Abstract
Background
Outbreaks of equine influenza (EI) in endemic populations cause disruption and economic loss.
Objectives
To identify (i) factors involved in the spread of EI (ii) virus strains responsible for outbreaks (iii) single radial haemolysis (SRH) antibody levels correlating with protection against current virus strains (iv) evidence of vaccination breakdown.
Methods
RT‐PCR, virus isolation and SRH were carried out on nasopharyngeal swabs and blood samples collected from horses, ponies and donkeys on affected premises. Data relating to 629 samples from 135 equidae were analysed.
Results and conclusions
Outbreaks were sporadic, self limiting and associated with the movement of horses. Vaccination status and age influenced clinical signs of disease while housing and fomites contributed to virus spread. Subclinical infection as defined as a horse which tested positive by one or more of the following; RT‐PCR, virus isolation and seroconversion in the absence of clinical signs, was identified in 9% of animals. Of the horses with up to date vaccination records 32% developed clinical signs. Vaccine breakdown occurred among horses vaccinated with all four commercially available vaccines. Analysis of HA1 sequence data generated for 26 viruses indicated that they all belonged to clade 2 of the Florida sublineage. Higher SRH antibody levels were required for both clinical and virological protection than reported in studies where vaccine strains were antigenically and genetically similar to those circulating in the field. The results of this study therefore support the OIE recommendations that vaccines be updated to include representatives of both clades of the Florida sublineage.
Keywords: Equine, influenza, investigations, Ireland, outbreak
Introduction
Equine influenza virus (EIV) a highly contagious respiratory pathogen of horses and other equidae is endemic in Europe and North America, and major outbreaks have occurred in Asia,1, 2, 3, 4, 5 Africa,6, 7 Australia8 and South America (http://www.oie.int/wahis/public.php). Two virus subtypes H7N7 and H3N8 have been isolated in horses, and for nearly two decades, both virus subtypes co‐circulated. It is now considered, however, that the H7N7 virus first identified in horses in 19569 is no longer in circulation.10 Since 1979, all outbreaks of equine influenza (EI) for which virus has been isolated have been caused by the H3N8 subtype. Both virus subtypes are included in the majority of commercially available vaccines despite the fact that the Office International des Epizooties (OIE) stipulate it is no longer a requirement for vaccines to include a H7N7 virus.11
Outbreaks of EI occur predominately among naïve and partially immune populations. During the 2‐year period 2010–2012, outbreaks were reported in Argentina, Brazil, Canada, Chile, China, Dominican Republic, France, Germany, India, Mongolia, Sweden, UAE, United Kingdom, USA and Uruguay (http://www.aht.org.uk/icc/linksicc.html; http://www.oie.int/wahis/public.php) in addition to those identified in Ireland during this study. At a minimum, such outbreaks result in suboptimal performance, severe disruption to training schedules and significant economic losses. The virus infects the ciliated epithelium and impairs mucociliary clearance which may take several weeks to recover, even if uncomplicated by secondary bacterial infection.12 Rest and restriction of horse movement are essential to the management of clinical cases.13 This can result in individual racehorses missing important racing fixtures which may impact on their potential value and that of their progeny. Where large populations are affected, betting revenue may decline as a result of the uncertainty relating to racing form. Continuing to exercise horses following infection has been demonstrated to exacerbate the severity of clinical disease and increase weight loss.14 In a study carried out in the UK, examinations of the network of contacts between racehorses suggest that the spread of infectious diseases such as EI may occur rapidly.15 This is largely due to the close proximity of horses within yards and horses from different yards on the training gallops. During an EI outbreak, which infiltrated the racing population in Newmarket in 2003, 21 training yards and over 1300 horses were affected during a 2‐month period.16 Equestrian events may also be affected and sometimes cancelled. During the outbreak in Uruguay in 2012, which affected over 2000 horses, race meetings were cancelled for several weeks and movement of horses out of the country was prohibited (http://www.oie.int/wahis/public.php). Equine influenza outbreaks also resulted in the cancellation of equestrian events in Brazil (http://www.oie.int/wahis/public.php). While mortality is very rarely associated with EI, more than 40 horses died during an outbreak affecting over 74 000 horses in Mongolia in 2011. Disconcertingly, several foal deaths were also reported during EI outbreaks in France during the same year (http://www.oie.int/wahis/public.php).
Vaccination against EI is an effective method of disease control and following the introduction of mandatory vaccination of racehorses in Ireland and the UK in 1981, no major equestrian event has been cancelled as a result of the disease.17 The racing authorities in Ireland and the UK stipulate that the first two doses of primary vaccination be administered between 21 and 92 days apart followed by a third vaccine dose 150–215 days after the second vaccination. Thereafter, annual booster vaccination is required (http://www.tuftclub.ie; http://www.thejockeyclub.co.uk). Outbreaks have occurred, however, when the virus circulating in the field was significantly different from that contained in the vaccines.16, 18, 19 The horse industry requires EI vaccines that prevent both clinical disease (clinical protection) and virus shedding. The role of subclinically infected vaccinated horses (horses that are not virologically protected) in the global spread of EIV is of major concern to the industry. The importation of such horses in conjunction with inadequate quarantine procedures has resulted in several major outbreaks with significant economic consequences. It is estimated that the eradication of EI from Australia following the importation of a subclinically affected horse cost in excess of one billion Australian dollars.8 Therefore, in order to maintain the effectiveness of vaccination, surveillance of EI viruses and prompt updating of vaccine strains is of fundamental importance. Ireland has an active EI surveillance programme as does the UK, but in many other countries, surveillance is inadequate predominately due to a lack of funding. The aims of this study were to identify: the factors involved in the spread of EI in a country where the virus is endemic, the virus strains responsible for the outbreaks, the single radial haemolysis (SRH) antibody levels correlating with protection against current virus strains circulating in the field and any evidence of vaccination breakdown as defined by disease and/or virus shedding.
Materials and methods
Sample collection and clinical histories
Nasopharyngeal swabs and clotted blood samples were submitted to the Irish Equine Centre by veterinary surgeons attending horses with acute respiratory disease. After collection, nasopharyngeal swabs were placed in 5 ml of viral transport medium as previously described.20 Clinical histories were obtained from the attending veterinary surgeon and personnel involved in the day‐to‐day management of the horses. On the premises where EI was confirmed, morbidity was defined as the presence of one or more of the three most common clinical signs associated with influenza, that is, pyrexia, nasal discharge and coughing. A confirmed case of EI was defined as a horse which tested positive by one or more of the following: RT‐PCR, virus isolation and seroconversion. Following diagnosis, the sampling regime on affected premises was dictated by the cooperation of the veterinary surgeon and the owner. The average time between consecutive samplings was 6·6 ± 0·95 days SE. Nasopharyngeal swabs and whole‐blood samples were collected on average 2·1 ± 0·04 SE and 2·6 ± 0·08 SE times, respectively, from all horses included in the study.
Real‐time RT‐PCR
RNA was extracted from 140 μl of nasal secretions using the QIAamp Viral RNA Mini kit (Qiagen Venlo, Limburg, Netherland) according to the manufacturer's instructions. One‐step RT‐PCR was performed using a LightCycler RNA Amplification SYBR Green I Kit (Roche, Burgess Hill, West Sussex, UK) on a LightCycler 2.0 platform (Roche) as previously described.21
Virus isolation
RT‐PCR positive samples (100 μl) were passaged up to five times in the allantoic cavities of 9‐ to 12‐day‐old embryonated hen's eggs. Eggs were incubated at 34°C (±1°C) for 72 hours and then placed at 4°C (±1°C) overnight before harvesting. The allantoic fluid was tested for haemagglutinating activity using 1% chicken erythrocytes in PBS according to standard procedure.22
Serological diagnosis
Antibodies against A/eq/Prague/56 (H7N7) and A/eq/Meath/07 (H3N8) were measured using SRH as previously described.23 Results were expressed in square millimeter, and a seroconversion was defined as an increase in SRH of 25 mm2 or 50% whichever was smaller between the paired serum samples.24 A difference of 25 mm2 or greater between the H3N8 and H7N7 antibody levels was considered significant.23
HAI gene sequencing and analysis
The HA1 sequence (1009 bp) of 26 viruses identified during this study was determined. This included nine viruses identified in 2010, two viruses identified in 2011 and 15 viruses identified in 2012. The sequence data for 24 of these viruses were derived from viral RNA extracted from nasopharyngeal swabs. Two viruses (A/eq/Carlow/11 and A/eq/Kilkenny/1/12) required amplification in embryonated hen's eggs prior to sequencing. The HA1 gene was amplified by RT‐PCR and sequenced in four separate reactions as previously described.25 The RT‐PCR products were analysed on a 1·2% agarose gel stained with SYBR safe DNA gel stain (Life Technologies, CA, USA) and purified using the QIAquick PCR Purification Kit (Qiagen). Sequencing was performed by Qiagen Sequencing Services (Hilden, Germany), and genetic analysis was undertaken with lasergene software (DNAstar, Madison, WI, USA).
HA1 gene sequence analysis
Multiple nucleotide and amino acid alignments were undertaken using the clustalw2 program26 from the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/clustalw2/). The HA1 gene sequences used in phylogenetic analysis, including the Irish viruses described here, are obtainable from the NCBI sequence database GenBank (Table S1). To determine the relationship between EI viruses, a maximum likelihood phylogenetic tree was constructed. The appropriate nucleotide model of substitution was determined using ModelGenerator.27 The optimum model was found to be HKY + I.28 One hundred bootstrap replicates were then carried out with the appropriate nucleotide model using the software program phyml (v3.0)29 and summarised using the majority‐rule consensus method.
Statistical analysis
IBM spss Statistics 21 (Armonk, NY, USA) was used to analyse the data. Difference in age between clinically affected horses and their healthy cohorts was examined using an independent t‐test. Difference in duration of clinical signs between vaccinated and unvaccinated horses was also examined using this test. A significance level of P < 0·05 was used for all statistical tests.
Results
Between November 2010 and October 2012, EI was diagnosed on nine premises located in five counties in Ireland (Table 1). Outbreaks occurred on a variety of premises among Thoroughbred horses, non‐Thoroughbred horses, ponies and donkeys (Table 1). Initial diagnosis on all premises was made by RT‐PCR. Following diagnosis, all clients in consultation with their veterinary surgeons agreed to participate in a disease investigation. Data relating to 629 samples from 135 equidae were analysed. Of these, at least 60 (44%) were described as clinically affected by the person responsible for the day‐to‐day management of the horses. A confirmed diagnosis of EI was made in 60 (44%) of the horses in the study. The vaccination status of the horses on each of the nine premises is summarised in Table 1.
Table 1.
Premises number, date, location, method of detection, type of premises, reported vaccination status and number of confirmed cases
| Premises number | Date | Location (county) | Method of detection | Type of premises | Vaccinated (%)a | Confirmed cases (%) |
|---|---|---|---|---|---|---|
| 1 | November 2010 | Kildare | RT‐PCR serology | Dealer's yard | 0 | 2/5 (40) |
| 2 | December 2010 | Kildare | RT‐PCR serology, VI | Racing yard | 44 | 9/18 (50) |
| 3 | June 2011 | Kilkenny | RT‐PCR serology | AI Stud Farm | 0 | 3/16 (19) |
| 4 | September 2011 | Carlow | RT‐PCR serology, VI | Private mixed yard (horses, pony and donkeys) | 0 | 4/6 (67) |
| 5 | January 2012 | Kilkenny | RT‐PCR serology, VI | Agricultural college | 23 | 21/43 (49) |
| 6 | August 2012 | Cork | RT‐PCR serology, VI | Racing yard | 38 | 3/16 (19) |
| 7 | September 2012 | Clare | RT‐PCR serology, VI | Stud Farm | 0 | 3/4 (75) |
| 8 | October 2012 | Kildare | RT‐PCR serology, VI | Racing yard | 0 | 4/13 (31) |
| 9 | October 2012 | Clare | RT‐PCR serology, VI | Dealers yard | 0 | 11/14 (79) |
VI, virus isolation; AI, artificial insemination.
Up to date vaccination records available.
Factors involved in the spread of EI
Spread between premises
Eight of the nine outbreaks occurred during the autumn (5) and winter (3) months. In all premises, EI was diagnosed following the movement of horses on or off the premises. Horses on five premises were affected following the introduction of a new arrival of unknown vaccination status (premises 3, 5, 6, 8, 9). On three premises, clinical signs were observed following the return of horses from an equestrian event including a local hunt (premises 1), a show jumping training show (premises 4) and a non‐TB sale (premises 7). In one racing yard (premises 2), horses were affected following the return of a horse from another premises. On six premises where it was presumed that the source of infection was identified, the incubation period for the index case ranged from 24 hours to 4 days.
Spread within premises
Housing
Accommodation details are presented in Table 2. Of the 135 equidae for which this information was available, 68 were housed in barns, 57 were housed in individual stables, and 10 were at grass. Twenty‐nine of the 68 horses (43%) housed in barns, 29 of the 57 horses (51%) housed in stables and two of the 10 (20%) horses at grass were described as clinically affected.
Table 2.
Vaccination status, housing type, morbidity, confirmatory diagnoses and subclinical infection
| Premise number | Vaccination (%)a | Housing type (no. equidae) | Morbidity (%) | Symptomatic/EI confirmed | Asymptomatic/EI confirmed |
|---|---|---|---|---|---|
| 1 | 0 | Stable block (5) | 2/5 (40) | 2/2 | 0/2 |
| 2 | 44 | Stable block (18) | 10/18 (56) | 6/9 | 3/9 |
| 3 | 0 |
Barn (6) Grass (9) Stable block (1) |
5/6 (83) 2/9 (22) 0/1 (0) |
3/3 | 0/3 |
| 4 | 0 | Barn (6) | 4/6 (68) | 4/4 | 0/4 |
| 5 | 23 | Multiple barns (43) | 17/43 (40) | 14/21 | 7/21 |
| 6 | 38 | Stable block (16) | 3/16 (19) | 2/3 | 1/3 |
| 7 | 0 |
Stable block (3) Grass (1) |
3/3 (100) 0/1 (0) |
3/3 | 0/3 |
| 8 | 0 | Barn (13) | 3/13 (23) | 3/4 | 1/4 |
| 9 | 0 | Stable block (14) | 11/14 (79) | 11/11 | 0/11 |
EI, equine influenza.
Up to date vaccination records available.
Fomites/personnel
There was evidence to suggest that horses were infected following exposure to contaminated fomites or personnel on two premises (1, 3). On premises 1, a 2‐year‐old filly that was isolated on her own in a separate barn following arrival on the premises exhibited clinical signs 4 days after the index case. This horse tested negative by RT‐PCR but subsequently seroconverted. The filly did not have direct contact with any of the other horses in the yard. On premises 3, two mares at grass exhibited mild clinical signs 10 days after the index case. Both mares were at grass full time and had no direct contact with any other horses on the premises. Both mares tested negative by RT‐PCR; however, one of the mares that was seronegative seroconverted. The mares had daily contact with handlers from the affected yard. Aerosol transmission was discounted as the source of infection as horses in neighbouring fields were not infected.
Clinical signs
Veterinary advice was sought from 48 hours to 14 days (average 4·9 ± 1·25 SE days) after first clinical signs were observed. Clinical signs most commonly reported were persistent coughing and nasal discharge. Pyrexia was also reported in several horses; however, temperature records were only available on one premises (premises 5). A significant difference (P = 0·000) in age between horses that were clinically affected (4·4 ± 0·44 SE years) and their healthy (7·6 ± 0·56 SE years) cohorts was observed. Evidence of subclinical infection in 12 horses (9%) on four premises was established (Table 2). Of these 12 horses, EI was confirmed in 2 (17%) by RT‐PCR and virus isolation, in 2 (17%) by RT‐PCR and serology and in 8 (67%) by serology only.
Serological status and clinical protection
The SRH results for acute serum samples collected from the 60 confirmed cases identified during this study are summarised in Table 3 as is the incidence of clinical signs. Five of the 60 confirmed cases had an up to date vaccination record.
Table 3.
SRH data of acute serum samples for confirmed equine influenza cases
| Premises number | No. confirmed cases | Seronegative H7N7 and H3N8 | Seronegative H7N7 Seropositive H3N8 | Seropositive H3N8 > H7N7 | Seropositive H3N8 = H7N7 |
|---|---|---|---|---|---|
| 1 | 2 | 1/2 | 1/2 | 0/2 | 0/2 |
| 2 | 9 | 2/9 | 4/9 | 0/9 | 3/9 |
| 3 | 3 | 3/3 | 0/3 | 0/3 | 0/3 |
| 4 | 4 | 4/4 | 0/4 | 0/4 | 0/4 |
| 5 | 21 | 7/21 | 6/21 | 1/21 | 7/21 |
| 6 | 3 | 1/3 | 1/3 | 1/3 | 0/3 |
| 7 | 3 | 2/3 | 1/3 | 0/3 | 0/3 |
| 8 | 4 | 3/4 | 1/4 | 0/4 | 0/4 |
| 9 | 11 | 5/11 | 2/11 | 3/11 | 1/11 |
| Total (%) | 60/135 (44) | 28/60 (47) | 16/60 (27) | 5/60 (8) | 11/60 (18) |
| Clinically affected (%) | 48/60 (80) | 28/28 (100) | 11/16 (69) | 4/5 (80) | 5/11 (46) |
H3N8 > H7N7: H3N8 antibody level >25 mm2 compared with H7N7 antibody level; H3N8 = H7N7: H3N8 antibody level <25 mm2 difference compared with H7N7 antibody level.
In order to study the level of protection afforded by pre‐existing antibodies, 68 horses were selected on the basis of having no serological evidence of recent exposure by natural infection. On examination of their acute serum samples, these horses were seronegative or had similar H3N8 to H7N7 antibody levels, that is, consistent with exposure to H3N8 by vaccination. Their clinical and serological status is illustrated in Figure 1. Of the 36 horses that were seronegative, 29 developed clinical signs. Twenty‐eight of these had a confirmed diagnosis of EI. Of the seven seronegative horses that did not develop clinical signs, five were at grass (premises 3) and had no direct contact with infected horses.
Figure 1.
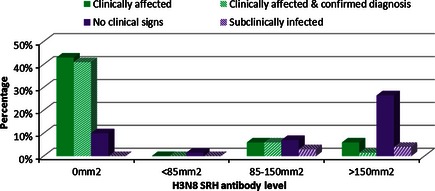
Percentage and H3N8 SRH level of horses which were clinically affected, clinically affected and equine influenza positive, developed no clinical signs or were subclinically infected.
Only one of the 32 seropositive horses had antibody levels <85 mm2. This horse did not develop clinical signs. Nine horses had antibody levels between 85 and 150 mm2 of which four became clinically affected and tested positive for EI. Two of the five horses with antibody levels 85–150 mm2 that did not develop clinical signs also tested positive for EI. Twenty‐two horses had antibodies levels >150 mm2 of which four were described as clinically affected. Only one of these tested positive for EI, and this horse had an SRH level of 151 mm2. Three of the remaining 18 horses with antibody levels >150 mm2 also tested positive.
Effectiveness of vaccination
Of the 60 horses described as clinically affected of which 80% tested positive for EI, 33 (55%) were reported unvaccinated, or of unknown vaccination status, 4 (7%) had out‐of‐date vaccination records, and the vaccination records of 15 (25%) horses were unavailable. On premises 2 where clinical signs were reported among both vaccinated and non‐vaccinated horses, the average duration of clinical signs among vaccinated horses (8·0 ± 0·63 SE days) was significantly less (P = 0·028) than that reported among unvaccinated horses or horses with unknown/out‐of‐date vaccination status (12·6 ± 1·60 SE days). On premises 5, booster vaccinations of Duvaxyn IE‐T Plus were given to 11 healthy horses 5 days after first clinical signs were observed. No new cases were observed post‐vaccination.
Twenty‐five horses in this study had up to date vaccination records, eight of these (32%) developed clinical signs and one subclinically affected horse shed virus (Table 4). This horse which was not virologically protected was a vaccinated 2‐year‐old in training (premises 2). Two nasopharyngeal swabs collected 72 hours apart from this horse were RT‐PCR and virus isolation positive despite the absence of clinical signs (Table 4). Of the 25 horses, four were vaccinated with ProteqFlu Te which contains H3N8 only and two of these developed clinical signs. Of the remaining 21 horses which were vaccinated with products which contained both H7N7 and H3N8 virus, 8 (38%) of these had comparable H3N8 and H7N7 antibody levels. All eight horses had H3N8 SRH levels >117 mm2, and six had antibody levels >150 mm2. Only one of these horses that had a H3N8 SRH level of 118 mm2 developed clinical signs. Five of the 21 horses (24%) had H3N8 antibody levels that where were >25 mm2 compared with their H7N7 antibody level. These horses had a mean H7N7 antibody level of 138 ± 24·8 mm2 and none developed clinical signs. Eight of the 21 horses (38%) were H7N7 seronegative but H3N8 seropositive, and four of these developed clinical signs. One horse which was seronegative for both subtypes was also clinically affected.
Table 4.
Vaccination breakdown
| Vaccinea | No. doses | Time since last vaccination (months) | Acute SRH H7N7 (mm2) | Acute SRH H3N8 (mm2) | Clinical signs | EI confirmed | EI detection method |
|---|---|---|---|---|---|---|---|
| Equilis Prequenza Te | 2 | 5 | 106 | 118 | Yes | Yes | RT‐PCR, seroconversion |
| Equilis Prequenza Te | 2 | 2 | Negative | Negative | Yes | Yes | RT‐PCR, seroconversion |
| Duvaxyn IE‐T Plus | 2 | 4 | Negative | 146 | Yes | Yes | RT‐PCR |
| ProteqFlu Te | 3 | 5 | Negative | 32 | Yes | Yes | RT‐PCR, seroconversion |
| ProteqFlu Te | 3 | 7 | Negative | 127 | Yes | No | N/A |
| Equilis Prequenza Te | 3 | 9 | 116 | 127 | No | Yes | RT‐PCR, VI, seroconversion |
| Equilis Prequenza Te | 3 | 5 | Negative | 203 | Yes | No | N/A |
| Equip FT | 5 | 5 | Negative | 313 | Yes | Yes | RT‐PCR, VI |
| Duvaxyn IE‐T Plus | 6 | 5 | Negative | 314 | Yes | No | N/A |
EI, equine influenza; VI, virus isolation.
Last vaccine dose.
Availability of vaccination records
The vaccination records of 41 horses on five premises (2, 3, 5, 6, 9) were unknown or unavailable. Nine of the 41 (22%) horses were seronegative for antibodies to H7N7 and H3N8 on initial sampling. Nine (22%) horses were seronegative for antibodies to H7N7 but seropositive for antibodies to H3N8 on initial testing. Ten of the horses (24%) had H3N8 antibody levels significantly higher (188 ± 18·2 mm2 SE) than that of their H7N7 antibody levels (109 ± 19·8 mm2 SE). Thirteen horses (32%) had H7N7 antibodies in acute samples (166 ± 9·5 mm2 SE), which were comparable to their H3N8 antibody level (163 ± 8·7 mm2 SE).
Genetic characterisation and phylogeny
The nucleotide sequence of the HA1 gene of nine viruses identified on two premises in 2010, two viruses identified on separate premises in 2011 and 15 viruses identified on five premises in 2012 was determined. Analysis of the sequence data generated for the 26 viruses indicated that they all belonged to clade 2 of the Florida sublineage of the American lineage (Figure 2). The HA1 nucleotide sequence of nine viruses identified on two premises in Kildare in November and December of 2010 was identical. The sequence of five viruses identified on a single premises in Kilkenny in January 2012 was identical. The sequence of a further 10 viruses identified on four premises in Cork, Clare and Kildare from August to October 2012 was also identical. These latter 10 viruses differed from the five viruses identified in January 2012 by a single nucleotide substitution which did not result in an amino acid change. There was no HA1 nucleotide sequence difference between RNA extracted directly from swab material and RNA extracted from allantoic fluid of infected eggs from samples taken on the same premises (data not shown).
Figure 2.
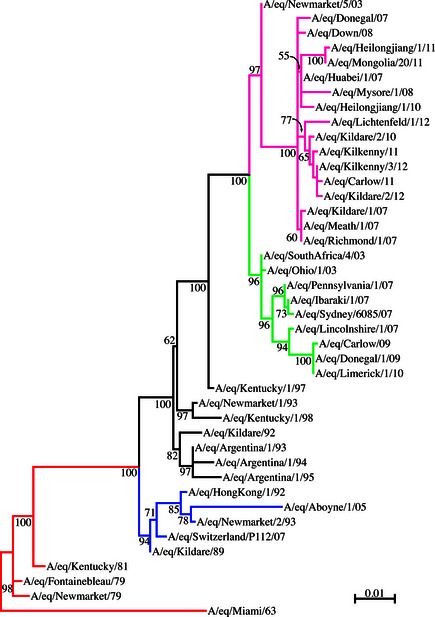
Phylogenetic tree of HA1 nucleotide sequences. Phylogenetic analysis of HA1 nucleotide sequences encoded by EIV, subtype H3N8. Bootstrap values obtained after 100 replicated are shown at the major nodes. Accession numbers for the genes reported in this manuscript are listed in Table S1. Red = pre‐divergent; blue = Eurasian; black = American; green = Florida sublineage clade 1; pink = Florida sublineage clade 2. A/eq/Kildare/2/10 is representative of A/eq/Kildare/1/10, A/eq/Kildare/3/10, A/eq/Kildare/4/10, A/eq/Kildare/5/10, A/eq/Kildare/6/10, A/eq/Kildare/7/10, A/eq/Kildare/8/10 and A/eq/Kildare/9/10; A/eq/Kilkenny/3/12 is representative of A/eq/Kilkenny/1/12, A/eq/Kilkenny/2/12, A/eq/Kilkenny/4/12, A/eq/Kilkenny/5/12; A/eq/Kildare/2/12 is representative of A/eq/Cork/1/12, A/eq/Cork/2/12, A/eq/Clare/1/12, A/eq/Clare/2/12, A/eq/Clare/3/12, A/eq/Kildare/1/12, A/eq/Kildare/3/12, A/eq/Kildare/4/12 and A/eq/Kildare/5/12.
Amino acid alignment
The HA1 amino acid sequence of EI viruses identified during this study were aligned with A/eq/Newmarket/5/03, a representative virus of the Florida sublineage clade 2, and any amino acid changes are summarised in Figure 3. The numbering of the HA1 amino acid sequence starts with the serine residue immediately downstream of the predicted signal peptide cleavage site.30 Negative numbers represent the predicted signal sequence. All Irish viruses identified during this study belonged to clade 2 of the Florida sublineage and did not have the two characteristic amino acid substitutions (V78A and N159S) in putative antigenic sites that distinguish viruses such as A/eq/Wisconsin/1/03 (clade 1) from the UK prototype clade 2 virus A/eq/Newmarket/5/03.16 All Irish viruses had the ‘FNF’ motif at position −11 to −9 first identified in a UK isolate (A/eq/Newmarket/1/07) in 2007.31 In addition, all Irish viruses had three amino acid substitutions (G7N, P103L and V111I) when compared to A/eq/Newmarket/5/03. G7N has previously been observed in viruses identified in Europe, China, Mongolia and India.4, 25, 31, 32, 33 Within the Irish viruses, a further three amino acid substitutions occurred, two of which were in antigenic sites. One of these, A144V, which occurred in A/eq/Carlow/11, A/eq/Kilkenny/3/12 and A/eq/Kildare/2/12, was in antigenic site A.34 A second amino acid change I242T that occurred in A/eq/Kilkenny/11 was in antigenic site D.34 To the best of our knowledge, this is the first published report regarding these two amino acid changes. The third amino acid change P169H occurred in A/eq/Carlow/11, but was not in an antigenic site.
Figure 3.

HA1 amino acid sequence alignment.A/eq/Kildare/2/10 is representative of A/eq/Kildare/1/10, A/eq/Kildare/3/10, A/eq/Kildare/4/10, A/eq/Kildare/5/10, A/eq/Kildare/6/10, A/eq/Kildare/7/10, A/eq/Kildare/8/10 and A/eq/Kildare/9/10; A/eq/Kilkenny/3/12 is representative of A/eq/Kilkenny/1/12, A/eq/Kilkenny/2/12, A/eq/Kilkenny/4/12, A/eq/Kilkenny/5/12; A/eq/Kildare/2/12 is representative of A/eq/Cork/1/12, A/eq/Cork/2/12, A/eq/Clare/1/12, A/eq/Clare/2/12, A/eq/Clare/3/12, A/eq/Kildare/1/12, A/eq/Kildare/3/12, A/eq/Kildare/4/12 and A/eq/Kildare/5/12.
Discussion
Equine influenza outbreaks identified during this study were sporadic and self‐limiting and occurred primarily among equidae which were unvaccinated or did not have up to date vaccination records. This study included the first‐confirmed case of EI affecting donkeys in Ireland (premises 4). In all cases, outbreaks occurred following the movement of horses. On the majority of premises, horses were clinically affected following the introduction of a new arrival of unknown vaccination status. The risk associated with failing to isolate new arrivals and establish their antibody status was highlighted previously.20, 31, 32 The seasonality of the outbreaks and the contribution of stabling and fomites/personnel to virus spread were also observed previously as was the influence of age on the clinical severity of disease.2, 6, 7, 19, 20, 35, 36 However, the rate of virus isolation was greater than that reported in a similar study from 2007 to 2010.20 In the current study, virus was isolated from 18 of the 45 (40%) RT‐PCR‐positive nasal swabs which were collected from 14 of the 135 horses. In the previous study, a delay in veterinary intervention was identified as a possible contributing factor to the low rate of virus isolation.20 As a similar delay was observed in the present study, it is likely that the increased rate of virus isolation was due to the fact that 10 of the 14 horses were seronegative on initial sampling. Seronegative horses rapidly amplify and shed large quantities of virus and are frequently the index case during outbreaks.37, 38, 39
A definite correlation between SRH antibody levels (H3N8) and protective immunity has been established following experimental challenge studies and observations in the field. Horses with antibody levels >85 mm2 are clinically protected, that is, protected against clinical disease, and horses with antibody levels >150 mm2 are virologically protected, that is, protected against infection, provided the vaccine strains are closely related to those circulating in the field.24, 38, 40 It is not possible to differentiate between H3N8 antibodies due to vaccination and natural infection. Other studies have defined pre‐infection antibody titres as those of blood samples taken the day after influenza was first diagnosed.16, 19 However, delayed veterinary intervention as observed in previous outbreaks20 affects interpretation of serological data, as at the time of sample collection, horses have often mounted an antibody response to the virus that is circulating on the premises. Thus, it is often not possible to determine the pre‐infection SRH titre that correlates with protection. In horses vaccinated with the majority of vaccines which contain the H7N7 subtype, the measurement of antibodies against H7N7 is a useful aid to the identification of those horses that have vaccinal rather than post‐infection titres as a higher H3N8 antibody level in comparison with H7N7 suggests exposure by natural infection.23 In this study, horses sampled pre‐infection were defined as those that were seronegative or had similar H7N7 and H3N8 antibody titres. These horses exhibited a strong correlation between their pre‐existing SRH antibody level and susceptibility to influenza as defined by clinical signs and confirmatory laboratory diagnosis. Seventy‐eight per cent of the seronegative horses, 44% of horses with antibody levels of between 85 and 150 mm2 and 5% of horses with antibody levels >150 mm2 developed influenza. No horse with a pre‐existing antibody level >151 mm2 exhibited clinical signs that were confirmed as EI. No horse with a pre‐existing antibody level >210 mm2 tested positive for EI, that is, they were virologically protected. Subclinical infection was identified in 12 horses on four different premises. Of the 12 horses, six had antibody titres consistent with vaccination. The mean H3N8 antibody titre of these six horses was 175 ± 14·7 mm2 SE. Virus was isolated from one of these horses on two occasions 72 hours apart. This horse which had a H3N8 SRH antibody level of 127 mm2 had received its third vaccine dose (Equilis Prequenza Te) 9 months prior to the outbreak.
Over 80% of the horses included in this study were unvaccinated, of unknown/unavailable vaccination status or their vaccination record was out of date. Furthermore, over 70% of horses that tested positive in this study were seronegative for H7N7, suggesting no vaccinal antibodies at the time of exposure. On three premises, it was reported that the majority of horses were vaccinated; however, vaccination records were only available for between 23% and 44% of horses. Nevertheless, examination of the H7N7 and H3N8 antibody level indicated that horses whose vaccination record was unknown or unavailable were not necessarily unvaccinated and highly susceptible to infection. Some had antibody levels in excess of 150 mm2.
Of the 25 horses with up to date vaccination records, a confirmed diagnosis of EI was made in 5 (20%) of 8 (32%) clinically affected horses. The average time since last vaccination for the eight clinically affected horses was 4·8 ± 0·49 SE months. These horses had been vaccinated with all four commercially available products, Duvaxyn IE‐T Plus (2), Equip FT (1), Equilis Prequenza Te (3) and ProteqFlu Te (2). Two of the horses had received their third vaccine dose of ProteqFlu Te on average 6 months prior to developing clinical signs. These horses had a mean H3N8 antibody level of 79 ± 47·3 mm2 SE on initial sampling. A third horse which had received its second dose of Equilis Prequenza Te 5 months prior to the outbreak had comparable H7N7 (106 mm2) and H3N8 (118 mm2) antibody levels, thus suggesting this horse had responded well to vaccination but was also inadequately protected. The five remaining horses were all vaccinated with products containing both virus subtypes. They had received on average 3·6 doses of vaccine and had last been vaccinated between 2 and 5 months prior to developing clinical signs. All were H7N7 seronegative and one was also H3N8 seronegative, suggesting these horses had responded poorly to vaccination. Poor response to vaccination is seen in some horses and is believed to play an important role in the transmission of virus among vaccinated horses.24 Two recent comparative vaccine studies have demonstrated that between 39% and 43% of Thoroughbred weanlings failed to seroconvert following first vaccination and that the incidence of poor responders was shown to be vaccine related41 Poor durability of antibody response between second and third vaccination and approximately 6 months following third vaccination have also previously been reported.37, 38, 41, 42, 43, 44
All virus strain identified as being responsible for outbreaks in Ireland during this study were from the Florida sublineage clade 2 and were similar to those identified during previous outbreaks in Ireland in 2007 and 200825 in Europe31, 32 and in Asia.4, 33 Experimental challenge studies and field data have demonstrated that protection against virus shedding correlates with the degree of antigenic relatedness between the vaccine and challenge strain.16, 19, 45 In this study, evidence of vaccine breakdown occurred in eight horses, six of which had been vaccinated with products not updated in line with the OIE recommendations of 2004 or 2010. These vaccines contained A/eq/Newmarket/1/93 (Duvaxyn IE‐T Plus, Equilis Prequenza Te) and A/eq/Kentucky/98 (Equip FT) as a representative of the American lineage. Evidence of vaccine breakdown occurred in a further two horses vaccinated with ProteqFlu Te. This vaccine has been updated in line with OIE recommendations from 2004 to include A/eq/Ohio/03 as a representative of clade 1 of the Florida sublineage; however, it does not include a representative of clade 2 as recommended by the OIE since 2010.
Cross‐protection is the ability of an antigen to elicit a protective immune response against infection with another antigen, for example a heterologous strain of virus. The findings of this study suggest that cross‐protectivity between the viruses included in the current vaccines and the circulating (field) strains has decreased. Forty‐four per cent and 18% of 68 horses that had pre‐infection antibody levels that correlate with clinical protection and virological protection with homologous virus developed clinical signs. Subclinical infection was detected in horses which had a mean H3N8 SRH antibody level of 175 ± 14·7 mm2 SE, and one horse had an SRH level of 210 mm2, that is, a level higher than that usually elicited by booster vaccination.46 The results of this study therefore suggest that vaccinal antibody levels indicative of clinical protection against homologous virus did not correlate with protection against the clade 2 viruses responsible for these outbreaks.
Equine influenza vaccines are updated to effect qualitative rather than quantitative improvements in the immune response. Comparative vaccine studies suggest that the level of antibodies elicited by current vaccines is related to adjuvant composition rather than antigenic load or virus strains.41, 42, 46 When there is mismatch between the circulating field strains and the vaccine strains, infection may result in clinical influenza and/or virus shedding regardless of a high SRH response to vaccination. Replacing an out‐of‐date virus in a vaccine with an epidemiologically relevant strain is unlikely to elicit a greater antibody response but should maximise protective immunity against EI. In order to ensure that mandatory vaccination programmes provide optimum protection, vaccines must contain strains that are antigenically similar to those circulating in the field. The findings of epidemiological investigations such as this should not be overlooked, and vaccine strains must be updated in a timely manner as recommended by the OIE.
Supporting information
Table S1. Equine influenza viruses included in phylogenetic analysis.
Acknowledgements
This study would not have been possible without the cooperation of the veterinary surgeons, owners and animal handlers to whom the authors are extremely grateful. All of the experimental work was funded by the Department of Agriculture under the National Development Plan and carried out at the Irish Equine Centre.
Gildea et al (2013) Epidemiological and virological investigations of equine influenza outbreaks in Ireland (2010–2012). Influenza and Other Respiratory Viruses 7 (Suppl. 4), 61–72.
References
- 1. Guo Y, Wang M, Zheng GS, Li WK, Kawaoka Y, Webster RG. Seroepidemiological and molecular evidence for the presence of two H3N8 equine influenza viruses in China in 1993–94. J Gen Virol 1995; 76(Pt 8):2009–2014. [DOI] [PubMed] [Google Scholar]
- 2. Powell DG, Watkins KL, Li PH, Shortridge KF. Outbreak of equine influenza among horses in Hong Kong during 1992. Vet Rec 1995; 136:531–536. [DOI] [PubMed] [Google Scholar]
- 3. Yamanaka T, Niwa H, Tsujimura K, Kondo T, Matsumura T. Epidemic of equine influenza among vaccinated racehorses in Japan in 2007. J Vet Med Sci 2008; 70:623–625. [DOI] [PubMed] [Google Scholar]
- 4. Virmani N, Bera BC, Singh BK et al Equine influenza outbreak in India (2008–09): virus isolation, sero‐epidemiology and phylogenetic analysis of HA gene. Vet Microbiol 2010; 143:224–237. [DOI] [PubMed] [Google Scholar]
- 5. Yondon M, Heil GL, Burks JP et al Isolation and characterization of H3N8 equine influenza A virus associated with the 2011 epizootic in Mongolia. Influenza Other Respi Viruses 2013; 5:659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guthrie AJ, Stevens KB, Bosman PP. The circumstances surrounding the outbreak and spread of equine influenza in South Africa. Rev Sci Tech 1999; 18:179–185. [DOI] [PubMed] [Google Scholar]
- 7. King EL, Macdonald D. Report of the Board of Inquiry appointed by the Board of the National Horseracing Authority to conduct enquiry into the causes of the equine influenza which started in the Western cape in early December 2003 and spread to the Eastern Cape and Gauteng. Aust Equine Vet 2004; 23:139–142. [Google Scholar]
- 8. Garner MG, Cowled B, East IJ, Moloney BJ, Kung NY. Evaluating the effectiveness of early vaccination in the control and eradication of equine influenza–a modelling approach. Prev Vet Med 2011; 99:15–27. [DOI] [PubMed] [Google Scholar]
- 9. Sovinova O, Tumova B, Pouska F, Nemec J. Isolation of a virus causing respiratory disease in horses. Acta Virol 1958; 2:52–61. [PubMed] [Google Scholar]
- 10. Webster RG. Are equine 1 influenza viruses still present in horses? Equine Vet J 1993; 25:537–538. [DOI] [PubMed] [Google Scholar]
- 11. OIE Bulletin . Expert surveillance panel of equine influenza vaccines – conclusions and recommendations. Mill Hill, London, United Kingdom, 2008; 42–45.
- 12. Willoughby R, Ecker G, McKee S et al The effects of equine rhinovirus, influenza virus and herpesvirus infection on tracheal clearance rate in horses. Can J Vet Res 1992; 56:115–121. [PMC free article] [PubMed] [Google Scholar]
- 13. Cullinane A, Newton JR. Equine influenza‐A global perspective. Vet Microbiol 2013; doi: 10.1016/j.vetmic.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 14. Gross DK, Hinchcliff KW, French PS et al Effect of moderate exercise on the severity of clinical signs associated with influenza virus infection in horses. Equine Vet J 1998; 30:489–497. [DOI] [PubMed] [Google Scholar]
- 15. Christley RM, French NP. Small‐world topology of UK racing: the potential for rapid spread of infectious agents. Equine Vet J 2003; 35:586–589. [DOI] [PubMed] [Google Scholar]
- 16. Newton JR, Daly JM, Spencer L, Mumford JA. Description of the outbreak of equine influenza (H3N8) in the United Kingdom in 2003, during which recently vaccinated horses in Newmarket developed respiratory disease. Vet Rec 2006; 158:185–192. [DOI] [PubMed] [Google Scholar]
- 17. Cullinane A. Equine influenza – a constantly evolving challenge. Equine Vet Educ 2009; 8:1–7. [Google Scholar]
- 18. Livesay GJ, O'Neill T, Hannant D, Yadav MP, Mumford JA. The outbreak of equine influenza (H3N8) in the United Kingdom in 1989: diagnostic use of an antigen capture ELISA. Vet Rec 1993; 133:515–519. [DOI] [PubMed] [Google Scholar]
- 19. Newton JR, Verheyen K, Wood JL, Yates PJ, Mumford JA. Equine influenza in the United Kingdom in 1998. Vet Rec 1999; 145:449–452. [DOI] [PubMed] [Google Scholar]
- 20. Gildea S, Arkins S, Cullinane A. Management and environmental factors involved in equine influenza outbreaks in Ireland 2007–2010. Equine Vet J 2011; 43:608–617. [DOI] [PubMed] [Google Scholar]
- 21. Quinlivan M, Dempsey E, Ryan F, Arkins S, Cullinane A. Real‐time reverse transcription PCR for detection and quantitative analysis of equine influenza virus. J Clin Microbiol 2005; 43:5055–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. OIE . Manual of Standards for Diagnostics Tests and Vaccines, Equine Influenza. Chapter 2.5.7, Paris: OIE , 2012; 1–14. [Google Scholar]
- 23. Gildea S, Arkins S, Cullinane A. A comparative antibody study of the potential susceptibility of Thoroughbred and non‐Thoroughbred horse populations in Ireland to equine influenza virus. Influenza Other Respi Viruses 2010; 4:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newton JR, Townsend HG, Wood JL, Sinclair R, Hannant D, Mumford JA. Immunity to equine influenza: relationship of vaccine‐induced antibody in young Thoroughbred racehorses to protection against field infection with influenza A/equine‐2 viruses (H3N8). Equine Vet J 2000; 32:65–74. [DOI] [PubMed] [Google Scholar]
- 25. Gildea S, Quinlivan M, Arkins S, Cullinane A. The molecular epidemiology of equine influenza in Ireland from 2007–2010 and its international significance. Equine Vet J 2012; 44:387–392. [DOI] [PubMed] [Google Scholar]
- 26. Larkin MA, Blackshields G, Brown NP et al Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 27. Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 2006; 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hasegawa M, Kishino H, Yano T. Dating of the human‐ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 1985; 22:160–174. [DOI] [PubMed] [Google Scholar]
- 29. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696–704. [DOI] [PubMed] [Google Scholar]
- 30. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X‐ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 2003; 309:209–218. [DOI] [PubMed] [Google Scholar]
- 31. Bryant NA, Rash AS, Russell CA et al Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet Microbiol 2009; 138:41–52. [DOI] [PubMed] [Google Scholar]
- 32. Bryant NA, Rash AS, Woodward AL et al Isolation and characterisation of equine influenza viruses (H3N8) from Europe and North America from 2008 to 2009. Vet Microbiol 2011; 147:19–27. [DOI] [PubMed] [Google Scholar]
- 33. Qi T, Guo W, Huang WQ et al Genetic evolution of equine influenza viruses isolated in China. Arch Virol 2010; 155:1425–1432. [DOI] [PubMed] [Google Scholar]
- 34. Lai AC, Chambers TM, Holland RE Jr et al Diverged evolution of recent equine‐2 influenza (H3N8) viruses in the Western Hemisphere. Arch Virol 2001; 146:1063–1074. [DOI] [PubMed] [Google Scholar]
- 35. Morley PS, Townsend HG, Bogdan JR, Haines DM. Risk factors for disease associated with influenza virus infections during three epidemics in horses. J Am Vet Med Assoc 2000; 216:545–550. [DOI] [PubMed] [Google Scholar]
- 36. Callinan I. The August 2007 outbreak in Australia: Commonwealth of Australia. Available at http://www.equineinfluenzainquiry.gov.au (Accessed 12th June 2008).
- 37. Mumford JA, Jessett D, Dunleavy U et al Antigenicity and immunogenicity of experimental equine influenza ISCOM vaccines. Vaccine 1994; 12:857–863. [DOI] [PubMed] [Google Scholar]
- 38. Mumford JA, Wilson H, Hannant D, Jessett DM. Antigenicity and immunogenicity of equine influenza vaccines containing a Carbomer adjuvant. Epidemiol Infect 1994; 112:421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wood JLN. A review of the history and epidemiology and a description of recent outbreak. MSc Dissertation. London: University of London, 1991. [Google Scholar]
- 40. Mumford JA. Biology, epidemiology and vaccinology of equine influenza. Proceedings of the International Symposium, Budapest, 10–11 December 2001; 7–12.
- 41. Gildea S, Arkins S, Walsh C, Cullinane A. A comparison of antibody responses to commercial equine influenza vaccines following primary vaccination of Thoroughbred weanlings–a randomised blind study. Vaccine 2011; 29:9214–9223. [DOI] [PubMed] [Google Scholar]
- 42. Gildea S, Quinlivan M, Murphy BA, Cullinane A. Humoral response and antiviral cytokine expression following vaccination of thoroughbred weanlings ‐ A blinded comparison of commercially available vaccines. Vaccine 2013; in press doi: 10.1016/j.vaccine.2013.08.083. [DOI] [PubMed] [Google Scholar]
- 43. Cullinane A, Weld J, Osborne M, Nelly M, McBride C, Walsh C. Field studies on equine influenza vaccination regimes in thoroughbred foals and yearlings. Vet J 2001; 161:174–185. [DOI] [PubMed] [Google Scholar]
- 44. Heldens JG, Kersten AJ, Weststrate MW, van den Hoven R. Duration of immunity induced by an adjuvanted and inactivated equine influenza, tetanus and equine herpesvirus 1 and 4 combination vaccine. Vet Q 2001; 23:210–217. [DOI] [PubMed] [Google Scholar]
- 45. Daly JM, Yates RJ, Browse G et al Comparison of hamster and pony challenge models for evaluation of effect of antigenic drift on cross protection afforded by equine influenza vaccines. Equine Vet J 2003; 35:458–462. [DOI] [PubMed] [Google Scholar]
- 46. Gildea S, Arkins S, Walsh C, Cullinane A. A comparison of antibody responses to commercial equine influenza vaccines following annual booster vaccination of National Hunt horses – a randomised blind study. Vaccine 2011; 29:3917–3922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Equine influenza viruses included in phylogenetic analysis.


