Abstract
Single-molecule approaches to solving biophysical problems are powerful tools that allow static and dynamic real-time observations of specific molecular interactions of interest in the absence of ensemble-averaging effects. Here, we provide detailed protocols for building an experimental system that employs atomic force microscopy and a single-molecule DNA tightrope assay based on oblique angle illumination fluorescence microscopy. Together with approaches for engineering site-specific lesions into DNA substrates, these complementary biophysical techniques are well suited for investigating protein–DNA interactions that involve target-specific DNA-binding proteins, such as those engaged in a variety of DNA repair pathways. In this chapter, we demonstrate the utility of the platform by applying these techniques in the studies of proteins participating in nucleotide excision repair.
1. INTRODUCTION
Experiments studying nucleotide excision repair (NER) proteins using optical imaging in our laboratories usually go through three distinct phases: biochemical analysis (Croteau, DellaVecchia, Perera, & Van Houten, 2008; Croteau et al., 2006), atomic force microscopy (AFM) (Wang et al., 2006), and fluorescence single-molecule imaging (Hughes et al., 2013; Kad, Wang, Kennedy, Warshaw, & Van Houten, 2010; Kong et al., 2016). First, proteins should be highly purified and exhibit excellent activity. Purification of these proteins often includes a size-exclusion chromatography step to ensure a homogenous preparation of non-aggregated protein, free of contaminating DNA, which is then examined by a variety of bulk biochemistry methods such as fluorescence anisotropy and electrophoretic mobility shift assays for DNA-binding affinities. These proteins are then imaged alone and complexed with DNA substrates using AFM to assess properties such as homogeneity, stability, stoichiometry (Ghodke et al., 2014; Yeh et al., 2012), specificity, and DNA bend angles (Kong et al., 2016). Finally, the dynamic interactions of these proteins with DNA are visualized with the DNA tightrope assay and fluorescence microscopy (Ghodke et al., 2014; Kad et al., 2010; Kong et al., 2016; Kong & Van Houten, 2016). This chapter first gives detailed protocols on preparing defined DNA substrates for analysis by AFM or our tightrope assay. We then discuss how AFM is used to determine specificity, stoichiometry, and DNA bend angles. Finally, we end with a description of our optical DNA tightrope flow cell setup with which we can observe quantum dot (Qdot or QD)-labeled proteins using oblique angle illumination on a total internal reflection fluorescence microscope.
2. PREPARATION OF DEFINED LESION SUBSTRATES FOR AFM AND DNA TIGHTROPE ASSAY
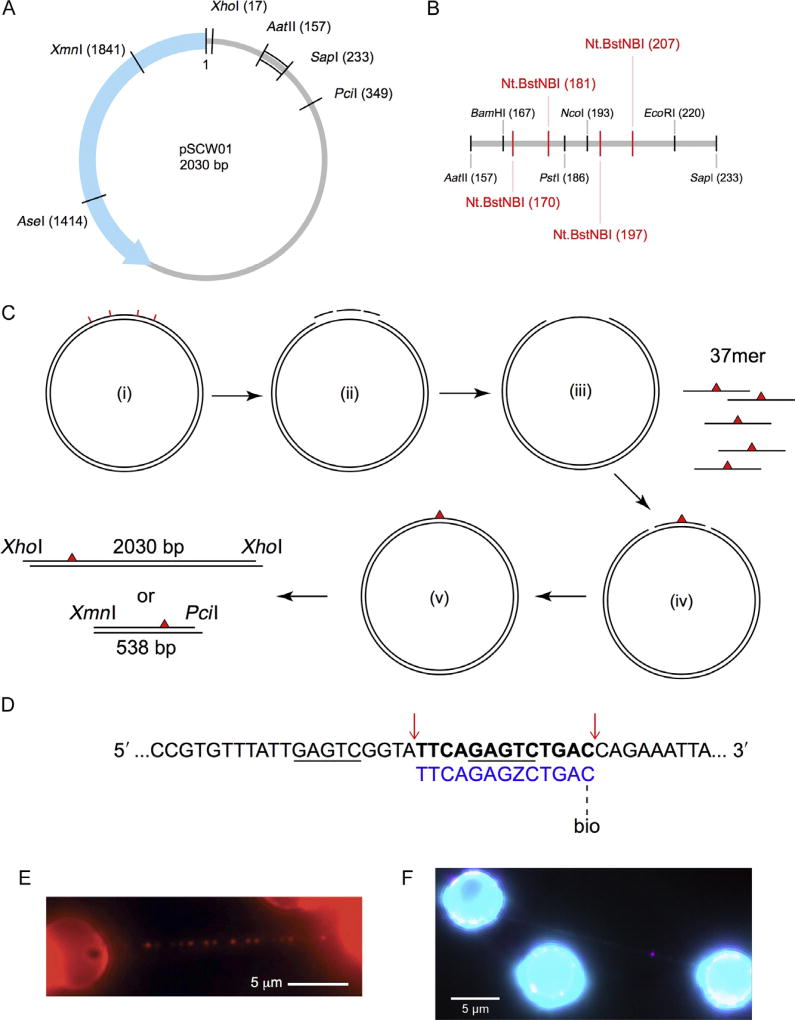
To characterize protein–DNA interactions involving proteins that recognize specific targets, DNA sequences or otherwise, it is important to ensure that an optimal number of target sites exist in the DNA substrate against a vast nonspecific background, such that binding events can be observed efficiently. For DNA repair proteins that carry out damage recognition, a common method to globally induce different types of lesions in a random manner is to subject commercially available λ-DNA to physical or chemical manipulations (Kad et al., 2010; Nelson, Dunn, Kathe, Warshaw, & Wallace, 2014). The number of total lesions can be estimated qualitatively for comparison purposes or, in the case of UV-induced photoproducts, explicitly calculated as an average lesion density through quantitative PCR (qPCR) (Furda, Bess, Meyer, & Van Houten, 2012; Meyer et al., 2007). It is also worth noting that UV irradiation of DNA generates 6,4-photoproducts as well as cyclobutane pyrimidine dimers, both of which contribute to the global average lesion density derived from qPCR. Compared to the random distributions of possibly more than one type of lesion generated as briefly described above, a DNA substrate containing site-specific lesion(s) of desired identity offers more control in the sequence context around the lesion site and leads to more predictable binding patterns that may correlate with specific binding events. To this end, we have developed two different strategies for making DNA substrates containing site-specific lesions, suitable for single-molecule AFM and DNA tightrope assays. The first approach, based on the plasmid pSCW01 (Fig. 1A and B) previously used to study DNA mismatch repair, places a 37mer lesion-containing oligonucleotide in a gap created in the plasmid via nicking at four Nt.BstNBI sites (Fig. 1C (i)–(iv)) (Geng et al., 2011; Ghodke et al., 2014). The oligonucleotide containing the defined lesion is sealed into the plasmid by T4 DNA ligase with high efficiency approaching 98%–99% (Fig. 1C (v)). The plasmid can be digested to yield a 538-bp lesion-containing fragment for AFM studies (Fig. 1C; Section 2.7). Alternatively, it is linearized and tandemly ligated (end to end) to form long DNA substrates suitable for the tightrope assay (Fig. 1C; Sections 2.1–2.6). These defined lesion damage arrays thus contain one site-specific lesion every 2030 bp (Fig. 1E). Another approach inserts an oligonucleotide containing a site-specific lesion into λ-DNA (Fig. 1D; Section 2.8). In this method, λ-DNA is first nicked by Nt.BstNBI at 61 different sites and the shortest single-stranded fragment, between bases 33,778 and 33,791, is then liberated and replaced with a lesion-containing oligonucleotide.
Fig. 1.
Design of defined lesion substrate. (A) Map of the pSCW01 plasmid and locations of restriction digest sites. (B) Detailed map of restriction digest and nicking sites for plasmid sequence between the AatII (157) and SapI (233) sites. Nt.BstNBI nicking sites are shown in red. (C) Strategy for generating defined lesion substrate based on the pSCW01 plasmid. The plasmid is first nicked by Nt.BstNBI at four different locations (i), which yields three short single-stranded fragments (ii) that are liberated from the plasmid via heating, resulting in a gapped plasmid (iii). 37mer oligonucleotides, each containing a site-specific lesion, are annealed to the gapped plasmids (iv) before the nicks on either side of the oligonucleotides are sealed by overnight ligation (v). The plasmids can now be linearized by XhoI and then tandem-ligated to form long DNA substrates, containing one site-specific damage per 2030 bp, for use in the DNA tightrope assay. Alternatively, the plasmids can be double digested by XmnI and PciI and gel purified to obtain 538 bp fragments, each containing one site-specific damage ~160 bp from the PciI site. (D) Strategy for inserting a damaged oligonucleotide with a biotin conjugate for quantum dot visualization in λ-DNA. The upper λ-DNA sequence is underlined at the binding sites for Nt.BstNBI. Cut sites are indicated by red arrows, leading to the release of the bolded segment. This is replaced by the lower 5′ phosphorylated oligonucleotide (blue) containing damage (Z = fluorescein-dT) and biotin-conjugated via TEG at the 3′ end. (E) An array of streptavidin-conjugated quantum dots on a DNA tightrope of a defined lesion substrate containing one site-specific abasic site analog per 2030 bp, each with a proximal biotin marking the site of the lesion. (F) DNA damage (magenta) visualized with 655 streptavidin-conjugated quantum dot on a λ-DNA tightrope stained with YOYO-1 (cyan). Panel E: Adapted with permission from Ghodke, H., Wang, H., Hsieh, C. L., Woldemeskel, S., Watkins, S. C., Rapic-Otrin, V., et al. (2014). Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proceedings of the National Academy of Sciences of the United States of America, 111(18), E1862–E1871. http://dx.doi.org/10.1073/pnas.1323856111 (fig. 4A).
2.1 Growing pSCW01 Plasmid
2.1.1 Equipment
37°C shaking incubator
Laboratory centrifuge
2.1.2 Buffers and Reagents
Escherichia coli transformed with pSCW01 on LB-Amp agar plates
LB media with 100 µg/mL ampicillin (LB-Amp)
2.1.3 Procedure
Pick a single colony from a freshly transformed plate.
Inoculate a 2-mL LB-Amp starter culture for 6 h at 37°C.
Inoculate 1 L LB-Amp with 1 mL starter culture. Grow for 18 h at 37°C.
Centrifuge at 6000 × g for 15 min at 4°C to harvest. Store each liter of culture as two pellets.
2.2 Maxiprep of Plasmid DNA
2.2.1 Equipment
Laboratory centrifuge
SpeedVac or other vacuum concentrator
2.2.2 Buffers and Reagents
pSCW01 E. coli pellets
QIAGEN Plasmid Maxi Kit
Isopropanol
70% ethanol
2.2.3 Procedure
Resuspend each pellet of culture in 25 mL of buffer P1.
Add 25 mL of buffer P2. Incubate at room temperature for 5 min.
Add 25 mL of prechilled buffer P3. Mix well.
Follow the manufacturer’s protocol.
Resuspend each DNA pellet in 500 µL of ddH2O.
Concentrate DNA in SpeedVac to ~1 µg/µL.
2.2.4 Notes
In step 5, DNA is resuspended in ddH2O instead of Tris or Tris–EDTA buffer so that samples can be concentrated without affecting concentrations of the buffer components.
2.3 Plasmid DNA Nicking and Oligo Displacement
2.3.1 Equipment
Heat block or thermocycler
2.3.2 Buffers and Reagents
Purified plasmid DNA (pSCW01)
Displacer oligonucleotides (Table 1, IDT)
Nickase (Nt.BstNBI, 10 U/µL, NEB)
10× NEBuffer 3.1 (NEB)
Table 1.
Sequences of Oligonucleotides Used in Preparation of Defined Lesion Substrates
| Oligonucleotide | Sequence |
|---|---|
| Displacer1 | ATTTGACTCC |
| Displacer2 | CATGGACTCGCTGCAG |
| Displacer3 | GAATGACTCGG |
| FL37 | CCGAGTCATTCCTGCAGCGAGTCCATGGGAGTCAAAT |
| FL37BiodT | CCGAGTCATTCCTGCAGCGAGTCCATGGGAGTCAAA/BiodT/ |
| FL13 | TTCAGAGTCTGAC/BioTEG/ |
T indicates an internal fluorescein-modified deoxythymidine. /BiodT/ indicates a biotin-modified deoxythymidine. /BioTEG/ indicates a biotin modification attached via a triethylene glycol (TEG) spacer.
2.3.3 Procedure
Prepare, in 1 × NEBuffer 3.1 (NEB), purified plasmid DNA (pSCW01) at the final concentration of 400 ng/µL, with 50-fold molar excess of each of the three displacer oligonucleotides (Table 1), and twice the number of units of nickase (Nt.BstNBI, 10 U/µL, NEB) as the amount of plasmid DNA in micrograms. Incubate the reaction at 55°C for 4 h. Before proceeding to the next step, save 1–2 µL of the nicking reaction for diagnostic tests.
Inactivate the nicking reaction at 85°C for 10 min before turning off the heat block. Let the heat block cool down to room temperature for approximately 3.5–4 h to allow annealing of displacer oligos with complementary short fragments liberated from plasmids through the nicking reaction. Before proceeding to the next step, save 1–2 µL of the gapped DNA for diagnostic tests.
2.3.4 Notes
Start the nicking reaction with at least 50 µg of plasmid DNA for better yield in the next step.
During cooling, the excess displacer oligonucleotides capture and anneal to those liberated from the nicking reaction, preventing them from reannealing to the plasmid. These short fragments and oligonucleotides are then removed in the next step.
2.4 PEG Purification of Gapped Plasmid DNA
2.4.1 Equipment
Heat block or thermocycler
Benchtop centrifuge
Nanodrop or other UV–vis spectrophotometer
Standard agarose gel electrophoresis equipment
2.4.2 Buffers and Reagents
2× PEG solution (26% polyethylene glycol, MW 8000 and 20 mM MgCl2)
70% ethanol
1× TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA)
Restriction enzymes (PstI and NcoI, NEB)
2.4.3 Procedure
Pool and transfer now gapped DNA plasmids to new Eppendorf tubes, each containing no more than 500 µL in volume. Add equal volume of 2× PEG solution to each tube and mix well.
Centrifuge at 4°C for 1 h at the maximum speed (14,800 rpm) on a benchtop centrifuge.
Carefully remove the supernatant from each tube. Precipitated DNA should have formed a thin film stuck on the side of the tube. Using a pipette, wash the side wall with 500 µL of 70% ethanol. The white film of DNA should peel off and settle to the bottom of the tube.
Centrifuge and collect the DNA pellet at 4°C for 15 min at the maximum speed (14,800 rpm) on a benchtop centrifuge.
Carefully remove the supernatant from each tube without disturbing the DNA pellet at the bottom.
Air dry the tube and the pellet before resuspending the pellet in 200 µL of ddH2O.
Dilute 1 µL of the purified gapped plasmid DNA in 20 µL of 1× TE buffer and measure the DNA concentration at A260 using a UV–vis spectrophotometer (NanoDrop 2000, Thermo Scientific).
Save 1–2 µL of the purified gapped plasmid DNA for diagnostic tests.
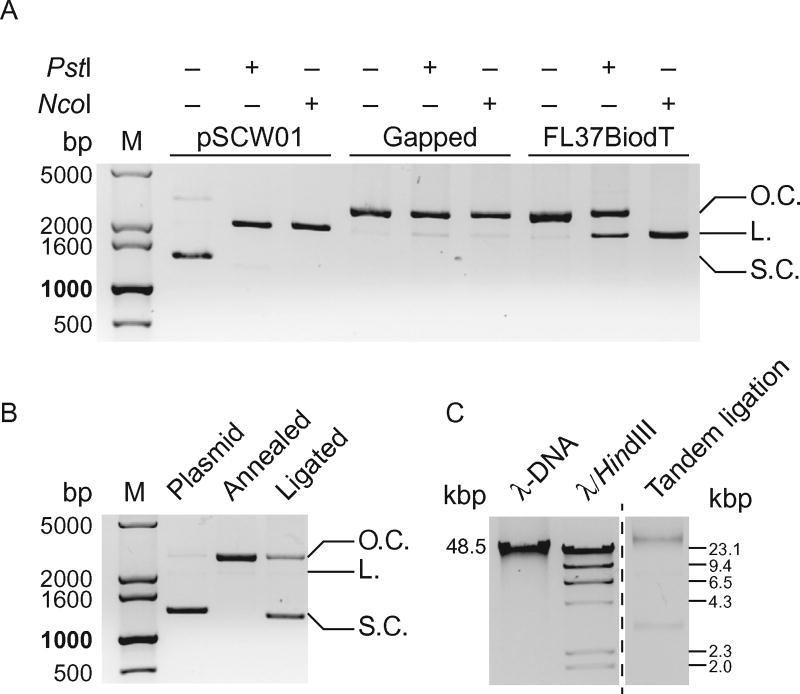
Test for completeness of nicking and gapping reactions by setting up restriction digests of samples saved previously after nicking and gapping reactions. Purified pSCW01 plasmids should be used as a positive control. Restriction enzymes (PstI and NcoI, NEB) target the sequence that is nicked and/or liberated after nicking, and therefore will not incise the gapped plasmid DNA. Typical reactions contain 100–200 ng of nicked or gapped plasmid DNA and 5 U of restriction enzyme in 20 µL of appropriate reaction buffer and are incubated at 37°C for 2 h. Run all digested reactions and undigested controls on a 1% agarose gel (Fig. 2A).
Fig. 2.
Diagnostic agarose gels for preparation of defined lesion substrates. (A) 1% agarose gel of diagnostic restriction digests of pSCW01 plasmid, gapped plasmid, and gapped plasmid with FL37BiodT annealed. PstI and NcoI, whose restriction sites are within the 37-base gap, do not linearize the gapped plasmid. With FL37BiodT annealed in the gap, NcoI linearizes the now nicked plasmid. Restriction digest by PstI on the FL37BiodT-annealed plasmid is hindered due to the presence of the fluorescein in the PstI restriction site. L., linearized plasmid; O.C., open circle, nicked or gapped plasmid; S.C., supercoiled plasmid. (B) 1% agarose gel of pSCW01 plasmid, FL37BiodT-annealed plasmid, and FL37BiodT-ligated plasmid. The reappearance of the supercoiled band in the ligated plasmid lane indicates completion of the ligation reaction. L., linearized plasmid; O.C., open circle, nicked or gapped plasmid; S.C., supercoiled plasmid. (C) 0.8% agarose gel of tandem-ligated the FL37-containing defined lesion substrate with full-length λ-DNA and λ-DNA HindIII digest fragments as size markers.
2.4.4 Notes
During the resuspension step, it may be helpful to heat the tube at 55°C for 10 min to help resolubilize the DNA.
2.5 Annealing and Ligation of 37mer Oligo
2.5.1 Equipment
Heat block
Thermocycler or heat block in cold room or fridge
Standard agarose gel electrophoresis equipment
Standard denaturing polyacrylamide electrophoresis equipment
Typhoon scanner (GE Healthcare)
2.5.2 Buffers and Reagents
10× NEBuffer 2.1 (NEB)
Lesion-containing 37mer oligonucleotides (Table 1, IDT)
Fluorescently labeled 37mer and 50mer oligonucleotides (IDT)
100 mM ATP solution
T4 DNA ligase (NEB)
Restriction enzymes (PstI, NcoI, EcoRI, and AatII, NEB)
2× denaturing sample loading buffer (NEB)
2.5.3 Procedure
Fill the gap by annealing a 37mer oligonucleotide that contains a lesion of choice. Always carry out the annealing and ligation steps for the lesion-containing oligo in parallel to the same experiments using a fluorescein-labeled 37mer (FL37, Table 1), which can be later used to check annealing and ligation reactions.
Set up annealing reactions in 1 × NEBuffer 2.1 (NEB), containing 400 nM gapped plasmids and threefold molar excess of 37mer lesion-containing oligonucleotides (and in parallel, FL37). Incubate at 85°C for 10 min before turning off the heat block. Let the heat block cool down to room temperature for approximately 3.5–4 h to allow annealing of 37mer oligonucleotides. Save 1–2 µL of the annealed plasmid DNA for diagnostic tests.
Set up ligation reaction to seal the 5′- and/or 3′-nicks that remain after annealing. To the annealing reaction, add ATP and T4 DNA ligase (2000 U/µL, NEB) to a final concentration of 8 mM and 20 U/µL, respectively. Incubate at 16°C for 18 h.
Inactivate the ligation reaction at 65°C for 10 min before turning off the heat block. Let the heat block cool down slowly to room temperature.
Save 1–2 µL of each ligation reaction for diagnostic tests.
To test for completeness of annealing reaction, set up restriction digest reactions of saved sample of annealed plasmid DNA with restriction enzymes that target the sequence in the annealed oligo. Incubate 100–200 ng of annealed plasmids with 5 U of restriction enzymes (PstI or NcoI, NEB) in 20 µL reaction volume in appropriate buffers for 2 h at 37°C and run with undigested control on 1% agarose gel (Fig. 2A).
To test for completeness of the ligation reaction, set up restriction digest reactions of the saved ligated plasmids with FL37 in the gap. Prepare single digestions of the sample with either EcoRI or AatII, as well as a double digestion with both enzymes. Incubate 20 µL reactions containing 100–200 ng plasmids and 5 U of restriction enzyme(s) in appropriate buffer at 37°C for 2 h. To each 5 µL of digested samples and undigested control, as well as 2 µL of 25 nM fluorescein-labeled oligonucleotides of appropriate lengths (37mer and 50mer), add equal volume of 2 × denaturing sample loading buffer. Heat all samples at 90°C for 5 min and chill on ice immediately. Load these samples on a prerun 10% denaturing polyacrylamide gel. Ensure that the gel runs hot to the touch to prevent reannealing of single-stranded DNA and image on a fluorescence scanner (Typhoon 9400, GE Healthcare). Lengths of the diagnostic restriction digests will vary depending on whether the 5′- or the 3′-nick was sealed. We normally observe >98% ligation of both ends of the modified 37mer.
2.5.4 Notes
Ideally, steps in the protocol from nicking plasmids to annealing of 37mer oligonucleotides should be completed in 1 day, with the 18-h ligation setup to take place overnight. This is so that the time that plasmids remain gapped, during which they are presumably the most fragile, is minimized. However, if necessary, purified gapped plasmids can be stored overnight at 4°C without significant adverse effects on the quality of the entire preparation.
Ligation reaction can also be confirmed by comparing overnight-ligated plasmids to those before ligation. A supercoiled band similar to that seen in purified plasmids should reappear after ligation (Fig. 2B).
2.6 Linearization and Tandem Ligation
2.6.1 Equipment
Heat block or thermocycler
Standard agarose gel electrophoresis equipment
2.6.2 Buffers and Reagents
10× NEBuffer 2.1 (NEB)
50 mM EDTA
Restriction enzyme (XhoI, NEB)
T4 DNA ligase (NEB)
2× Quick Ligation Reaction Buffer (NEB)
Dry ice
λ-DNA and λ-DNA HindIII digest fragments (NEB)
2.6.3 Procedure
Linearize ligated plasmids by incubating them with twice the number of units of XhoI (20 U/µL, NEB) as the amount of DNA in micrograms at 37°C for 2 h. Adjust the final concentration of NEBuffer 2.1 (NEB) to 1 × with 10 × stock if necessary.
Heat inactivate XhoI by incubating at 80°C for 20 min. Turn off heat block and allow it to cool down slowly to room temperature. Linearized plasmids can be stored at −20°C.
For tandem (end-to-end) ligation of linearized plasmids: incubate 1 µg of plasmids with 2 µL of T4 DNA ligase (2000 U/µL, NEB) in 1 × Quick Ligation Reaction Buffer (NEB) in a total reaction volume of 20 µL at room temperature for 15 min.
At the end of the ligation, save 2 µL of the reaction and stop the reaction by adding 1 µL of 50 mM EDTA. Stop the rest of the reaction (18 µL) by placing the ligation reaction tube on dry ice till frozen. Ligation products can be kept for short-term storage at −20°C.
To check the efficiency of tandem ligation, run the saved sample from the step above on 0.8% agarose gel with full-length λ-DNA (NEB) and λ-DNA HindIII digest fragments (NEB) as standards (Fig. 2C). Tandem-ligation products should be at least the same length as the longest λ-DNA HindIII digest fragment (~23,000 bp), preferably equal to or longer than λ-DNA (~48,000 bp).
2.7 Preparation of DNA Substrate for AFM
2.7.1 Equipment
Heat block or thermocycler
Standard agarose gel electrophoresis equipment
UV transilluminator
Benchtop centrifuge
Nanodrop or other UV–vis spectrophotometer
SpeedVac or other vacuum concentrator
2.7.2 Buffers and Reagents
Restriction enzymes (XmnI and PciI, NEB)
10× NEBuffer 2.1 (NEB)
Agarose gel purification kit (Wizard SV Gel and PCR Clean-Up System, Promega)
PCR purification kit (QIAquick PCR Purification Kit, Qiagen)
AFM water: autoclaved nuclease-free ddH2O, 0.02 µm filtered
2.7.3 Procedure
Set up a double digest with restriction enzymes (XmnI and PciI, NEB) in appropriate buffer (1 × NEBuffer 2.1, NEB). Use twice the number of units of each restriction enzyme as the amount of annealed and ligated lesion-containing plasmid DNA in micrograms. Incubate the reaction at 37°C for 4 h.
Inactivate the digestion reaction at 80°C for 20 min before turning off the heat block. Let the heat block cool down slowly to room temperature. Run a small sample of the digested product on 1% agarose gel toensure that digestion was complete.
Run the rest of digestion reaction on 1% agarose gel. Excise the band of appropriate size from gel and extract DNA with a commercial gel purification kit per manufacturer’s protocol. See Section 2.7.4 for notes on avoiding UV damage.
Purify gel-extracted DNA one more time with a commercial PCR purification kit (QIAquick PCR Purification Kit, Qiagen) per manufacturer’s protocol to ensure complete removal of restriction enzymes from the desired DNA fragments. The final elution of DNA should be carried out in AFM water. Measure DNA concentration at A260 using a UV–vis spectrophotometer (NanoDrop 2000, Thermo Scientific).
In a vacuum concentrator (SpeedVac DNA120, Thermo Scientific), concentrate DNA sample to desired concentration appropriate for AFM-binding experiments (~200–300 nM). DNA can be kept at 4°C for immediate use, or −80°C for long-term storage.
2.7.4 Notes
When excising gel bands on the UV transilluminator, it is important to minimize the bands’ exposure toUV as UV light could induce additional undesired photoproducts in DNA. To do so, load in a separate lane a small amount of digested DNA for visualization purpose only and shield the bulk of the DNA sample in gel from UV with aluminum foil.
Two-step purification (gel extraction and PCR purification kits) should remove all DNA-bound restriction enzymes from the sample. However, if proteins are found bound to DNA upon quality check under AFM, additional rounds of PCR purification may be needed at the cost of slight loss of DNA sample. Additionally, it may be necessary to do a phenol–chloroform extraction and ethanol precipitation to get rid of stubborn proteins.
It may be desirable to aliquot purified DNA sample into single-use tubes and store at −80°C to avoid repeated freeze–thaw cycles.
2.8 Defined Lesion Substrates Based on λ-DNA
2.8.1 Equipment
Heat block
2.8.2 Buffers and Reagents
Nickase (Nt.BstNBI, 10 U/µL, NEB)
10× NEBuffer 3.1 (NEB)
λ-DNA (NEB)
T4 DNA ligase (1 U/µL, NEB)
100 mM ATP solution
1 µM 13mer oligonucleotide with site-specific damage at position 8 and a 3′ biotin modification via a TEG linker (Table 1)
Qdot Streptavidin Conjugate (Thermo Scientific)
YOYO-1 dye (Thermo Scientific)
2.8.3 Procedure
Prepare the nicking reaction using NEBuffer 3.1, 5 µg of λ-DNA and 2 U of enzyme; incubate at 55°C for 2 h.
Digestion of λ-DNA with the single-stranded nickase will create numerous nicks with which only one pair will be close enough together to generate an oligonucleotide fragment with a near room temperature melting point, regions 33,778–33,791 of λ-DNA (Fig. 1D).
Incubate with a 10-fold excess of damage-containing oligonucleotide (FL13, Table 1) at 55°C for 10 min.
Allow the solution to cool to room temperature.
Perform the ligation with 1 U of T4 DNA ligase and 1 mM ATP at room temperature overnight.
Removal of DNA ligase can be achieved using phenol:chloroform extraction (Sambrook, Fritsch, & Maniatis, 1989).
The lesion-containing DNA is ready to be used for DNA tightropes.
The DNA can be stored at 4°C for use within a day or two, for longer storage −20°C is preferred.
To visualize the damage site located 5 bases from the biotin, add 10 nM streptavidin-conjugated Qdots into a flow cell and incubate for 15 min. This can be combined with 100 nM YOYO-1 dye to visualize the DNA simultaneously (Fig. 1F).
2.8.4 Notes
For longer tightropes, DNA can be concatemerized (Springall, Inchingolo, & Kad, 2016).
This procedure is based on the method of Tafvizi, Huang, Fersht, Mirny, and van Oijen (2011).
3. ATOMIC FORCE MICROSCOPY
AFM provides a topographical view of protein–DNA interactions (Fig. 3). Three major sets of data can be obtained from a single protein–DNA experiment: protein specificity for site-specific lesions, as determined by its binding position on a DNA substrate (Fig. 4B and F); the bend angle of DNA at points of specific and nonspecific protein binding or otherwise (Fig. 4C and G); and the stoichiometry of protein binding to DNA substrates as determined by the volume of the complex (Fig. 4A, D, and E). The steps needed to acquire these data are outlined below. The overall process involves setting up a protein–DNA-binding reaction and depositing the sample onto atomically smooth mica (Section 3.1), imaging with an atomic force microscope (Section 3.2), and analyzing data (Section 3.3).
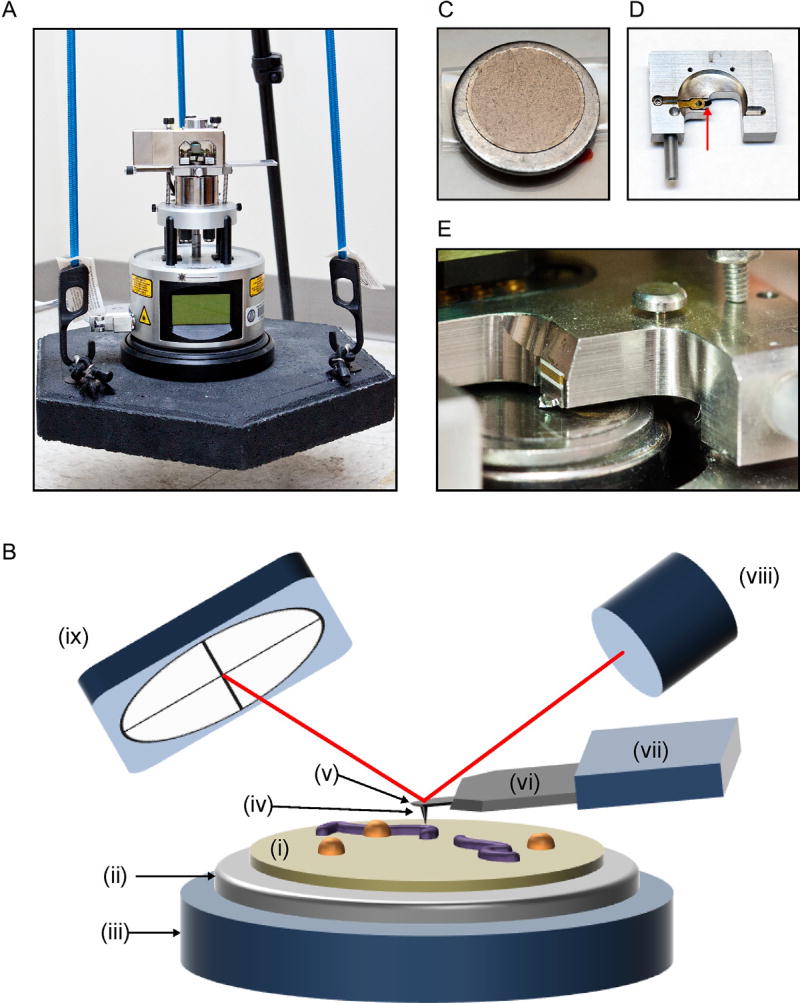
Fig. 3.
Atomic force microscopy setup. (A) diMultiMode V atomic force microscope by Veeco. For noise isolation, the AFM is placed on a heavy platform suspended by elastic bungee cords that are secured to a tripod. (B) Schematic of AFM tapping mode in air (not to scale). A protein–DNA sample on mica (i) glued to a metal disc (ii) is placed on the AFM scanner (iii). The probe tip (iv) scans across the sample to generate AFM data. The tip is located at the end of a cantilever (v), which is attached to a support chip (vi) and held by the probe holder (vii). A laser (viii) is reflected off the cantilever and onto a photodetector (ix). Deflection of the cantilever induced by the sample surface changes the path of the laser beam and provides topographical information about the sample (not shown). (C) 12-mm mica chip glued to metal disc. (D) Probe holder with probe (red arrow) for tapping in air. (E) Close-up of probe holder and probe installed above mica on scanner.
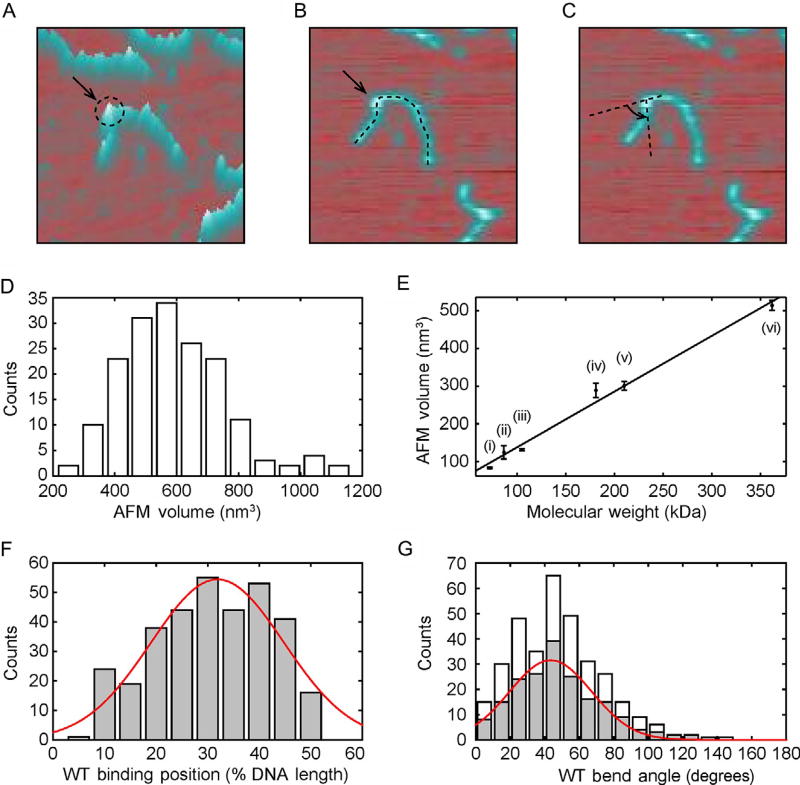
Fig. 4.
AFM imaging of protein volume, position, and bend angle. (A)–(C) AFM images of each protein-binding event on DNA can be used to extract information on the protein volume, binding position, and DNA bend angle, respectively. (D) Histogram of UV-DDB K244E mutant volumes on DNA (n = 171). (E) Calibration curve relating the molecular weight of a complex to its measured AFM volume, mean ± SD of three separate determinations. The curve was generated using the following proteins in solution: (i) Pot1 monomer (65 kDa), (ii) PcrA monomer (86.4 kDa), (iii) UvrA monomer (105 kDa), (iv) Taq MutS dimer (181 kDa), (v) UvrA dimer (210 kDa), and (vi) Taq MutS tetramer (362 kDa). Linear fit to the data yields V(nm3) = 1.471 MW (kDa) − 7.294 with R2 = 0.9886. (F) Histogram and Gaussian fitting (red curve) of wild-type Rad4–Rad23-binding positions (32% ± 13%, n = 335) on 538 bp DNA fragment in terms of percentage of total contour length measured from one end. (G) Histogram of DNA bend angles at all internal wild-type Rad4–Rad23-binding sites (white, n = 335). The histogram (gray) and Gaussian fitting (red curve) show DNA bend angles (43 ± 24°, n = 189) at specific binding events (proteins bound between 20% and 40%). Panel (D): Adapted with permission from Ghodke, H., Wang, H., Hsieh, C. L., Woldemeskel, S., Watkins, S. C., Rapic-Otrin, V., et al. (2014). Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proceedings of the National Academy of Sciences of the United States of America, 111(18), E1862–E1871. http://dx.doi.org/10.1073/pnas.1323856111 (fig. 5E). Panel (E): Adapted with permission from Ghodke, H., Wang, H., Hsieh, C. L., Woldemeskel, S., Watkins, S. C., Rapic-Otrin, V., et al. (2014). Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proceedings of the National Academy of Sciences of the United States of America, 111(18), E1862–E1871. http://dx.doi.org/10.1073/pnas.1323856111, fig. S6D. Panel (F): Adapted with permission from Kong, M., Liu, L., Chen, X., Driscoll, K. I., Mao, P., Bohm, S., et al. (2016). Single-molecule imaging reveals that Rad4 employs a dynamic DNA damage recognition process. Molecular Cell, 64(2), 376–387. http://dx.doi.org/10.1016/j.molcel.2016.09.005 (fig. 5A). Panel (G): Adapted with permission from Kong, M., Liu, L., Chen, X., Driscoll, K. I., Mao, P., Bohm, S., et al. (2016). Single-molecule imaging reveals that Rad4 employs a dynamic DNA damage recognition process. Molecular Cell, 64(2), 376–387. http://dx.doi.org/10.1016/j.molcel.2016.09.005 (fig. 5B).
Binding reactions are set up using purified proteins and DNA substrates 500–600 bp in length. The process described in Section 2.7 produces a 538-bp DNA duplex with a single site-specific lesion, positioned at 30% the contour length. Empirically, substrates of this size are ideal for AFM because they are long enough to allow for precise assessment of protein-binding positions and DNA bend angles, but short enough such that a large number of molecules can be captured in a single 1 × 1 µm field without excessive overlap and convolution.
Many protocols for AFM take advantage of the chemical properties of mica. First, mica exists in sheets that can be easily cleaved. Freshly cleaved mica is an atomically smooth surface, ideal for AFM imaging, as it will not contribute to the landscape being imaged. Second, the surface of freshly cleaved mica has a negative charge, which may be useful for studying certain positively charged particles. However, when studying protein–DNA interactions, mica can be treated with divalent and/or monovalent cations; we use a combination of sodium and magnesium salts in our deposition buffer. This confers a positive charge to the mica surface that will attract the negative phosphate backbone of DNA and enhance sample adhesion (Hansma & Laney, 1996; Vesenka et al., 1992).
Finally, the atomic force microscope scans the samples on mica to produce topographical data. Suspension of the microscope with bungee cords provides some protection from interfering vibrations (Fig. 3A). In AFM tapping mode (Fig. 3B), a cantilever (with probe tip at the end) is driven to oscillate vertically near its resonance frequency. The AFM scanner allows the probe to track a sample field in the X–Y dimensions. In tapping mode, the oscillation amplitude is kept constant, but interaction with the sample surface causes deflection of the cantilever and alters the path of the reflected laser beam. Three-dimesional images are captured and can be analyzed using various computer programs.
3.1 Binding Reaction and Sample Preparation
3.1.1 Equipment
Heat block
Mica (SPI) fixed to metal disks with low-melt glue (SPI)
Forceps
Compressed nitrogen gas
Scotch tape (3M)
3.1.2 Buffers and Reagents
Purified protein, ~5 µM
DNA substrate, ~ 200 nM (Section 2.7)
Protein-binding buffer (Note 2)
Deposition buffer: 25 mM HEPES, pH 7.5, 25 mM NaOAc, 10 mM Mg(OAc)2, 0.02 µm filtered
AFM water: autoclaved nuclease-free ddH2O, 0.02 µm filtered
3.1.3 Procedure
Set up binding reaction. This will vary depending on the protein and DNA being studied. In general, a 10-µL reaction can be prepared with 100 nM DNA substrate and 500 nM protein in binding buffer. Allow the reaction to proceed for 30 min at room temperature.
During reaction, heat the required volume (~200 µL) of deposition buffer at 65°C for 15–20 min. After heating, vortex buffer and spin down briefly. Allow to cool back to room temperature before setting up dilutions in step 4.
During reaction, and while deposition buffer is preheating, cleave mica using scotch tape. A razor blade may be used to make a shallow cut across the edge of the mica as a starting point for peeling. Smooth tape over the surface of the mica and, gripping the metal disk with forceps, pull the tape back. Check surface for uneven cleavage and repeat if necessary. A mica chip glued on a metal disc is shown in Fig. 3C.
When steps 1–3 are complete, set up depositions one at a time, in order to minimize time sample is spent in deposition buffer (Note 3). Add 1 µL of the reaction to 24 µL of deposition buffer and mix gently. Transfer all 25 µL of the diluted reaction onto the mica (Note 4); be careful not to touch the surface with the pipette tip. Immediately after depositing the droplet, gently rock the mica back and forth and swirl to distribute the sample evenly on the surface. Do this for 30 s and immediately begin step 5.
Aspirate 1000 µL of AFM water in a micropipette, dispense approximately 200 µL onto the mica surface, and flick water into sink. Repeat until you have used all the water (~5 washes total).
Dry the mica under a gentle stream of N2 gas. Push the liquid off the mica and onto a paper towel. Be careful of the air stream such that water droplets run down and off the surface, but are not allowed to come back up.
3.1.4 Notes
It is important that the purified protein is very clean and is stored in a buffer that does not include BSA, as this will interfere with imaging and analysis in the following sections.
Protein-binding buffer will vary depending on the specific reaction being studied. 50 mM HEPES (pH 7.5) and 150 mM NaCl are a good starting point.
These steps would be easiest with three hands, but they are manageable with a little forethought. We recommend setting up the “wash station” prior to beginning the depositions: aspirate 1000 µL of AFM water and leave pipette by N2 tank, along with some paper towels laid on the bench to collect runoff. Then, make the dilution and aspirate the sample. Carefully set the pipette on the bench while picking up the mica with forceps. Then, the operator can transfer the forceps/mica to their nondominant hand and dispense the sample using their dominant hand.
Sample concentration and volume both affect distribution on the mica. Typically, depositing 25 µL of a 1:25 dilution (for a final concentration of 20 nM protein and 4 nM DNA) of the reaction results in favorable sample distribution without overcrowding. However, some optimization may be required. We suggest setting up multiple depositions to test these factors.
3.2 Imaging With AFM
3.2.1 Equipment
Atomic force microscope (Veeco, diMultiMode V, or other/newer models)
AFM controller (Bruker, NanoScope V)
NanoScope 9.0 software (earlier versions will also work)
Tripod with bungees or air table to protect AFM from environmental vibrations
Probes for tapping in air (Nanosensors PointProbe® Plus)
3.2.2 Procedure
Turn on the AFM controller.
Open the NanoScope software and begin a Tapping in Air protocol. Select a Capture Directory for files to be saved. Basic steps of the procedure are outlined on the left of the window.
Begin with Setup. Enter probe information if desired.
On the microscope itself, switch the mode to AFM/LFM. Insert a fresh probe into the probe holder (Fig. 3D). Adjust the laser and mirror positions for the maximum signal intensity.
Remove the AFM head by releasing the springs on either side and place the mica onto the magnetic sample pedestal. Carefully, replace the AFM head and reattach the springs. The probe is now positioned over the sample on the scanner (Fig. 3E).
Lower the tip to ~50–100 µm above the mica surface. When using the NanoSensors PointProbe® Plus, this can be estimated as roughly half the length of the cantilever that is visible.
Use the AFM knobs to adjust the laser position on the detector. The display on the AFM should read as close to 0 as possible for both vertical and horizontal differences.
Switch the AFM to Tapping/TFM mode and carefully transfer the AFM to the bungee setup (Fig. 3A).
Still on the Setup step in NanoScope, press Auto Tune. Verify that the Drive Amplitude is less than 100 mV. It may be necessary to use Manual Tune to achieve appropriate settings.
Click on the next step in NanoScope: Check Parameters. Begin with the following settings: scan size: 0.00 nm; aspect ratio: 1.00; X offset: 0.00 nm; Y offset: 0.00 nm; scan rate: 3.26 Hz; Samples/Line: 512; Lines: 512.
Click on the next step in NanoScope: Engage. The tip will lower toward the mica until it engages and begins tracking the sample. Ensure you are scanning in the height channel. Because the scan size is set to 0, the surface should appear completely flat. Verify that the Trace and Retrace curves are both sufficiently flat.
Click on the Withdraw step in NanoScope and then return to Setup. Repeat steps 9–11.
Increase the scan size to 1000 nm. Press the Frame Up or Frame Down arrows to begin at the bottom or top of the field, respectively.
While scanning, capture the current field by pressing the camera button (Capture). The status bar will read “Capture: On” for the duration of the scan, and “Capture: Done” when it is complete and the file has been saved to the Capture Directory. The status bar will read “Capture: Next” if parameters have been changed within the current scan, such as scan size or offset.
After each image, change the X and Y offsets to move to a new area on the mica and capture a new image. We suggest moving by 1.1 µm each time to account for drift and avoid redundancy.
Open raw 001 files in NanoScope Analysis. Flatten the images using the Flatten tool. Select first- (line by line) or second (to correct for bowing effect)-order flattening and press Execute. Adjust data scales and colors as desired. Save changes.
To export BMP files of the height images, select the desired files in the Browse Menu. Right-click > Export… > bmp.
When imaging is complete, withdraw the trip (press Withdraw several times) and remove the AFM from the bungee setup. Close the NanoScope software and then shut off the controller.
3.2.3 Notes
When inserting probes and samples, be careful to keep the tip position high off the surface to prevent accidental damage.
Analysis can be done on any of the following file formats: BMP, TIFF, and JPEG.
3.3 Data Analysis
3.3.1 Equipment
ImageJ (https://imagej.nih.gov/ij/, NIH)
Image SXM
Microsoft Excel
GraphPad Prism 7 or other data analysis program
3.3.2 Procedure
Open the image file (Section 3.2.2, step 17) with ImageJ.
- Label protein–DNA complexes using the Text Tool. Use the following criteria to ensure that all data points will provide reliable information:
- The protein–DNA complex is larger than unbound DNA (typically in both height and area).
- Entire DNA molecule is visible in the image. It must not continue past the edge of the image nor overlap with other molecules.
- DNA is the correct length. This can be judged initially by eye, and again when measuring the contour length (step 3). DNA molecules within 10% of the expected length can be counted.
Measure DNA contour length and protein-binding position (Fig. 4B). In Image J, use the Segmented Line Tool (right-click on Straight Line to select Segmented Line) to measure the contour length and binding position. Left-click to begin the line and add vertices, along the length of the DNA; right-click to end the line. Select Analyze > Measure (or use shortcut “m”) to add the current length measurement in units of pixels to the Results window. If images were captured as above, the conversion factor 1000 nm/512 pixels should be used to calculate length in appropriate units. Measure total contour length of the DNA molecule, as well as the length from the bound protein to the closest DNA end. Protein-binding position can be reported as percent from one end of the total DNA contour length, P = (100 × length from DNA end to protein)/total DNA contour length. Repeat for all labeled complexes.
Measure DNA bend angle (Fig. 4C). Use the Angle Tool in ImageJ to measure the DNA bend angle at the bound protein. Left-click to create the three points of the angle; these may be adjusted by dragging the points as desired. Place the middle point at the center of the bound protein, such that the angle measures the bend in the DNA immediately adjacent on either side. Select Analyze > Measure (or use shortcut “m”) to add the angle measurement (α) to the Results Window. DNA bend angles are typically reported as β = 180 − α. Repeat for all labeled complexes.
Measure complex volume (Fig. 4A). Open the height channel from the flattened .001 file in Image SXM. Select Analyze > Show Histogram to display the distribution of heights in the image; record the mean (histogram peak) as the background for the image. Locate protein–DNA complexes (identified in step 2) and draw a line around the footprint with the eraser tool to demarcate it from naked DNA. Then, select Options > Density Slice to define the thresholds for analysis; set the upper threshold to its maximum and drag the lower threshold such that all particles (DNA and proteins) are highlighted with minimal background noise. To count particles, select Analyze > Analyze Particles and choose the following settings: Min Particle Size (pixels): 15; Max Particle Size (pixels): 999,999; Label Particles; Ignore Particles Touching Edge; Include Interior Holes; Reset Measurement Counter. Then, select Analyze > Measure, followed by Analyze > Show Results. This will open a new Results window, which can be copied into an Excel spreadsheet. “Mean” and “Area” are the average height of the particle and the area of its footprint, respectively; ensure that these values are reported in nm. Volumes (nm3) are calculated as V = (mean − background) × area.
Generate histograms of the binding positions (P), DNA bend angles (β), and complex volumes (V) using GraphPad Prism. Create a column table of the data and, in the Analysis toolbox, select Analyze > Column analyses > Frequency distribution. Adjust bin centers and widths as appropriate for the sample size (typically, the number of bins should be approximately √n). Under the New graph heading, select Create a new graph of the results and change the graph type to XY graph, Histogram spikes.
Viewing the histogram, select Fit a curve with nonlinear regression from the Analysis toolbox. Under the Fit tab, select Gaussian > Gaussian. This will generate a new page showing the parameters for the Gaussian fit to the histogram data. The best-fit values for mean and standard deviation can be used to describe the properties of the bound proteins.
Volume data can be further processed to infer molecular weights, and thus binding stoichiometry. Because AFM volumes are directly proportional to MW for most globular proteins (Ratcliff & Erie, 2001; Schneider, Larmer, Henderson, & Oberleithner, 1998), a standard curve can be generated and used for all experiments with the same probe type and mode of data collection (Fig. 4E). Using the center of the fitted Gaussian as the mean volume, calculate the experimental MW to determine stoichiometry.
3.3.3 Notes
It may be useful to have the file open in the NanoScope Analysis software as well. The 3D view is helpful when identifying proteins on DNA, particularly in the case of smaller proteins.
In step 5, the minimum particle size may vary depending on the protein being studied and the threshold settings applied.
Sample data from different NER proteins are shown in Fig. 4. Wild-type Rad4–Rad23 binding to a 538-bp DNA substrate was analyzed for protein-binding position (Fig. 4F) and DNA bend angle (Fig. 4G) (Kong et al., 2016). A mutant form of UV-DDB (127 kDa) binding to a 538-bp DNA substrate was analyzed for protein volumes (Fig. 4D); the mean volume corresponds to a MW of 388.6 kDa, which suggests that the protein was bound as a dimer (Ghodke et al., 2014).
4. SINGLE-MOLECULE DNA TIGHTROPE ASSAY
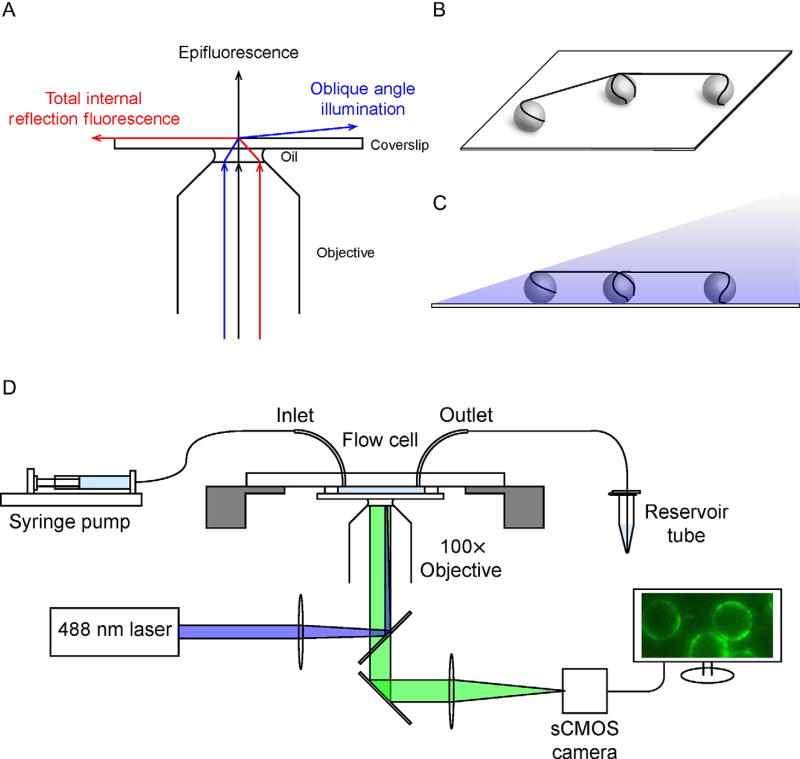
To eliminate the need for constant flow and the potential of surface interactions, we have developed a unique optical platform, based on the ability to anchor both ends of a long DNA molecule on two nearby micronsized poly-l-lysine-coated silica beads via electrostatic interaction, with the rest of the DNA suspended in between them, forming DNA tightropes (Fig. 5) (Kad et al., 2010). While the procedure involved does not offer the degree of precision and control afforded by the nanofabrication process used in constructing flow cells for DNA curtain assays (Gorman et al., 2007; Graneli, Yeykal, Robertson, & Greene, 2006; Lee et al., 2015; Sternberg, Redding, Jinek, Greene, & Doudna, 2014), its implementation is relatively straightforward. The DNA tightrope assay also elevates the DNA molecules, and therefore protein–DNA interactions, away from the coverslip, allowing complete access to elongated DNA in space and minimizing any potential adverse surface effects. To illuminate protein–DNA interactions taking place microns above the surface, a subcritical, oblique angle must be used to maximize the signal-to-noise ratio (Konopka & Bednarek, 2008; Tokunaga, Imamoto, & Sakata-Sogawa, 2008). Since its inception, we and others have utilized the DNA tightrope platform extensively to characterize proteins involved in prokaryotic and eukaryotic nucleotide and base excision repair, as well as telomere shelterin complex components TRF1 and TRF2 (Dunn, Kad, Nelson, Warshaw, & Wallace, 2011; Ghodke et al., 2014; Hughes et al., 2013; Kong et al., 2016; Lin et al., 2014, 2016; Nelson et al., 2014). Due to the oblique angle illumination, the tightrope platform requires the use of Qdots to label proteins and provide sufficient fluorescence for visualization. These fluorescently stable and brilliant nanoparticles allow continuous imaging at rates of 10–100 frames per second for collection periods of minutes without any photobleaching. Preparation of the flow cell begins with precoating clean coverslips with polyethylene glycol (Sections 4.1 and 4.2) and assembling predrilled microscope slides with inlet and outlet tubing (Section 4.3). Flow cells are constructed by attaching the coverslip to the slide assembly via a double-sided tape spacer (Section 4.5). Microspheres are simply flowed in such that they are distributed randomly but uniformly throughout the imaging area. Following deposition of the silica beads, tightropes are set up by continuously flowing DNA back and forth inside the flow cell for 40–60 min at the rate of 0.3 mL/min (Section 4.6). This step allows one end of the negatively charged DNA molecule to anchor to a positively charged bead, while the rest of the molecule is elongated by hydrodynamic force in the flow. With bead density optimized for length of DNA substrate used, the free end of the DNA molecule can attach to another bead in the vicinity. Proteins are visualized by Qdot labeling (Section 4.7), which is achieved either by conjugating a streptavidin-coated Qdot to a biotinylated antibody that recognizes the affinity tag on the protein (Ghodke et al., 2014; Kong et al., 2016) or through an antibody sandwich approach that utilizes a primary antibody against the affinity tag on the protein combined with a secondary antibody-coated Qdot (Kad et al., 2010; Wang, Tessmer, Croteau, Erie, & Van Houten, 2008). Data are collected, exported, and analyzed with a combination of software and scripts (Sections 4.8 and 4.9).
Fig. 5.
Schematics of the DNA tightrope assay. (A) Schematic ray diagram of incident laser light paths for epifluorescence (black), total internal reflection fluorescence (TIRF) at the critical angle (red), and oblique angle illumination (blue). (B) Schematic of 5 µm poly-l-lysine-coated microspheres deposited on a glass coverslip with DNA tightropes suspended between them. (C) Schematic of DNA tightropes in the flow cell under oblique angle illumination. (D) Schematic of the experimental setup for the DNA tightrope assay. The flow cell is connected on the one end (inlet) to a syringe mounted on a syringe pump, while the other end (outlet) is connected to an Eppendorf tube reservoir. DNA tightropes in the flow cell are illuminated by a 488 nm laser (blue) under oblique angle through a 100× objective. Fluorescence signal (green) is imaged on a sCMOS camera connected to a computer.
4.1 Cleaning Coverslips
4.1.1 Equipment
Ultrasonic cleaning bath (Branson)
Glass or plastic staining jars
4.1.2 Buffers and Reagents
20% Liquinox (ALCONOX)
100% acetone
100% ethanol
1 M potassium hydroxide (KOH)
Coverslips (No. 1½, 24 × 40 mm, Corning)
4.1.3 Procedure
Load coverslips into staining jars and fill with 20% Liquinox detergent solution. Sonicate for 60 min.
Dump out detergent solution and rinse coverslips under deionized water until suds no longer form. Then fill staining jars with deionized water and sonicate for 5 min.
Replace deionized water in staining jars with acetone. Sonicate for 15 min.
Pour off acetone and rinse coverslips thoroughly under deionized water. Then fill staining jars with deionized water and sonicate for 5 min.
Replace deionized water in staining jars with 1 M KOH solution. Sonicate for 15 min.
Pour off and save KOH solution. Rinse coverslips thoroughly under deionized water. Then fill staining jars with 100% ethanol. Sonicate for 15 min.
Pour off ethanol and rinse coverslips thoroughly under deionized water. Then fill staining jars with 1 M KOH solution saved from the previous step. Sonicate for 15 min.
Pour off KOH solution and rinse coverslips thoroughly under deionized water. Then fill staining jars with deionized water and sonicate for 15 min.
Replace the deionized water in staining jars. Slides can be stored in water until they are to be used.
4.1.4 Notes
Do not allow coverslips to sit in 1 M KOH solution for prolonged time as they can be slowly etched by the solution.
4.2 PEGylation of Coverslips
4.2.1 Equipment
Ultrasonic cleaning bath (Branson)
Glass or plastic staining jars
4.2.2 Buffers and Reagents
Aminosilane solution (for eight coverslips, 1.0 mL (3-aminopropyl) triethoxysilane, 2.5 mL glacial acetic acid, and 50 mL methanol, scale up if needed)
10 mM NaHCO3, adjusted to pH ~8.5
PEG solution (25 mg mPEG-succinimidyl valerate, MW 5000 (Laysan Bio) dissolved in 96 µL of NaHCO3 solution)
Compressed nitrogen gas
4.2.3 Procedure
Dry the cleaned coverslips completely with compressed nitrogen gas.
Let the coverslips sit in aminosilane solution for 20 min total. After 10 min, sonicate for 1 min and then sit for the remaining 9 min.
Pour off aminosilane solution and rinse the coverslips thoroughly under deionized water and dry with compressed nitrogen.
Prepare an empty tip box: fill it with deionized water up to a depth of ~1 cm and soak a piece of paper towel in the water.
Take a dry coverslip, mark the side that is not to be PEGylated with marker. Lay the coverslip marked-side-down on the tip rack. Take another coverslip, mark the side that is not to be PEGylated with marker. Set it aside, marked-side-down.
To create a coverslip “sandwich,” deposit 20 µL of the PEG solution in the middle of the coverslip on the tip rack. Lay the other coverslip on top, marked-side-up. The liquid should spread out evenly between the two coverslips without forming any bubbles.
Repeat steps 5 and 6 for the remaining coverslips.
Shield the tip box from light with aluminum foil and place it in a dark place at room temperature overnight.
Disassemble the coverslip “sandwiches,” place the coverslips in staining jars, and rinse them thoroughly under deionized water.
Blow dry coverslips with nitrogen gas and place them back on the tip rack, PEGylated side up (marked-side-down).
Cover the tip box with aluminum foil. PEGylated coverslips can be stored at 4°C for 2 weeks.
4.2.4 Notes
NaHCO3 and PEG solutions can be prepared during step 2. PEG is especially light sensitive when in solution and should be protected from light.
After step 3, coverslips may be stored in methanol if the PEG solution is not ready.
Steps 6 through 10 should be carried out in a dark environment.
PEGylation (step 8) can be as short as 3 h, but overnight is preferred.
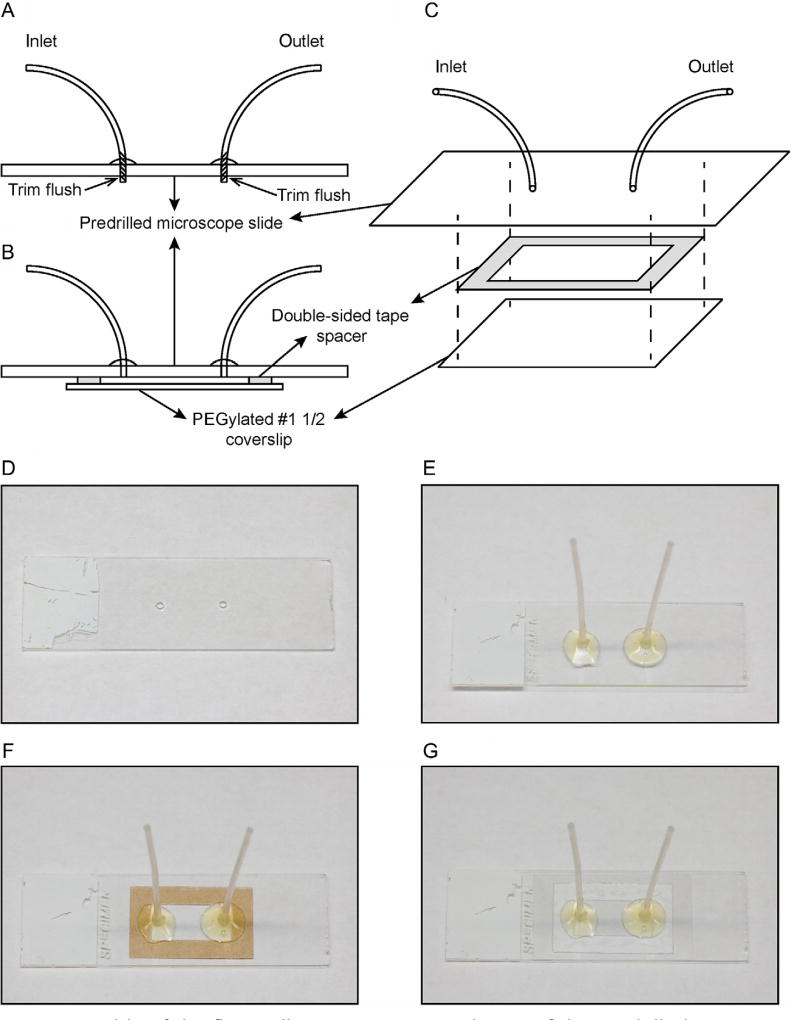
4.3 Assembly and Disassembly of Slides With Tubing (See Fig. 6)
Fig. 6.
Assembly of the flow cell. (A) Cross-sectional view of the predrilled microscope slide with inlet and outlet tubing attached. (B) Cross-sectional view of the assembled flow cell, where the double-sided tape spacer is sandwiched between the slide assembly and the PEGylated coverslip. (C) Exploded view of the flow cell assembly. (D) Microscope slide with predrilled holes. (E) Microscope slide with inlet and outlet tubing attached. (F) Microscope slide with inlet and outlet tubing and rectangular double-sided tape spacer (brown). (G) Complete flow cell assembly with glass coverslip attached to the microscope slide.
4.3.1 Equipment
Ultrasonic cleaning bath (Branson)
Glass or plastic staining jars
Benchtop drill press and 1.25 mm diamond drill bit
Extra fine grit sanding sponge (3M)
4.3.2 Buffers and Reagents
Microscope slides (25 × 75 × 1 mm, Thermo Scientific)
Teflon PFA tubing (1/16″ OD × 0.030″ ID, IDEX)
Adhesive (BONDiT B-45TH, RELTEK)
Slides cleaning solution (1 M HCl and 20% ethanol)
100% acetone
100% ethanol
4.3.3 Procedure
Drill two holes, 15–16 mm apart horizontally, in the center of a microscope slide (Fig. 6A and D). The precise distance between the holes is dependent on the desired size of the usable flow cell area.
Cut two pieces of the Teflon tubing to size, ~3 cm each. Rough up one end (~5 mm) of each piece of tubing with the sanding sponge for better adhesion.
Thread the roughed-up ends of the tubing through the holes in the slide. Apply adhesive around the base. Allow the ends to protrude ~1 mm from the other (bottom) side of the slide (Fig. 6A and E). This ensures that should some adhesive seeps through, it will not block the tubing.
Set the assembled slides aside at room temperature for at least 24–48 h to allow the adhesive to cure completely.
Drilled slides and Teflon tubing may be reused. For disassembly, submerge the flow cell (see below) in acetone for 1–2 days until it falls apart. Keep the slide and tubing and discard everything else. Remove any residual adhesive from the slide with a razor blade or KimWipe soaked in acetone.
In a staining jar, submerge used slides in acetone and sonicate for 1 h.
Discard acetone, rinse the slides thoroughly under deionized water, and fill the staining jar with the slides cleaning solution (1 M HCl and 20% ethanol). Sonicate for 1 h.
Discard the cleaning solution, rinse the slides thoroughly under deionized water, and fill the staining jar with 100% ethanol.
Wipe dry slides with KimWipes. Any remaining adhesive on the slides should be rubbed off with KimWipes and 100% ethanol.
4.3.4 Notes
It may be helpful to drill holes in the slide while it is submerged in water in order to help reduce the probability of slides cracking.
Some adhesives may cure faster (i.e., overnight) if the assembled slides are left in a 37°C incubator.
4.4 Preparation of Poly-l-Lysine-Coated Beads
4.4.1 Equipment
Benchtop centrifuge
Vertical rotators
4.4.2 Buffers and Reagents
5 µm silica microspheres (Polysciences)
Poly-l-lysine powder (Waco Chemicals)
4.4.3 Procedure
Resuspend 100 µL of beads in 500 µL of ddH2O. Centrifuge at 4°C for 4 min at 12,000 rpm.
Remove supernatant and resuspend beads in 400 µL of 2.5 mg/mL poly-l-lysine solution.
Rotate end to end at 4°C overnight on a vertical rotator.
4.4.4 Notes
2.5 mg/mL poly-l-lysine solution is made in ddH2O and can be stored at −20°C.
Poly-l-lysine-coated beads can be stored at 4°C.
4.5 Flow Cell Assembly (See Fig. 6)
4.5.1 Equipment
Benchtop centrifuge
Low-magnification light microscope
4.5.2 Buffers and Reagents
Assembled predrilled slide with tubing
Double-sided tape spacer
PEGylated coverslip
200 µL gel-loading tips
Blocking buffer (10 mM HEPES, pH 7.5, 50 mM NaCl, 1 mg/mL bovine serum albumin (Roche))
Poly-l-lysine-coated silica beads
4.5.3 Procedure
Take a clean slide and use a razor blade to cut the protruding ends of tubing flush with the slide. Scrape back and forth to ensure that the bottom side of the slide is flat and smooth.
Cut out a double-sided tape spacer with a razor blade. Peel one side and paste it to the slide, using fingernail to firmly press the sticky tape (Fig. 6C and F).
Take one PEGylated coverslip from 4°C storage. Make sure that there is no excessive condensation or water on the treated (unmarked) surface. Hold the coverslip on its edges with fingers so that any condensation on the treated side evaporates quickly. Wipe the untreated (marked) surface dry with KimWipes.
Peel off the adhesive backing, make sure that the coverslip is completely dry, and place the PEGylated coverslip over the sticky tape spacer. Make sure the edges of the coverslip do not extend beyond those of the slide underneath it. Again, using the thumbnail, gently press around the outline of the spacer (Fig. 6B, C, and G).
With a 200-µL gel-loading tip, fill the flow cell with ~100 µL of the blocking buffer (10 mM HEPES, pH 7.5, 50 mM NaCl, 1 mg/mL BSA). Block the flow cell for 10 min.
After 10 min of initial blocking, examine the flow cell to ensure that no leakage has occurred, and then prepare the beads while blocking continues. First, vortex and resuspend the stock of beads in poly-l-lysine solution.
Add 13–15 µL of bead stock to 400 µL of ddH2O. Resuspend again by vortexing and then centrifuge at 12,000 rpm for 4 min at 16°C. Carefully discard the supernatant without disturbing the pellet.
Repeat the washing step above with another 400 µL of ddH2O. This time, after centrifugation, take out 300 µL of ddH2O and then resuspend beads in the remaining ~110 µL.
Immediately after mixing, pipette ~110 µL of the suspension slowly into the flow cell with a gel-loading tip. Collect the bead flow-through and recirculate once if necessary.
Check the distribution and density of deposited beads in the flow cell with a low-magnification light microscope. Add more beads if necessary.
Allow the beads to settle for 10 min, and then flow 200 µL of ddH2O through the flow cell to wash away any free beads.
4.5.4 Notes
Spacers can be prepared by folding a piece of double-sided tape (3M) onto itself to double the thickness and create two adhesive sides with removable backing. A nested-rectangle design pattern is then cut from the tape to make spacers. The outer rectangle should be slightly less than the size of the coverslip. The size of the inner rectangle corresponds to the usable flow cell area and should be large enough to encompass the predrilled holes in the microscope slide.
Poly-l-lysine-coated beads settle and clump together easily if left unperturbed. To ensure reproducible results, any pipetting should be done immediately after resuspension and vortexing. This is especially important in step 10.
It is useful to keep in mind the length of DNA tightropes to be used in the system when checking bead distribution and density. In order to determine whether enough beads have been deposited on the coverslip, compare the expected DNA tightrope length to inter-bead distances, which can be estimated based on known bead diameters.
The amounts of beads required may need to be further optimized with respect to the person carrying out this protocol.
4.6 Preparation of DNA Tightropes
4.6.1 Equipment
Syringe pump (WPI)
5-mL glass syringe or plastic syringe (Hamilton)
21G hypodermic needle (BD)
Teflon PFA tubing (1/16″ OD × 0.030″ ID, IDEX)
Union assembly (0.020 through hole, for 1/16″ OD, IDEX)
Flangeless ferrule (for 1/16″ OD, IDEX)
4.6.2 Buffers and Reagents
Assembled flow cell
1× TR buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 3 mM MgCl2)
Long DNA substrate (λ-DNA or defined lesion substrates)
4.6.3 Procedure
Assemble the syringe, needle, tubing, and all fitting pieces. Wash the system by flowing ~5–6 mL of 1× TR buffer through it. Leave ~1 mL in the syringe.
Set up and secure the syringe on the syringe pump. Set the flow rate to 0.3 mL/min, volume = 100 µL.
Connect the flow cell to the syringe by first pushing the TR buffer in the system through until the solution starts to drip from the female fitting piece that is to be connected. Quickly attach the fitting piece on the flow cell to that on the tubing.
Attach the outlet tubing to the other side of the flow cell and set up a predrilled Eppendorf tube as the reservoir. Add 500 µL of TR buffer to the reservoir tube and withdraw until there is only 1–2 µL left.
Thaw out DNA tightrope substrate, make up the volume to 100 µL with 1× TR buffer. Vortex to resuspend well and spin down briefly to collect. Add DNA to reservoir tube and withdraw all.
Add 250 µL of 1× TR buffer. Withdraw 100 µL to push the DNA from the outlet tubing into the flow cell. Set up the program to the continuous push–pull cycle (infusion followed by withdrawal) at the rate of 0.3 mL/min for a total volume of 100 µL in each direction.
Pause the syringe pump after 40–60 min of the continuous cycle.
If using ligated defined lesion damage arrays, wash the flow cell with 200 µL of 1× high-salt TR buffer containing 1 M NaCl to remove DNA-bound ligase carried over from the ligation reaction. Then equilibrate the flow cell with 400 µL of protein binding buffer.
4.6.4 Notes
Introduction of air bubbles during step 3 is a common cause of failure. Attaching the flow cell to the tubing in a swift and smooth manner usually leads to better results. It is important to inspect the flow cell after step 3 for the presence of air bubbles. Small air bubbles trapped in the tubing that is attached to the flow cell can be backed out into the syringe and will not cause any issues downstream. A large column of air pushed into the flow cell will displace deposited beads, rendering the flow cell unusable.
The combination of the flow rate (0.3 mL/min) and time (40–60 min) of the continuous cycle employed to string up DNA tightropes has been shown to not overstretch DNA (Kad et al., 2010). Different combinations can also be explored for potential effects on DNA tightrope conformations and protein binding.
4.7 Protein Conjugation
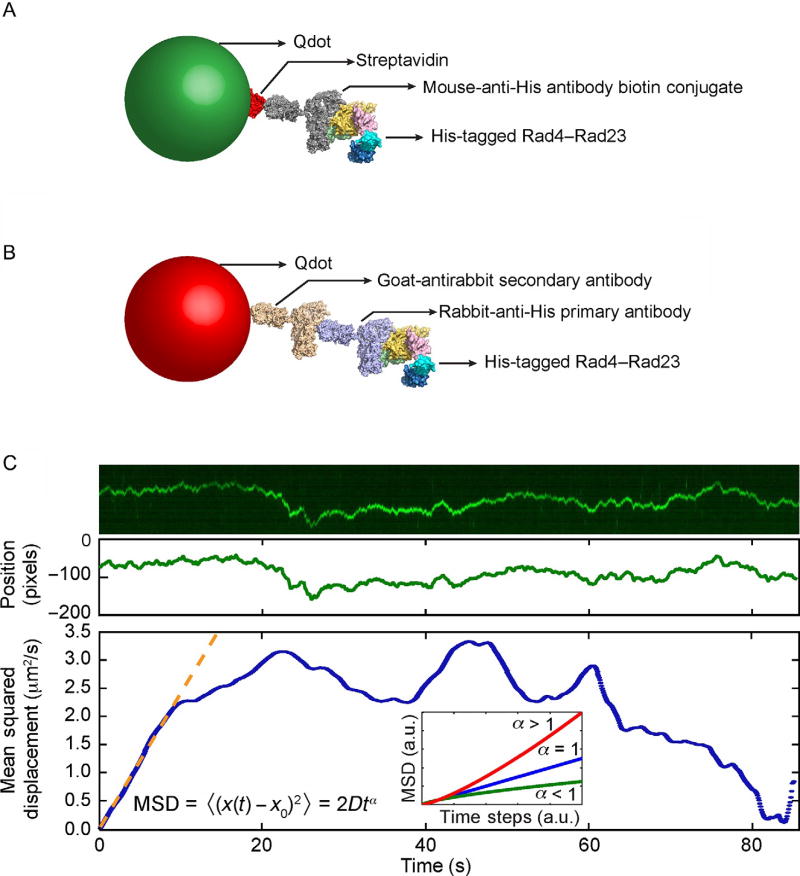
The use of oblique angle illumination for probing of protein–DNA interactions on tightropes that are suspended 5 µm above the surface requires the use of fluorescent probes that are exceptionally bright. Bioconjugated Qdots or QDs are commercially available and possess characteristics such as broad excitation spectrum and narrow size-dependent emission spectrum, as well as excellent brightness and photostability, all of which are highly beneficial to single-particle tracking (Bruchez, 2011). We have developed several approaches to label affinity purified proteins with Qdots for imaging on the tightrope platform, two of which are shown in Fig. 7. The first strategy takes advantage of the highly specific streptavidin–biotin interaction by conjugating streptavidin-coated Qdots with biotinylated antibodies against the affinity tag used in the purification of the protein of interest (Fig. 7A). Under certain circumstances, the placement of a relatively large Qdot close to the protein of interest may interfere with its ability to interact with other proteins or DNA. To prevent potential steric hindrance, we also developed the antibody sandwich approach, where a primary antibody against the affinity tag on the protein serves as the linker between a secondary antibody-coated Qdot and the affinity-tagged protein of interest (Wang et al., 2008) (Fig. 7B). Both approaches are straightforward to implement in one-color imaging of protein on λ-DNA or defined lesion substrates without biotin in the damage-containing oligonucleotide. However, to image more than one protein, it is essential to ensure that the Qdots on those proteins cannot exchange. The antibody sandwich approach can be easily adapted to this situation by using an orthogonal set of species of antibodies, i.e., goat-antimouse secondary antibody Qdots paired with mouse-anti-6×His primary antibody, and goat-antirabbit secondary antibody Qdots paired with rabbit-anti-6×His primary antibody. We have had great success at imaging two colors using this approach (Hughes et al., 2013), and depending on the optical setup with appropriate splitters and the number of protein tags, as many as six uniquely Qdot-labeled proteins could be feasibly imaged simultaneously.
Fig. 7.
Qdot conjugation strategies and data analysis. (A) Streptavidin (red)-coated quantum dot (green) is conjugated to His-tagged Rad4–Rad23 via the biotin-conjugated mouse-anti-His antibody (gray). (B) His-tagged Rad4–Rad is labeled by goat-antirabbit secondary antibody (wheat)-conjugated quantum dot (red) via a rabbit-anti-His primary antibody (purple). (C) Top: Representative kymograph of a diffusing particle. Middle: Plot of position, in the units of pixels (1 pixel = 46 nm), vs time, after fitting the light intensity profile at each time point in the kymograph with a one-dimensional Gaussian. Bottom: Plot of mean squared displacement (MSD), calculated from Gaussian-fitted positions, vs time steps. Orange dashed line is the result of fitting the initial portion of the MSD curve to the equation MSD = 2Dtα. Inset: three types of one-dimensional diffusion characterized by different α values: superdiffusion (red), random diffusion (blue), and subdiffusion (green). Based on Movie 1 in the online version at http://dx.doi.org/10.1016/bs.mie.2017.03.027. Adapted with permission from Kong, M., & Van Houten, B. (2016). Rad4 recognition-at-a-distance: Physical basis of conformation-specific anomalous diffusion of DNA repair proteins. Progress in Biophysics and Molecular Biology. http://dx.doi.org/10.1016/j.pbiomolbio.2016.12.004 (fig. 2C).
4.7.1 Equipment
Benchtop centrifuge
4.7.2 Buffers and Reagents
1 µM Qdot (streptavidin- or secondary antibody-conjugated, Invitrogen)
1 µM biotin-conjugated anti-His antibody (Qiagen) or other anti-His primary antibody
His-tagged protein of interest
Protein storage buffer
4.7.3 Procedure
- For streptavidin-conjugated Qdots (SAQD): incubate 1 µL of 1 µM of SAQD with 5 µL of 1 µM of biotin-conjugated anti-His antibody (HisAb) so that the molar ratio of SAQD:HisAb is 1:5. Allow the binding reaction to proceed at 4°C for 1 h.
- For secondary antibody-conjugated Qdots (IgGQD): incubate 1 µL of 1 µM of anti-His primary antibody with 1 µL of 1 µM of the His-tagged protein of interest and make up the volume with protein storage buffer to 5 µL. The molar ratio of protein:antibody is 1:1. Allow the binding reaction to proceed at 4°C for 1 h.
- For SAQD: incubate 1 µL of the mixture prepared in step 1(a) with 1 µL of 1/6 µM of the protein of interest, such that the molar ratio of SAQD:HisAb:protein is 1:5:1 and the final concentration of the protein is ~83 nM. Allow the binding reaction to proceed at 4°C for 1 h.
- For IgGQD: incubate 1 µL of the mixture prepared in step 1(b) with 1 µL of 1 µM of IgGQD with the appropriate secondary antibody, such that the molar ratio of IgGQD:HisAb:protein is 5:1:1 and the final concentration of the protein is ~100 nM. Allow the binding reaction to proceed at 4°C for 1 h.
4.7.4 Notes
Depending on the stability of the protein of interest, conjugation steps may be carried out at room temperature to speed up the reaction.
Agarose gel-based electrophoretic mobility shift assays should be carried out with short DNA substrate, protein of interest, and each intermediate step of the Qdot conjugation protocol (i.e., protein with primary antibody, protein with primary antibody and Qdot) to ensure that DNA-binding activity is not lost due to conjugation of Qdot (Ghodke et al., 2014).
4.8 Data Collection
4.8.1 Equipment
Benchtop centrifuge
Inverted fluorescence microscope (Nikon Ti) with 100 × oil-based high-NA objective for TIRF-M, appropriate filter set for the wavelengths of Qdots used (optional), and high-speed sCMOS camera (Andor).
Microscope user interface and image collection software (NIS-Elements Ar, Nikon)
4.8.2 Buffers and Reagents
1 × TR buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 3 mM MgCl2)
1 × high-salt TR buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 3 mM MgCl2, 1 M NaCl)
Protein binding buffer
QD-conjugated protein of interest
Immersion oil
4.8.3 Procedure
Set up and secure the flow cell in the holder of the translational stage on the microscope, using immersion oil with appropriate 100 × objective lens. Focus the objective on the beads that have been deposited on the coverslip. Turn on any focus drift compensation if applicable (Perfect Focus System, Nikon).
Equilibrate the flow cell by passing through 4 volumes (400 µL) of protein binding buffer from the reservoir tube.
Dilute 1 µL of QD-conjugated protein of interest in 100 µL of protein binding buffer. Pipette the diluted protein solution into the reservoir tube and withdraw all. The final concentration of QD-labeled protein is ~1 nM.
Pipette 100 µL of protein binding buffer into the reservoir tube and withdraw all, such that the protein solution is pushed into the flow cell.
Turn on the excitation laser and find the critical TIRF angle where QD fluorescence can just begin to be observed. Then increase the angle slightly to optimize for signal-to-noise ratio.
Move the translational stage to look for binding events in the live-view window.
After locating a region of interest, set up recording frame rate (~10 fps), time (~5–15 min), and file directory. If more than one Qdot wavelength is used, configure the emission filter as needed. Record the time series.
Refresh QD-labeled proteins at least every 2 h, depending on the stability of protein while under the microscope.
4.8.4 Notes
When using tandem-ligated plasmid DNA substrates, wash the flow cell with 200 µL of 1 × high-salt TR buffer containing 1 M NaCl prior to equilibration with protein-binding buffer to remove DNA-bound ligase carried over from the ligation reaction.
It is important to perform negative controls with QD–HisAb complexes only, in the absence of protein conjugation, to confirm that they do not stick to DNA in a nonspecific manner.
If DNA binding is rare, consider increasing the concentration of QD-labeled protein in the flow cell. The empirical maximum concentration of fluorescent Qdots, including both free and protein-conjugated, is ~10 nM. Background fluorescence from freely diffusing Qdot could become overwhelming above this limit.
4.9 Data Analysis
4.9.1 Equipment
Image processing software (NIS-Elements Ar or NIS-Elements Viewer, Nikon)
ImageJ (https://imagej.nih.gov/ij/, NIH)
Data processing and fitting software (Matlab, MathWorks)
4.9.2 Procedure
Convert manufacturer-specific proprietary image stack file format (.nd2, Nikon) to a time series of individual TIFF files. Separate the channels if multiple Qdot emission wavelengths are used.
Import the time series of TIFF files as an Image sequence in ImageJ. Save the image stack in ImageJ as a single TIFF file. For an example of a time series, see Movie 1 in the online version at http://dx.doi.org/10.1016/bs.mie.2017.03.027.
Using the Straight line tool in the tool bar, trace the linear trajectory of one-dimensional diffusion of one QD-labeled particle. Ensure that the length of the line covers the entire range of motion.
Press the “/” key or go to Image > Stacks > Reslice. In the Reslice window that pops up, check the box Rotate 90 degrees. Click OK to generate a kymograph that displays the particle position (on the vertical axis) over time (on the horizontal axis). Save the kymograph as a TIFF file.
Fit the fluorescence intensity in the kymograph with a one-dimensional Gaussian fitting algorithm in ImageJ. Save the Gaussian-fitted peak positions (Fig. 7C).
- In Matlab, or other appropriate data processing software, import the Gaussian-fitted peak positions and calculate the one-dimensional mean square displacement (MSD) as a function of time steps
where N is the total number of frames in the time series, n is the number of frames for different time steps, xi is the Gaussian-fitted peak position in the ith frame, and Δt is the unit time step between consecutive frames, i.e., the inverse of the frame rate. - Extract diffusion coefficient D and anomalous diffusion exponent α from the MSD by fitting the equation
Begin the fitting process by using all available data points in the MSD curve. In each round of fitting, reduce the number of data points used by one, taken from the end of the MSD curve, until desired goodness of fit is achieved (Fig. 7C). For an example of Matlab script, see MSD_main.m in the supplementary file (http://dx.doi.org.10.1016/bs.mie.2017.03.027).
4.9.3 Notes
It is important to first establish the systematic noise level of the platform, in terms of the one-dimensional diffusion coefficient value of stably bound nonmotile Qdots on the tightrope.
By analyzing the component of diffusive motion that is along the direction of the tightrope (longitudinal), an implicit assumption is made that particle motion perpendicular to the tightrope (transverse) is at the background noise level. This assumption can be verified by observing that the particle of interest, motile or nonmotile, does not exhibit any kind of “wobble” on the tightrope, whose direction is in general parallel to that of the hydrodynamic flow expected in the flow cell. Quantitatively, two-dimensional tracking of the particle can be employed to determine its x and y positions. In practice, particles that exhibit any “wobble” on the tightrope should be excluded from further analysis as the behavior indicates that the tightrope itself is not anchored properly on beads or has structural defects.
In the case of multiple binding events on one DNA tightrope, it is beneficial to extract the kymographs of all particles on the tightrope, motile and nonmotile, by drawing one straight line through all the particles. Kymographs of individual particles can be cropped out and analyzed independently.
An ImageJ script for one-dimensional Gaussian fitting is available for download at http://kadlab.mechanicsanddynamics.com/images/Downloads/Gaussian_Fit.txt (Kad et al., 2010).
Resolution of the system can be characterized by the positional accuracy (Thompson, Larson, & Webb, 2002) and localization precision (Arnspang, Brewer, & Lagerholm, 2012). Calculations of these quantities relevant to the tightrope platform have been detailed elsewhere (Ghodke et al., 2014).
5. CONCLUSIONS
In summary, we have established a complete laboratory workflow from bulk biochemistry to single-molecule biophysics. The experimental platform detailed in this chapter is well suited for characterization of not just proteins involved in NER, but protein–DNA interactions in general. Specifically, the DNA tightrope assay is straightforward to implement and its versatility allows the technique to be applied to investigate repair pathways such as base excision repair and mismatch repair, as well as the target search process of telomere shelterin complex proteins (Lin et al., 2014). Tightropes have also been constructed from actin filaments to study the cooperative activation of thin filaments (Desai, Geeves, & Kad, 2015). In addition to the dynamic and transient behavior observable on DNA tightropes, the use of AFM allows independent snapshot measurements of specific and nonspecific binding in the absence of any labeling fluorescent probes and visualization of any mechanical changes in DNA conformation that can be induced through protein binding. Both complementary techniques benefit greatly from the utilization of defined lesion substrates such that specific binding events can be more readily differentiated from nonspecific ones. In the future, the challenges ahead lie in the development of incorporating nucleosomes (Lee & Greene, 2011; Visnapuu & Greene, 2009) in the defined lesion damage arrays, as well as complete reconstitutions of repair pathways at the single-molecule level with efficient real-time multicolor imaging capabilities.
Supplementary Material
Acknowledgments
This work was made possible through funding from the National Institutes of Health 5R01ES019566 to B.V.H., and 2P30CA047904 to University of Pittsburgh Cancer Institute.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/bs.mie.2017.03.027.
References
- Arnspang EC, Brewer JR, Lagerholm BC. Multi-color single particle tracking with quantum dots. PLoS One. 2012;7(11):e48521. doi: 10.1371/journal.pone.0048521. http://dx.doi.org/10.1371/journal.pone.0048521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchez MP. Quantum dots find their stride in single molecule tracking. Current Opinion in Chemical Biology. 2011;15(6):775–780. doi: 10.1016/j.cbpa.2011.10.011. http://dx.doi.org/10.1016/j.cbpa.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, DellaVecchia MJ, Perera L, Van Houten B. Cooperative damage recognition by UvrA and UvrB: Identification of UvrA residues that mediate DNA binding. DNA Repair (Amst) 2008;7(3):392–404. doi: 10.1016/j.dnarep.2007.11.013. http://dx.doi.org/10.1016/j.dnarep.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. The Journal of Biological Chemistry. 2006;281(36):26370–26381. doi: 10.1074/jbc.M603093200. http://dx.doi.org/10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Geeves MA, Kad NM. Using fluorescent myosin to directly visualize cooperative activation of thin filaments. The Journal of Biological Chemistry. 2015;290(4):1915–1925. doi: 10.1074/jbc.M114.609743. http://dx.doi.org/10.1074/jbc.M114.609743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Research. 2011;39(17):7487–7498. doi: 10.1093/nar/gkr459. http://dx.doi.org/10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furda AM, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods in Molecular Biology. 2012;920:111–132. doi: 10.1007/978-1-61779-998-3_9. http://dx.doi.org/10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Du C, Chen S, Salerno V, Manfredi C, Hsieh P. In vitro studies of DNA mismatch repair proteins. Analytical Biochemistry. 2011;413(2):179–184. doi: 10.1016/j.ab.2011.02.017. http://dx.doi.org/10.1016/j.ab.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke H, Wang H, Hsieh CL, Woldemeskel S, Watkins SC, Rapic-Otrin V, et al. Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(18):E1862–E1871. doi: 10.1073/pnas.1323856111. http://dx.doi.org/10.1073/pnas.1323856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Molecular Cell. 2007;28(3):359–370. doi: 10.1016/j.molcel.2007.09.008. http://dx.doi.org/10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graneli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(5):1221–1226. doi: 10.1073/pnas.0508366103. http://dx.doi.org/10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma HG, Laney DE. DNA binding to mica correlates with cationic radius: Assay by atomic force microscopy. Biophysical Journal. 1996;70(4):1933–1939. doi: 10.1016/S0006-3495(96)79757-6. http://dx.doi.org/10.1016/S0006-3495(96)79757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CD, Wang H, Ghodke H, Simons M, Towheed A, Peng Y, et al. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Research. 2013;41(9):4901–4912. doi: 10.1093/nar/gkt177. http://dx.doi.org/10.1093/nar/gkt177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Molecular Cell. 2010;37(5):702–713. doi: 10.1016/j.molcel.2010.02.003. http://dx.doi.org/10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M, Liu L, Chen X, Driscoll KI, Mao P, Bohm S, et al. Single-molecule imaging reveals that Rad4 employs a dynamic DNA damage recognition process. Molecular Cell. 2016;64(2):376–387. doi: 10.1016/j.molcel.2016.09.005. http://dx.doi.org/10.1016/j.molcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M, Van Houten B. Rad4 recognition-at-a-distance: Physical basis of conformation-specific anomalous diffusion of DNA repair proteins. Progress in Biophysics and Molecular Biology. 2016 doi: 10.1016/j.pbiomolbio.2016.12.004. http://dx.doi.org/10.1016/j.pbiomolbio.2016.12.004. [DOI] [PMC free article] [PubMed]
- Konopka CA, Bednarek SY. Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. The Plant Journal. 2008;53(1):186–196. doi: 10.1111/j.1365-313X.2007.03306.x. http://dx.doi.org/10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- Lee JY, Greene EC. Assembly of recombinant nucleosomes on nanofabricated DNA curtains for single-molecule imaging. Methods in Molecular Biology. 2011;778:243–258. doi: 10.1007/978-1-61779-261-8_16. http://dx.doi.org/10.1007/978-1-61779-261-8_16. [DOI] [PubMed] [Google Scholar]
- Lee JY, Terakawa T, Qi Z, Steinfeld JB, Redding S, Kwon Y, et al. DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases. Science. 2015;349(6251):977–981. doi: 10.1126/science.aab2666. http://dx.doi.org/10.1126/science.aab2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Countryman P, Buncher N, Kaur P, Longjiang E, Zhang Y, et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Research. 2014;42(4):2493–2504. doi: 10.1093/nar/gkt1132. http://dx.doi.org/10.1093/nar/gkt1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Countryman P, Chen H, Pan H, Fan Y, Jiang Y, et al. Functional interplay between SA1 and TRF1 in telomeric DNA binding and DNA-DNA pairing. Nucleic Acids Research. 2016;44(13):6363–6376. doi: 10.1093/nar/gkw518. http://dx.doi.org/10.1093/nar/gkw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biology. 2007;8(5):R70. doi: 10.1186/gb-2007-8-5-r70. http://dx.doi.org/10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SR, Dunn AR, Kathe SD, Warshaw DM, Wallace SS. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(20):E2091–E2099. doi: 10.1073/pnas.1400386111. http://dx.doi.org/10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff GC, Erie DA. A novel single-molecule study to determine protein–protein association constants. Journal of the American Chemical Society. 2001;123(24):5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schneider SW, Larmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflügers Archiv. 1998;435(3):362–367. doi: 10.1007/s004240050524. http://dx.doi.org/10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- Springall L, Inchingolo AV, Kad NM. DNA-protein interactions studied directly using single molecule fluorescence imaging of quantum dot tagged proteins moving on DNA tightropes. Methods in Molecular Biology. 2016;1431:141–150. doi: 10.1007/978-1-4939-3631-1_11. http://dx.doi.org/10.1007/978-1-4939-3631-1_11. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. http://dx.doi.org/10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafvizi A, Huang F, Fersht AR, Mirny LA, van Oijen AM. A single-molecule characterization of p53 search on DNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(2):563–568. doi: 10.1073/pnas.1016020107. http://dx.doi.org/10.1073/pnas.1016020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophysical Journal. 2002;82(5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. http://dx.doi.org/10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nature Methods. 2008;5(2):159–161. doi: 10.1038/nmeth1171. http://dx.doi.org/10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- Vesenka J, Guthold M, Tang CL, Keller D, Delaine E, Bustamante C. Substrate preparation for reliable imaging of DNA molecules with the scanning force microscope. Ultramicroscopy. 1992;42–44(Pt B):1243–1249. doi: 10.1016/0304-3991(92)90430-r. [DOI] [PubMed] [Google Scholar]
- Visnapuu ML, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nature Structural & Molecular Biology. 2009;16(10):1056–1062. doi: 10.1038/nsmb.1655. http://dx.doi.org/10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. The Journal of Biological Chemistry. 2006;281(22):15227–15237. doi: 10.1074/jbc.M601476200. http://dx.doi.org/10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- Wang H, Tessmer I, Croteau DL, Erie DA, Van Houten B. Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Letters. 2008;8(6):1631–1637. doi: 10.1021/nl080316l. http://dx.doi.org/10.1021/nl080316l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JI, Levine AS, Du S, Chinte U, Ghodke H, Wang H, et al. Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(41):E2737–E2746. doi: 10.1073/pnas.1110067109. http://dx.doi.org/10.1073/pnas.1110067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.