] [PubMed] [Google Scholar]
- 105.Vuong LM, Dhahbi J, Sladek FM. SAT-273: expression of nuclear receptor HNF4a in mouse embryonic stem cells is sufficient to inhibit cell proliferation and drive differentiation. 2015. [Google Scholar]
- 106.Lin S-J, Lin C-Y, Yang D-R, Izumi K, Yan E, Niu X, Chang H-C, Miyamoto H, Wang N, Li G. The differential effects of anti-diabetic thiazolidinedione on prostate cancer progression are linked to the TR4 nuclear receptor expression status. Neoplasia. 2015;17(4):339–347. doi: 10.1016/j.neo.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghoneim RH, Ngo Sock ET, Lavoie J-M, Piquette-Miller M. Effect of a high-fat diet on the hepatic expression of nuclear receptors and their target genes: relevance to drug disposition. Br J Nutr. 2015;113(03):507–516. doi: 10.1017/S0007114514003717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nuclear receptors are differently expressed in human and mouse tissues. A: representative tissue mRNA distribution profile of housekeeping gene ARHGDIA in humans and Ldha in mice. See “Experimental Procedures” for details. B: mRNA distribution profiles of 26 nuclear receptors in 21 human tissues. C: MRNA distribution profiles of 15 nuclear receptors in 17 mouse tissues. The statistical significance was defined as when gene expression was larger than the upper limit of the confidence interval. Gene symbols are listed in Tables 1, 2, and 3. (PDF 296 kb)
Forty-eight nuclear receptors are associated with SAH and SAM/SAH ratio in mouse tissues. A: shows the correlation of 48 nuclear receptors associated with SAH levels (hypomethylation status) in mouse tissues. B. shows the correlation of 48 nuclear receptors associated with SAM/SAH ratio in mouse tissues. (PDF 648 kb)
Data Availability Statement
Experimentally verified mRNA expressions of NRs in various tissues were obtained from databases of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) UniGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene). The microarray datasets that were used in this study were retrieved from NIH-GEO database (https://www.ncbi.nlm.nih.gov/gds), and the numbers of datasets that were used are as follows: GSE55235, GSE6088, GSE6054, GSE23561, GSE13670, GSE55100, GSE48964, GSE15773, GSE9490, GSE43292, GSE13139, GSE18443, GSE24342, GSE21324, GSE3920, GSE19240, GSE40838, GSE10939, GSE46150, GSE42267, GSE33101, GSE2489, GSE41802, GSE57002, GSE56670, and GSE49694. MouseMine database (http://www.mousemine.org/mousemine/begin.do) was used to analyze how the deficiency of NRs lead to development of metabolic and cardiovascular pathologies.


