Abstract
Background
Since the influenza A(H1N1)pdm09 virus was first introduced to the Norwegian pig population in September 2009, it has repeatedly been detected in pigs in Norway. No other subtypes of influenza virus are circulating in Norwegian pigs.
Objective
To follow the diversity of A(H1N1)pdm09 viruses circulating in pigs in Norway and to investigate the relationship between viruses circulating in Norwegian pigs and in humans.
Methods
Between January 2011 and January 2013, nasal swabs from 507 pigs were tested for A(H1N1)pdm09 virus by real‐time RT‐PCR. The hemagglutinin (HA) gene of virus‐positive samples was sequenced and compared with publically available sequences from viruses circulating in humans at the time.
Results
Sequencing and phylogenetic analysis of the HA gene showed that the A(H1N1)pdm09 virus circulating in Norwegian pigs early in 2011 resembled the A(H1N1)pdm09 virus circulating in humans during this time. Viruses detected in pigs by the end of 2011 had acquired four characteristic amino acid substitutions (N31D, S84I S164F, and N473D) and formed a distinct phylogenetic group.
Conclusions
A(H1N1)pdm09 virus detected in Norwegian pigs by the end of 2011 formed a distinct genetic lineage. Also, our findings indicate that reverse‐zoonotic transmission from humans to pigs of the A(H1N1)pdm09 virus is still important.
Keywords: Influenza A(H1N1)pdm09 virus, Swine, Public health, Norway
Introduction
Influenza A viruses of three subtypes (H1N1, H1N2, and H3N2) occur endemic within most pig populations throughout the world. However, these swine influenza subtypes differ in origin and genetic characteristics in different continents and regions. In Norway, a swine influenza surveillance program based on serological screening of blood samples has been in place since 1997. Results from this program have shown that influenza A virus was not present in the Norwegian pig population prior to 2009. However, following the worldwide spread of the pandemic influenza A(H1N1) strain (A(H1N1)pdm09) in humans, influenza A virus was detected in Norwegian pigs for the first time in the autumn of 2009.1 Serological testing of slaughter pigs showed that 2 years after the virus was first introduced, about 60% of the tested pigs had antibodies against the influenza A(H1N1)pdm09 virus,2 and by the end of 2012, 49% of the pigs included in the program tested positive.3 Together, this indicates that the pandemic subtype has established itself within the Norwegian pig population.
The introduction of the A(H1N1)pdm09 virus to commercial swine herds was first described from Canada in April 2009 and was most likely caused by human‐to‐pig transmission.4 The virus was subsequently isolated from pigs throughout the world.1, 5, 6, 7, 8, 9 It is efficiently transmitted between pigs, as confirmed by experimental studies,10, 11 and is now co‐circulating with endemic swine influenza viruses in most countries. Different reassortant viruses containing genes from A(H1N1)pdm09, and other swine influenza viruses are regularly isolated, and the new viruses are typically also efficiently transmitted between pigs.4, 6, 7, 8 Some of these reassortant viruses have also caused infections in humans.12
In order to follow the diversity of the A(H1N1)pdm09 virus in the Norwegian pig population, where no other influenza subtype is circulating, a limited target surveillance at a boar testing station was carried out. In addition, diagnostic samples submitted to the Norwegian Veterinary Institute were included in the study. The hemagglutinin (HA) gene of virus‐positive samples was sequenced, and their relationship to contemporary viruses circulating in humans examined.
Material and methods
Study material
Nasal swab (Copan Innovation LTD, Brescia, Italy) samples from a total of 316 pigs were collected from pigs at a boar testing station at eight different time points between April and November 2011. The test station receives piglets aged 10–12 weeks from closed nucleus herds located in different parts of Norway every week. The test station recruits boars from a total of 46 closed nucleus herds. Sampling was performed based on suspicion of influenza virus infection at the station, but the symptoms were generally mild, and the majority of sampled pigs did not display any symptoms of respiratory illness. Furthermore, lung tissue was collected from seven pigs with clinical symptoms at the station in November 2011.
In addition, 191 nasal swabs from 17 herds with suspected influenza virus infection submitted to the Norwegian Veterinary Institute for diagnostic purposes between January 2011 and January 2013 were available for the study. The nasal swab samples were placed in 1 ml of transport media (EMEM 2% IBFS/Tris) before they were shipped to the laboratory.
Virological investigation
RNA was extracted from lung tissue samples and nasal swabs. Approximately 20 mg of lung tissue in 650 μl of NucliSens lysis buffer (bioMériux, Norge AS, Oslo, Norway) was homogenized using Qiagen tissue lyser (Qiagen, Valencia, CA) at 25 Hz for 10 min. 200 μl of lung tissue homogenate or transport media from nasal swabs was used for RNA extraction using the automatic extraction instrument NucliSens easyMag (bioMériux, Norge AS, Oslo, Norway) according to the manufacturer's instructions. Detection of influenza A virus by real‐time RT‐PCR was performed as described by the World Health Organization Collaborating Centre for the Surveillance, Epidemiology, and Control of Influenza at CDC, Atlanta, USA.13 Specific detection of the HA gene of the A(H1N1)pdm09 subtype was carried out on influenza‐A‐positive samples, also by real‐time RT‐PCR.14 Amplification was performed on a Stratagene Mx3500P (LaJolla, CA, USA) using the SuperScript® III Platinum® One‐Step qRT‐PCR Kit (Invitrogen, Paisley, UK).
Sequencing and phylogenetic analysis
Altogether, 10 positive samples, including nasal swabs and lung tissue samples, from five different time points (April, July, October, November, and December) in 2011 and one positive nasal swab sample from 2013 (January) were selected for HA gene sequencing. From 2012, no samples were available for sequencing. The HA gene was reverse transcribed and amplified using the OneStep RT‐PCR Kit (Qiagen).15 RT‐PCR products were excised from agarose gel and purified using the Qiaquick Gel extraction Kit (Qiagen). DNA was sequenced using the Prism BigDye Terminator v3.1 Cycle Sequencing Kit on 3130 Genetic Analyzer (Applied Biosystem, Warrington, USA) according to manufacturer's instructions. Sequence assembly and multiple sequence alignment were performed using Sequencher version 4.5 (Gene codes Corporation, Ann Arbor, MI; http://www.genecodes.com), and MEGA version 5.0. Sequences obtained in this study have been submitted to the EpiFlu database provided by the Global Initiative on Sharing All Influenza Data (GISAID) (accession numbers are provided in Table 1 and Figure 1). For comparison, the HA sequences from one pig virus from Norway 2009, human viruses occurring in Norway and obtained by the Norwegian Institute of Public Health, as well as a selection of representative international reference strains and the closest matching sequences available in the publicly accessible EpiFlu database, were included in the analysis. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5.0.
Table 1.
Substitutions in the HA gene of A(H1N1)pdm09 viruses detected in humans and pigs in Norway between October 2009 and January 2013
| HA0 Amino acid position | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 31 | 56 | 83 | 84 | 97 | 141 | 143 | 164 | 185 | 197 | 203 | 205 | 216 | 321 | 374 | 451 | 473 | |
|
A/California/7/2009*
(EPI176620) |
N | N | P | S | D | A | S | S | S | T | S | R | I | I | E | S | N |
|
A/Norway/3721/09 Nord‐Trondelag October 2009 (EPI381831) |
‐ | ‐ | S | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | A | T | ‐ | ‐ | V | ‐ | ‐ | ‐ |
|
A/swine/Norway/02‐11342/2009
1
October 2009 (EPI352385) |
‐ | ‐ | S | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | A | T | ‐ | ‐ | V | ‐ | ‐ | ‐ |
|
A/Norway/324/2011 Akershus January 2011 (EPI304461) |
‐ | ‐ | S | ‐ | ‐ | ‐ | G | ‐ | T | A | T | ‐ | ‐ | V | K | ? | ? |
|
A/swine/Hedmark/P249_1/2011
2
April 2011 (EPI 378462) |
‐ | ‐ | S | ‐ | ‐ | ‐ | G | ‐ | T | A | T | ‐ | ‐ | A | K | N | ‐ |
|
A/Norway/1404/2011 Vest‐Agder April 2011 (EPI381832) |
‐ | ‐ | S | ‐ | N | ‐ | ‐ | ‐ | ‐ | A | T | K | V | V | K | ? | ? |
|
A/swine/Hedmark/A64/2011 April 2011 (EPI378461) |
‐ | ‐ | S | ‐ | N | ‐ | ‐ | ‐ | ‐ | A | T | K | V | V | K | ‐ | ‐ |
|
A/swine/Hedmark/P249_3/2011
1
April 2011 (EPI378464) |
‐ | ‐ | S | ‐ | N | ‐ | ‐ | ‐ | ‐ | A | T | K | V | V | K | ? | ? |
|
A/swine/Hedmark/A66_1/2011
1
April 2011 (EPI378466) |
‐ | ‐ | S | ‐ | N | ‐ | ‐ | ‐ | ‐ | A | T | K | V | V | K | ‐ | ‐ |
|
A/swine/Hedmark/A66_2/2011
1
April 2011 (EPI378467) |
‐ | ‐ | S | ‐ | N | T | ‐ | ‐ | ‐ | A | T | K | V | V | K | ‐ | ‐ |
|
A/Norway/231/2011 Oslo January 2011 (EPI304457) |
‐ | ‐ | S | ‐ | N | ‐ | ‐ | ‐ | T | A | T | ‐ | ‐ | V | K | N | ‐ |
|
A/swine/Hedmark/A114/2011
1
July 2011 (EPI378468) |
‐ | S | S | ‐ | N | ‐ | ‐ | ‐ | T | A | T | ‐ | ‐ | V | K | N | ‐ |
|
A/swine/Oppland/A2/2013
1
January 2013 (EPI440999) |
‐ | ‐ | S | ‐ | N | ‐ | ‐ | S | T | A | T | ‐ | ‐ | V | K | N | ‐ |
|
A/Norway/331/2011 Akershus January 2011 (EPI304465) |
‐ | ‐ | S | I | N | ‐ | ‐ | ‐ | T | A | T | ‐ | ‐ | V | ? | ? | ? |
|
A/swine/Nord_Trøndelag/A148/2011
1
October 2011 (EPI378469) |
D | ‐ | S | I | N | ‐ | ‐ | F | T | A | T | ‐ | ‐ | V | K | N | D |
|
A/swine/Hedmark/P907/2011
2
November 2011 (EPI378470) |
D | ‐ | S | I | N | ‐ | ‐ | F | T | A | T | ‐ | ‐ | V | K | N | D |
|
A/swine/Hedmark/A161/2011
1
November 2011 (EPI378471) |
D | ‐ | S | I | N | ‐ | ‐ | F | T | A | T | ‐ | ‐ | V | K | N | D |
|
A/swine/Hedmark/A173/2011
1
December 2011 |
D | ‐ | S | I | N | ‐ | ‐ | F | T | A | T | ‐ | ‐ | V | K | N | D |
*Reference strain,1nasal swab,2lung tissue, (‐) amino acids identical to California/7/2009, (?) virus sequence not determined, (green) amino acid changes characteristic for the distinct lineage seen in Norwegian pigs in late 2011.
Figure 1.
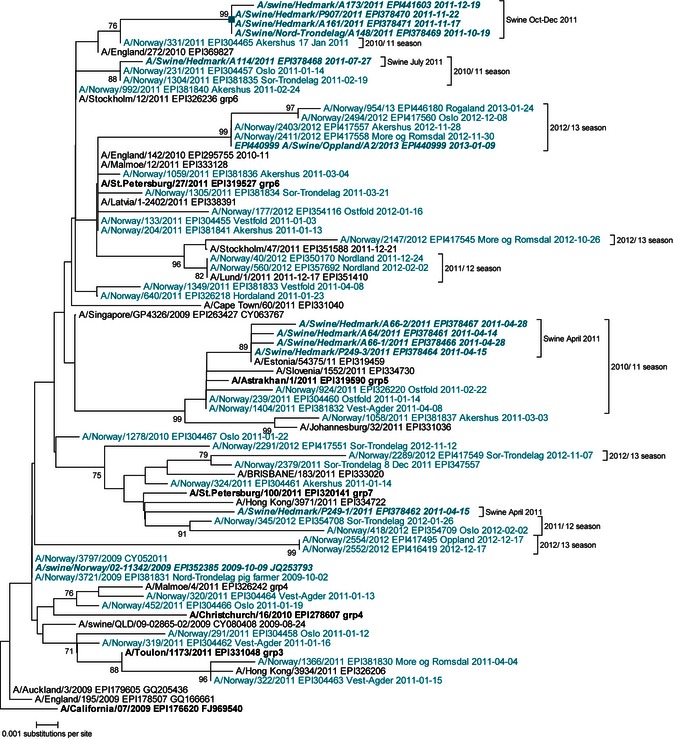
Phylogenetic reconstruction of the hemagglutinin gene (HA1 subunit) of influenza A(H1N1)pdm09 viruses from pigs in Norway, together with viruses from humans in Norway and elsewhere during the corresponding period. An alignment of the 981 nucleotides encoding the HA1 subunit was used. The evolutionary history was inferred using the neighbor‐joining method, from pairwise evolutionary distances computed using the Kimura 2‐parameter method. Bootstrap scores of 70 or higher (1000 replicates) are shown next to the branches. Norwegian isolates are in color. Viruses from pigs are in bold italics. International reference strains are in bold. The clade emerging in Norwegian pigs in late 2011 is indicated with a square.
Results
Detection of virus
At the boar testing station, 160 of the 316 nasal swabs and all seven lung tissue samples collected in 2011 were positive for influenza A virus. Of the 191 diagnostic samples submitted to Norwegian Veterinary Institute between January 2011 and January 2013, 59 samples from seven herds tested positive for influenza A virus. In 2012, no samples were found to be positive. All positive samples were confirmed to be A(H1N1)pdm09 virus positive by the HA subtype‐specific real‐time RT‐PCR.
Characterization of A(H1N1)pdm09 in Norwegian pigs
Results from sequencing analysis of the HA gene of A(H1N1)pdm09 viruses detected in this study, together with one Norwegian pig isolate from 2009, and selected human isolates are summarized in Table 1. Up until now, the A(H1N1)pdm09 viruses in humans have evolved and diversified to some extent, but are not yet shown to have changed substantially in antigenic or other important characteristics. The HA sequences of the swine viruses detected between April and July 2011 were similar to the virus sequence found in 20091 and showed only minor nucleotide substitutions which did not result in any amino acid changes of known importance (Table 1). The changes that were observed corresponded to changes seen in human viruses and phylogenetic analysis showed that the swine viruses closely clustered with contemporary A(H1N1)pdm09 viruses circulating in humans in Norway (Figure 1). However, in October, November, and December 2011, viruses detected in pigs on two well‐separated production sites in Norway were forming a distinct phylogenetic group (Figure 1). No sequences from human‐derived viruses fell into this clade. The most closely related available human virus was a Norwegian isolate from January 2011. The distinct group is characterized by amino acid substitutions N31D, S84I, S164F, and N473D in the viral HA0 protein, with the substitutions at positions 31, 164, and 473 (position 146 in the HA2 subunit) appearing to be unique to this group (Table 1). Amino acid position 164 is within a known antigenic site,16 but antigenic characterization of one of the isolates showed that the virus was not distinguishable from viruses lacking the S164F substitution (data not shown). The HA gene of the virus detected in pigs in January 2013 was again similar to the viruses detected in humans during the 2012/2013 season.
Discussion
In Norway, pigs are free of endemic swine influenza viruses of other subtypes than A(H1N1)pdm09. Therefore, the Norwegian pigs may serve as a unique population for studying the developing characteristics of an influenza strain where the chance of coinfections with other swine influenza viruses is negligible. Here, we report on the emergence of a distinct clade of influenza A(H1N1)pdm09 viruses in Norwegian swine. Sequence analysis shows that the emergence of the new clade most likely occurred during 2011 and that human viruses circulating in Norway in early 2011 are the most closely related viruses from which sequence is available. The HA gene of the new clade contains few amino acid substitutions as compared to other A(H1N1)pdm09 viruses. One of the substitutions, S164F, is located in the HA globular head subunit (HA1) and has not been reported elsewhere. The N473D substitution, located in the HA2 stalk subunit (position 146), has subsequently been found in another A(H1N1)pdm09 genetic lineage. This linage was first seen in some African countries but has also been observed in European countries including Norway during the influenza season 2012/201317 and is represented by the viruses A/Norway/2252/2012 and A/Norway/2254/2012 in Figure 1. During the 2011/12 influenza season in Norway, A(H1N1)pdm09 viruses occurred only sporadically in humans, presumably because the immunity in the human population following the widespread infection and vaccination in 2009 remained high. Viruses resembling the pig viruses from the new group have not been detected in the human influenza surveillance. This may indicate that the few amino acid changes in the HA of this swine lineage have not caused significant antigenic drift and immune evasion enabling the virus to spread more easily in humans.
Viruses representing the new phylogenetic clade originated from swine at three well‐separated pig farms, where there were no known contact between at least two of the farms. However, no thorough epidemiological work was performed to determine the source of the viruses in each case, and transmission of virus between the farms cannot be completely ruled out. To determine how commonly viruses that fall into the new group occur, extensive sampling from influenza infected pigs is needed. Even though the serological surveillance shows that infection with influenza viruses of the pandemic subtype frequently occurs in Norwegian pigs, most infections seem to be subclinical or only induce mild symptoms. This, together with the fact that influenza virus only is shed during the initial phase of infection1, 10, 11 makes sampling of influenza virus‐positive material from Norwegian pigs difficult.
The virus sampled from pigs in January 2013 did not fall into the new group of viruses. It was more closely related to viruses isolated from humans during the 2012/2013 influenza season and is likely to have originated from a new introduction of the virus from humans. Consistent with this, influenza caused by the A(H1N1)pdm09 virus was confirmed in persons who had been in contact with the pigs. The distinct lineage observed in pigs in late 2011 was not detected during the winter of 2012/2013, but as only a few positive samples were available for sequencing, it cannot be ruled out that it is still circulating. New introductions of the A(H1N1)pdm09 virus from humans to swine occur at a higher frequency than what is seen with seasonal influenza viruses.12 The phylogenetic analysis performed in this study indicates that also the situation in the Norwegian pig population is driven by constant spillover from humans.1, 2 As influenza A(H1N1)pdm09 in general seems to cause only modest or no symptoms at all in Norwegian pigs, it is notable that the herd sampled in January 2013 that most likely got infected by humans, presented with typical clinical symptoms of influenza virus infection. This suggests that new introductions of the virus from humans to pigs could cause more severe infection in pigs, but this needs to be studied in more details.
Pigs can serve as a major reservoir for influenza viruses. Once a herd is infected, the virus is likely to persist through continuous introduction of young susceptible pigs. The selection pressure toward antigenic drift (i.e., amino acid substitutions affecting antigenicity) is lower as compared to humans due to short life span of the pigs.18 Although molecular data indicate that A(H1N1)pdm09 viruses from humans have continued to seed the pig population, the wide distribution of this virus in Norwegian pigs suggests that A(H1N1)pdm09 also has been transmitting extensively among pigs, suggestive of endemicity. The demonstration of endemic, all‐year circulation of a distinct A(H1N1)pdm09 lineage in the farmed pig population in Norway in 2011 has some animal and public health implications. Firstly, a novel natural virus reservoir has arisen, from which the human population can be seeded with virus at any time of year. Hitherto, the direction of cross‐species transmission appears to have been primarily humans to animals. However, sporadic cases of humans infected with other swine influenza viruses have been documented in other parts of the world,19 and as long as the virus remains fully human transmissible, pig‐to‐human transmission is likely to occur. This may lead to more frequent out‐of‐season influenza cases in humans. Secondly, reproduction and transmission of the virus in another mammalian species may impose distinct evolutionary selection pressures on the virus, with unpredictable effects on the biological properties of the virus. Further analysis of the virus with full‐genome sequencing will reveal whether additional mutations in other gene segments have been acquired. More detailed studies may also reveal whether the mutations described here has any impact on viral fitness.
This study shows the high importance of continuing the surveillance of A(H1N1)pdm09 in pigs, to monitor the circulation and further evolution of this and other genetic groups, as well as to further elucidate the possible impact of this novel lineage for public and animal health.
Acknowledgements
The authors gratefully acknowledge Bjørn Lium and Peer‐Ola Hofmo for assisting in collection of samples. This work was supported by a grant from the Norwegian Research Council (NFR 207836). We further thank the contributors in the Human Influenza Surveillance and the staff of the influenza laboratory, Department of Virology, Norwegian Institute of Public Health. We also gratefully acknowledge the contribution of submitting and original laboratories for the reference virus sequences accessed in the GISAID sequence database.
Forberg et al (2013) Swine influenza in Norway: a distinct lineage of influenza A(H1N1)pdm09 virus. Influenza and Other Respiratory Viruses 7 (Suppl. 4), 21–26.
References
- 1. Hofshagen M, Gjerset B, Er C et al Pandemic influenza A(H1N1)v: human to pig transmission in Norway? Euro Surveill 2009; 14:5. [DOI] [PubMed] [Google Scholar]
- 2. Gjerset B, Er C, Løtvedt S et al Experiences after twenty months with pandemic influenza A (H1N1) 2009 infection in the naive Norwegian pig population. Influenza Res Treat 2011; 2011:206975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lium B, Zerihun A, Er C. The surveillance and control programme for specific viral infections in swine herds in Norway 2012. Surveillance and control programmes for terrestrial and aquatic animals om Norway Annual report 2012.
- 4. Howard WA, Essen SC, Strugnell BW et al Reassortant Pandemic (H1N1) 2009 virus in pigs, United Kingdom. Emerg Infect Dis 2011; 17:1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Welsh MD, Baird PM, Guelbenzu‐Gonzalo MP et al Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet Rec 2010; 166:642–645. [DOI] [PubMed] [Google Scholar]
- 6. Moreno A, Di TL, Faccini S et al Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet Microbiol 2011; 149:472–477. [DOI] [PubMed] [Google Scholar]
- 7. Vijaykrishna D, Poon LL, Zhu HC et al Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 2010; 328:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kitikoon P, Sreta D, Nuntawan Na AS et al Brief report: molecular characterization of a novel reassorted pandemic H1N1 2009 in Thai pigs. Virus Genes 2011; 43:1–5. [DOI] [PubMed] [Google Scholar]
- 9. Deng YM, Iannello P, Smith I et al Transmission of influenza A(H1N1) 2009 pandemic viruses in Australian swine. Influenza Other Respi Viruses 2012; 6:e42–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brookes SM, Nunez A, Choudhury B et al Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non‐immune pigs. PLoS ONE 2010; 5:e9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lange E, Kalthoff D, Blohm U et al Pathogenesis and transmission of the novel swine‐origin influenza virus A/H1N1 after experimental infection of pigs. J Gen Virol 2009; 90:2119–2123. [DOI] [PubMed] [Google Scholar]
- 12. Nelson MI, Vincent AL, Kitikoon P, Holmes EC, Gramer MR. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009‐2011. J Virol 2012; 86:8872–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. World Health Organization . CDC protocol of realtime RTPCR for influenza A (H1N1). Available at https://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/index.html.
- 14. Schulze M, Nitsche A, Schweiger B, Biere B. Diagnostic approach for the differentiation of the pandemic influenza A(H1N1)v virus from recent human influenza viruses by real‐time PCR. PLoS ONE 2010; 5:e9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization . Sequencing Primers and Protocols. Available at http://www.who.int/csr/resources/publications/swineflu/sequencing_primers/en/index.html.
- 16. Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982; 31:417–427. [DOI] [PubMed] [Google Scholar]
- 17. Daniels R, Gregory V, McCauley J. Influenza virus characterisation, Summary Europe February 2013. ECDC Surveillance Report.
- 18. Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 2000; 74:29–46. [DOI] [PubMed] [Google Scholar]
- 19. Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis 2007; 44:1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]


