Abstract
Background
Swine influenza A virus (IAV) reassortment with 2009 H1N1 pandemic (H1N1pdm09) virus has been documented, and new genotypes and subclusters of H3N2 have since expanded in the US swine population. An H3N2 variant (H3N2v) virus with the H1N1pdm09 matrix gene and the remaining genes of swine triple reassortant H3N2 caused outbreaks at agricultural fairs in 2011–2012.
Methods
To assess commercial swine IAV vaccines' efficacy against H3N2 viruses, including those similar to H3N2v, antisera to three vaccines were tested by hemagglutinin inhibition (HI) assay against contemporary H3N2. Vaccine 1, with high HI cross‐reactivity, was further investigated for efficacy against H3N2 virus infection in pigs with or without maternally derived antibodies (MDA). In addition, efficacy of a vaccine derived from whole inactivated virus (WIV) was compared with live attenuated influenza virus (LAIV) against H3N2.
Results
Hemagglutinin inhibition cross‐reactivity demonstrated that contemporary swine H3N2 viruses have drifted from viruses in current swine IAV vaccines. The vaccine with the highest level of HI cross‐reactivity significantly protected pigs without MDA. However, the presence of MDA at vaccination blocked vaccine efficacy. The performance of WIV and LAIV was comparable in the absence of MDA.
Conclusions
Swine IAV in the United States is complex and dynamic. Vaccination to minimize virus shedding can help limit transmission of virus among pigs and people. However, vaccines must be updated. A critical review of the use of WIV in sows is required in the context of the current IAV ecology and vaccine application in pigs with MDA.
Keywords: Antigenic drift, H3N2v, hemagglutinin inhibition, influenza A virus, maternally derived antibody, vaccines
Introduction
Transmission of influenza A virus (IAV) between people and pigs results in a similar disease outcome. Three swine IAV subtypes, H1N1, H3N2, and H1N2 are endemic in the United States and world's pig population.1 In the last two decades, the emergence of H3N2 virus in 1998 and a number of human‐to‐swine IAV spillover events in the United States contributed to increased genetic diversity of swine IAV and multiple co‐circulating hemagglutinin (HA) gene subclusters.1 North American swine H3N2 viruses are characterized by the triple‐reassorted internal gene (TRIG) constellation of human, swine, and avian lineages.2 The H3N2‐TRIG viruses were classified as H3 clusters I to III, a result of three separate human seasonal H3 gene introductions in 1995 (H3‐I), 1997 (H3‐II), and 1996 (H3‐III), respectively,3 with low serological cross‐reactivity among the clusters.4, 5, 6 Swine H3‐IV evolved from cluster III and was reported in 2005,7 but remained relatively stable until 2010. From 1998 to 2009, the majority, if not all endemic H1 and H3 swine IAV in the United States contained the TRIG backbone.8 The TRIG was also found in an increased number of swine‐to‐human cases in the United States. Six human cases of classical swine H1N1 viruses were reported between 1990 and 1995, whereas the number increased to 21 from 1998‐2010, and all of these cases had the TRIG backbone (14 H1 and 7 H3 viruses).9 The introduction of the 2009 H1N1 pandemic virus (H1N1pdm09) and reassortment with H3N2‐TRIG viruses (rH3N2p) altered both the genotype and phenotype of the previously stable swine H3‐IV viruses. At least 10 H3N2 genotype patterns emerged in the United States, with subsequent HA evolution and emergence of H3‐IV subclusters with inconsistent HI and serum neutralizing antibody cross‐reactivity among them.4, 10 The H3N2 genotype that inherited only the matrix gene from the H1N1pdm09 virus in particular has raised public health concerns. This virus caused over 340 H3N2v cases across 13 US states, with 16 cases hospitalized and 1 death.11 Infections mostly occurred in children and were associated with direct exposure to pigs at agricultural fairs. The H3 genes of the H3N2v in people and closely related rH3N2p swine viruses fall into a monophyletic cluster, IV‐A.10 Along with the unique genotype and HA cluster, the N2 gene is derived from a human seasonal virus from 2002 instead of the N2‐1998 lineage associated with the original swine H3N2‐TRIG virus.12 Data show that current trivalent human influenza vaccine generates minimal HI cross‐reactivity and would likely provide no protection for this virus.13, 14 Similar data evaluating swine vaccines or population immunity in swine herds against contemporary H3N2 have not been reported previously. Additionally, data generated from the US Department of Agriculture (USDA)‐National Animal Health Laboratory Network (NAHLN) voluntary swine IAV surveillance system during 2009–2012 (with over 1000 HA and NA sequences) indicate that swine H3N2 isolates increased in detection from under 25% in 2009–2010 to 33% in 2012 (T.K. Anderson, in press). This increase suggests that these antigenically diverse rH3N2p may not be adequately controlled by current approaches.
Control of IAV in pigs through vaccination is a common practice in the USA. Current swine IAV vaccines are available as licensed commercial products with whole inactivated virus (WIV) preparations with oil‐based adjuvants.15 The vaccine strains vary in different regions of the world but usually consist of two or more isolates of the H1 and H3 subtypes (bivalent or multivalent vaccines). Vaccine efficacy of WIV correlates well with the antigenic distance between the vaccine strains and the circulating virus strains.16 As recent swine IAV have become increasingly diverse, updating vaccine strains in commercial vaccines has fallen behind, and in response, autogenous vaccines have become common among US swine producers. Vaccines are used primarily in sow herds to induce maternally derived antibody (MDA) that can pass to piglets through colostrum. Maternally derived antibody from vaccinated sows can last up to 13–14 weeks.17 The presence of swine IAV‐specific MDA may protect pigs through the nursery period, but not the growing phase where, although less common, vaccination is also used to control IAV.18 A drawback associated with using WIV vaccine is the presence of MDA at vaccination causes vaccine failure.19 Additionally, using a mismatched vaccine/infection strain of the same subtype or when heterologous MDA is present at the time of vaccination with mismatched vaccine/field strain can cause IAV vaccine‐associated enhanced respiratory disease (VAERD).20, 21
Our study assessed cross‐reactive hemagglutinin inhibition (HI) antibody following vaccination with currently available commercial swine IAV vaccines and contemporary US swine H3N2 viruses (including swine rH3N2p viruses similar to the human H3N2v viruses) against contemporary swine H3N2 viruses with genetically diverse H3 genes. A vaccine that induced the highest HI cross‐reactivity to tested viruses was selected for further investigation to evaluate efficacy when delivered in the presence or absence of heterologous MDA (animal trial 1). Experimental WIV and live attenuated influenza virus (LAIV) vaccines were also tested against contemporary H3N2 challenge (animal trial 2). Vaccine efficacy was based on the ability to decrease pneumonia, virus replication in lungs, and shedding in pigs following rH3N2p virus infection.
Materials and methods
Antisera production and hemagglutinin inhibition cross‐reactivity test
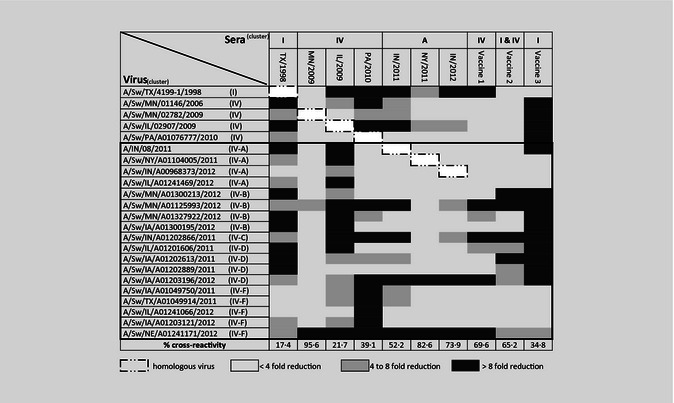
Twenty‐three H3N2 viruses including one 1998 isolate, four isolates from 2006 to 2010, and 18 isolates from 2011 to 2012 (including a human H3N2v and three nearly identical swine viruses associated with 2011–2012 county fair cases) were selected as HI test antigens and/or antigens for swine IAV‐antisera production. The hemagglutinin genes of all viruses in this study were characterized according to established criteria as either H3 cluster I or IV‐A to F.10 An H3 phylogeny demonstrating cluster and subcluster relationships is presented in Figure S1. In addition, three US commercial swine IAV vaccines were used for the generation of antisera to determine their cross‐reactivity against the selected swine isolates. Viruses and vaccines included are listed in Table 1.
Table 1.
Swine H3N2 viruses and commercial swine influenza A virus (IAV)‐inactivated vaccines used for the generation of antisera* and test antigen in the hemagglutination inhibition assay
| Viruses and vaccines | HA cluster | HA accession# |
|---|---|---|
| Viruses | ||
| A/swine/Texas/4199‐2/1998 | H3‐I | CY095675 |
| A/swine/Minnesota/01146/2006 | H3‐IV | CY099035 |
| A/swine/Minnesota/02782/2009* | H3‐IV | CY099103 |
| A/swine/Illinois/02907/2009 | H3‐IV | KF739387 |
| A/swine/Pennsylvania/A01076777/2010 | H3‐IV | JF263535 |
| A/Indiana/08/2011** | H3‐IV (A) | JN638733 |
| A/swine/New York/A01104005/2011*, *** | H3‐IV (A) | JN940422 |
| A/swine/Indiana/A00968373/2012*, *** | H3‐IV (A) | JX534982 |
| A/swine/Illinois/A01241469/2012 | H3‐IV (A) | JX422497 |
| A/swine/Minnesota/A01300213/2012 | H3‐IV (B) | JX657030 |
| A/swine/Minnesota/A01125993/2012 | H3‐IV (B) | JX422257 |
| A/swine/Minnesota/A01327922/2012 | H3‐IV (B) | JX422521 |
| A/swine/Iowa/A01300195/2012 | H3‐IV (B) | JX657018 |
| A/swine/Indiana/A01202866/2011 | H3‐IV (C) | JX092535 |
| A/swine/Illinois/A01201606/2011 | H3‐IV (D) | CY107066 |
| A/swine/Iowa/A01202613/2011 | H3‐IV (D) | JX092307 |
| A/swine/Iowa/A01202889/2011 | H3‐IV (D) | JX092542 |
| A/swine/Iowa/A01203196/2012 | H3‐IV (D) | JQ739697 |
| A/swine/Iowa/A01049750/2011 | H3‐IV (F) | JN652493 |
| A/swine/Texas/A01049914/2011 | H3‐IV (F) | JN652507 |
| A/swine/Illinois/A01241066/2012 | H3‐IV (F) | JX422557 |
| A/swine/Iowa/A01203121/2012 | H3‐IV (F) | JX092555 |
| A/swine/Nebraska/A01241171/2012 | H3‐IV (F) | JX422575 |
| Vaccines* | ||
| Vaccine 1 (FluSure® XP, Zoetis Animal Health) | ||
| A/swine/North Carolina/031/2005 | H1‐Delta 1 | |
| A/swine/Iowa/110600/2000 | H1‐Gamma | |
| A/swine/Oklahoma/0726H/2008 | H1‐Delta 2 | |
| A/swine/Missouri/069/2005 | H3‐IV | |
| Vaccine 2 (MaxiVacExcell® 5.0, Merck Animal Health) | H1‐Beta | |
| H1‐Gamma | ||
| H1‐Delta | ||
| H3‐I | ||
| H3‐IV | ||
| Vaccine 3 (PneumoStar, Novartis) | H1‐Alpha | |
| H3‐I | ||
HA, hemagglutinin.
H3N2 variant (H3N2v) virus from human case.
H3N2 viruses genetically similar to H3N2v.
Three‐week‐old cross‐bred pigs were obtained from a herd free of IAV, porcine reproductive and respiratory syndrome virus (PRRSV), and Mycoplasma hyopneumoniae (MHYO). For each virus, two pigs were immunized with 256 hemagglutinin units (HAU) of ultraviolet (UV)‐inactivated IAV combined with 20% commercial adjuvant (Emulsigen D; MVP Technologies, Omaha, NE, USA) by intramuscular (IM) route. Two or three doses of inactivated vaccines were given approximately 2–3 weeks apart. Commercial swine IAV vaccines were administered according to the manufacturers' guidance with the exception of vaccine 3 where an additional dose was administered. When HI titers to homologous virus reached at least 1:160, pigs were humanely euthanized with pentobarbital sodium (Fatal Plus; Vortech Pharmaceuticals, Dearborn, MI, USA) for blood collection. Sera were collected and stored at −80°C.
Prior to HI testing, sera were treated with receptor‐destroying enzyme (Sigma‐Aldrich, St. Louis, MO, USA), heat inactivated at 56°C for 30 minutes, and adsorbed with 50% turkey red blood cells (RBCs). Hemagglutinin inhibition assays were performed with four HAU of virus antigen and 0·5% turkey RBC according to standard techniques.22 Geometric mean titers were used to calculate the fold change between heterologous and homologous reactions. Fold changes were then used to generate a heat map indicating level of cross‐reactivity. The percentage of cross‐reactivity for each antiserum was calculated by dividing the number of virus antigens that cross‐reacted with antisera (<4‐fold reduction) by the total number of viruses tested. The commercial vaccine that provided the highest HI cross‐reactivity was used in subsequent experiments.
In vivo experimental design
Two separate animal trials were conducted to (i) investigate the role IAV‐specific MDA have on commercial vaccine immunogenicity and efficacy; and (ii) compare vaccine efficacy between WIV and LAIV against an rH3N2p virus genetically similar to the human H3N2v viruses reported in 2011–2012. Ten 3‐week‐old conventional pigs with swine IAV‐specific MDA were obtained from previously naïve sows that were vaccinated with an experimental WIV at 128 HAU [A/swine/Texas/4199‐2/1998 (TX98, H3N2 cluster I, MDA strain)] derived as described in Antisera production and hemagglutinin inhibition cross‐reactivity test and 33, and 3‐week‐old conventional pigs without MDA were procured from a high‐health status herd free of IAV, PRRSV, and MHYO. Upon arrival, pigs without IAV‐specific MDA were confirmed seronegative by Flock Check AI MultiS‐Screen diagnostic kit (IDEXX Laboratories, Westbrook ME, USA).23 To reduce bacterial contaminants, all pigs were treated with ceftiofur crystalline‐free acid (Excede; Zoetis Animal Health, Florham Park, NJ, USA) and enrofloxacin (Baytril 100; Bayer HealthCare AG, Monheim, Germany). The experimental protocol is summarized in Table 2. The eight pig groups were housed in individual isolation rooms and cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center.
Table 2.
Animal study design
| Group | MDA to TX98 | Vaccine status | Challenge with NY11 | n |
|---|---|---|---|---|
| Trial 1 | ||||
| MDA/C | Yes | No | Yes | 5 |
| MDA/V/C | Yes | Yes (vaccine 1) | Yes | 5 |
| V/C | No | Yes (vaccine 1) | Yes | 5 |
| C | No | No | Yes | 5 |
| Trial 2 | ||||
| WIV/C | No | Yes (OH04) | Yes | 6 |
| LAIV/C | No | Yes (OH04) | Yes | 6 |
| C | No | No | Yes | 6 |
| Negative control | ||||
| NEG | No | No | No | 5 |
C, challenged; MDA, maternal derived antibody; V, vaccinated; NEG, non‐vaccinated, non‐challenged (negative control); WIV, whole virus inactivated vaccine; LAIV, live attenuated influenza vaccine; TX98, A/swine/Texas/4199‐2/1998 (H3N2); NY11, A/swine/New York/A01104005/2011 (H3N2).
FluSure® XP (Vaccine 1; Zoetis Animal Health) was used in trial 1 to study the effect of IAV‐specific MDA on commercial vaccine efficacy. Pigs in groups MDA/V/C and V/C were vaccinated at 4 weeks of age and boosted 3 weeks later according to manufacturers' recommendation. In trial 2, two vaccine platforms were used, WIV and LAIV, with the vaccine strain of both from a cluster IV swine triple reassortant H3N2 A/turkey/Ohio/313053/2004 (OH04). Whole inactivated virus was prepared as described in Antisera production and hemagglutinin inhibition cross‐reactivity test and administered IM to the WIV/C group at approximately 4 weeks of age and boosted at 7 weeks of age. The LAIV H3N2 OH04,24 contains modifications to the viral polymerase genes to decrease virus polymerase activity and impair virus growth in normal core body temperatures.25 Pigs in the LAIV/C group were vaccinated and boosted with 1 ml of 106 50% tissue culture infective dose (TCID50) per ml virus intranasally (IN) at the same timing as the WIV group. In trials 1 and 2, five and six pigs, respectively, served as non‐vaccinated, challenged positive control groups (C). In addition, one group of five pigs served as non‐vaccinated, non‐challenged negative controls (NEG).
Pigs in both trials were challenged intratracheally (2 ml) and IN (1 ml) with 105·5 TCID50 per ml of virus A/swine/New York/A01104005/2011 (NY11), a swine rH3N2p similar to H3N2v, approximately 2 weeks following booster (0 days post‐infection, dpi). Viral inocula were back‐titrated in Madin–Darby canine kidney (MDCK) cells. Inoculation was performed under anesthesia administered IM, using an anesthetic cocktail of ketamine (8 mg/kg), xylazine (4 mg/kg), and Telazol (6 mg/kg, Zoetis Animal Health, USA). Pigs were observed daily for clinical signs of respiratory disease. Rectal temperatures were taken at −1 dpi and daily until 5 dpi. All pigs were humanely euthanized with a lethal dose of pentobarbital and necropsied at 5 dpi.
Genetic similarity of the HA between MDA vaccine strain, antisera vaccine strains, and challenge strain was calculated in MEGA v5.05 (Table S1). The MDA strain (TX98) had 91·3% amino acid identity with the challenge strain (NY11), and the LAIV strain (OH04) had 97% identity with the challenge strain.
Macroscopic and microscopic examination of lungs
At necropsy, lungs were removed and evaluated for the percentage of the surface affected with pneumonia for each lung lobe, and a total percentage for the entire lung was calculated based on weighted proportions of each lobe to the total lung volume.26
Tissue samples from the distal trachea and right cardiac lung lobe and/or affected lobes were fixed in 10% buffered formalin for histopathologic examination. A single pathologist blinded to the treatment groups scored all tissues.27 Bronchoalveolar lavage fluid (BALF) was collected at necropsy by lavaging lungs with 50 ml minimal essential medium (MEM) were screened for additional respiratory pathogens including PRRSV, porcine circovirus type 2 (PCV2), and MHYO nucleic acid by real‐time RT‐PCR test with VetMax NA and EU PRRSV, VetMax PCV2 (custom assay), and VetMax MHYO kits, respectively (Life Technologies, Carlsbad, CA, USA).
Virus titer in lungs and nasal swabs
To evaluate virus shedding, nasal swabs (NS) were collected on −1, and 1, 3, and 5 dpi from both trials and placed in 2 ml MEM. To assess IAV titers in lungs, BALF was collected from all pigs at necropsy. All NS and BALF were stored at −80°C until study completion.
For virus titration, NS and BALF samples were serially diluted (10‐fold) with serum‐free Opti‐MEM® reduced serum medium (Life Technologies) supplemented with TPCK trypsin and antibiotics. Each dilution was plated in triplicate (100 μl per well) onto PBS‐washed confluent MDCK cells in 96‐well tissue culture plates. At 48 hour, plates were fixed with 4% phosphate‐buffered formalin and stained using immunocytochemistry with an anti‐influenza A nucleoprotein monoclonal antibody.20 Virus titer was calculated for each sample using the Reed and Muench method.28
Hemagglutination inhibition assay
The HI assays were performed as described in Antisera production and hemagglutinin inhibition cross‐reactivity test to measure MDA titers and detect systemic antibody response to vaccination and infection. Serum samples were collected prior to vaccinations and challenge (−35, −14, and 0 dpi), and at necropsy (5 dpi). H3N2 virus antigens utilized in the HI assay included the challenge virus (NY11), MDA strain (TX98), a putative commercial vaccine 1 strain (A/Swine/Missouri/069/2005; vaccine 1), and laboratory WIV and LAIV strain (OH04).
ELISA for mucosal influenza A virus‐specific antibody detection
Prior to testing, BALF was incubated at 37°C for 1 hour with an equal amount of 10 mM dithiothreitol (DTT; Sigma‐Aldrich) to disrupt mucus. ELISAs for mucosal IAV antibodies to challenge virus (NY11) were performed on each BALF sample in duplicate.21 Antibody levels were reported as the mean optical density (OD), and the mean OD of each treatment group was compared.
Statistical analysis
The percent macroscopic pneumonia, microscopic lesion scores, log10 transformations of BALF and NS virus titers, and log2‐transformed HI titers were analyzed using analysis of variance (anova). Significant differences between treatment groups were evaluated using the Tukey–Kramer HSD multiple comparison test, with a P‐value of ≤0·05 considered significant. All data analyses were performed using GraphPad Prism 5.03 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Hemagglutination inhibition cross‐reactivity
Pairwise HI cross‐reactivity is presented as a heat map, indicating the fold reduction obtained by dividing the log2‐transformed reciprocal titer to heterologous antigen by the homologous antigen titer (Table 3). Sera from pigs vaccinated with the H3‐I (TX98) had limited cross‐reactivity (17·4%) with contemporary H3‐IV isolates. The percentages of HI cross‐reactivity detected between antisera produced against the two 2009 viruses, MN/2009 (95·6%) and IL/2009 (21·7%), were highly variable. Overall percentages of cross‐reactivity of antisera against 2010–2012 viruses ranged from 39·1% to 82·6%. Serum from pigs exposed to H3 cluster IV(A) viruses, including H3N2v (IN/2011) and swine IV(A) rH3N2p (NY/2011 and IN/2012), had a range in reactivity to recent H3‐IV viruses (52·2–82·6%). Vaccine 1, containing a 2005 H3‐IV strain, showed limited cross‐reactivity to H3‐I virus, but an overall 69·6% had cross‐reactivity to H3‐IV viruses. Vaccine 2, containing H3‐I and ‐IV strains, cross‐reacted with H3‐I virus but had 65·2% cross‐reactivity to H3‐IV viruses. Vaccine 3 containing an H3‐I virus cross‐reacted with the tested H3‐I virus but had low cross‐reactivity (34·8%) to H3‐IV viruses. Antisera to vaccines 1 and 2 cross‐reacted well with H3N2v and all three IV‐A rH3N2p swine isolates, while vaccine 3 had minimal cross‐reactivity to the H3N2v isolate but cross‐reacted to the swine IV‐A rH3N2p. In general, all three vaccines appeared to be more cross‐reactive with H3 IV‐A and IV‐F viruses but had reduced cross‐reactivity to H3 IV‐B and IV‐D viruses. Vaccine 1 had the highest level of HI cross‐reactivity among the three vaccines and was selected for further analysis of vaccine efficacy.
Table 3.
Heat map indicating hemagglutinin inhibition (HI) titer reduction between homologous and heterologous titers. Percentage of cross‐reactivity for each antisera was calculated by dividing the number of virus antigens with high cross‐reactivity (<4‐fold) by the total number of viruses tested
Clinical signs, macroscopic and microscopic findings
Clinical signs
None of the non‐infected control pigs or the infected animals in either trial displayed IAV‐associated respiratory signs. Maternally derived antibody alone reduced fever, as only the positive control pigs (Group C) in trial 1 spiked a fever ≥104°F at 1 and 2 dpi (Figure S2). Rectal temperatures of all animals in trial 2 remained below the febrile cutoff during the trial period (data not shown).
Macroscopic and microscopic lesions
Back titration from challenge inocula indicated that pigs in trial 1 were infected with 105·5 TCID50/ml, while pigs in trial 2 received 104 TCID50/ml virus. Mean macroscopic lesions were not statistically different between challenge groups in either trial. One pig vaccinated in the presence of MDA had enhanced macroscopic lesions of 18·6% (Figure 1A), while the remaining pigs in the same group had <6% lung lesions.
Figure 1.
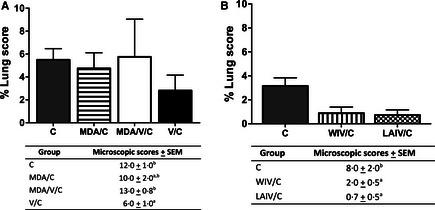
Percentage of macroscopic lesion from MDA/C, MDA/V/C, and V/C pigs from trial 1 (A) and C, WIV/C, and LAIV/C pigs in trial 2 (B) at 5 days post‐infection. Average microscopic lesion scores ± SEM of each treatment group are indicated in table. Means with different letters within a column are statistically different (P ≤ 0·05). Non‐challenged pigs had no macroscopic and microscopic lesion scores and are not included. C, challenged; MDA, maternal derived antibody; V, vaccinated; WIV, whole virus inactivated vaccine; and LAIV, live attenuated influenza vaccine.
Statistical differences were detected in the IAV‐associated microscopic pneumonia scores (Figure 1A, B). In trial 1, V/C pigs had significantly reduced scores compared with C pigs and MDA/V/C pigs. Maternally derived antibody alone did not reduce microscopic lesions as the scores were similar to groups C and MDA/V/C. In trial 2, pigs vaccinated with both experimental vaccine platforms had significantly lower pneumonia scores compared with non‐vaccinated, challenged pigs.
Virus in lungs and nasal swabs
All vaccinated pigs had a significantly reduced amount of virus in the lungs on 5 dpi compared with non‐vaccinated, challenged positive control groups (Figure 2A, B). Maternally derived antibody alone did not reduce virus replication in lungs. In trial 2, both WIV and LAIV significantly reduced virus in the lungs, and the level of virus was not different between the vaccinated groups.
Figure 2.
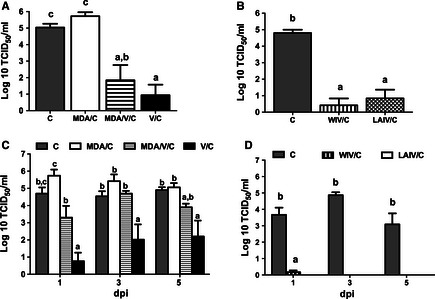
Average virus titers in lungs (A, B) at 5 days post‐infection (dpi) and nasal swabs (C, D) at 1, 3, and 5 dpi from C, MDA/C, MDA/V/C, and V/C pigs from trial 1 (A and C) and from C, WIV/C, and LAIV/C in trial 2 (B and D) as mean log10 TCID50/ml ± SEM. Different superscript letters or * within the figure are significant difference between the values (P ≤ 0·05). Virus was not isolated from any non‐challenged pigs, and results are not included. C, challenged; MDA, maternal derived antibody; V, vaccinated; WIV, whole virus inactivated vaccine; and LAIV, live attenuated influenza vaccine.
Pigs in the V/C group shed significantly reduced levels of virus in nasal secretions compared with positive controls in trial 1 on 1, 3, and 5 dpi (Figure 2C). Maternally derived antibody alone or MDA with vaccination did not reduce nasal shedding. In trial 2, no virus was detected in LAIV/C pigs, while one pig in WIV/C group shed virus at 1 dpi (Figure 2D). No virus was isolated from negative control pigs at any time point (data not shown).
Serum hemagglutinin inhibition antibody
In trial 1, MDA pigs had average reciprocal HI titers of 160 to MDA strain (TX98) prior to the first vaccination, and the level decreased approximately 2‐fold by the end of the study (Figure 3A). Pre‐vaccination MDA appeared to have moderate cross‐reactivity to the challenge strain (NY11) and less cross‐reactivity to the commercial vaccine strain (Figure 3B, C). Piglets derived from non‐vaccinated sows did not have HI titers to any tested virus prior to vaccination, and all negative control pigs remained seronegative throughout the study (data not shown). V/C pigs had HI titers to vaccine 1 (1:640) after two vaccinations and high cross‐reactivity to the challenge strain. Maternally derived antibody significantly reduced vaccine immunogenicity as reflected by lower HI titers to both vaccine 1 and challenge viruses in MDA/V/C pigs compared with V/C pigs. In trial 2 (Figure 3D, E) after the booster dose, WIV/C pigs had significantly higher HI titers to both vaccine strain (OH04) and challenge virus compared with LAIV/C vaccinated pigs, yet both vaccines were highly efficacious in non‐MDA vaccinated pigs.
Figure 3.
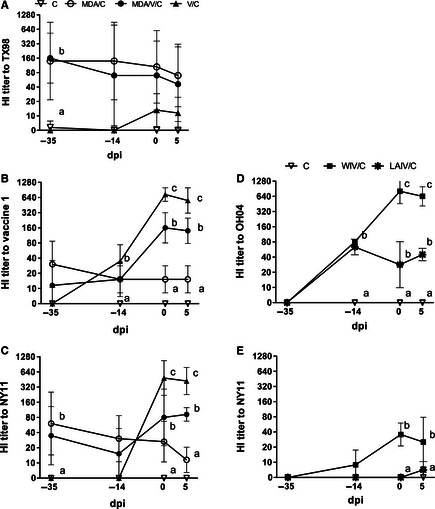
Average hemagglutinin inhibition (HI) titers against maternal derived antibody strain, TX98 (A), commercial vaccine 1 strain (B), experimental vaccine strain, OH04 (D), and challenge virus strain, NY11 (C and E). Sera were collected prior to first and second vaccination (−35 and −14 days post‐infection; dpi), prior to influenza A virus (IAV) infection (0 dpi) and at 5 dpi. Sera were collected from C, MDA/C, MDA/V/C and V/C pigs from trial 1 (A–C) and from C, WIV/C, and LAIV/C pigs in trial 2 (D, E). Means with different letters at the same time point are statistically different (P ≤ 0·05). Non‐challenged pigs had no HI titers, and results are not included. C, challenged; MDA, maternal derived antibody; V, vaccinated; WIV, whole virus inactivated vaccine; and LAIV, live attenuated influenza vaccine.
Mucosal SIV‐specific antibody
At 5 dpi, there was detectable IgG specific to challenge virus (NY11) in the lungs of vaccinated pigs regardless of MDA and vaccine platform (WIV or LAIV). Post‐challenge V/C pigs had significantly higher levels of NY11‐specific IgA in the lungs only if the vaccine was administered in the absence of MDA (Figure 4A). The LAIV/C pigs had significantly higher levels of NY11‐specific IgA in the lungs compared with all other groups (Figure 4B).
Figure 4.
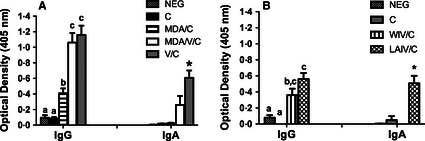
Average O.D. ± SEM of NY11 (challenge virus) ‐specific IgG and ‐specific IgA from lower airways of NEG, C, MDA/C, MDA/V/C, and V/C pigs in trial 1 (A) and from NEG, C, WIV/C, and LAIV/C pigs in trial 2 (B) measured by ELISA on 5 dpi. Means with different letters or * within a column are statistically different (P ≤ 0·05). NEG, non‐vaccinated, non‐challenged; C, challenged; MDA, maternal derived antibody; V, vaccinated; WIV, whole virus inactivated vaccine; and LAIV, live attenuated influenza vaccine.
Discussion
Here, we demonstrated that contemporary swine H3N2 viruses selected from subclusters of H3‐IV have drifted antigenically, affecting the efficacy of currently available commercial swine‐inactivated vaccines in the United States. These genetically distinct clusters of viruses continue to diversify and represent a challenge for IAV control. One commercial vaccine had limited cross‐reactivity to over half the contemporary viruses tested, although it was not unexpected as the vaccine strain was H3‐I, while the majority of the viruses tested were H3‐IV. The other two vaccines containing H3‐IV vaccine strains appeared to cross‐react with the H3N2v and similar rH3N2p viruses, but elicited moderate to significantly reduced cross‐reactivity to many of the ten representative 2012 viruses tested. Data from human influenza vaccines indicate that vaccine updates are required when a >8‐fold reduction HI antibody (>4‐fold of log2‐transformed HI antibody) is detected.29 Results of HI cross‐reactivity using antisera generated with three commercial vaccines suggest a reduction in vaccine efficacy against contemporary swine H3N2 strains.
To date, all commercially available swine IAV vaccines are inactivated bivalent or multivalent formulations including H1 and H3 subtype strains or a combination of strains representing the distinct subclusters.15 The HA is the primary immune target responsible for WIV‐mediated protection against IAV infection. Limited cross‐reactive antibodies to different HA subtypes, or clusters within the same subtype, is one of the major obstacles for optimal WIV vaccine efficacy. The presence of IAV‐specific MDA in weanling pigs at the time of vaccination is another factor that further limits WIV vaccine performance. Piglets born to sows that have been repeatedly vaccinated can have serum antibodies to H1N1 and H3N2 viruses until 13–14 and 9–10 weeks of age, respectively.17 While IAV‐specific MDA can reduce clinical disease in piglets, MDA‐positive piglets infected with IAV shed virus longer than pigs without MDA,30 and a negative correlation was observed between swine IAV‐specific MDA levels at vaccination and the development of primary antibody response to the vaccine.17 Consistent with previous studies, we report that the efficacy of a commercial inactivated swine IAV vaccine administered to pigs in the presence of MDA was markedly reduced. Maternally derived antibody‐negative pigs given two doses of vaccine had antibodies cross‐reactive to the challenge virus and had significantly reduced levels of rH3N2p virus in lungs and nasal cavities following challenge. The presence of MDA at vaccination interfered with the development of humoral immunity and subsequently reduced vaccine efficacy. This occurred despite the low HI cross‐reactivity level between MDA and the vaccine strain (<1:40) at vaccination. Reduced pneumonia is used as a parameter to assess IAV vaccine protection in pigs, but pneumonia was not different between any groups tested here. H3N2v was shown to cause fewer lung lesions when compared with endemic swine H3N2‐TRIG viruses,31 and consequently may not be a valid measurement, particularly with viruses that do not induce high percentages of pneumonia. One caveat is that our study had a limited number of animals, which may have reduced the power to detect small differences in lung lesion scores. However, there was one striking result, one pig vaccinated in the presence of MDA had a percentage of macroscopic lung lesions three times over the group mean detected in non‐vaccinated positive control pigs. Previous data using an inactivated adjuvanted IAV vaccine reported the occurrence of VAERD in vaccinated, MDA‐positive pigs where the MDA vaccinated and challenge viruses were heterologous strains of the same subtype.20, 21 These findings reflect events in the field where mis‐matching of MDA, vaccine, and circulating strains is expected to be common.
Recent studies with vectored and LAIV vaccines have suggested that they improve immunity to heterologous IAV strains,32 with a reduction in MDA interference33, 34 and avoidance of MDA‐VAERD.21 This study compared the capability of experimental LAIV and WIV platform vaccines (with the identical vaccine strain, OH04). The difference in lung lesions between the infected groups was not definitive because pneumonia was relatively mild. Both vaccine platforms were comparable in limiting virus replication in lungs, demonstrating the performance of WIV is effective in the absence of MDA. However, the LAIV completely prevented nasal shedding. Mucosal immunity likely played a major role as the LAIV‐vaccinated pigs had significantly higher mucosal IgA than WIV pigs but barely detectable challenge virus‐specific HI antibodies in serum. It is known that secretory IAV‐specific IgA is important in defending both the upper and lower respiratory tract, while serum IAV‐specific IgG protects the lower respiratory tract.16 Although not measured here, cell‐mediated immunity (CMI) must have a role in protection: a previous study using similar LAIV modified by replacing the H3 and N2 of the attenuated OH04 virus with H1 and N1 from a H1N1pdm09 strain demonstrated higher CMI response with increased IFN‐gamma secreting cells and T cell populations.35 Following infection, the LAIV vaccine in that study induced sterilizing immunity, while pigs vaccinated with WIV had virus in lungs and nasal cavities.25
Influenza continues to be a major health problem for swine, and new reassortant strains periodically cause public health threats. Vaccination remains an essential tool in controlling IAV in pigs. However, the increasing antigenic diversity of IAV makes development of adequate WIV vaccines for swine challenging. This study indicates that contemporary swine H3N2 viruses have drifted from the currently available swine vaccines. The rH3N2p virus similar to H3N2v used in this study caused mild clinical disease, but replicated and shed well from the nasal cavities. This is an advantage to the virus in spreading among immunologically naive pigs and people. Commercial WIV in non‐MDA pigs reduced virus in lungs and nasal cavities, but the response was diminished when vaccinated in the presence of heterologous MDA. Although not tested in pigs with MDA here, LAIV was previously shown to be more effective than WIV,21 and LAIV should be considered as an improved vaccine platform over WIV under appropriate conditions. Our vaccine study underscores the need for improving the process of swine IAV vaccine strain updates as well as consideration of a variety of appropriate vaccine platforms. Similarly, the extensive use of WIV in sows to increase MDA that is mismatched to the circulating strains to which piglets are exposed should be re‐evaluated.
Supporting information
Table S1. Percent nucleotide identity of the hemagglutinin (HA) gene compared between H3N2 isolates used in the animal trials.
Figure S1. Phylogeny of representative and study H3N2 influenza A isolates using the hemagglutinin (HA) gene.
Figure S2. Average rectal temperature of nonvaccinated, challenged pigs (C), MDA+ ‐nonvaccinated, challenged pigs (MDA/C), MDA+ ‐vaccinated, challenged pigs (MDA/V/C), and MDA− ‐vaccinated, challenged pigs (V/C) before infection (0 days post infection; dpi) and after infection (1–5 dpi) with NY11 (H3N2v‐like virus) at 7 weeks of age. Results of nonchallenged pigs are not included.
Acknowledgements
Authors wish to thank Michelle Harland, Gwen Nordholm, Katie Gibson, and Alicia Janas‐Martindale for excellent technical assistance and Jason Huegal, Jason Crabtree, and Tyler Standley for animal care and handling assistance. Thank you to Dr. Matthew Sandbulte for providing MDA‐positive piglets. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. Funding was provided by USDA‐ARS and USDA‐APHIS. USDA is an equal opportunity provider and employer.
Kitikoon et al (2013) Swine influenza virus vaccine serologic cross‐reactivity to contemporary US swine H3N2 and efficacy in pigs infected with an H3N2 similar to 2011–2012 H3N2v. Influenza and Other Respiratory Viruses 7(Suppl. 4), 32–41.
References
- 1. Vincent A, Awada L, Brown I et al Review of Influenza A Virus in Swine Worldwide: a Call for Increased Surveillance and Research. Zoonoses Public Health 2013; doi: 10.1111/zph.12049. [DOI] [PubMed] [Google Scholar]
- 2. Zhou NN, Senne DA, Landgraf JS et al Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 1999; 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Webby RJ, Swenson SL, Krauss SL et al Evolution of swine H3N2 influenza viruses in the United States. J Virol 2000; 74:8243–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hause BM, Oleson TA, Bey RF, Stine DL, Simonson RR. Antigenic categorization of contemporary H3N2 Swine influenza virus isolates using a high‐throughput serum neutralization assay. J Vet Diagn Invest 2010; 22:352–359. [DOI] [PubMed] [Google Scholar]
- 5. Leuwerke B, Kitikoon P, Evans R, Thacker E. Comparison of three serological assays to determine the cross‐reactivity of antibodies from eight genetically diverse U.S. swine influenza viruses. J Vet Diagn Invest 2008; 20:426–432. [DOI] [PubMed] [Google Scholar]
- 6. Richt JA, Lager KM, Janke BH et al Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 Swine influenza viruses cocirculating in the United States. J Clin Microbiol 2003; 41:3198–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsen CW, Karasin AI, Carman S et al Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis 2006; 12:1132–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorusso A, Vincent AL, Gramer MR, Lager KM, Ciacci‐Zanella JR. Contemporary epidemiology of North American lineage triple reassortant influenza A viruses in pigs. Curr Top Microbiol Immunol 2013; 370:113–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shu B, Garten R, Emery S et al Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology 2012; 422:151–160. [DOI] [PubMed] [Google Scholar]
- 10. Kitikoon P, Nelson MI, Killian ML et al Genotype patterns of contemporary reassorted H3N2 virus in US swine. J Gen Virol 2013; 94:1236–1241. [DOI] [PubMed] [Google Scholar]
- 11. CDC . Influenza A (H3N2) variant virus‐related hospitalizations — Ohio, 2012. MMWR Morb Mortal Wkly Rep 2012; 61(39):802. [PubMed] [Google Scholar]
- 12. Nelson MI, Vincent AL, Kitikoon P, Holmes EC, Gramer MR. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J Virol 2012; 86:8872–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Houser KV, Katz JM, Tumpey TM. Seasonal trivalent inactivated influenza vaccine does not protect against newly emerging variants of influenza A (H3N2v) virus in ferrets. J Virol 2013; 87:1261–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skowronski DM, De Serres G, Janjua NZ et al Cross‐reactive antibody to swine influenza A(H3N2) subtype virus in children and adults before and after immunisation with 2010/11 trivalent inactivated influenza vaccine in Canada, August to November 2010. Euro Surveill 2012; 17:1852–61. [DOI] [PubMed] [Google Scholar]
- 15. Van Reeth K, Ma W. Swine influenza virus vaccines: to change or not to change‐that's the question. Curr Top Microbiol Immunol 2012; 370:173–200. [DOI] [PubMed] [Google Scholar]
- 16. Gasparini R, Amicizia D, Lai PL, Panatto D. Live attenuated influenza vaccine–a review. J Prev Med Hyg 2011; 52:95–101. [PubMed] [Google Scholar]
- 17. Markowska‐Daniel I, Pomorska‐Mol M, Pejsak Z. The influence of age and maternal antibodies on the postvaccinal response against swine influenza viruses in pigs. Vet Immunol Immunopathol 2011; 142:81–86. [DOI] [PubMed] [Google Scholar]
- 18. Beaudoin A, Johnson S, Davies P, Bender J, Gramer M. Characterization of influenza a outbreaks in Minnesota swine herds and measures taken to reduce the risk of zoonotic transmission. Zoonoses Public Health 2012; 59:96–106. [DOI] [PubMed] [Google Scholar]
- 19. Allerson M, Deen J, Detmer SE et al The impact of maternally derived immunity on influenza A virus transmission in neonatal pig populations. Vaccine 2013; 31:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitikoon P, Nilubol D, Erickson BJ et al The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet Immunol Immunopathol 2006; 112:117–128. [DOI] [PubMed] [Google Scholar]
- 21. Vincent AL, Ma W, Lager KM et al Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine‐associated enhanced respiratory disease. J Virol 2012; 86:10597–10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. World Health Organization . 2002. Available at WHO_manual_on_animal-diagnosis_and_surveillance_2002_5.pdf (Accessed June 2012).
- 23. Ciacci‐Zanella JR, Vincent AL, Prickett JR, Zimmerman SM, Zimmerman JJ. Detection of anti‐influenza A nucleoprotein antibodies in pigs using a commercial influenza epitope‐blocking enzyme‐linked immunosorbent assay developed for avian species. J Vet Diagn Invest 2010; 22:3–9. [DOI] [PubMed] [Google Scholar]
- 24. Tang Y, Lee CW, Zhang Y et al Isolation and characterization of H3N2 influenza A virus from turkeys. Avian Dis 2005; 49:207–213. [DOI] [PubMed] [Google Scholar]
- 25. Pena L, Vincent AL, Ye J et al Modifications in the polymerase genes of a swine‐like triple‐reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol 2011; 85:456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Halbur PG, Paul PS, Frey ML et al Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 1995; 32:648–660. [DOI] [PubMed] [Google Scholar]
- 27. Gauger PC, Vincent AL, Loving CL et al Kinetics of lung lesion development and pro‐inflammatory cytokine response in pigs with vaccine‐associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol 2012; 49:900–912. [DOI] [PubMed] [Google Scholar]
- 28. Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg 1938; 27:493–497. [Google Scholar]
- 29. Cai Z, Zhang T, Wan XF. A computational framework for influenza antigenic cartography. PLoS Comput Biol 2010; 6:e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loeffen WLA, Heinen PP, Bianchi ATJ, Hunneman WA, Verheijden JHM. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet Immunol Immunopathol 2003; 92:23–35. [DOI] [PubMed] [Google Scholar]
- 31. Kitikoon P, Vincent AL, Gauger PC et al Pathogenicity and transmission in pigs of the novel A(H3N2)v influenza virus isolated from humans and characterization of swine H3N2 viruses isolated in 2010–2011. J Virol 2012; 86:6804–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kappes MA, Sandbulte MR, Platt R et al Vaccination with NS1‐truncated H3N2 swine influenza virus primes T cells and confers cross‐protection against an H1N1 heterosubtypic challenge in pigs. Vaccine 2012; 30:280–288. [DOI] [PubMed] [Google Scholar]
- 33. Wesley RD, Lager KM. Overcoming maternal antibody interference by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of swine influenza virus. Vet Microbiol 2006; 118:67–75. [DOI] [PubMed] [Google Scholar]
- 34. Gorres JP, Lager KM, Kong WP et al DNA vaccination elicits protective immune responses against pandemic and classic swine influenza viruses in pigs. Clin Vaccine Immunol 2011; 18:1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loving CL, Vincent AL, Pena L, Perez DR. Heightened adaptive immune responses following vaccination with a temperature‐sensitive, live‐attenuated influenza virus compared to adjuvanted, whole‐inactivated virus in pigs. Vaccine 2012; 30:5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Percent nucleotide identity of the hemagglutinin (HA) gene compared between H3N2 isolates used in the animal trials.
Figure S1. Phylogeny of representative and study H3N2 influenza A isolates using the hemagglutinin (HA) gene.
Figure S2. Average rectal temperature of nonvaccinated, challenged pigs (C), MDA+ ‐nonvaccinated, challenged pigs (MDA/C), MDA+ ‐vaccinated, challenged pigs (MDA/V/C), and MDA− ‐vaccinated, challenged pigs (V/C) before infection (0 days post infection; dpi) and after infection (1–5 dpi) with NY11 (H3N2v‐like virus) at 7 weeks of age. Results of nonchallenged pigs are not included.


