Abstract
Objectives
An epidemiological survey was carried out in order to obtain a better understanding of the role of wild boars in the epidemiology of the influenza virus.
Design
The samples were submitted to Real‐Time PCR testing for gene M of the swine influenza virus (SIV), and virus isolation was performed from the positive PCR samples. Genome sequence analysis was performed on the isolates. Additionally, 1,977 boar sera samples were analyzed using ELISA and hemoagglutination inhibition.
Setting
Over recent years, the wild boar population has greatly increased in Italy, including in areas of high‐density industrial pig farming, where the influenza virus is widespread. From July to December 2012, wild boar lung samples were collected in the Parma and Piacenza area, in the Emilia Romagna region.
Sample
354 wild boar lung samples were collected.
Main outcome measures
Wild‐boar influenza A virus infection should be studied more broadly in order to obtain a better understanding of the epidemiological role played by this species.
Results
Three SIV strains were isolated out of 12 samples that resulted positive using PCR analysis and they were identified as avian‐like SIV subtype H1N1. Phylogenetic analysis of the sequences obtained from isolate A/wild boar/291320/2012 showed that it clustered with recent Italian avian‐like H1N1 SIVs isolated from domestic pigs. Sixty‐eight sera samples showed a positive titer to the isolate A/wild boar/291320/2012.
Conclusions
This study suggests that SIV actively circulates in the wild boar population in the investigated. area.
Keywords: influenza, wild boar, virus, antibodies
Introduction
Influenza viruses are segmented RNA viruses belonging to the Orthomixoviridae family, which includes three different Influenza virus genera: A, B, C. Among these, influenza A viruses (IAVs) are the most common and can infect humans and several animal species, both wild and domestic, such as pigs, dogs, horses, and birds. Wild birds play a key role in the ecology of IAV, generally being considered its reservoir. Due to their viral genome segmentation, all IAVs, including pig influenza viruses, undergo continuous genetic evolution. Swine has very important role in influenza virus ecology, being sensitive to both avian and mammalian (including human) influenza viruses.
Nowadays, there are three principal subtypes of IAVs circulating in the Italian pig population: H1N1, H3N2, H1N2, and more recently, H1N1 pdm 2009 has also been described.1 In Italy, SIV is responsible for 12% of respiratory diseases on intensive pig farms.2
While many studies have been carried out on domestic animals, there is still much to investigate in wild mammals, especially with regards to influenza virus evolutionary dynamics.
Epidemiological surveys are essential for better understanding the role of wild pigs and boars (both belonging to the Sus scrofa species) in influenza virus epidemiology. SIV infection in wild boar has been reported in several European countries with varying prevalence such as 0·7% in Poland,3 4% 4 and 6·4% 5 in Spain, but it has not been reported in Italy until now. A survey in Slovenia did not demonstrate serological response for the influenza virus.6 In Germany, a survey was carried out over two consecutive years and showed 7·8% and 5·2% of positive samples, respectively, and also an H3N2 subtype was isolated.7, 8 In Croatia, a recent investigation revealed 9·7% positive response.9 Serological positive response was also shown in the USA.10, 11 Variable rates of positivity, related to the pig farm density, ranging from 0% to 91% were shown in different geographical areas of South and North Carolina.12 Recently in Texas, subtype H1N1 pdm 2009 circulation was demonstrated in feral pigs.13 In France, no evidence of influenza virus circulation was found in wild boars living in an area shared with migratory waterfowl,14 even if in some regions detection of antibodies in wild boars is reported.15 In Italy, only two serological surveys have been carried out in the northern part of the country in 1992 16 and 1996–1998,17 revealing no circulation of the influenza virus in the wild boar population.
The aim of this study is to provide a preliminary updating overview of the circulation of SIV in the wild boar population of the neighboring Apennine areas of the provinces of Parma and Piacenza (3449 and 2589 km2, respectively), in the Emilia Romagna region, in Northern Italy, where this species is widespread.
Materials and methods
During an active surveillance program carried out from July to December 2012 (as part of the 2012–2013 Emilia Romagna Regional Wildlife Control Plan), 354 lung samples and 1977 serum samples were collected in wild boars. The lung samples were tested for the influenza A virus using a real‐time RT‐PCR.18 To isolate the virus from positive samples, they were inoculated onto MDCK and CACO‐2 cells and into 11‐day‐old SPF chicken embryonated eggs.19 The cell‐culture supernatant and allantoic fluid were tested by hemagglutination assay (HA) with chicken erythrocytes, using the standard procedure.20 To reveal the presence of IAV, a double‐antibody sandwich ELISA (NPA‐ELISA) was used.21 The positive samples were tested by multiplex RT‐PCR 22 for virus subtyping. Antigenic characterization was performed by hemagglutination inhibition (HI).20
When it was not possible to isolate the virus from them, the positive samples were tested directly using multiplex RT‐PCR to reveal the SIV subtype present in the lung.
Full genome sequence analysis 23 was carried out on one isolated virus (A/wild boar/Italy/291320/2012) and partial genome sequencing, HA1 region for HA gene and a 514 base pairs region of the NA gene,22 was carried out on A/wild boar/Italy/324480‐4/2012 and A/wild boar/Italy/324486‐3/2012, resolving the sequences using an ABI 3130 DNA automatic sequencer (Applied Biosystems, Foster City, CA, USA). The Lasergene sequencing analysis software package (DNASTAR, Madison, WI) was applied to combine and edit DNA sequences. ClustalW was used to make multiple sequence alignments, and MEGA5 software 24 was used to generate distance‐based phylogenetic trees.
Phylogenetic analysis was conducted using neighbor‐joining,25 following the method used by Tamura–Nei.26
Serum samples collected from wild boars (n = 1977) were examined using homemade ELISA test for influenza A NP antibodies according to de Boer et al.27 Next, positive samples for NP antibodies were tested using HI 20, 28 to detect the presence of antibodies against the main SIV subtypes circulating in Italy, that is, hu‐like H1N2 (A/swine/Italy/284922/2009), av‐like H1N1 (A/swine/Italy/267505/2010), human‐derived H3N2 (A/swine/Italy/312583/2009), H1N1 pdm 2009 (A/swine/Italy/290271/2009), and the virus isolated from wild boar A/wild boar/Italy/291320/2012. A serum sample was considered positive if the HI titer was ≥1/20. If a sample scored positive for more than one antigen, due to sera cross‐reaction, it was assigned to the subtype showing an HI titer at least twofold higher than to the other viral types.
Moreover, we assessed whether HI titers obtained from A/wild boar/Italy/291320/2012 and from A/swine/Italy/267505/2010 are significantly different by fitting a generalized linear model (GLM) with binomial error distribution for every dilution of the HI test. Statistical analyses were performed with R 2.15.0.29
Results
During the study, a total of 354 samples of lung tissue were tested using real‐time RT‐PCR targeting the M gene: 12 lung samples (3·38%) gave positive results. Virological examination performed on the PCR positive samples led, after one passage on the substrates, to the isolation of 3 IAVs (A/wild boar/Italy/291320/2012, A/wild boar/Italy/324480‐4/2012, A/wild boar/Italy/324486‐3/2012). RT multiplex PCR testing and serological antigenic characterization identified the isolates as avian‐like H1N1 SIVs. The RT multiplex PCR test performed on the 9 lung homogenates, which were negative for viral isolation, showed the presence of avian‐like H1N1 SIVs. Full sequences of the strain A/wild boar/Italy/291320/2012 were obtained and then they were registered in the GenBank (KC984942, KC984943, KC984944, KC984945, KC984946, KC984947, KC984948, KC984949). The gene sequences of the wild boar strain were compared with swine, avian, and human influenza virus sequences, retrieved from the NCBI Influenza Virus Resource as well as from our collection from SIV diagnostic activity. Sequences of HA and NA genes of A/wild boar/Italy/291320/2012 clustered with sequences of H1N1 SIVs isolated recently in Italy (Figure 1). In particular, the sequences of all eight gene segments were closely related (with 99–100% of nucleotide identity) with the strain A/swine/Italy/218884‐2/2012 (GenBank accession numbers KC984938, KC984941, KF000002, KF000003, KF000004, KF000005, KF000006, KF000007) isolated during a respiratory outbreak in a pig farm located in the same area (Piacenza province) of the wild boar sampling.
Figure 1.
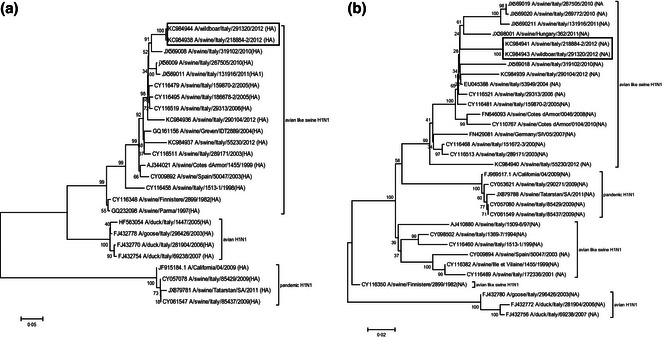
Phylogenetic tree of HA (A) and NA (B) genes of SIVs and A/wild boar/291320/2012. The evolutionary history was inferred using the neighbor‐joining method with bootstrap test (1000 replicates). The evolutionary distances were computed using the Tamura–Nei method. The rate variation among sites was modeled with a gamma distribution [shape parameter = 0·38 for (A) and 0·43 for (B)]. Evolutionary analyses were conducted in MEGA5.
The analysis of the nucleotide sequences of the fragments derived from HA and NA genes of the other two isolates (A/wild boar/Italy/324480‐4/2012, A/wild boar/Italy/324486‐3/2012), revealed that the sequences of these gene fragments were identical (100%) to the sequences of the strain A/wild boar/Italy/291320/2012.
Upon the examination of 1977 sera from wild boar shot in 2012 in the area considered, antibodies against NP of IAV were detected using ELISA in 78 samples (3·9%). Seventy‐eight positive samples were subjected to HI tests, 58 sera showed antibodies against the avian‐like H1N1 A/swine/Italy/267505/2010 and A/wild boar/Italy/291320/2012, 10 more samples showed antibodies only against the homologous A/wild boar/Italy/291320/2012 while 9 samples resulted negative against the strains tested. One serum sample showed antibodies only against H3N2 SIV subtype with 1:640 titer. There were no positive results for SIV H1N2 subtype, while only some cross‐reactions were observed using the H1N1 pdm 2009 subtype.
It is interesting to highlight that the titers obtained using HI, with the influenza virus isolated from wild boar, were higher compared with HI titers obtained from the A/swine/Italy/267505/2010 strain, and the differences were statistically significant (Table 1).
Table 1.
Comparison between serological titers obtained with HI reaction using H1N1 SIV and H1N1 wild boar virus
| SIV Strain | No. of positives | Titer | ||||||
|---|---|---|---|---|---|---|---|---|
| 20 | 40 | 80 | 160 | 320 | 640 | >1280 | ||
| A/swine/Italy/267505/2010 | 58 | 21 | 21 | 12 | 1 | 3 | 0 | 0 |
| A/wild boar/Italy/291320/2012 | 68 | 8* | 16*** | 15*** | 17*** | 8* | 2 | 2 |
Significance of the Chi‐square tests obtained from GLM with binomial error distributions: *P < 0·05; ** P < 0·01; *** P < 0·001.
Discussion
Swine are very important to influenza virus ecology, being sensitive to both avian and mammal (including human) influenza viruses, whereas the role of wild boar is not clear. In particular, it is necessary to clarify its significance in maintaining and spreading this virus within wild animal populations. SIV infection in wild boar has been reported in several European countries with varying prevalence, but it has not been reported in Italy until now.
The wild boar population in Italy has increased greatly over recent years, both in number and in geographical distribution,30 and today, in Emilia Romagna (22124 km2), around 21000 wild boar are hunted each year; moreover, intensive pig farming is practiced in this region and thus SIV actively circulates. In particular, the SIV H1N1 subtype was found to be responsible for clinical respiratory outbreaks in intensive pig farms in 47% of cases.2
This study revealed the circulation of SIV H1N1 for the first time in the Italian boar population, in an area highly populated by this species. The rate of lung samples positive for SIV gene M by biomolecular assay was 3·38% (12/354). The phylogenetic analysis of the isolate showed that the wild boar strain was closely related to a pig SIV strain isolated 3 months before from pigs reared in the same area. It can be hypothesized that pigs are a source of SIV for wild boars, as observed by Corn in North Carolina.12
Active circulation of SIV H1N1 subtype in the wild population of the examined area is confirmed from the high HI titers observed during the serological investigation performed using both the SIV pig strain and the wild boar strain. In the wild boar population considered, the circulation of IAV H1N1 pdm 2009 and H1N2 subtypes was not shown. Moreover, it is important to highlight the serological positivity at 1:640 titer for the H3N2 SIV strain in a boar serum, even if it only occurred in one isolated case.
It was investigated whether there were any significant habitat data that could have affected the epidemiology of the SIV infection reported. The number and the location of swine farms in the study area were analyzed, and the presence of small family‐run pig and boar farms with low biosecurity levels was observed, as they allow contact between wild and domestic populations. Once infected, the high density of the population, the gregarious habits of boars, mainly nearby food and water sources, could play a role in spreading and maintaining SIV within the boar population and possibly in other wildlife species.
The virological and serological data collected suggest the need for carrying out other investigations to clarify further which SIV subtypes, besides H1N1 subtype, circulate among wild boars and also to obtain a better understanding of the role that this wild species has in the epidemiology of IAVs within wild animal populations.
Usually, SIVs do not cause zoonotic infections; however, sporadic, dead‐end infection of humans has been reported with most SIV subtypes or lineages31, 32 although in the wild, human contact would not be very common. In any case, the frequency of antigenic variation of IAVs and the possibility of cross‐species transmission highlights the importance of monitoring influenza virus circulation, not only in domestic species, but also in wildlife, including mammals.
Acknowledgements
The authors thank Dr. Luca Bolzoni for statistical analysis, Mrs. Roberta Manfredi and Mrs. Daniela Bresciani for technical assistance, and Dr. Steans Nuala for English language revision.
Foni et al (2013) Epidemiological survey of swine influenza A virus in the wild boar population of two Italian provinces. Influenza and Other Respiratory Viruses 7(Suppl. 4), 16–20.
References
- 1. Moreno A, Di Trani L, Alborali L et al First pandemic H1N1 outbreak from a pig farm in Italy. Open Virol J 2010; 4:52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Foni E, Chiapponi C, Sozzi E et al Characterization of swine influenza viruses circulating in Italy in 2008‐2009; in SIPAS eds. Atti del XXXVI meeting annuale della Società Italiana di patologia ed Allevamento dei suini. Montichiari, Brescia, Italy: Società Italiana di Patologia ed Allevamento dei suini, 2010; 159–166. [Google Scholar]
- 3. Markowska‐Daniel I. Monitoring of swine influenza in Poland in the season 2001/2002; in: Proceedings of the Fourth International Symposium on Emerging and Re‐emerging pig diseases. Rome, Italy, 2003; 277–278. [Google Scholar]
- 4. Vicente J, Leon‐Vizcaino L, Gortazar C, Jose Cubero M, Gonzalez M, Martin‐Atance P. Antibodies to selected viral and bacterial pathogens in European wild boars from southcentral Spain. J Wildl Dis 2002; 38:649–652. [DOI] [PubMed] [Google Scholar]
- 5. Closa‐Sebastià F, Casas‐Diaz E, Cuenca R, Lavin S, Mentaberre G, Marco I. Antibodies to selected pathogens in wild boar (sus scrofa) from Catalonia (NE spain). Eur J Wildl Res 2011; 57:977–981. [Google Scholar]
- 6. Vengust G, Valencak Z, Bidovec A. A serological survey of selected pathogens in wild boar in Slovenia. J Vet Med B Infect Dis Vet Public Health 2006; 53:24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaden V, Lange E, Hänel A et al Retrospective serological survey on selected viral pathogens in wild boar populations in Germany. Eur J Wildl Res 2009; 55:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaden V, Lange E, Starick E, Bruer W, Krakowski W, Klopries M. Epidemiological survey of swine influenza A virus in selected wild boar populations in Germany. Vet Microbiol 2008; 131:123–132. [DOI] [PubMed] [Google Scholar]
- 9. Roic B, Jemersic L, Terzic S, Keros T, Balatinec J, Florijancic T. Prevalence of antibodies to selected viral pathogens in wild boars (sus scrofa) in Croatia in 2005‐06 and 2009‐10. J Wildl Dis 2012; 48:131–137. [DOI] [PubMed] [Google Scholar]
- 10. Saliki JT, Rodgers SJ, Eskew G. Serosurvey of selected viral and bacterial diseases in wild swine from Oklahoma. J Wildl Dis 1998; 34:834–838. [DOI] [PubMed] [Google Scholar]
- 11. Gipson PS, Veatch JK, Matlack RS, Jones DP. Health status of a recently discovered population of feral swine in Kansas. J Wildl Dis 1999; 35:624–627. [DOI] [PubMed] [Google Scholar]
- 12. Corn JL, Cumbee JC, Barfoot R, Erickson GA. Pathogen exposure in feral swine populations geographically associated with high densities of transitional swine premises and commercial swine production. J Wildl Dis 2009; 45:713–721. [DOI] [PubMed] [Google Scholar]
- 13. Clavijo A, Nikooienejad A, Esfahani MS et al Identification and analysis of the first 2009 pandemic H1N1 influenza virus from U.S. feral swine. Zoonoses Public Health 2013; 60:327–335. [DOI] [PubMed] [Google Scholar]
- 14. Vittecoq M, Grandhomme V, Simon G et al Study of influenza A virus in wild boars living in a major duck wintering site. Infect Genet Evol 2012; 12:483–486. [DOI] [PubMed] [Google Scholar]
- 15. Payne A, Rossi S, Lacour SA et al Bilan sanitaire du sanglier vis‐à‐vis de la trichinellose, de la maladie d'Aujeszky, de la brucellose, de l'hépatite E et des virus influenza porcins en France. Bulletin Epidémiologique Santé Animale et Alimentation Anses‐DGAl 2011; 44:2–8. [Google Scholar]
- 16. Cordioli P, Callegari S, Berlinzani A, Foni E, Candotti P, Barigazzi G. Indagine sierologica su cinghiali selvatici dell'Appennino Parmense In SISVET eds. Atti del meeting annuale della Società Italiana delle Scienze Veterinarie.Riccione, Italy: Società italiana delle Scienze Veterinarie, 1993. 1159–1162. [Google Scholar]
- 17. Ferroglio E, Acutis PL, Masoero L, Gennero S, Rossi L. Indagine sierologica su una popolazione di cinghiali nelle alpi occidentali. J Mt Ecol 2003; 7:225–228. [Google Scholar]
- 18. Slomka MJ, Densham AL, Coward VJ et al Real time reverse transcription (RRT)‐polymerase chain reaction (PCR) methods for detection of pandemic (H1N1) 2009 influenza virus and European swine influenza A virus infections in pigs. Influenza Other Respi Viruses 2010; 4:277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiapponi C, Zanni I, Garbarino C, Barigazzi G, Foni E. Comparison of the usefulness of the CACO‐2 cell line with standard substrates for isolation of swine influenza A viruses. J Virol Methods 2010; 163:162–165. [DOI] [PubMed] [Google Scholar]
- 20. OIE . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 7th ed OIE, 2012. Chapter 2.8.8, Swine Influenza. Available at http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.08.08_SWINE_INFLUENZA.pdf (Accessed 5 May 2013). [Google Scholar]
- 21. Foni E, Candotti P, Raffo A, Gamba D, Cataldi M, Barigazzi G. Utilizzo di ELISA sandwich nella diagnosi di influenza suina a confronto con tecniche di isolamento virale; in SISVET (eds): Atti del XLIX meeting annuale della SISVET: Società Italiana delle Scienze Veterinarie, 1995; 543–544. [Google Scholar]
- 22. Chiapponi C, Moreno A, Barbieri I, Merenda M, Foni E. Multiplex RT‐PCR assay for differentiating European swine influenza virus subtypes H1N1, H1N2 and H3N2. J Virol Methods 2012; 184:117–120. [DOI] [PubMed] [Google Scholar]
- 23. Moreno A, Chiapponi C, Boniotti MB et al Genomic characterization of H1N2 swine influenza viruses in italy. Vet Microbiol 2012; 156:265–276. [DOI] [PubMed] [Google Scholar]
- 24. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saitou N, Nei M. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 26. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993; 10:512–526. [DOI] [PubMed] [Google Scholar]
- 27. De Boer GF, Back W, Osterhaus AD. An ELISA for detection of antibodies against influenza A nucleoprotein in humans and various animal species. Arch Virol 1990; 115:47–61. [DOI] [PubMed] [Google Scholar]
- 28. Kendal AP, Pereira MS, Skehel JJ. Hemagglutination inhibition; in Kendal AP, Pereira MS, Skehel JJ. (eds): Concepts and Procedures for Laboratory‐Based Influenza Surveillance. Atlanta, GA: Centers for Disease Control and Prevention, 1982. [Google Scholar]
- 29. R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2012. Available at http://www.R-project.org/ [Google Scholar]
- 30. Carnevali L, Pedrotti L, Riga F, Toso S. Banca dati ungulati: status, distribuzione, consistenza, gestione e prelievo venatorio delle popolazioni di ungulati in Italia. Rapporto 2001‐2005. Biol Cons. Fauna 2009; 117:1–168. [Google Scholar]
- 31. Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis 2007; 44:1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Reeth K. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet Res 2007; 38:243–260. [DOI] [PubMed] [Google Scholar]


