Abstract
Invariant NKT (iNKT) cells bridge innate and adaptive immunity by rapidly secreting cytokines and lysing targets following TCR recognition of lipid antigens. Based on their ability to secrete IFN-γ, IL-4 and IL-17A, iNKT-cells are classified as NKT-1, NKT-2 and NKT-17 subsets, respectively. The molecular pathways regulating iNKT-cell fate are not fully defined. Recent studies implicate Rictor, a required component of mTORC2, in the development of select iNKT-cell subsets, however these reports are conflicting. To resolve these questions, we used Rictorfl/fl CD4cre+ mice and found that Rictor is required for NKT-17 cell development and normal iNKT-cell cytolytic function. Conversely, Rictor is not absolutely required for IL-4 and IFN-γ production as peripheral iNKT-cells make copious amounts of these cytokines. Overall iNKT-cell numbers are dramatically reduced in the absence of Rictor. We provide data indicating Rictor regulates cell survival as well as proliferation of developing and mature iNKT-cells. Thus, mTORC2 regulates multiple aspects of iNKT-cell development and function.
Keywords: Natural killer T cell, thymus, development, cytotoxicity, differentiation, signal transduction
Introduction
Natural Killer T (NKT) cells play a central role in innate and adaptive immune responses. These cells share characteristics with conventional T cells but are distinguished by their restricted TCR usage, unique MHC restriction and expression of cell surface markers typically found on NK cells. The most abundant NKT-cell type is Type I or “invariant” NKT (iNKT) cells. In mice, these cells express an invariant Vα14-Jα18 TCRα chain that pairs with a limited number of Vβ chains [1]–[4]. Similar to conventional T cells, NKT-cells are thought to go through selection during their development [5]–[8]. However, whereas conventional T cells are restricted by peptide antigens presented by classical MHC I or MHC II molecules, iNKT-cells are selected based on reactivity to glycolipid antigens presented by the non-classical MHC I molecule, CD1d [9]–[11]. Indeed, identification of iNKT-cells is greatly aided by CD1d tetramers that when loaded with glycolipid antigens, such as PBS57, an analogue of the iNKT-cell agonist α-galactosylceramide (α-GalCer), can distinguish this rare population [12],[13].
iNKT-cells acquire an innate-like or activated phenotype during development in the thymus. This phenotype is associated with expression of receptors typically found on NK cells or activated T cells, and the ability to quickly secrete cytokines upon antigen exposure. The ability to respond rapidly, allows iNKT-cells to be early participants in immune responses and to direct the activation of adaptive immune cells.
Within the iNKT-cell population there is maturational, phenotypic, and functional diversity. Immature iNKT-cells are CD24+, which is rapidly lost as iNKT-cells mature [14],[15]. Initially, CD24− iNKT-cells were further “staged” based on their expression of CD44 and NK1.1 [16]. These stages (stages 1–3) are associated with iNKT-cell maturation and differentiation. More recently, transcription factor expression was described as a means to functionally delineate mature iNKT-cell subsets [17]. This method results in grouping iNKT-cells by their cytokine potential and is reminiscent of how CD4+ T helper (TH) cells are delineated. In fact, many transcription factors that define CD4+ TH cell subsets also define iNKT-cells and the cytokines they produce. For example, T-bet marks IFN-γ producing TH1 and NKT-1 subsets, GATA3 drives IL-4 production in TH2 and NKT-2 subsets, and retinoic acid receptor-related orphan nuclear receptor γt (RORγt) regulates IL-17 production in TH17 and NKT-17 cells [18].
Many studies have explored the signaling events that drive CD4+ TH cell fate. In recent years, much of this work has focused on the role of mammalian target of rapamycin (mTOR) and its two signaling complexes, mTOR complex 1 and 2 (mTORC1 and mTORC2), key components of the PI3K/Akt pathway [19],[20]. Activation of PI3K leads to the membrane recruitment and activation of Akt. Activated Akt inhibits TSC1/TSC2, negative regulators of mTORC1, resulting in mTORC1 activation and increased cell survival, proliferation and growth. In contrast to mTORC1, mTORC2 phosphorylates Akt and contributes to actin remodeling, survival and proliferation. For CD4+ TH cell differentiation, studies have shown that mTORC1 is required for TH1 and TH17 differentiation, whereas mTORC2 is necessary for TH2 responses, and in some cases, TH1 differentiation [21],[22].
mTOR complexes are also important for iNKT-cell development [23]–[25], and two reports have described different roles for mTORC2 in the development of specific iNKT-cell subsets [23],[26]. One report suggested mTORC2 was required for NKT-17 development (and not NKT-1 or NKT-2), while the other described a selective defect in NKT-2 development with normal NKT-17 development. To resolve these discordant findings, we used Rictorfl/fl CD4cre+ mice (RictorcKO) to show that NKT-17 cell development, defined both transcriptionally and functionally, requires mTORC2. We also clarify that although IL-4 production is diminished among Rictor deficient thymic iNKT-cells, peripheral iNKTs do produce this cytokine. These data suggest that NKT-17 cells are highly dependent on Rictor and although the NKT-2 axis may be altered, it is not abrogated. We extended our studies to another functional readout of iNKT-cells and uncovered a novel role for mTORC2 in iNKT-cell mediated killing of tumor targets. Lastly, we identify Rictor as a key regulator of TCR/CD28 induced proliferation in developing and mature iNKT-cells.
Results
Rictor is required for iNKT-cell development
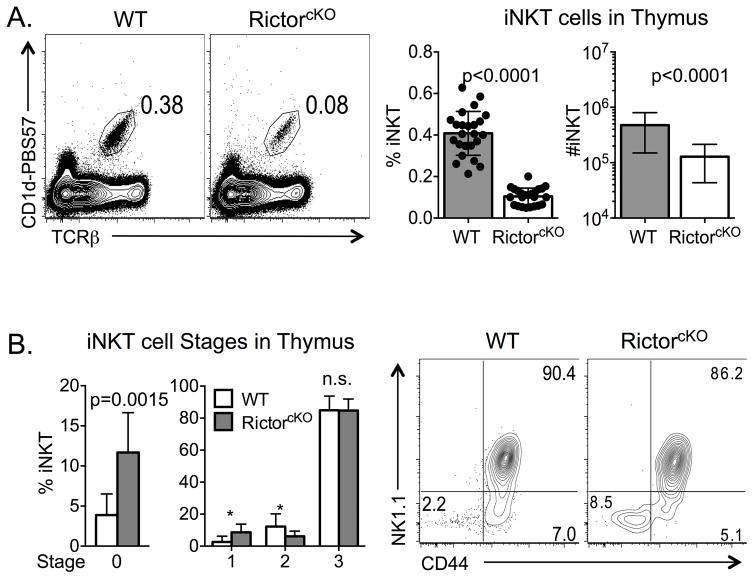
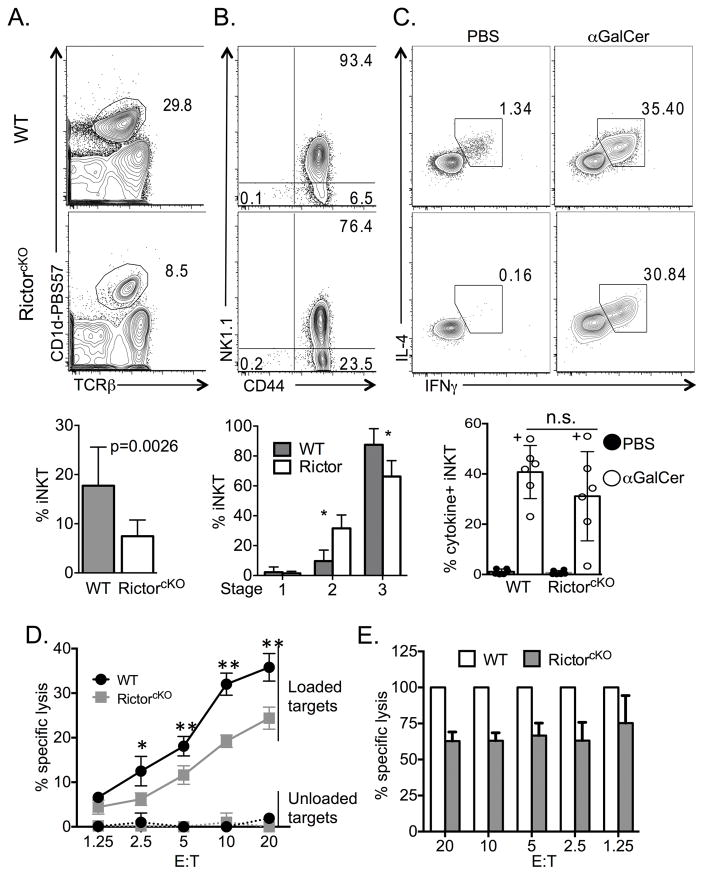
mTORC2 is important for proper differentiation of conventional CD4+ T cells into IL-4 secreting TH2 cells [21],[22]. To determine whether mTORC2 plays a similar role in iNKT-cell development, we analyzed thymocytes from WT and Rictorfl/fl CD4cre+ mice (RictorcKO), and found a marked decrease in frequency and absolute number of iNKT-cells in the absence of Rictor (Figure 1A and Supporting Information Figure 1 gating strategy). Further characterization revealed an increased frequency of CD24+ immature iNKT-cells (Stage 0). Beyond this stage, RictorcKO mice had a higher frequency of Stage 1 iNKT-cells (CD44− NK1.1−) and a diminishment of Stage 2 cells (CD44+ NK1.1−). Stage 3 (CD44+ NK1.1+) cell frequencies were similar between genotypes (Figure 1B). Consistent with a previous report [26], the failure of RictorcKO mice to generate normal numbers of iNKT-cells was cell intrinsic. In mixed bone marrow chimeras (equal contribution of WT and RictorcKO bone marrow), the iNKT-cell population was derived overwhelmingly from WT cells despite similar representation within the TCRβlow CD4−CD8− (DN) precursor thymocyte population (Supporting Information Figure 2).
Figure 1.
Rictor is required for iNKT-cell maturation. A. Frequency of iNKT-cells from WT and RictorcKO thymi was determined by flow cytometry. Gates were set on live, singlets, TCRβ+ CD1d-PBS57+ cells. Compiled frequency and absolute number of iNKT-cells is graphed. Data are from 12 independent experiments with 2–6 mice per experiment, n=24 mice per genotype, unpaired t-test mean ± S.D. B. Compiled frequency of Stage 0 iNKT (CD24+ iNKT) cells from 5 experiments with 2–4 mice per experiment, n=8 mice total per genotype. Frequencies of cells in Stages 1–3 (CD24−) are graphed using compiled data from 9 experiments with 2–6 mice per experiment, n=16 mice per genotype, multiple t-test mean ± S.D. Representative CD44 and NK1.1 profile of mature iNKT-cells from WT and RictorcKO mice is shown. * p ≤ 0.009.
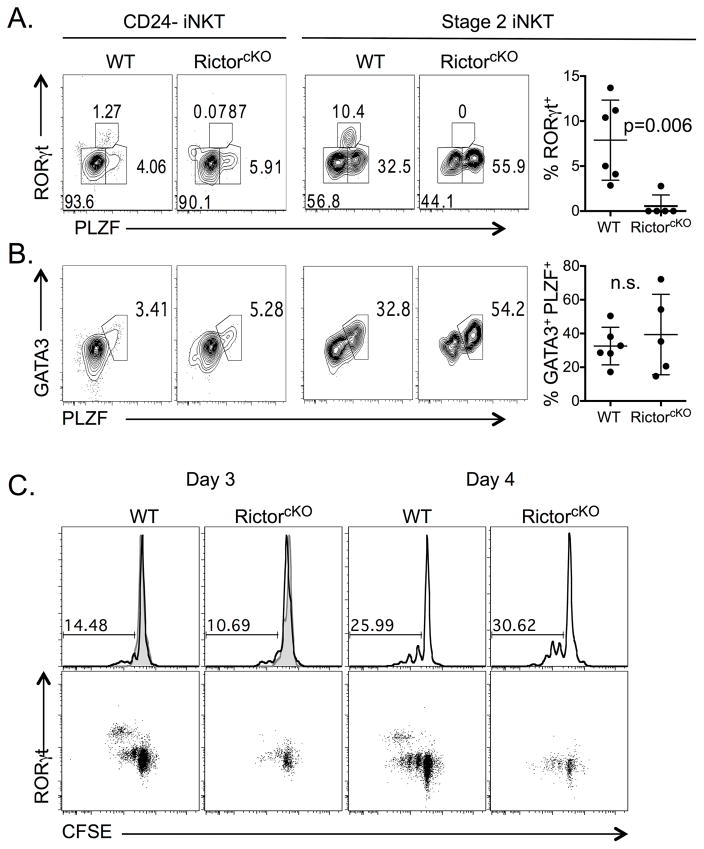
iNKT-cell subsets can also be identified based on their transcription factor expression [17]. Staining cells for promyelocytic leukemia zinc finger (PLZF) and RORγt allows for the resolution of three subsets: NKT-1 (PLZFlow RORγt−), NKT-2 (PLZFhi RORγt−), and NKT-17 (PLZFint RORγt+). These subsets primarily produce IFN-γ, IL-4 and IL-17, respectively. Among mature (CD24−) iNKT-cells, the frequencies of NKT-1 and NKT-2 cells were similar in WT and RictorcKO mice, but the frequency of NKT-17 cells was dramatically decreased in RictorcKO mice (Figure 2A, left). By focusing on Stage 2 iNKT-cells (the stage in which NKT-17 cells are most prevalent) [17],[27], the loss of the NKT-17 subset was even more apparent (Figure 2A, right). We also interrogated the NKT-2 population by analyzing the expression of GATA3, a transcription factor that is co-expressed with PLZF in NKT-2 cells. In both CD24− iNKT-cells and Stage 2 cells, where the majority of NKT-2 cells reside [15]-[17], we found no difference in the frequency of PLZF+ GATA3+ co-expressing cells (Figure 2B). Similarly, T-bet expression in mature iNKT-cells and NKT-1 cells was normal (Supporting Information Figure 3A).
Figure 2. Rictor is required for the development of RORγt+ iNKT-cells.
RORγt+ iNKT (A) or GATA3+ PLZF+ iNKT (B) cells in the thymus of WT and RictorcKO mice were analyzed among CD24− iNKT-cells (left plots) or Stage 2 iNKT-cells (right plots) by flow cytometry. Frequency of RORγt+ (A) or GATA3+ PLZF+ (B) cells among Stage 2 iNKT-cells was compiled over 4 experiments with 2–5 mice per experiment, n=5–6 mice total per genotype, unpaired t-test mean ± S.D. C. WT and RictorcKO thymocytes were cultured with or without IL-7 for 3 or 4 days. Top row: Numbers indicate the percent of divided iNKT-cells gated from live, singlet gates. Bottom row: CFSE versus RORγt profile from the cultures depicted in Top row. Data representative of 3 experiments, 2 mice per experiment, n=3 mice total per genotype for day 3; 2 experiments with n=2 mice per genotype for day 4. Filled histograms represent cells cultured without IL-7.
RORγt− iNKT-cells maintain homeostasis by utilizing IL-15 or IL-7 whereas the RORγt+ NKT-17 population is uniquely dependent on IL-7 [28]. Since this population was most affected in RictorcKO mice, we tested whether failure to respond to IL-7 might contribute to their decreased numbers. iNKT-cells from WT and RictorcKO mice persisted and divided similarly in response to IL-7 (Figure 2C, top row and Supporting Information Figure 3B gating stragegy). As expected, RORγt+ NKT-17 cells divided more rapidly than RORγt− iNKT-cells in the presence of IL-7. However, despite robust proliferation of RORγt− iNKT-cells from RictorcKO mice, IL-7 did not lead to an accumulation of RORγt+ NKT-17 cells (Figure 2C, bottom row). These data suggest IL-7 signaling is largely intact in RictorcKO iNKT-cells, and point to a role for Rictor in directing early signals required for RORγt+ iNKT-cell development.
Rictor is required for the development of IL-17A producing iNKT-cells
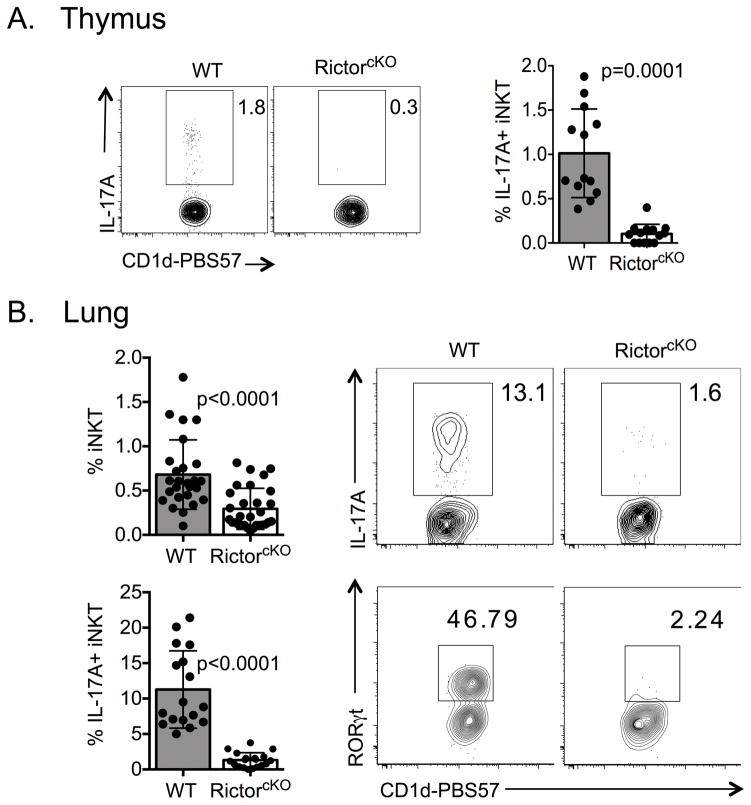
Given the near absence of RORγt+ iNKT-cells in the thymus of RictorcKO mice, we predicted iNKT-cells in these mice would be deficient in IL-17A production. Indeed, we detected little IL-17A from RictorcKO thymic iNKT-cells following ex vivo PMA/ionomycin stimulation (Figure 3A). We also evaluated iNKT-17 cells in the lung, a tissue enriched for NKT-17 cells [27],[29]. Similar to the thymus, RictorcKO mice had a reduced frequency of iNKT-cells in the lung and produced significantly less IL-17A compared to WT iNKT-cells (Figure 3B). This finding was consistent with a dramatic reduction in the frequency of lung iNKT-cells expressing RORγt (Figure 3B). Together, these data clearly demonstrate that Rictor is required for the development of RORγt+ IL-17A producing NKT-17 cells.
Figure 3. Rictor regulates NKT-17 development.
A. IL-17A production from live, singlet, TCRβ+ CD1d-PBS57 thymocytes from WT and RictorcKO mice following stimulation with PMA/ionomycin was assessed by flow cytometry. Plots are representative of 6 experiments with 3–6 mice per experiment; n=13–14 mice per genotype and the compiled percent of IL-17A+ iNKT-cells is shown in the graph, unpaired t-test mean ± S.D. B. iNKT-cell frequency of lung lymphocytes. Data compiled from 15 experiments with 2–7 mice per experiment, n>20 mice total per genotype (top graph), unpaired t-test mean ± S.D. The frequencies of IL-17A+ cells among lung iNKT-cells compiled from 8 experiment with 3–8 mice per experiment, n=17–19 mice total per genotype (bottom graph), unpaired t-test mean ± S.D. Flow plots: Frequency of IL-17A+ iNKT-cells from lungs of WT and RictorcKO mice (top) and frequency of RORγt+ iNKT-cells (bottom). RORγt plot is representative of 4 experiments with 2–3 mice per experiment, n=4–6 mice total per genotype.
Rictor regulates the balance of cytokine producing iNKT-cells
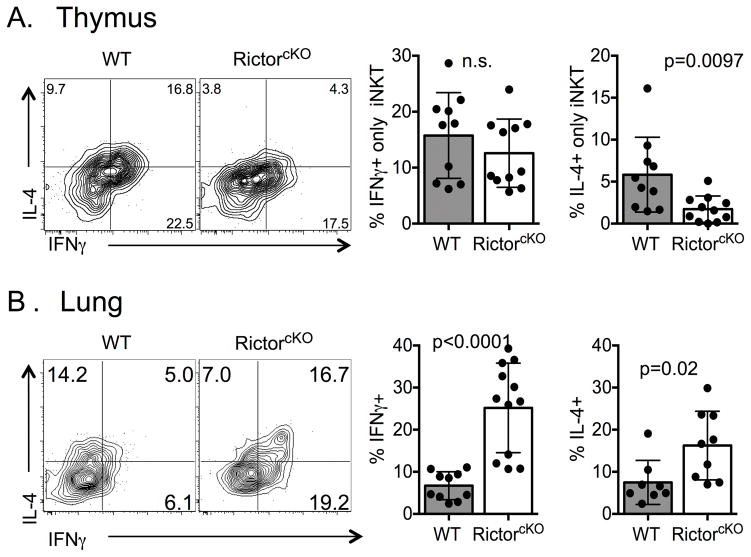
To determine whether Rictor also impacts NKT-1 and NKT-2 function, we assessed IFN-γ and IL-4 production. Both cytokines were produced by WT and RictorcKO iNKT-cells with a similar frequency of IFN-γ+IL-4− single producing cells (Figure 4A). However, despite similar GATA3 and PLZF expression (Figure 2), we noted a defect in IL-4+IFN-γ− producers as well as IFN-γ+IL-4+ double producing cells (Figure 4A and data not shown). These data suggest that Rictor contributes to IL-4 production.
Figure 4. Rictor regulates cytokine production from thymic iNKT-cells.
Thymi and lungs of WT and RictorcKO mice were analyzed by flow cytometry. Frequency of IFN-γ+ and IL-4+ from thymi (A) and lungs (B) live, singlet, TCRβ+ CD1d-PBS57+ cells following stimulation with PMA/ionomycin. A. Representative flow plots and compiled data from 5 experiments with 3–6 mice per experiment; n=10–11 mice per genotype, unpaired t-test mean ± S.D. B. Representative flow plots and compiled data are from 5 (IFN-γ) or 4 (IL-4) experiments with 2–8 mice per experiment, n=8–12 mice total per genotype, unpaired t-test mean ± S.D.
Some iNKT-cells complete their maturation in peripheral tissues [15],[16]. Thus, to better understand how mTORC2 shapes iNKT-cell development, we analyzed IFN-γ and IL-4 secretion from lung iNKT-cells. We found RictorcKO lung iNKT-cells were capable of both IFN-γ and IL-4 production. On average there were enhanced frequencies of total IFN-γ+ cells, total IL-4+ cells (Figure 4B) and IFN-γ+IL-4+ double producing NKT-cells (data not shown) in RictorcKO mice. To evaluate the in vivo responsiveness of RictorcKO iNKT-cells, we challenged mice with glycolipid antigens and analyzed their liver iNKT-cells, as this organ harbors a high frequency of iNKT-cells that have the potential to produce IFN-γ and IL-4. Similar to the thymus and lung, RictorcKO mice have a decreased frequency of iNKT-cells among liver lymphocytes compared to WT mice (Figure 5A). While the proportion of CD44+ iNKT-cells was similar between WT and Rictor deficient mice, the proportion of NK1.1− versus NK1.1+ (Stage 2 versus Stage 3, respectively) was skewed (Figure 5B). Despite this difference, following injection with the iNKT-cell ligands, α-GalCer or PBS44, WT and RictorcKO mice produced similar levels of IFN-γ and IL-4 on a per cell basis indicating that Rictor is not absolutely required for production of either of these cytokines (Figure 5C).
Figure 5. Rictor is required for development and cytolytic function of liver iNKT-cells.
A. Representative live singlet, TCRβ+ CD1-PBS57 profile of WT and RictorcKO liver lymphocytes and compiled frequencies of iNKT liver lymphocytes from 8 experiments, with 2–3 mice per experiment, n=8–9 mice total per genotype, unpaired t-test mean ± S.D. B. CD44 and NK1.1 profile of liver live singlet, TCRβ+ CD1-PBS57, CD24− cells and compiled frequencies of Stage 1–3 iNKT-cells from 3 experiments, n=4–6 mice per genotype, multiple t-test mean ± S.D. * p<0.001. C. IFN-γ and IL-4 production from liver iNKT-cells following in vivo challenge. Representative cytokine plots and graphs of compiled frequencies of IFN-γ and IL-4 producing iNKT-cells from mice injected with PBS (filled histograms and bars) or α-GalCer (open histograms and bars). “+” denotes data from mice injected with PBS44. Compiled data from 6 independent experiments with n=6 mice per genotype, paired t-test mean ± S.D. D and E. Line graph of percent specific lysis of unloaded or PBS44-loaded targets by NKT-cells sorted from WT or RictorcKO mice. Analysis by two-way ANOVA/Bonferroni post-test (* p<0.05, ** p<0.01) mean ± S.D of triplicate wells. Line graph (D) is representative of 2 independent experiments and bar graph (E) shows combined data of the specific lysis from RictorcKO iNKT-cells relative to WT; paired t-test mean ± S.D. of all replicate wells, p=0.0001. For each experiment liver lymphocytes from 3 (WT) or 8 (RictorcKO) mice were pooled and sorted for NK1.1+ TCRβ+ cells.
Optimal NKT-cell cytotoxicity is dependent on Rictor
In addition to cytokine release, iNKT-cells can lyse cells bearing lipid-antigen loaded CD1d molecules [30]–[32]. To test whether Rictor impacts the cytolytic function of NKT-cells, we sorted NK1.1+ TCRβ+ NKT-cells from the livers of WT and RictorcKO mice and compared their ability to lyse EL4 cells loaded with PBS44. Using a number of effector to target ratios, we found that liver NKT-cells lacking Rictor were less cytolytic (Figure 5D and E). This defect was not attributable to differences in the proportion of NKT-cells capable of recognizing lipid-loaded targets, as approximately 75–80% of NK1.1+ TCRβ+ liver lymphocytes from WT and RictorcKO mice are also CD1d-PBS57+ (Supporting Information Figure 4). Taken together, these data indicate that Rictor is required for optimal NKT-cell killing.
Rictor contributes to iNKT-cell expansion and survival
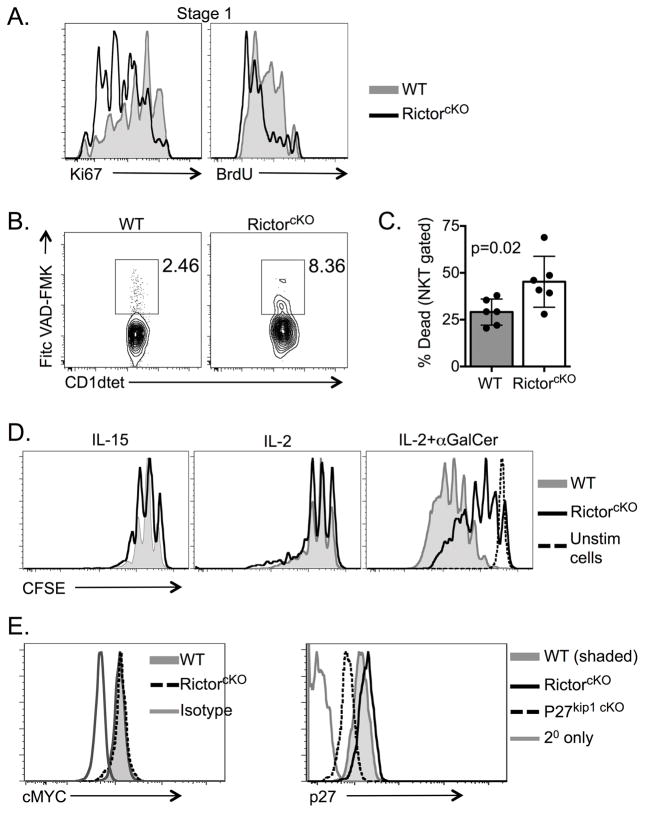
To address why RictorcKO mice have fewer iNKT-cells than WT mice we evaluated division and cell death. Ki67 staining of total iNKT-cells was similar between WT and RictorcKO mice, but analyses of individual iNKT-cell stages revealed a significant decrease in the frequency of Ki67+ Stage 1 cells in RictorcKO mice (Figure 6A and Supporting Information Figure 5A). This finding was supported by in vivo BrdU labeling in which a significant decrease in BrdU uptake was seen in RictorcKO Stage 1 iNKT-cells (Figure 6A and Supporting Information Figure 5B). To assess cell survival, we measured caspase expression using labeled Fitc-VAD-FMK and found increased staining in RictorcKO iNKT-cells compare to WT (Figure 6B). Consistent with this finding, we observed that when cultured overnight in media alone, the viability of RictorcKO iNKT-cells was decreased compared to WT cells (Figure 6C and Supporting Information Figure 5C).
Figure 6. Rictor regulates iNKT-cell proliferation and survival.
A. WT and RictorcKO iNKT-cells in Stage 1 (CD24− NK1.1− CD44−) after Ki76 staining and BrdU incorporation. Data are representative of 2 (BrdU) or 3 (Ki67) independent experiments with 3–7 mice per experiment with a total of 6–7 mice per genotype. B. The frequency of total CD24− iNKT-cells that were caspase+ was determined. Representative of 3 independent experiments with 2–4 mice per experiment and total of 4 mice per genotype, p=0.0114 ratio paired t-test. C. iNKT-cell enriched thymocytes were cultured in media overnight then evaluated for cell survival. Representative plots show dead cells (LIVE/DEAD+) gated on all iNKT-cells after exclusion of doublets. Gating strategy included in Supporting Information. Compiled data are from 6 experiments with 2 mice per experiment total of n=6 mice per genotype, paired t-test mean ± S.D. D. CFSE dilution of live, singlet, iNKT-cells from unfractionated thymocytes cultured for 4 days with the indicated stimuli. Plots representative of 3 independent experiments with 2 mice per experiment. E. Myc and p27kip1 staining of CD24− thymic iNKT-cells from WT, RictorcKO or p27kip1(fl/fl)CD4cre+ (p27kip1 cKO) mice as indicated. Plots are representative of 2 (Myc) or 3 (p27) independent experiments with 2–4 mice per experiment.
In addition to IL-7, iNKT-cells divide in response to IL-2 and IL-15 and exhibit robust proliferation following TCR ligation in combination with cytokine stimulation. To determine whether the response to these signals were compromised in the absence of Rictor, we cultured thymocytes with IL-2 or IL-15 alone or in combination with α-GalCer + anti-CD28. iNKT-cells from WT and RictorcKO proliferated similarly in response to cytokines, however, proliferation of RictorcKO iNKT-cells was greatly blunted upon addition of α-GalCer/CD28 in the presence of either cytokine (Figure 6D and data not shown). Taken together, these data show that Rictor is important for driving TCR/CD28 induced iNKT-cell proliferation.
Myc is required for the proliferative burst following agonist selection of iNKT-cells [33],[34]. Since mTORC2 can regulate Myc in other cell types, we hypothesized Myc levels may be decreased in the absence of Rictor [35],[36]. Although we were able to detect the expected changes in Myc expression during thymocyte development (Supporting Information Figure 6) [33], we saw no change in Myc expression between WT and RictorcKO iNKT-cells (Figure 6E). Thus, we investigated other mTORC2 targets that regulate cell division [37],[38]. To that end we found expression of the cyclin-dependent kinase, p27kip1, was consistently elevated in RictorcKO iNKT-cells compared to WT. This finding warrants further investigation to determine whether this pathway contributes to the proliferative defects observed in RictorcKO iNKT-cells (Figure 6E).
Discussion
Accumulating data support a role for the PI3K/Akt/mTOR pathway in regulating iNKT-cell development [39]–[43]. Several reports show mTORC1 is required for iNKT-cell proliferation and development of NKT-1 and NKT-2 subsets. However, the role of mTORC2 in iNKT-cell development is less clear. Although there is agreement that deletion of Rictor results in diminished iNKT-cell numbers (reported here and previously [23],[26]), the mechanism(s) underlying this observation are discordant, pointing to either diminished proliferation [26] or decreased survival [23]. More strikingly, a requirement for Rictor in iNKT-cell subsets has been ascribed to regulate either NKT-2 [26] cells or NKT-17 [23], with normal development of the remaining iNKT subtypes. Our studies affirm that Rictor is required for NKT-17 development but its requirement for NKT-2 cell development and IL-4 production is more nuanced. We also provide clear evidence that TCR/CD28 driven iNKT-cell proliferation is diminished in the absence of Rictor, and suggest this defect contributes to the reduced frequency of iNKT-cells found in RictorcKO mice.
Few iNKT-cells are present in either the thymus or peripheral organs of RictorcKO compared to WT controls ([23],[26] and herein). Similar to one report, our data revealed decreased proliferation among stage 1 iNKT-cells, while division of other iNKT-cells was normal (data not shown). A second study finding no role for Rictor in iNKT-cell proliferation evaluated BrdU incorporation and Ki67 staining in bulk iNKT-cells, which likely accounts for the disparate conclusion. How mTORC2 regulates iNKT-cell proliferation is unclear. Taking a candidate approach, we noted levels of p27kip1 were elevated in RictorcKO iNKT-cells. Studies in other cell types show mTORC2 can regulate p27kip1 levels by upregulating expression of proteins involved in its degradation, or transcriptionally through FOXO1 inhibition [37],[38],[44]. Understanding whether either of these mechanisms play a role in iNKT-cell development will be important to address. Importantly, our in vitro proliferation studies suggest that signals downstream of cytokines important for iNKT-cell homeostasis are not affected by the loss of Rictor, but rather point to a role for Rictor in TCR/CD28 mediated proliferation. While defective proliferation may account for the decreased numbers of iNKT-cells present in RictorcKO mice, we did observe some increased cell death in the absence of Rictor, which may be a contributing factor.
In addition to regulating the size of the iNKT-cell pool, Rictor is differentially required for iNKT-cell subset development. In the absence of Rictor, NKT-17 numbers are dramatically reduced. NKT-17 cells are defined by expression of RORγt and ability to produce IL-17. Both of theses features were greatly diminished in the absence of Rictor. Whereas these results are concurrent with an earlier report, it is unclear why this phenotype was not observed in a second report [23],[26]. Possible explanations could be differences in the source of RictorcKO mice or age of mice used for these studies. However, the original source of mice appears to be the same, and we have noted decreased IL-17A production from iNKT-cells over a wide range of ages. To probe why iNKT-17 cells were affected by the loss of Rictor, we investigated whether Rictor was required for IL-7 driven expansion of this subset. We found that IL-7 (as well as IL-2 and IL-15) driven proliferation was normal in RictorcKO iNKT-cells suggesting Rictor is important for the very early stages of NKT-17 development and/or RORγt expression.
Our findings shed light on previous reports that evaluated the role of Rictor in the development of NKT-2 cells and which appear to be contradictory. Both prior studies focused on the thymus with one reporting normal NKT-2 cell development as assessed by the evaluation of PLZF and GATA3 expression, with no measurement of IL-4 [23]. The other reported reduced IL-4 expression [26]. In the thymus, we did note a decrease in IFN-γ+IL-4+ double producing and IL-4+ IFN-γ− producing iNKT-cells. However, evaluation of peripheral iNKT-cells revealed that RictorcKO iNKT-cells can produce this cytokine following either in vitro or in vivo stimulation. In fact, IL-4 production, as well as IFN-γ production, was increased in the lungs of RictorcKO mice. We suspect these increases might be a compensatory reflection of the loss of NKT-17 cells that normally comprise a substantial proportion of the iNKT-cells in this organ. Taken together, our data indicate that Rictor contributes to the proper complement of IL-4 producing iNKT-cells in the thymus, but that the IL-4 axis within the iNKT-cell compartment is not absent as RictorcKO mice harbor IL-4 producing iNKT-cells in peripheral organs that are capable of responding to in vivo challenge.
In addition to early cytokine release, iNKT-cells are important participants in cytolytic responses. Several reports show iNKT-cells can lyse targets that express glycolipid-loaded CD1d molecules [30]–[32]. Using liver derived iNKT-cells, we show that the iNKT-cells present in RictorcKO mice do not lyse glycolipid-loaded targets as effectively as WT derived iNKT-cells. How Rictor contributes to iNKT-cell mediated killing is unknown, and both the cellular and molecular mechanisms governing this process in WT mice are still being elucidated. Indeed, recent reports have suggested that several factors, both iNKT-cell intrinsic as well as extrinsic, may cooperate to regulate iNKT cytotoxicity [30],[32]. A better understanding of iNKT-cell cytotoxicity will help guide our studies of how Rictor contributes to the establishment of an efficient cytolytic iNKT-cell population.
mTORC1 and mTORC2 play important and distinct roles in regulating cytokine production in CD4+ TH cell subsets, and it is interesting that these two complexes do not appear to regulate the same cytokines in corresponding iNKT-cell subsets [21],[22]. However, these findings are not unlike reports describing differences in signaling requirements between conventional T cells and T cells that share characteristics with either innate or memory T cells. Indeed, both NKT-17 and other innate-like T cell lineages (such as TCRγδ cells) differ from conventional CD4+ TH cells in their requirements for becoming IL-17A producing cells [45]–[47]. Together with these studies, our work highlights the importance of defining the contribution of distinct signaling pathways to the differentiation of both conventional and non-conventional T cell subsets, as therapeutic targeting of such pathways may have opposing outcomes depending on cell lineage.
Materials and Methods
Mice
Rictorfl/fl mice were originally generated in M. Magnuson’s laboratory, Vanderbilt University School of Medicine and Rictorfl/fl CD4cre+ (RictorcKO) mice were obtained with permission from J. Powell, John’s Hopkins School of Medicine. p27kip1(fl/fl) CD4cre+ mice were a gift from A. Wells, The Children’s Hospital of Philadelphia; p27kip1(fl/fl) mice originally made by M. Fero, Fred Hutchinson Cancer Research Center. C57BL/6J mice were from Jackson Laboratories or bred in the University of Pennsylvania facility. Animals were housed at the University of Pennsylvania. All experiments were performed in accordance with guidelines provided by the University of Pennsylvania Institutional Animal Care and Use Committee under the supervision of the University Laboratory Animal Resources.
Antibodies
The following antibodies were used: APC labeled PBS57 loaded CD1d-tetramers from the NIH tetramer facility, TCRβ APC-e780 (H57-597, eBiosciences) or Pacific Blue (H57-597, Biolegend), CD44 AlexaFluor(AF)700 (IM7, Biolegend), CD45.2 Pacific Blue (104, Biolegend), Thy1.1 phycoerythrin (PE) (A85-1, BD Pharmingen), Thy1.2 FITC (53-2.1, BD Pharmingen), NK1.1 PE-Cy7 or PE (PK136, BD Pharmingen), or Pacific Blue (PK136, Biolegend), or NK1.1-biotin (PK136, eBiosciences) followed by streptavidin-PE Texas Red, CD24 PerCPCy5.5 (M1/69, eBiosciences) or FITC or PE (M1/69, BD Pharmingen), IL-17A PE (17B7, eBiosciences), IL-4 PE-Cy7 (BVD6-24G2, eBiosciences), IFN-γ PerCPCy5.5 (XMG1.2, Biolegend), cMyc (Y69, abcam) or p27kip1 (D69C12; Cell Signaling Technology) followed by anti-rabbit IgG PE (Life Sciences), Ki67-PeCy7 (B56, BD Pharmingen), rabbit IgG isotype (Life Technologies) and BrdU (BrdU flow kit; BD Pharmingen). For transcription factor staining, cells were incubated with antibody to PLZF (D-9, Santa Cruz), followed by anti-mouse IgG1 PE (A85-1, eBiosciences), and then T-bet PE-Cy7 (4B10, eBiosciences), GATA3 PerCpCy5.5 (TWAJ, eBiosciences), and RORγt-BV421 (Q31-378, BD Biosciences).
Flow Cytometry and BrdU injection
Cells were stained with LIVE/DEAD Fixable Aqua Dead Cell (Life Technologies), as per manufacturer’s instructions, washed and stained with surface antibodies in FACS buffer (PBS 2% FBS and .05% sodium azide). Foxp3 Staining Buffer (eBiosciences) was used per manufacturer’s instructions except for detection of BrdU for which BD Pharmingen kit was used according to manufacturer’s instructions (BD Pharmingen). Prior to BrdU detection, mice were injected with 3 doses of 300μg BrdU every 4 hours. For caspase staining, cells were incubated with FITC-VAD-FMK or DMSO vehicle (Promega) for 20min at 37°C before surface staining. All incubations were performed in the dark at 4°C. Cells were acquired on LSRII Flow Cytometer (BD Biosciences). Analysis was performed using FlowJo software (TreeStar).
Flow cytometry gating strategy
Unless otherwise noted, iNKT-cells were analyzed using the following gating strategy: lymphocyte gate, excluded doublets, negative for LIVE/DEAD aqua, TCRβ+ CD1d-PBS57+. Analysis of mature iNKT-cells included further exclusion of CD24+ (stage 0) cells.
Bone Marrow Chimeras
C57BL/6-SJL (CD45.1) mice were irradiated with 950 rads and injected intravenously with a 1:1 mixture of T cell-depleted (Magnetic bead depletion, Qiagen) bone marrow from WT C57BL/6 (CD45.2/Thy1.2) and RictorcKO (CD45.2/Thy1.1) mice. Recipients were analyzed 6–7 weeks post transplantation.
Isolation of Lung and Liver Hematopoietic Cells
Lungs were perfused by flushing PBS into the right ventricle of the heart using a 27½-gauge needle. Lungs were diced into fragments and digested in RPMI 1640, 10% P/S/G, 0.065U/ml Liberase TM (Roche), 0.05% DNAse I (Sigma), and 20mM HEPES for 1hr in a 37°C shaker. Cells were filtered through a 70um cell strainer and washed prior to staining. Livers were removed and made into single-cell suspensions. Lymphocytes were isolated using density centrifugation with a Percoll(TM) (GE Healthcare) gradient.
Stimulation of thymic iNKT-cells
For experiments involving ex vivo stimulation for cytokine assessment, lymphocytes were cultured in the presence of PMA (50 ng/ml), ionomycin (500 ng/ml), GolgiStop protein transport inhibitor with monensin (BD Pharmingen) and/or 1 mg/ml brefeldin A for 2 hours at 37°C and then analyzed by flow cytometry. For in vivo stimulation of liver iNKT-cells, mice were injected i.p. with PBS or 4μg/ml α-GalCer. Where indicated, thymocytes were enriched for iNKT-cells by using anti-CD8 MACS microbeads per manufacturer’s instructions (Miltenyi Biotech).
CFSE CellTrace
Thymocytes were labeled with CFSE according to the CellTraceTM CFSE Cell Proliferation Kit (Life Technologies) protocol and plated in the presence or absence of IL-7 (10ng/ml), IL-2 (50U/ml), IL-15 (50ng/ml), αGalCer (125ng/ml) and anti-CD28 (5μg/ml). Cells were harvested on Day 3 and/or Day 4.
Cell lines and reagents
The EL4 cell line was from American Type Culture Collection (Manassas, VA). The αGalCer analogue, PBS44, was a kind gift from Dr. Paul B. Savage (Brigham Young University; Provo, UT).
In vitro cytotoxicity assay
Hepatic mononuclear cells were isolated and stained with anti-NK1.1 and anti-TCRβantibodies and sorted using a BD FACS Aria (BD Biosciences). Cells were >97% NK1.1+TCRβ+. iNKT-cell cytotoxicity was evaluated using a 51Cr-release assay. Briefly, EL4 target cells (T, 1×106) were labeled with 100μCi 51Cr (Na2CrO4; Perkin Elmer) for 2 h at 37°C and extensively washed. 51Cr-labeled targets were loaded with PBS44 (100 ng/ml) or left untreated, washed and then cultured in triplicate with effector cells (E) at varying E:T ratios. After 16 h, experimental supernatants were collected, applied to LumaPlates (Perkin Elmer) and radioactivity (experimental counts per minute [CPM]) was measured using a TopCount scintillation counter (Perkin Elmer). Cultures of target cells alone were assayed in a similar fashion to determine spontaneous release of 51Cr (Spont CPM). Maximal CPM was assessed by complete lysis of target cells by using 0.1% IGEPAL CA-630 in water. Percent specific lysis was calculated using the following formula: %Specific lysis = 100 × [Experimental CPM – Spont CPM]/[Total CPM – Spont CPM]. Not significant (n. s.) differences are indicated.
Statistics
Statistical analyses were performed using GraphPad software (GraphPad PRISM). Data points for the in vitro cytotoxicity assays represent mean of triplicates and error bars indicate SD. Percent specific lysis at different E:T between the groups were compared using two-way ANOVA/Bonferroni post-test. All other compiled data were analyzed using a t-test unless otherwise noted. Error bars represent SD unless otherwise indicated.
Supplementary Material
Acknowledgments
The authors thank Dr. Gary Koretzky and Dr. Shannon Carty for critical reading of this manuscript and Dr. Andrew Wells for p27kip1(fl/fl) CD4Cre+ mice. We acknowledge the NIH Tetramer Core Facility for CD1d tetramers. Support was from a NIH grant awarded to M. S. J. and A. H. (R21AI105046) and NIH K22 grant (1 K22 CA188149-01) awarded to R. D. Other support from R01AI082292.
Abbreviations
- α-GalCer
α-galactosylceramide
- cKO
conditional knockout
- CPM
counts per minute
- DN
double negative (CD4−CD8−)
- iNKT
invariant NKT
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- n.s
not significant
- PLZF
promyelocytic leukemia zinc finger
- RORγt
retinoic acid receptor-related orphan nuclear receptor γt
- TH
T helper
- WT
wild type C57BL/6
Footnotes
Conflict of interest
The authors declare no commercial or financial conflict of interest.
References
- 1.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohteki T, MacDonald HR. Stringent V beta requirement for the development of NK1.1+ T cell receptor-alpha/beta+ cells in mouse liver. J Exp Med. 1996;183:1277–1282. doi: 10.1084/jem.183.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makino Y, Kanno R, Ito T, Higashino K, Taniguchi M. Predominant expression of invariant V alpha 14+ TCR alpha chain in NK1.1+ T cell populations. Int Immunol. 1995;7:1157–1161. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 4.Bendelac A, Killeen N, Littman DR, Schwartz RH. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 5.Chun T, Page MJ, Gapin L, Matsuda JL, Xu H, Nguyen H, Kang H-S, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med. 2003;197:907–918. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 8.Pellicci DG, Uldrich AP, Kyparissoudis K, Crowe NY, Brooks AG, Hammond KJL, Sidobre S, et al. Intrathymic NKT cell development is blocked by the presence of alpha-galactosylceramide. Eur J Immunol. 2003;33:1816–1823. doi: 10.1002/eji.200323894. [DOI] [PubMed] [Google Scholar]
- 9.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MCJ, Pellicci DG, Koh R, Besra GS, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 10.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 11.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellicci DG, Hammond KJL, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(−)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 17.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013 doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das R, Sant'Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei J, Yang K, Chi H. Cutting Edge: Discrete Functions of mTOR Signaling in Invariant NKT Cell Development and NKT17 Fate Decision. The Journal of Immunology. 2014 doi: 10.4049/jimmunol.1402042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin J, Wang S, Deng W, Wu J, Gao J, Zhong X-P. Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc Natl Acad Sci US A. 2014;111:E776–83. doi: 10.1073/pnas.1315435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Tschumi BO, Corgnac S, Rüegg MA, Hall MN, Mach J-P, Romero P, et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. The Journal of Immunology. 2014;193:1759–1765. doi: 10.4049/jimmunol.1400769. [DOI] [PubMed] [Google Scholar]
- 26.Prevot N, Pyaram K, Bischoff E, Sen JM, Powell JD, Chang C-H. Mammalian target of rapamycin complex 2 regulates invariant NKT cell development and function independent of promyelocytic leukemia zinc-finger. The Journal of Immunology. 2015;194:223–230. doi: 10.4049/jimmunol.1401985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci US A. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webster KE, Kim H-O, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, Brink R, et al. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol. 2014 doi: 10.1038/mi.2013.122. [DOI] [PubMed] [Google Scholar]
- 29.Michel M-L, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong C-H, et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingender G, Krebs P, Beutler B, Kronenberg M. Antigen-specific cytotoxicity by invariant NKT cells in vivo is CD95/CD178-dependent and is correlated with antigenic potency. The Journal of Immunology. 2010;185:2721–2729. doi: 10.4049/jimmunol.1001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das R, Bassiri H, Guan P, Wiener S, Banerjee PP, Zhong M-C, Veillette A, et al. The adaptor molecule SAP plays essential roles during invariant NKT cell cytotoxicity and lytic synapse formation. Blood. 2013;121:3386–3395. doi: 10.1182/blood-2012-11-468868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassiri H, Das R, Guan P, Barrett DM, Brennan PJ, Banerjee PP, Wiener SJ, et al. iNKT cell cytotoxic responses control T-lymphoma growth in vitro and in vivo. Cancer Immunol Res. 2014;2:59–69. doi: 10.1158/2326-6066.CIR-13-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci US A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mycko MP, Ferrero I, Wilson A, Jiang W, Bianchi T, Trumpp A, MacDonald HR. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. The Journal of Immunology. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 35.Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, Iwanami A, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo Y, Huang H, Cai T, Wang T. Target of Rapamycin Complex 2 regulates cell growth via Myc in Drosophila. Sci Rep. 2015;5:10339. doi: 10.1038/srep10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanmugasundaram K, Block K, Nayak BK, Livi CB, Venkatachalam MA, Sudarshan S. PI3K regulation of the SKP-2/p27 axis through mTORC2. Oncogene. 2013;32:2027–2036. doi: 10.1038/onc.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang Z, Zhang T, Dizeyi N, Chen S, Wang H, Swanson KD, Cai C, et al. Androgen Receptor Enhances p27 Degradation in Prostate Cancer Cells through Rapid and Selective TORC2 Activation. J Biol Chem. 2012;287:2090–2098. doi: 10.1074/jbc.M111.323303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limón P, Kaech SM, Nakayama M, et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 2013;38:984–997. doi: 10.1016/j.immuni.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, Yang J, Yang K, Wang H, Gorentla B, Shin J, Qiu Y, et al. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J Clin Invest. 2014;124:1685–1698. doi: 10.1172/JCI69780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishimoto H, Ohteki T, Yajima N, Kawahara K, Natsui M, Kawarasaki S, Hamada K, et al. The Pten/PI3K pathway governs the homeostasis of Valpha14iNKT cells. Blood. 2007;109:3316–3324. doi: 10.1182/blood-2006-07-038059. [DOI] [PubMed] [Google Scholar]
- 42.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finlay DK, Kelly AP, Clarke R, Sinclair LV, Deak M, Alessi DR, Cantrell DA. Temporal differences in the dependency on phosphoinositide-dependent kinase 1 distinguish the development of invariant Valpha14 NKT cells and conventional T cells. The Journal of Immunology. 2010;185:5973–5982. doi: 10.4049/jimmunol.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birkenkamp KU, Coffer PJ. FOXO transcription factors as regulators of immune homeostasis: molecules to die for? J Immunol. 2003;171:1623–1629. doi: 10.4049/jimmunol.171.4.1623. [DOI] [PubMed] [Google Scholar]
- 45.Rachitskaya AV, Hansen AM, Horai R, Li Z, Villasmil R, Luger D, Nussenblatt RB, et al. Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J Immunol. 2008;180:5167–5171. doi: 10.4049/jimmunol.180.8.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doisne JM, Soulard V, Becourt C, Amniai L, Henrot P, Havenar-Daughton C, Blanchet C, et al. Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1- invariant NKT cells to bacteria. J Immunol. 2011;186:662–666. doi: 10.4049/jimmunol.1002725. [DOI] [PubMed] [Google Scholar]
- 47.Powolny-Budnicka I, Riemann M, Tanzer S, Schmid RM, Hehlgans T, Weih F. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in gammadelta T cells. Immunity. 2011;34:364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.