Abstract
Accurate potential energy models are necessary for reliable atomistic simulations of chemical phenomena. In the realm of biomolecular modeling, large systems like proteins comprise very many noncovalent interactions (NCIs) that can contribute to the protein’s stability and structure. This work presents two high-quality chemical databases of common fragment interactions in biomolecular systems as extracted from high-resolution Protein DataBank crystal structures: 3380 sidechain-sidechain interactions and 100 backbone-backbone interactions that inaugurate the BioFragment Database (BFDb). Absolute interaction energies are generated with a computationally tractable explicitly correlated coupled cluster with perturbative triples [CCSD(T)-F12] “silver standard” (0.05 kcal/mol average error) for NCI that demands only a fraction of the cost of the conventional “gold standard,” CCSD(T) at the complete basis set limit. By sampling extensively from biological environments, BFDb spans the natural diversity of protein NCI motifs and orientations. In addition to supplying a thorough assessment for lower scaling force-field (2), semi-empirical (3), density functional (244), and wavefunction (45) methods (comprising 1M interaction energies), BFDb provides interactive tools for running and manipulating the resulting large datasets and offers a valuable resource for potential energy model development and validation.
I. INTRODUCTION
Computational chemical models are being more routinely applied to large biological systems with the aid of increased computational resources, specialized hardware, and efficient algorithms. Examples of modeling successes in the recent literature are quite exciting: long-time scale molecular dynamics (MD) simulations of fast-folding proteins have yielded insights into how some proteins fold,1,2 Markov state models have been used to construct models of protein dynamics from parallel simulations distributed through a crowd-sourced network of personal computers and gaming consoles,3,4 human brain power itself was utilized to help solve protein folding problems through online video games,5 free energy perturbation calculations were used to guide the design of some of the most potent known non-nucleoside HIV reverse transcriptase inhibitors,6 MD simulations of protein-ligand systems have been used to reconstruct the complete dynamics of the protein-ligand binding process,7,8 and semiempirical,9,10 density functional,11,12 and ab initio13–16 quantum chemical methods are being more routinely applied to larger and larger systems, particularly to elucidate noncovalent interactions17 (NCIs).
While reports such as these are very encouraging, a different perspective can be obtained from analyzing the results of blind prediction challenges. These benchmarks are important for measuring our modeling abilities without the influences of retrospective ad hoc corrections or pressure to publish positive results. For example, in last year’s SAMPL5 competition, poor correlation and high magnitudes of error were present in a variety of methods used to make blind predictions of host-guest complexation free energies.18 Similarly the CSAR 2014 exercise, in which participants were asked to predict relative or absolute binding affinities of given ligand poses in protein receptor structures, showed that it is still very difficult to accurately score ligands across different protein targets.19 Analogously in the protein folding community, results from the Critical Assessment of Protein Structure Prediction (CASP) experiments show that current methodologies are only very newly able to make practicable inter-residue contact predictions.20,21 Thus it seems that our ability to routinely make reliable predictions concerning biomolecular systems still needs to be developed further.
The molecular mechanics methods that are the mainstays of protein dynamics modeling are very fast and tuned to their task. But difficulties can arise for new chemical environments or be masked by seemingly good collective results. For example, fragmenting the 1HSG-indinavir docking site into contact pairs yields errors whose sum could easily overwhelm the magnitude of the overall binding energy.22 In contrast, ab initio approaches that are the mainstay of small molecule modeling are far more robust for new regions of chemical space but can scarcely approach the thousand-atom scale.
A necessary resource for creating, optimizing, and validating potential energy models is a large dataset of biologically relevant fragment interactions and their associated reference interaction energies (IEs). Several such databases exist,22–26 though they most commonly are organic rather than biological in origin and include only dozens of systems, and many are available through user-friendly web portals.27,28 Several of them utilize “gold standard” quantum chemical methods such as coupled-cluster singles and doubles with perturbative triples, CCSD(T),29 at the complete basis set (CBS) limit to estimate reference energies at equilibrium geometries. Non-equilibrium geometries, which are often encountered in composite biomolecular systems, are less often included in benchmark databases. Some geometry variation was introduced with the S22x5 and S66x8 datasets, for which previously optimized fragments were translated along their intermolecular axes,25,30 with S66a8, which systematically scanned interfragment angles,31 with NBC10 and HBC6, which provided frozen momomers and relaxed potential energy curves,24,32 and with ACHC, which examined nucleobase pair degrees of freedom.33 Hobza and co-workers recently assembled a database of 4014 protein-nucleic acid complexes from the Protein DataBank (PDB) and analyzed them by contact; however, that work used B3LYP-D3/def2-TZVPP as a reference for molecular mechanics.34 Those authors also collected 272 sidechain and nucleic acid contacts that they benchmarked at CCSD(T)/[aTQZ; :aDZ].35,36 Collections of databases have also been assembled, such as by Goerigk and Grimme37 (GMTKN30) and by Mardirossian and Head-Gordon.38 As the cost of quantum chemical computations has diminished, even for reference-quality methods, the quantity that comprises a large-data study has grown commensurately, even an order of magnitude from Ref. 39 to the present work. At this level, data management and indexing are just as important as generating calculations.
In order to provide a large, diverse, and more complete dataset of biologically relevant bimolecular fragments and their interaction energies, we have begun the construction of the BioFragment Database (BFDb). We have selected high-resolution protein structures from the PDB and fragmented them with in-house code to produce a very large () set of biomolecular fragment complexes. The fragments are clustered into sets depending on their origin (e.g., backbone or sidechain) and interaction type (e.g., nonpolar or anionic). The resulting sets are analyzed in terms of gas-phase absolute electronic interaction energy with several potential models, including force fields (FFs), semiempirical (SE) methods, density functional theory (DFT), wavefunction (WFN) methods, and a benchmark-quality reference approximating the CCSD(T)/CBS gold-standard model. The database is hosted online (accessible via http://vergil.chemistry.gatech.edu/bfdb) in order to provide reference results for potential energy model development, validation, and error estimation methods. Herein is described the construction of BFDb, general trends observed, and example usage.
II. COMPUTATIONAL METHODS
A. Construction of BFDb
Initial protein structures were hand-selected from the PDB according to specific criteria: (i) no nonstandard amino acid residues, (ii) no bound ligand, nucleic acid, or metal, and (iii) crystal resolution better than 2 Å. This resulted in a set of 47 proteins, which had a range of crystal structure resolution from 0.62 to 2.0 Å and a range of chain lengths from 9 to 333. The selected PDBIDs are as follows: 1BN6, 1DF4, 1G9W, 1LU4, 1NG6, 1R7J, 1UCS, 1UKF, 1YU5, 1YZM, 1ZVA, 2BV9, 2BVV, 2ESK, 2JLJ, 2PMR, 2VQ4, 2WJ5, 2WLW, 2WWE, 2X4L, 2X5Y, 2XGV, 3A2Z, 3A7L, 3DVW, 3EAZ, 3EY6, 3HNX, 3I35, 3ICH, 3JQU, 3JVE, 3K0M, 3K0N, 3KB5, 3KSN, 3KZD, 3LAA, 3M9J, 3MSI, 3N11, 3NE0, 3NGP, 3NR5, 3O1Z, and 3O3T. The set of 47 protein structures was processed with LEAP in AMBER.40 Water molecules were removed, hydrogen atoms were added, and the proteins were energy minimized with ff99SB41 in a generalized Born implicit solvent model42 with tight restraints (force constant = 1000 kcal/mol Å2) on non-hydrogen atoms. The relaxed structures were then processed by our fragmentation code.
The fragmentation algorithm is based on detected intramolecular interactions. First, noncovalent interactions are detected by measuring distances between heavy atoms from different amino acid residues. A threshold of 4 Å is set for detecting nonpolar interactions and 3.5 Å for polar interactions. Corresponding atom pairs within these distances are chosen and labeled as “starters” for the program to initialize each fragment. The program searches for atoms adjacent to every starter and labels them as “seekers.” When all of the seekers are selected, a new round of searching starts in which atoms adjacent to previous seekers are regarded as new seekers. The searching procedure ends only if (i) the seeker is a sp3-hybridized carbon atom, (ii) the seeker is a terminal atom, or (iii) all adjacent atoms to the current seeker have already been chosen. Finally, starters and seekers for both interacting moieties are chosen as members of a fragment pair. Hence, this fragmentation program keeps all interacting atoms and cuts protein molecules at the nearest non-interacting sp3 carbons. For example, a particular histidine and aspartic acid contact becomes 4-methylimidazole and formate ion fragments, while an isoleucine and cysteine contact becomes ethane and ethanethiol fragments. After fragmentation, hydrogen atoms are added to the terminal sp3 carbons to replace the cut atoms. Each hydrogen atom is positioned according to the position vector to the cut atom and the resulting C–H bond length set to 1.1 Å. All modeling and quantum chemical methods employed these geometries without further optimization or adjustment to hydrogen positions.
The fragmentation procedure was applied to the set of 47 proteins, which resulted in an unfiltered dataset of 10 972 biomolecular fragment interactions observed in protein crystal structures. The set was divided into 4640 interactions between protein backbone moieties (e.g., from -helices and -sheets), 2774 backbone-sidechain interactions (BBI), and 3558 sidechain-sidechain interactions. The distribution of various interacting sidechains is shown as a heatmap in Fig. 1(a), with the increased frequency for cation-anion and nonpolar-nonpolar contacts reflecting natural abundance. The size distribution of sidechain-sidechain complexes from 12 to 41 atoms is shown as a histogram in Fig. 1(b). Most of the backbone-backbone type interactions (uniformly 24 atoms in size) were the same dimer complexes in different geometries [see insets of Fig. 2(d) for representatives of the two binding motifs], so from this group, 100 were randomly selected to become the set of backbone-backbone interactions called BBI. 3380 of the sidechain-sidechain fragments were collected into the dataset SSI.43 A selection of the sidechain-backbone interactions makes up the dataset SBI. Table I summarizes the available datasets.
FIG. 1.
Composition of the SSI database. (a) The number of intermolecular contacts by amino acid as a heatmap, (b) the size of interacting complex as a histogram, and (c) the nature of intermolecular contacts by amino acid as presented by plotting each database member individually and coloring it according to SAPT2+/aug-cc-pVDZ decomposition.
FIG. 2.
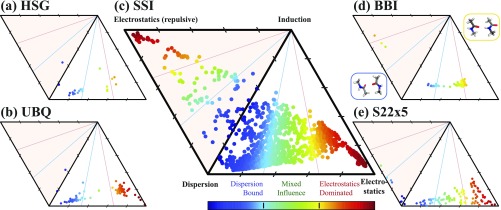
SAPT interaction energy decomposition of the four BFDb databases. The relationship between the attractive components—electrostatics, dispersion, and induction—of a SAPT analysis of each member of (a) HSG, (b) UBQ, (c) SSI, and (d) BBI is plotted on a ternary diagram, where proximity to a vertex indicates an accordant fraction of attraction which is due to that component. Database members are colored to differentiate binding motif: hydrogen-bonded (red), mixed-influence (green), and dispersion-dominated (blue). The shaded upper triangle contains complexes supporting a repulsive electrostatics component. Comparison to scanned-coordinate-sampling database S22x5 shown in (e).
TABLE I.
Databases of the BFDb.
| Database | Source | Size | Reference level | Citation |
|---|---|---|---|---|
| HSG | HIV-II protease/indinavir (PDBID:1HSG) | 21 | Gold | 22 and 44 |
| UBQ | Ubiquitin (PDBID:1UBQ) | 94 | UBQREFa | 45 |
| BBI | Backbone-backbone (all PDB) | 100 | Silver | This work |
| SSI | Sidechain-sidechain (all PDB) | 3380 | Silver | This work |
| SBIb | Sidechain-backbone (all PDB) | 2774 |
UBQREF comprises a combination of MP2/CBS estimates from aug-cc-pVTZ and aug-cc-pVQZ basis set extrapolation and CCSD(T)/CBS estimations for aromatic complexes.
SBI not included in this work.
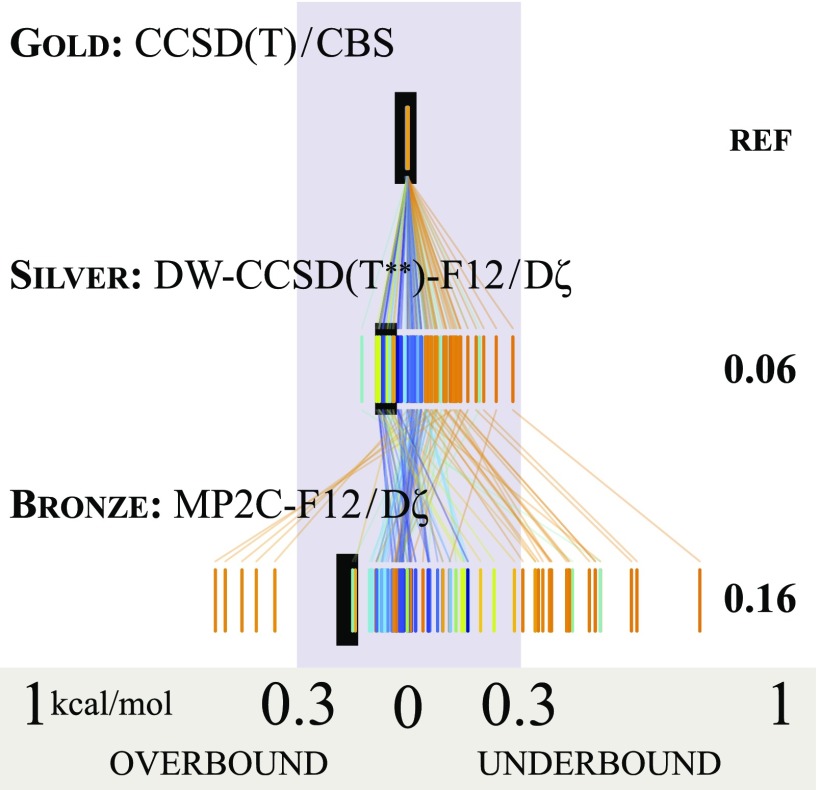
Truly high quality estimates of the complete basis set limit of gold standard CCSD(T) for NCI can be achieved using focal point estimates based on basis set extrapolations of second-order Møller–Plesset perturbation theory (MP2) and a [CCSD(T) − MP2] correction evaluated in at least a triple- basis set augmented by diffuse functions. Benchmarks of this quality or better have been evaluated for several databases of noncovalent interactions, including HSG,22 S22,46 NBC10,24 and HBC6.32,44 The high-quality reference energies available for the 345 bimolecular complex geometries in those four databases enable wavefunction methods with an excellent balance between error and computational cost to be identified.47 These are designated “silver” and “bronze” standards, and their comparison with gold is shown in Fig. 3. The DW-CCSD(T**)-F12 method of Marshall and Sherrill48 is employed with the aug-cc-pV(D+d)Z basis set as the silver standard, achieving mean absolute error (MAE) among the four databases of 0.06 kcal/mol while remaining computationally tractable for a nucleobase pair on a typical workstation. For orientation, levels of theory such as MP2/aug-cc-pV5Z, CCSD(T)/(heavy-)aug-cc-pVTZ, and B3LYP-D3/aug-cc-pVTZ yield MAE of 0.35–0.59 kcal/mol, again with respect to the databases and gold standard reference described above. By employing an explicitly correlated and dispersion-weighted methodology, DW-CCSD(T**)-F12/aug-cc-pV(D+d)Z succeeds for all broad classes of NCI. MP2C-F12/aug-cc-pVDZ is designated as the bronze standard. This approach corrects the MP2-F12 interaction energy with a coupled dispersion term derived from TDDFT and thus provides quite accurate interaction energies (IEs) for dispersion-dominated systems, though errors for hydrogen-bonding average 0.2 kcal/mol. Full technical details associated with computing reference IE can be found in Ref. 47.
FIG. 3.
Levels of reference interaction energy. The performance of silver and bronze standards is shown for 121 bimolecular complexes (from S22, NBC10, HBC6, and HSG databases) for which gold standard reference is available. Errors for individual complexes are presented as thread plots, discussed in the penultimate paragraph of Sec. II B, and the mean absolute error summary statistic is to the right in kcal/mol.
B. Computational and graphical details
DFT and wavefunction methods were evaluated with the correlation-consistent basis sets of Dunning, augmented by diffuse functions on all atoms.49,50 These are notated as aug-cc-pVDZ (aDZ), aug-cc-pVTZ (aTZ), and aug-cc-pVQZ (aQZ) throughout. Occasionally, results for unaugmented basis sets such as cc-pVQZ (QZ) and def2-QZVP (QZVP)51 are also reported. Reference interaction energies for DW-CCSD(T**)-F12 employed the modified aug-cc-pV(D+d)Z52 basis to better treat sulfur-containing residues. All interaction energies were computed using the counterpoise (CP) correction scheme of Boys and Bernardi,53 unless explicitly stated otherwise as unCP. Density fitting was employed wherever possible. For wavefunction calculations, CCSD(T)-F12,54 MP2,55 and MP2C56,57 (the TDDFT and MP2 parts) were performed within Molpro58 2009.1 and 2010.1; MP2,55 SAPT0 (which also goes into MP2C interaction energies), and SAPT2+59 have been accessed through the quantum chemistry code Psi4.60 Details of computational methodology stated in Refs. 47 (WFN) and 61 (SAPT) also apply to this work unless contradicted; this includes levels of SAPT theory, MP2 spin-component-scaling (SCS) variants, auxiliary basis sets for density fitting, triples scaling in F12 computations, and equations for basis set extrapolations36,62 and focal point analysis. DFT computations were performed in Psi4 (PBE,63 BP86,64,65 BLYP,64,66 B97,67 PBE0,68,69 B3LYP,70,71 M05-2X,72 LC-PBE,73 B97X-D,74 and B2PLYP;75 density-fitted) and Q-Chem76 4.3 (B97X-V77 and B97M-V38) employing a Lebedev grid with 100 radial and 302 angular points and the SG-1 grid78 where relevant for VV10. Following developer recommendations for best B97M-V performance, a (99,590) (BBI and SSI500) or (75,590) (for remainder of SSI) primary grid was used. DFT dispersion corrections D2,67 D3 (zero-damping),79 D3(BJ),80 and revised D3M and D3M(BJ)39 were accessed through external program dftd3.81 The PBEh-3c method82 was run in Psi4 and by design uses basis set def2-mSVP and no explicit counterpoise correction (an effective geometrical counterpoise correction (gCP) is applied among the three corrections of “3c”). The performance of a method and basis set (together, a theoretical model chemistry83) for a given database is evaluated by comparing the interaction energy (always in kcal/mol) for each constituent system to that of its reference CBS value and is summarized by the quantity mean absolute error ( ).
Fragment-specific CHARMM atom types, bonded parameters, and charges were assigned using version 1.0.0 of the CHARMM General Force Field (CGenFF) program.84,85 Vacuum interaction energies were calculated with CHARMM version 39b2,86 reading in the aforementioned fragment-specific parameters on top of version 3.0.1 of the CHARMM general force field.87,88 No optimization was performed on the fragment pair for this purpose, i.e., CHARMM’s INTEraction command was applied directly to the PDB geometry, with the distance cutoffs for all nonbonded interactions set to 997 Å in order to ensure recovery of the full interaction energy. GAFF results were generated with Amber,40 while semi-empirical methods Austin Model 1 (AM1)89 and Parameterized Model 6 with empirical corrections for dispersion and hydrogen-bonding interactions and 2nd-generation (PM6-DH2)90 were run in MOPAC2009.91
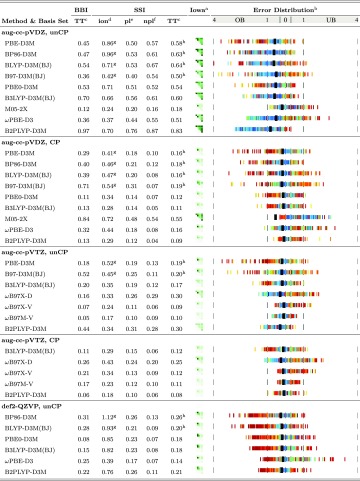
While summary statistics are valuable, a model chemistry can be more completely assessed by a visual representation of errors from all database members. Results for several of the best methods examined in this work are illustrated by “thread” and “Iowa” plots in, respectively, Tables II–IV (rightmost column) and Figs. 4 and 5. The conventions for these graphs are outlined here. For thread plots, each horizontal strip represents the results for the database (or multiple databases in Fig. 3) with a given model chemistry. Thin vertical lines plot the error in interaction energy (kcal/mol) for each member of the database in either the underbound (right of the zero-error line) or overbound (left of the zero-error line) sector of the chart. Individual quantities lying beyond the range of the graph are omitted without annotation. The vertical lines are colored according to SAPT decomposition (vide infra Sec. III A); typically the member is red if hydrogen-bonded or blue if dispersion-bound. A black rectangular marker indicates the MAE over all the databases, with its position being fixed in the “overbound” sector of the graph for ease of comparison to the zero line, regardless of the preponderance of individual subset markings.
TABLE II.
Interaction energy (kcal/mol) MAE statistics by force-field and semi-empirical methods.
SSI MAE (dark by 1.0 kcal/mol) for subsets in residue classes cation, anion, polar, aliphatic, and aromatic (L to R).
SSI Errors with respect to benchmark within 4.0 kcal/mol. Guide lines are at 0.0, 0.3, 1.0, and 4.0 overbound (−) and underbound (+).
Total database.
Charged block: union of +/+, +/−, −/−.
Polar-containing: union of +/pl, −/pl, pl/pl, pl/al, pl/ar.
Nonpolar block: union of al/al, al/ar, ar/ar.
Missing 1 of 230 interactions.93
Missing 1 of 1696 interactions.93,94
Missing 5 of 3380 interactions.93,94
Missing 2 of 230 interactions.94
TABLE III.
Interaction energy (kcal/mol) MAE statistics by wavefunction methods, CP-corrected.
SSI MAE (dark by 1.0 kcal/mol) for subsets in residue classes cation, anion, polar, aliphatic, and aromatic (L to R).
SSI Errors with respect to benchmark within 4.0 kcal/mol. Guide lines are at 0.0, 0.3, 1.0, and 4.0 overbound (−) and underbound (+).
Total database.
Charged block: union of +/+, +/−, −/−.
Polar-containing: union of +/pl, −/pl, pl/pl, pl/al, pl/ar.
Nonpolar block: union of al/al, al/ar, ar/ar.
TABLE IV.
Interaction energy (kcal/mol) MAE statistics by DFT methods.
SSI MAE (dark by 1.0 kcal/mol) for subsets in residue classes cation, anion, polar, aliphatic, and aromatic (L to R).
SSI Errors with respect to benchmark within 4.0 kcal/mol. Guide lines are at 0.0, 0.3, 1.0, and 4.0 overbound (−) and underbound (+).
Total database.
Charged block: union of +/+, +/−, −/−.
Polar-containing: union of +/pl, −/pl, pl/pl, pl/al, pl/ar.
Nonpolar block: union of al/al, al/ar, ar/ar.
Missing 2–7 of 230 interactions.95
Missing 2–7 of 3380 interactions.95
FIG. 4.
Performance of [(a)–(e)] force field and semiempirical, [(f)–(k)] density functional, and (continuing in Fig. 5) [(l)–(p)] wavefunction methods.
FIG. 5.
Continuing from Fig. 4, [(l)–(p)] performance of wavefunction methods.
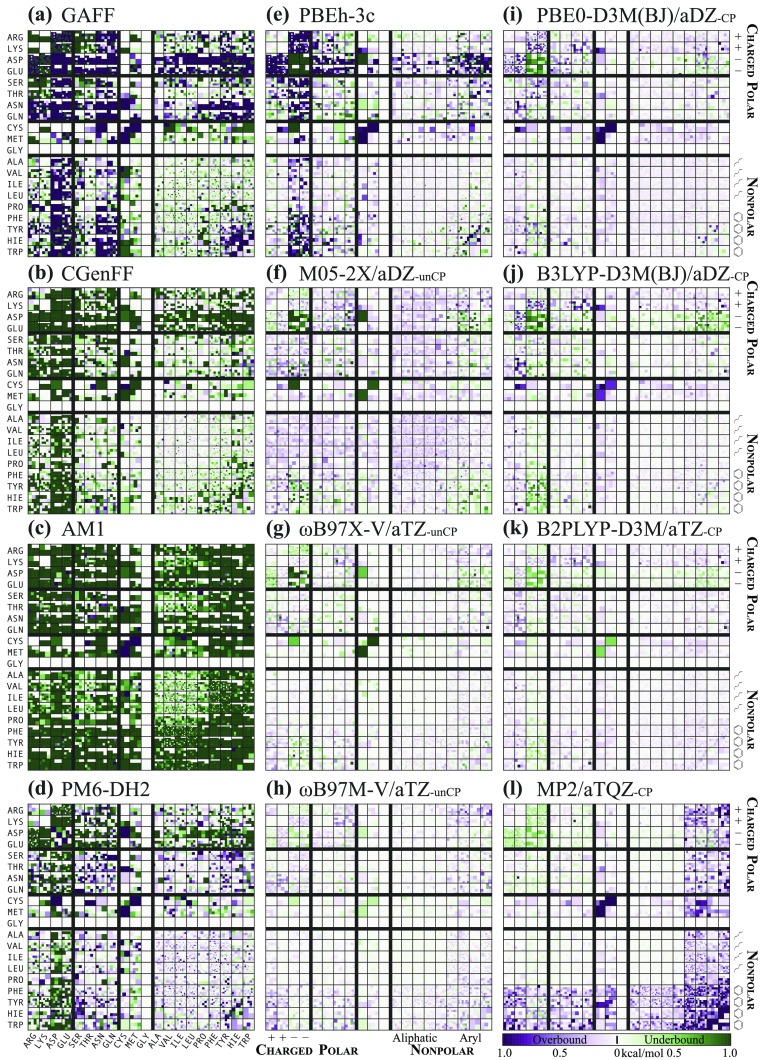
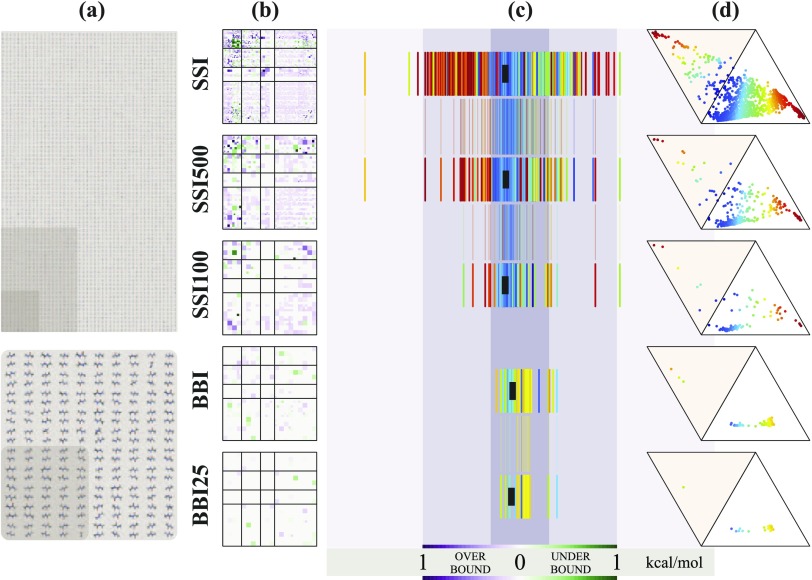
An Iowa plot (e.g., Fig. 4) also depicts the performance of a given model chemistry for an entire database, albeit only for databases composed of inter-residue interactions. The model chemistry error for each database member is depicted by a color scale, where intensity of green reflects the degree to which the system is underbound, intensity of purple reflects the degree to which the system is overbound, white reflects no error, and intensities are saturated at kcal/mol. The colored squares for the subset of database members drawn from contacts between two given amino acids are tiled to form a larger square (blank slots are left when subset is not a square number). Amalgamated squares are arranged in a 20 × 20 array indexed by amino acids ordered roughly by polarity. The resulting plot is symmetric across a diagonal axis (in information if not, strictly, visually), easily examined by subsets of interactions (e.g., anionic or polar/aromatic), and, since an individual database member’s square is depicted as larger or smaller depending on the frequency of that interaction type (i.e., PRO/TRP) in the database, resembles an aerial view of farmland in the American Midwest. Together, thread and Iowa plots provide a good visual representation of the performance of a model chemistry for a protein-based database. Where thread plots require comparisons between several to reveal subset differences, Iowa plots show subsets side-by-side. Where thread plots obscure database member identity except as thread color, Iowa plots sort by residue contacts. Where Iowa plots obscure the worst outliers by saturating the color scale at kcal/mol, thread plots show the full error range.
III. RESULTS AND DISCUSSION
A. SAPT components
SAPT decompositions of fragment interactions from each of the BFDb databases are shown as ternary representations in Figs. 2(a)–2(d). SSI and BBI computations are SAPT2+/aug-cc-pVDZ,92 though ternary representations are particularly insensitive to SAPT level of theory.61 In Fig. 2, the position of a database member reflects the relative contributions of its three attractive physical components (electrostatics, induction, and dispersion), and color reflects the relative contributions of its electrostatics and dispersion components. This color is used in Figs. 1(c), 3, 6(c), and 6(d) and Tables II–IV (rightmost column), as it provides a quick visual distinction between polar/hydrogen-bonded complexes in red and nonpolar/dispersion-dominated complexes in blue. For BBI in Fig. 2(d), the two binding motifs of backbone-backbone interactions, single hydrogen-bonded (SHB) and unaligned (UA), are apparent in the separate clusters. In contrast, the SSI database in Fig. 2(c) spans a significant fraction of the space of noncovalent interactions. This is especially clear in comparison with a general-purpose organic database such as S22, even extended to dissociation curves by S22x5, in Fig. 2(e). Though sharing many binding motifs with S22, SSI is far more comprehensive through its sampling of a large number of interactions and its inclusion of charged systems (which tend to have a stronger induction component) and non-equilibrium systems (which often have a repulsive electrostatics component).
FIG. 6.
Representative subsets of BBI and SSI. The SSI500, SSI100, and BBI25 subsets are shown in relation to their parent databases in size (a), in breadth of SAPT range (d), and in error range and distribution for a specific model chemistry [PBE0-D3(BJ)/aDZ-CP] (b) and (c).
B. Modeling performance
The BBI and SSI databases have been surveyed by a broad range of approaches to molecular modeling, from force-fields to coupled-cluster wavefunction theory. Web portal access to the full results by individual bimolecular complex is discussed in Sec. III D. Full model chemistry summary results (including most of the numbers quoted in the discussion below) are reported in the supplementary material. Abridged (by model chemistries and statistics reported) results are given in Table II (FF/SE), Table III (WFN), and Table IV (DFT). Summary Iowa plots for SSI with all force-field and semiempirical methods and selected DFT and wavefunction methods are presented in Figs. 4 and 5. Since more intense colors reflect larger errors with respect to the benchmark, it is clear from Figs. 4 and 5 that the expected progression in accuracy from force-field [(a) and (b)], to semi-empirical [(c)–(e)], to quantum chemical [(f)–(o)], to benchmark (p) holds. Iowa plots are not useful for BBI since the identity of the sidechain has little to do with the nature of the backbone fragment interaction.
It is helpful to discuss subsets of NCI binding motifs, and a convenient surrogate within SSI is amino acid sidechain classification. (Note that this surrogate is not rigorous since the current fragmentation procedure may truncate the residue’s “characteristic” functional group.) This work uses + -charged residues (ARG and LYS), − -charged residues (ASP and GLU), polar residues (SER, THR, ASN and GLN), aliphatic nonpolar residues (ALA, VAL, ILE, LEU and PRO), and aromatic nonpolar residues [PHE, TYR, HIE (neutral form with hydrogen at epsilon N), and TRP], with the remainder unclassified [CYS, GLY (absent since sidechain is trivial), and MET]. Designations can be made for subsets of interactions: pl, where one monomer is polar; −/al, where one monomer is aliphatic and the other negatively charged (intersection of al and −); the nonpolar block (union of al/al, ar/ar, and al/ar); and the charged block (union of +/+, −/−, and +/−). These five primary classes (+, −, pl, al, and ar) form the axes of the small plot in the “Iowa” column of Tables II–IV which illustrates the subset MAE of the 15 intersections (e.g., the upperrightmost square is +/ar); see Fig. S-2 of the supplementary material for a more explicit mapping. Meanwhile, BBI is discussed in terms of single hydrogen bond (SHB) and unaligned (UA) subsets determined by SAPT classification. Every number in Sec. III B and Tables II–IV, unless explicitly labeled otherwise, refers to an overall or subset MAE in kcal/mol. Note that the average IE varies considerably between contact types (see Tables S-3 and S-4 for figures of the supplementary material), with subset mean IE ranging from −73 to +69 kcal/mol. Naturally, the largest absolute errors (for ions) accompany the largest IE.
1. Force-fields and semi-empirical
Figure 4 shows strong colors for force-fields (a) GAFF and (b) CGenFF and semi-empirical (c) AM1 and (d) PM6-DH2, indicating large, widespread errors. Partially mitigating this assessment, which references in vacuo IE, is that force field parameterization roughly takes into account solvation by water. Nevertheless, large errors indicate that significant improvement in GAFF is possible. GAFF is at its best for the nonpolar block and other non-anion aliphatics (0.22–0.67), while the remaining BBI and SSI subsets have MAE 1, leading to overall 5.48 and 2.72, respectively. Overbinding of anions is the force field’s greatest weakness, yielding 15.24 for +/− and 94.24 for −/−. CGenFF does considerably better in overall statistics (0.51 for BBI and 1.30 for SSI). In addition to having less extreme outlier subsets (6.92 for +/− and 7.94 for −/−; generally underbinding), non-charged polar and SHB join the GAFF good performers as reasonably well-treated, leading to 0.22–0.84 for the polar/nonpolar block (+/al also) and 0.13 for al/al itself.
The semiempirical AM1 is almost uniformly underbinding, as seen from Fig. 4(c), where the only pale region is al/al at 0.50. That its overall BBI/SSI at 4.04/2.54 are comparable to GAFF is due to significant improvement in −/− to 3.07 since very many of the subsets, especially nonpolar-containing, are treated worse by AM1 than by either force field. PM6-DH2 yields significant errors for anions, +/pl, and UA (3.63 for +/−, others 0.95–1.45) but otherwise (0.7) is practicable over wide regions of NCI space at low cost, returning overall 0.70 for BBI and 0.65 for SSI. Its best performance is for the same nonpolar block and non-anion aliphatics (0.17–0.34) at which GAFF succeeded (though roughly 40% better). PBEh-3c in Fig. 4(e), though properly a DFT method, is discussed here as it is highly prescriptive and seeks (and succeeds at 0.46/0.41 overall BBI/SSI) to provide decent accuracy at low cost. Its weakness is − (excepting −/al), where it actually does worse than PM6-DH2 for −/−. Otherwise, those methods compare very much in favor of the former, with ten subsets’ error reduced to at least half. Apart from the already described − subsets, all but SHB and +/pl have MAE .
2. SAPT and wavefunction
One SAPT, 33 MP2-level, and 12 post-MP2 model chemistries are assessed. SAPT2+/aDZ (a SAPT Pauling point in Ref. 61) as a method itself [Fig. 5(o)] is very good for aliphatic interactions, with BBI UA and SSI pl/al, al/al, al/ar having 0.06. Its interactions involving anions (0.43; generally overbinding), particularly with other anions (1.29), are a clear shortcoming. The bronze standard MP2C-F12/aDZ in Fig. 5(p) (and its co-standard47 MP2C/[aTQZ; :aDZ]36) has greatest discrepancy from the (silver) reference for −/− and +/− in terms of the absolute error (MAE 0.32–0.43, though MA%E within 1.3%). The MP2 basis set limit—MP2-F12/aDZ and MP2/aTQZ [Fig. 4(l)]—equals or surpasses bronze in performance for pl and al (0.04–0.14) and anion complexes (0.07–0.33), always excepting combinations with ar, though the last reflects weakness in bronze as well. Aromatics (ar) are handled very poorly (ar/ar in particular 0.7 as expected96 for MP2/CBS) such that BBI, containing exclusively polar and non-polarizable dispersion interactions, has overall MAE 0.10 (half that of bronze) whereas SSI with abundant contacts has overall 0.21 (double that of bronze).
The basis-set convergence behavior of MP2 variants (convergence analysis excludes the un-augmented QZ basis) is best seen in Table S-7 or S-21 of the supplementary material. For MP2 itself, aliphatic and +/+ are converged by aTZ and polar by aDTZ, all to 0.15, excepting combinations with ar and −/pl that converges slower. Within the charged block, subsets −/− and +/− converge to 0.3 by aTQZ. Not unexpectedly, aromatic interactions are a weakness, particularly +/ar and ar/ar (0.33–0.73 for aTZ) which get worse with respect to basis treatment, particularly for extrapolations. For SCS-MP2, the nonpolar block does not improve beyond 0.45 by aDTZ. Otherwise the method is generally poor (0.5–3.0, including BBI). For SCS(N)-MP2, aromatic interactions not involving anions are quite good by aTZ as is SHB (0.11–0.31), while other subsets do no better than 0.3–0.5 and UA and −/− are poor (0.6). For SCS(MI)-MP2, ar/ar converge by aDTZ (0.16) and −/− is universally poor (0.8), but otherwise aTZ is the best basis choice [Fig. 5(m)], yielding all subsets (excepting −/−) 0.10–0.37 and overall BBI 0.15 and SSI 0.22. Considering the ar/ar high-error hotspot of converged MP2 and the −/− hotspot of every other MP2-based model chemistry, DW-MP2 is the only method to present an even error level across binding motifs, as shown by uniform color in the Iowa column of Table III for aTZ. Among these, extrapolated basis treatments are the best, with overall BBI/SSI 0.42/0.37 for aDTZ [Fig. 5(n)] and 0.32/0.35 for aTQZ. While the −/− subset is challenging even for augmented-triple- (apart from MP2 and DW-MP2 at 0.3–0.5, methods yield 0.8–1.4), QZ at 2.2–3.0 shows lack of diffuse functions to be far worse.
3. DFT
Twelve density functionals are assessed that lead to 244 model chemistries after basis set (aDZ, aTZ, and QZVP), BSSE treatment (unCP and CP), and dispersion [None, D2, D3, D3(BJ), D3M, and D3M(BJ)] combinatorics. Generalizations about a functional consider only the four D3 variants unless otherwise specified. “D3” itself means the original zero-damping variant. Similarly, when the shorthand is used with basis-BSSE treatments, the underlying ordering is aDZ-unCP, aDZ-CP, aTZ-unCP, aTZ-CP, QZVP-unCP, and QZVP-CP.
Considering all DFT model chemistries mingled together, DFT at its best rivals the benchmark error tolerance for certain NCI motifs. In particular, all BBI subsets and amongst SSI all al, non-anionic ar, pl/pl, and +/+ subsets have the best MAE, 0.03–0.07 kcal/mol. At the other end of the gamut, −/− contacts are such a challenge that best is 0.15 and only 19% of (DFT) model chemistries have MAE 0.5. Of the remaining, anion subsets (+/−, −/pl, and −/ar) and +/pl have 0.09–0.14 at best (only 39% 0.5 for +/−). The best result for BBI overall is 0.05 and for SSI overall is 0.08. In discussions below of particular model chemistries, the phrase “among the best DFT” means that the MAE for the subset in question is within 0.05 (0.10 for the charged block, where IE are so much greater in magnitude) of the best value listed above among the 244 DFT model chemistries. Most subsets have plenty of good performers by absolute error, even outside the “best” labeling, but −/− is so challenging that many rating “best −/−” are overall poor performers, with anions their sole strong point; only B97M-V and certain B3LYP- and B2PLYP-D3 variants, all unCP with aTZ, incorporate good anion and good general results.
DFT success is unsystematic—a rung 4 functional is not always better than a rung 2, a QZVP basis is not reliably better than an aDZ, D3(BJ) is not assuredly better than D2 (though it almost always is and any D is always better than None), and CP is not necessarily better than unCP. For example, overall SSI averaged for all QZVP-unCP is 0.23 vs. 0.19 for all aDZ-CP and the respective best for both is 0.12, yet neither best functional is uncommonly good upon basis-BSSE exchange with the other. Thus, the best DFT choice depends on the chemical system; the following discussion and elaborate tables in the supplementary material provide some guidance.
Among the simplest functionals (GGAs), aDZ and QZVP results are available for all, while PBE and B97 analyses additionally covers aTZ. PBE always has MAE 0.8 for at least one of −/− and +/−, making its overall utility low. Nevertheless, D3(BJ)-CP obtains satisfactory results, with aDZ and aTZ best, producing overall (0.21–0.23) and subsets (0.15–0.45, excepting −/−). Refitting the damping function39 improves results for nearly all subsets so that PBE-D3M(BJ) lower bounds decline to 0.19 overall and 0.11 subset over the same aDZ and aTZ with CP. For BP86, despite occasional good figures for al/al, al, and UA, poor performance for −/− and +/− and mediocre for other subsets makes BP86 unrecommendable. Refitting the dispersion damping function improves overall SSI by 0.04 on average. BLYP does quite well for the nonpolar block with basis set treatments aDZ-CP, especially D3(BJ) at 0.08–0.18 (among the best DFT for al/ar) and 0.13–0.28 for UA in BBI, yielding 0.19–0.21 for SSI overall. The refit D3M(BJ) additionally merits best DFT labels for pl/al and al/al. Yet −/− and +/− are so bad (0.46) that BLYP is not recommended in general. B97 similarly does well for the nonpolar block, +/+, and UA at aDZ-CP, particularly with D3 at 0.10–0.23 [and, after refitting, D3M(BJ) at 0.03–0.22], leading to 0.21–0.26 (0.17–0.20) for overall SSI. Despite its promise for conventional organic NCI, certain charged and polar subsets (SHB, +/−, −/−, −/pl, and pl/pl) have substantial errors (often 0.5) even at the highest basis treatments; these are computed much better at aDZ-unCP, suggesting an unbalanced parameterization.
As the functionals’ general habits for NCI are known,38,77,97 M05-2X is computed only with aDZ and B97X-D, B97X-V, and B97M-V only with aTZ. For M05-2X, aDZ-unCP [Fig. 4(f)] is remarkably good at overall BBI 0.12 and SSI 0.18. Apart from −/− at 0.86, subsets are 0.10–0.31, and SHB, +/+, +/− are among the best DFT. It should be noted that other studies24,97,98 have found this functional weak at non-equilibrium geometries, especially short-range (the PDB contact identification procedure precludes long-range NCI in BBI/SSI). With counterpoise correction, M05-2X aDZ-CP is uniformly bad, having systems strongly skewed toward underbound and only certain pl and al subsets 0.5. For B97X-D, unCP is better for SHB, −, pl/pl, while CP is better for UA and the remainder. Both are poor for −/− (1.0), weak for +/+, +/ar, −/ar (0.37–0.57), and middling for other SSI (0.18–0.39), averaging to about the same: unCP at 0.16/0.30 BBI/SSI and CP at 0.26/0.25. Although expensive, B97X-V is among the best and most consistent functionals examined. For unCP [Fig. 4(g)], all subsets (excepting −/− at 0.58) are 0.03–0.23 and all of those but +/ar (0.15) and ar/ar (0.11) are among the best of DFT. CP in comparison computes most of the SSI subsets slightly worse, while BBI subsets are considerably worse. Overall statistics are 0.07/0.09 BBI/SSI for unCP and 0.21/0.12 for CP. More expensive still due to the kinetic energy density dependence and larger grid, B97M-V with aTZ-unCP in Fig. 4(h) gives good performance for the greatest weakness of B97X-V— −/− at 0.24—while also improving −, pl/pl, and BBI subsets by up to 0.09. In turn, its own weakness is aromatic systems, for which subset values are 0.02–0.06 worse (excepting −/ar as previous) than the non-meta functional. The resulting overall metrics are about the same at 0.05/0.10 BBI/SSI. CP likewise is comparable or worse than unCP for all but +/ar.
Basis sets aDZ and QZVP are available for others in the PBE family. For PBE0, aDZ-unCP is remarkably evenhanded with D2. Though no subset excels (0.31–0.67), the worst (+/−) is better than the worst of M05-2X/aDZ-unCP (−/− at 0.86). For aDZ-CP, all binding motifs except charged interacting with −, pl, or ar perform well, especially using CP and D3(BJ), D3M, or D3M(BJ). By QZVP, +/− and −/− are quite bad, so D3M(BJ) with aDZ-CP is the optimal PBE0 model chemistry [Fig. 4(i)], producing 0.08 BBI and 0.13 SSI which are among the best DFT overall as well as for subsets SHB, UA, +/+, most al, and non-ionic ar, despite its weakness at −/− (0.58). PBE is best with D3 or D3M, and aDZ-CP yields overall BBI 0.19–0.33 (SHB and UA generally underperforming for this functional) and SSI 0.14–0.16. Although the usual weakness of −/− (especially severe at 1.4) and the particular weakness of −/ar (0.40–0.52) are present, strengths (0.03–0.21) include the nonpolar block, al, and certain additional cation (+/+ and +/pl) subsets, many of which are among the best DFT. The performance of aDZ-CP is nearly identical to QZVP-CP, and QZVP-unCP is somewhat superior to both.
For B3LYP, basis treatments aDZ-CP are roughly equal and quite good (excluding D3 with unCP and D3M in general) overall (0.10–0.20), but aDZ-CP and all aTZ avoid the severe −/− and +/− errors that arise by QZVP. D3/D3M is a bright spot for −/− (and anions in general), producing statistics at least 0.15 lower than D3(BJ)/D3M(BJ) counterparts and achieving 0.23–0.25 with aTZ-unCP, which is among the best DFT (actually aDZ-unCP earns this rating with D3M, but as its sole advantage). However, at the optimal aDZ-CP treatment, all subsets but SHB and − are treated better by D3M(BJ) [Fig. 4(j); 0.03–0.22] than by D3 (0.11–0.32), so the choice depends on the targeted NCI. D3 has −/− at 0.41, SHB and −/al among the best DFT, and overall 0.13/0.15 BBI/SSI, while D3M(BJ) has larger −/− at 0.55 yet many aliphatic and aromatic subsets (pl/al, pl/ar, al/al, al/ar, ar/ar, and overall) among the best DFT, and overall 0.13/0.11 BBI/SSI.
B2PLYP at aDZ-CP performs creditably (overall SSI 0.12–0.27) among non-refitted -D, and with D2 frequently as good as D3 and D3(BJ) (D2 aTZ-CP overall BBI/SSI 0.21/0.13, having most subsets 0.25, many among the best DFT). For good anion performance (for once including −/−), aTZ-unCP with any un-refitted -D treatment is to be recommended (overall 0.14–0.27), though D3 is best, producing − at 0.06–0.21, though with attendant poor (0.17–0.35) aromatic results. QZVP-unCP is notably worse for anions (especially +/−, −/−, −/pl) but otherwise good (all subsets 0.04–0.25). Dispersion refitting yielded excellent CP functionals, the best of which is D3M/aTZ-CP [Fig. 4(k)], for which every subset (excepting −/− at 0.37 and −/ar at 0.18) is either 0.15 or among the best DFT. Less costly, D3M/aDZ-CP also maintains (−/− at 0.50 excepted) all but SHB and non-−/al anion subsets within or among the best DFT. Their weaknesses are UA and certain aromatics (+/ar and ar/ar) for aTZ (0.11–0.13) and SHB and anions for aDZ (0.16–0.31), but their overall BBI/SSI figures show aTZ (0.06/0.08) and aDZ (0.13/0.09) to be outstanding performers.
4. Reappraisals
As BBI and SSI survey the space of NCI more extensively than earlier works and partition into more subsets, it is worthwhile to compare results and recommendations. From several databases comprising 3547 systems, Mardirossian and Head-Gordon reported RMSE for B97-D3(BJ) (1.73 kcal/mol), B97X-V (0.63), and B97M-V (0.49) across their subset of difficult NCI with unCP def2-QZVPPD.38 This relative ordering, the disappointing B97-D, and the excellent performance of the nonlocal dispersion functional is consistent with results in this work, although this work also finds subsets where X-V outperforms M-V. Goerigk et al. evaluate the S66 and S66x8 databases using unCP def2-QZVP, yielding M05-2X (0.58 MAE kcal/mol for the former), PBE-D3(BJ) (0.40), B97-D3(BJ) (0.29), B3LYP-D3(BJ) (0.28), B2PLYP-D3(BJ) (0.21), BLYP-D3(BJ) (0.19), and PBE-D3(BJ) (0.19) for DFT and SCS-MP2 MP2 SCS(MI)-MP2 DW-MP2 for MP2-level WFN, where inequalities refer to performance judged by the MAE.98 Such figures and ordering are not unlike what the present work finds for a conventional binding motif like pl, but considerably larger and considerably smaller MAEs are possible across the full diversity of SSI. In Hobza and co-workers’ survey of PDB contacts,35 they computed B3LYP- and BLYP-D3/QZVP-CP for some general categories of interaction, e.g., anion, neutral, polar, and aromatic. Loose mapping of subsets to the current project yields differences in the MAE usually well within 0.1 kcal/mol, though they find a much greater difference between B3LYP and BLYP performance (in the former’s favor) and less extreme ion subset errors. Regarding earlier recommendations from some of the authors based upon S22, NBC10, HBC6, and HSG databases,97 the following (overlapping) functionals were recommended for general NCI with aTZ-unCP: B97-D3 B3LYP-D3 B97X-D. While the latter two are, in this work, satisfactory functionals, B97-D does badly for several subsets, and the development of both dispersion (D3M) and functional family (B97) means that several attractive alternatives are available. Regarding wavefunction recommendations, DW-MP2 fulfills its role of balanced treatment of NCI subsets, if undistinguished overall, while SCS(MI)-MP2 parameterization is not able to cope with some of the new subsets (−/− and −/ar). For SAPT2+/aDZ, +/+ and +/− errors are much higher than predicted from previous work,61 while other subsets (UA, al/al, and al/ar) are as or better than expected.
C. Representative subsets
Since method development often involves separate fitting and validation stages and also since SSI can be unwieldy in its entirety, subsets of the BBI and SSI databases have been defined. These are BBI25, SSI100, and SSI500 (middle is a subset of last) shown in Fig. 6. Selection was made by generating dozens of random subsets from the whole, winnowing by minimizing the difference in the MAE and maximum error between the whole and the subset for several wavefunction and DFT model chemistries, and choosing one that is representative and diverse as gauged by ternary and Iowa plots. Gauges are shown in Fig. 6, and statistics for these subsets are reported for all model chemistries in Tables S-5–S-19 of the supplementary material. Among the nearly 300 DFT and wavefunction model chemistries, BBI25 and SSI500 differ by no more than 0.03 kcal/mol in the MAE from their respective whole databases. Force fields and semiempirical methods are more variable with MAE differences up to 0.11 kcal/mol. The much smaller SSI100 subset exhibits correspondingly larger variance: MAE within 0.05, 0.11, and 0.19 kcal/mol for wavefunction, DFT, and FF/SE classes, respectively. Naturally, the statistical subsets are less effective models of the whole when considering individual binding motif subsets.
D. Web portal
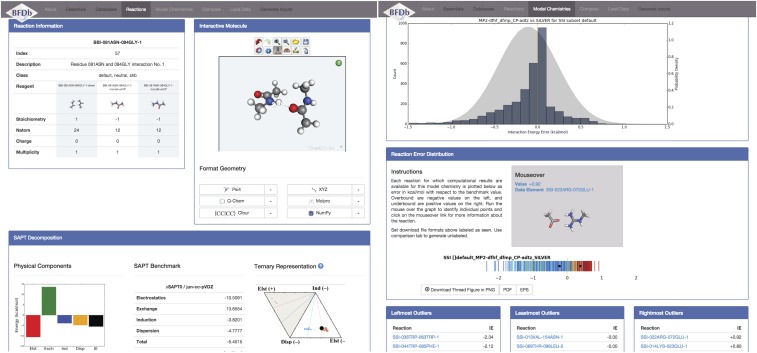
This work, which explores dozens of model chemistries for a database with thousands of members, has yielded large quantities of data (1 IE) whose usefulness is in en masse comparison rather than in simple look-up. Additionally, considerable metadata are associated with both the databases (charge, multiplicity, geometry, subset classification, SAPT decomposition, etc.) and the computational approach (density-fitting, counterpoise correction, dispersion variant, basis treatment, etc.). The sizeable new infrastructure required to manage and use the BFDb databases is encoded into a Python module, Quantum Chemistry Common Driver and Databases (QCDB).99 The BFDb project is the first major (second actual39) use of the database features of QCDB. These tools are made available to the scientific community through the BFDb portal at http://vergil.chemistry.gatech.edu/bfdb/. The portal consists of several pages for different stages of research. For a selected model chemistry, a page [Fig. 7(right)] presents its histogram and thread error distribution and statistics with respect to a reference, as well as Iowa plots if relevant, all restrictable by subset. For a selected database member, a page [Fig. 7(left)] presents its manipulatable geometry, SAPT decomposition, and tabular and thread error distribution for all available model chemistries. For a selected database, a page generates thread error distributions for any number of model chemistries or subsets, as configurable by the user. Whenever possible, DOI references to the published literature are supplied. At a further level of interaction, any user can upload new interaction energy datasets to the portal, which can then be analyzed with all the usual tools and alongside preloaded modeling results. Additionally, input geometries for a number of popular quantum chemistry packages can be produced.
FIG. 7.
BFDb web portal at http://vergil.chemistry.gatech.edu/bfdb. Screen shots sample website contents for a given bimolecular complex (left) and database model chemistry (right).
The flexibility of the BFDb portal is possible because all pages and figures are generated on-the-fly from the QCDB Python module storing the reaction energy results and facilitating chemical database manipulations. Throughout, generated plots and geometry and input files can be downloaded in different formats. Indeed, Figs. 3–5 and parts of Fig. 6 are composed of figures downloaded from the portal and slightly marked up. Additionally, the geometries and reference IE of SSI and BBI are available in Psi4 through the database () feature to simplify job execution and result collection. Finally, for those preferring command-line access, a Jupyter100 notebook sufficient to produce components of all tables and figures is included in the supplementary material. Currently loaded onto the BFDb portal are computational chemistry results from this work and Refs. 47, 61, 97, and 101 encompassing databases from BFDb (BBI, HSG, and SSI) and others (A24, HBC6, NBC10, and S22).
IV. SUMMARY AND CONCLUSIONS
This work has harvested distinct backbone/backbone and sidechain/sidechain intermolecular contact geometries from the Protein DataBank (PDB) and collected them alongside high-quality DW-CCSD(T**)-F12/aug-cc-pV(D+d)Z reference interaction energies into two chemical databases. The resulting SSI and BBI manifest several contrasts in construction compared to conventional organic databases. Namely, instead of near-equilibrium or scanned-coordinate geometries, BFDb collects 3000 bifragment contacts directly from biological systems. Instead of curating a limited number of “representative” complexes, BFDb samples many variations on a NCI binding motif. Instead of assigning broad classifications, BFDb provides SAPT decompositions and subsetting by residue/residue contacts. Instead of exerting great effort on a few heroic gold-standard computations, BFDb uses a tractable but accurate silver-standard coupled-cluster benchmark.
Having defined the core geometries and reference energies, a number of efforts have been made to package BFDb for the computational community. Dozens of model chemistries from all classes of computational chemistry are assessed. Subsets of convenient size for any application have been assembled to reflect the statistics and composition of the whole. A web portal is available to view error statistics, outliers, and visualizations by database and NCI class for very many model chemistries and references. Future work will look at the effect of geometry optimization and larger datasets that have a broader range of interaction types.
Surveying nearly 300 model chemistries necessitates only broad analysis, so those seeking advice for a known system type are best served by running a finger down the Iowa column of Tables II–IV [or their expanded (by model chemistry) and far more detailed (by binding motif subset) counterparts in the supplementary material]. Nevertheless, conclusions as to generally commendable functionals follow. Unsurprisingly, the balanced and successful performers were among those encoding the most physics in the most expensive methods: the double-hybrids and the non-local dispersion functionals. In no particular order, the five best all-around model chemistries are B2PLYP-D3M/aTZ-CP, B3LYP-D3M(BJ)/aDZ-CP, PBE0-D3M(BJ)/aDZ-CP, B97M-V/aTZ-unCP, B97X-V/aTZ-unCP, with overall MAE 0.05–0.13 kcal/mol. Best performance by one of the previous model chemistries taking computational cost into account goes to B3LYP-D3M(BJ)/aDZ-CP; best performance by a GGA goes to BLYP-D3M/aDZ-CP; by an unCP GGA, PBE-D3M/aTZ-unCP; by an unCP, B97X-V/aTZ-unCP; by an aDZ, B2PLYP-D3M; by an unCP aDZ, … is not recommended; by an unCP aTZ without VV10, B2PLYP-D3(BJ)/aTZ-unCP; for the contacts most difficult to compute, −/−, B2PLYP-D3M/aTZ-unCP; for ar/ar contacts, B2PLYP-D3M or -D3M(BJ)/aDZ-CP; for the most difficult to get right simultaneously, anion and aromatic, B97M-V/aTZ-unCP and B2PLYP-D3M/aTZ-CP; for contacts classified as electrostatically dominated by SAPT, B97M-V/aTZ-unCP; for contacts classified as dispersion dominated by SAPT, B2PLYP-D3M/aDZ-CP; for contacts classified as mixed influence by SAPT, B2PLYP-D3M/aTZ-CP. Best performance by a method with formal scaling less than O(N3), DH2-PM6; by an MP2 derivative, DW-MP2/aDTZ-CP; by an MP2 derivative without contrivance, MP2/aDTZ-CP; by a method available in Psi4 1.0, B2PLYP-D3M/aTZ-CP; by a method available in Psi4 1.2, B97M-V/aTZ-unCP.
SUPPLEMENTARY MATERIAL
See supplementary material for Table S-1 defining summary error statistics; Fig. S-2 illustrating how to read Iowa plots; Tables S-3 and S-4 containing summary statistics for BBI/SSI IE ranges over various subsets; Tables S-5–S-19 extending manuscript Tables II–IV to all model chemistries surveyed; and Tables S-20–S-39 presenting MAE statistics for all amino acid contact subsets. A Jupyter notebook to access the data and instructions for use is present as a separate file.
ACKNOWLEDGMENTS
This work in the Sherrill group was performed under the auspices of grants provided by the United States National Science Foundation (Grant Nos. ACI-1449723 and CHE-1566192). The Merz and MacKerell groups acknowledge financial support from the United States National Institutes of Health (Merz: R01s GM044974 and GM066859; MacKerell: GM070855, GM072558, and R44GM109635). The authors thank Dr. Narbe Mardirossian for suggesting the testing of the B97M-V functional and for advising how best to run it. The authors acknowledge the PACE advanced computing center at Georgia Tech for providing fantastic amounts of computer time for this project.
REFERENCES
- 1.Lindorff-Larsen K., Piana S., Dror R. O., and Shaw D. E., Science 334, 517 (2011). 10.1126/science.1208351 [DOI] [PubMed] [Google Scholar]
- 2.Huang J. and A. D. MacKerell, Jr., Biophys. J. 107, 991 (2014). 10.1016/j.bpj.2014.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman G. R., Voelz V. A., and Pande V. S., Curr. Opin. Struct. Biol. 21, 4 (2011). 10.1016/j.sbi.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zagrovic B., Snow C. D., Shirts M. R., and Pande V. S., J. Mol. Biol. 323, 927 (2002). 10.1016/s0022-2836(02)00997-x [DOI] [PubMed] [Google Scholar]
- 5.Cooper S., Khatib F., Treuille A., Barbero J., Lee J., Beenen M., Leaver-Fay A., Baker D., Popović Z., and players F., Nature 466, 756 (2010). 10.1038/nature09304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollini M., Domaoal R. A., Thakur V. V., Gallardo-Macias R., Spasov K. A., Anderson K. S., and Jorgensen W. L., J. Med. Chem. 54, 8582 (2011). 10.1021/jm201134m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buch I., Giorgino T., and Fabritiis G. D., Proc. Natl. Acad. Sci. U. S. A. 108, 10184 (2011). 10.1073/pnas.1103547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shan Y., Kim E. T., Eastwood M. P., Dror R. O., Seeliger M. A., and Shaw D. E., J. Am. Chem. Soc. 133, 9181 (2011). 10.1021/ja202726y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon S. L. and K. M. Merz, Jr., J. Chem. Phys. 107, 879 (1997). 10.1063/1.474386 [DOI] [Google Scholar]
- 10.van der Vaart A., Gogonea V., Dixon S. L., and K. M. Merz, Jr., J. Comput. Chem. 21, 1494 (2000). [DOI] [Google Scholar]
- 11.Yang W. and Lee T. S., J. Chem. Phys. 103, 5674 (1995). 10.1063/1.470549 [DOI] [Google Scholar]
- 12.Grimme S., Hansen A., Brandenburg J. G., and Bannwarth C., Chem. Rev. 116, 5105 (2016). 10.1021/acs.chemrev.5b00533 [DOI] [PubMed] [Google Scholar]
- 13.Fukuzawa K., Kitaura K., Uebayasi M., Nakata K., Kaminuma T., and Nakano T., J. Comput. Chem. 26, 1 (2005). 10.1002/jcc.20130 [DOI] [PubMed] [Google Scholar]
- 14.He X. and K. M. Merz, Jr., J. Chem. Theory Comput. 6, 405 (2010). 10.1021/ct9006635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesh V., Dongare R. K., Balanarayan P., and Gadre S. R., J. Chem. Phys. 125, 104109 (2006). 10.1063/1.2339019 [DOI] [PubMed] [Google Scholar]
- 16.Riplinger C., Sandhoefer B., Hansen A., and Neese F., J. Chem. Phys. 139, 134101 (2013). 10.1063/1.4821834 [DOI] [PubMed] [Google Scholar]
- 17.Hobza P. and Řezáč J., Chem. Rev. 116, 4911 (2016). 10.1021/acs.chemrev.6b00247 [DOI] [PubMed] [Google Scholar]
- 18.Yin J., Henriksen N. M., Slochower D. R., Shirts M. R., Chiu M. W., Mobley D. L., and Gilson M. K., J. Comput.-Aided Mol. Des. 31, 1 (2017). 10.1007/s10822-016-9974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson H. A., Smith R. D., Damm-Ganamet K. L., Stuckey J. A., Ahmed A., Convery M. A., Somers D. O., Kranz M., Elkins P. A., Cui G., Peishoff C. E., Lambert M. H., and Dunbar J. B., J. Chem. Inf. Model. 56, 1063 (2016). 10.1021/acs.jcim.5b00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moult J., Fidelis F., Kryshtafovych A., Schwede T., and Tramontano A., Proteins: Struct., Funct., Bioinf. 84, 4 (2016). 10.1002/prot.25064 [DOI] [Google Scholar]
- 21.Monastyrskyy B., D’Andrea D., Fidelis K., Tramontano A., and Kryshtafovych A., Proteins: Struct., Funct., Bioinf. 84, 131 (2016). 10.1002/prot.24943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faver J. C., Benson M. L., He X., Roberts B. P., Wang B., Marshall M. S., Kennedy M. R., Sherrill C. D., and K. M. Merz, Jr., J. Chem. Theory Comput. 7, 790 (2011). 10.1021/ct100563b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuzuki S., Honda K., Uchimaru T., Mikami M., and Tanabe K., J. Am. Chem. Soc. 124, 104 (2002). 10.1021/ja0105212 [DOI] [PubMed] [Google Scholar]
- 24.Sherrill C. D., Takatani T., and Hohenstein E. G., J. Phys. Chem. A 113, 10146 (2009). 10.1021/jp9034375 [DOI] [PubMed] [Google Scholar]
- 25.Řezáč J., Riley K. E., and Hobza P., J. Chem. Theory Comput. 7, 2427 (2011). 10.1021/ct2002946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Řezác J. and Hobza P., Chem. Rev. 116, 5038 (2016). 10.1021/acs.chemrev.5b00526 [DOI] [PubMed] [Google Scholar]
- 27.Řezáč J., Jurečka P., Riley K. E., Černý J., Valdes H., Pluháčková K., Berka K., Řezáč T., Pitoňák M., Vondrášek J., and Hobza P., Collect. Czech. Chem. Commun. 73, 1261–1270 (2008), http://www.begdb.com. 10.1135/cccc20081261 [DOI] [Google Scholar]
- 28.Grimme S., https://www.chemie.uni-bonn.de/pctc/mulliken-center/software/GMTKN/gmtkn.
- 29.Raghavachari K., Trucks G. W., Pople J. A., and Head-Gordon M., Chem. Phys. Lett. 157, 479 (1989). 10.1016/s0009-2614(89)87395-6 [DOI] [Google Scholar]
- 30.Gráfová L., Pitoňák M., Řezáč J., and Hobza P., J. Chem. Theory Comput. 6, 2365 (2010). 10.1021/ct1002253 [DOI] [PubMed] [Google Scholar]
- 31.Řezáč J., Riley K. E., and Hobza P., J. Chem. Theory Comput. 7, 3466 (2011). 10.1021/ct200523a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thanthiriwatte K. S., Hohenstein E. G., Burns L. A., and Sherrill C. D., J. Chem. Theory Comput. 7, 88 (2011). 10.1021/ct100469b [DOI] [PubMed] [Google Scholar]
- 33.Parker T. M. and Sherrill C. D., J. Chem. Theory Comput. 11, 4197 (2015). 10.1021/acs.jctc.5b00588 [DOI] [PubMed] [Google Scholar]
- 34.Jakubec D., Hostaš J., Laskowski R. A., Hobza P., and Vondrášek J., J. Chem. Theory Comput. 11, 1939 (2015). 10.1021/ct501168n [DOI] [PubMed] [Google Scholar]
- 35.Hostaš J., Jakubec D., Laskowski R. A., Gnanasekaran R., Řezáč J., Vondrášek J., and Hobza P., J. Chem. Theory Comput. 11, 4086 (2015). 10.1021/acs.jctc.5b00398 [DOI] [PubMed] [Google Scholar]
- 36.As described in Ref. 47, the basis set treatment aDTZ (aTQZ) is shorthand for aug-cc-pVDTZ (aug-cc-pVTQZ) that itself represents a two-point extrapolation62 of the correlation energy between the named basis set -levels atop an un-extrapolated HF energy at the larger basis. The further focal-point basis set treatment notated post-MP2/[aTQZ; :aDZ] expands to .
- 37.Goerigk L. and Grimme S., J. Chem. Theory Comput. 7, 291 (2011). 10.1021/ct100466k [DOI] [PubMed] [Google Scholar]
- 38.Mardirossian N. and Head-Gordon M., J. Chem. Phys. 144, 214110 (2016). 10.1063/1.4952647 [DOI] [PubMed] [Google Scholar]
- 39.Smith D. G. A., Burns L. A., Patkowski K., and Sherrill C. D., J. Phys. Chem. Lett. 7, 2197 (2016). 10.1021/acs.jpclett.6b00780 [DOI] [PubMed] [Google Scholar]
- 40.Case D. A., Darden T. A., Cheatham I. T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., Goetz A. W., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wolf R. M., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., and Kollman P. A., AMBER 12, University of California, San Francisco, 2012. [Google Scholar]
- 41.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C., Proteins: Struct., Funct., Bioinf. 65, 712 (2006). 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Still W. C., Tempczyk A., Hawley R. C., and Hendrickson T., J. Am. Chem. Soc. 112, 6127 (1990). 10.1021/ja00172a038 [DOI] [Google Scholar]
- 43.The remaining (3358 less 3380) mostly sulfur-containing systems were set aside. As seen in Fig. 1(a), SSI has a low population of sulfur-containing contacts which is due to the parameters of fragmentation and omission of disulfide bonds. While some CYSand METsystems are present, they are not grouped into any of the discussed sidechain subsets: +, −, pl, al, ar.
- 44.Marshall M. S., Burns L. A., and Sherrill C. D., J. Chem. Phys. 135, 194102 (2011). 10.1063/1.3659142 [DOI] [PubMed] [Google Scholar]
- 45.Faver J. C., Benson M. L., He X., Roberts B. P., Wang B., Marshall M. S., Sherrill C. D., and K. M. Merz, Jr., PLoS ONE 6, e18868 (2011). 10.1371/journal.pone.0018868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurečka P., Šponer J., Černý J., and Hobza P., Phys. Chem. Chem. Phys. 8, 1985 (2006). 10.1039/b600027d [DOI] [PubMed] [Google Scholar]
- 47.Burns L. A., Marshall M. S., and Sherrill C. D., J. Chem. Phys. 141, 234111 (2014). 10.1063/1.4903765 [DOI] [PubMed] [Google Scholar]
- 48.Marshall M. S. and Sherrill C. D., J. Chem. Theory Comput. 7, 3978 (2011). 10.1021/ct200600p [DOI] [PubMed] [Google Scholar]
- 49.Dunning T. H., J. Chem. Phys. 90, 1007 (1989). 10.1063/1.456153 [DOI] [Google Scholar]
- 50.Kendall R. A., Dunning T. H., and Harrison R. J., J. Chem. Phys. 96, 6796 (1992). 10.1063/1.462569 [DOI] [Google Scholar]
- 51.Weigend F. and Ahlrichs R., Phys. Chem. Chem. Phys. 7, 3297 (2005). 10.1039/b508541a [DOI] [PubMed] [Google Scholar]
- 52.Dunning T. H., Peterson K. A., and Wilson A. K., J. Chem. Phys. 114, 9244 (2001). 10.1063/1.1367373 [DOI] [Google Scholar]
- 53.Boys S. F. and Bernardi F., Mol. Phys. 19, 553 (1970). 10.1080/00268977000101561 [DOI] [Google Scholar]
- 54.Knizia G., Adler T. B., and Werner H.-J., J. Chem. Phys. 130, 054104 (2009). 10.1063/1.3054300 [DOI] [PubMed] [Google Scholar]
- 55.For MP2 (always density fitted in both SCF and MP2), aDZ was run in Molpro with aDZ JK and RI auxiliary basis sets. aTZ was run in Molpro with aQZ auxiliary bases. QZ was run in Psi4 with QZ auxiliary. aQZ was run in Molpro with a5Z auxiliary.
- 56.Hesselmann A., J. Chem. Phys. 128, 144112 (2008). 10.1063/1.2905808 [DOI] [PubMed] [Google Scholar]
- 57.Pitonak M. and Hesselmann A., J. Chem. Theory Comput. 6, 168 (2010). 10.1021/ct9005882 [DOI] [PubMed] [Google Scholar]
- 58.Werner H.-J., Knowles P. J., Manby F. R., Schütz M., Celani P., Knizia G., Korona T., Lindh R., Mitrushenkov A., Rauhut G., Adler T. B., Amos R. D., Bernhardsson A., Berning A., Cooper D. L., Deegan M. J. O., Dobbyn A. J., Eckert F., Goll E., Hampel C., Hesselmann A., Hetzer G., Hrenar T., Jansen G., Köppl C., Liu Y., Lloyd A. W., Mata R. A., May A. J., Tarroni R., Thorsteinsson T., Wang M., and Wolf A., molpro, version 2010.1, a package of ab initio programs, 2010, see http://www.molpro.net.
- 59.Whereas Ref. 61 recommends and uses exchange scaling factor , consensus on this issue has changed and is used in this work.
- 60.Parrish R. M. et al. , J. Chem. Theory Comput. 13(7), 3185 (2017). 10.1021/acs.jctc.7b00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker T. M., Burns L. A., Parrish R. M., Ryno A. G., and Sherrill C. D., J. Chem. Phys. 140, 094106 (2014). 10.1063/1.4867135 [DOI] [PubMed] [Google Scholar]
- 62.Halkier A., Helgaker T., Jørgensen P., Klopper W., Koch H., Olsen J., and Wilson A. K., Chem. Phys. Lett. 286, 243 (1998). 10.1016/s0009-2614(98)00111-0 [DOI] [Google Scholar]
- 63.Perdew J. P., Burke K., and Ernzerhof M., Phys. Rev. Lett. 77, 3865 (1996). 10.1103/physrevlett.77.3865 [DOI] [PubMed] [Google Scholar]
- 64.Becke A. D., Phys. Rev. A 38, 3098 (1988). 10.1103/physreva.38.3098 [DOI] [PubMed] [Google Scholar]
- 65.Perdew J. P., Phys. Rev. B 33, 8822 (1986). 10.1103/physrevb.33.8822 [DOI] [PubMed] [Google Scholar]
- 66.Lee C., Yang W., and Parr R. G., Phys. Rev. B 37, 785 (1988). 10.1103/physrevb.37.785 [DOI] [PubMed] [Google Scholar]
- 67.Grimme S., J. Comput. Chem. 27, 1787 (2006). 10.1002/jcc.20495 [DOI] [PubMed] [Google Scholar]
- 68.Adamo C. and Barone V., J. Chem. Phys. 110, 6158 (1999). 10.1063/1.478522 [DOI] [Google Scholar]
- 69.Ernzerhof M. and Scuseria G. E., J. Chem. Phys. 110, 5029 (1999). 10.1063/1.478401 [DOI] [Google Scholar]
- 70.Becke A. D., J. Chem. Phys. 98, 5648 (1993). 10.1063/1.464913 [DOI] [Google Scholar]
- 71.Stephens P. J., Devlin F. J., Chabalowski C. F., and Frisch M. J., J. Phys. Chem. 98, 11623 (1994). 10.1021/j100096a001 [DOI] [Google Scholar]
- 72.Zhao Y., Schultz N. E., and Truhlar D. G., J. Chem. Theory Comput. 2, 364 (2006). 10.1021/ct0502763 [DOI] [PubMed] [Google Scholar]
- 73.Vydrov O. A. and Scuseria G. E., J. Chem. Phys. 125, 234109 (2006). 10.1063/1.2409292 [DOI] [PubMed] [Google Scholar]
- 74.Chai J. and Head-Gordon M., Phys. Chem. Chem. Phys. 10, 6615 (2008). 10.1039/b810189b [DOI] [PubMed] [Google Scholar]
- 75.Grimme S., J. Chem. Phys. 124, 034108 (2006). 10.1063/1.2148954 [DOI] [PubMed] [Google Scholar]
- 76.Shao Y. et al. , Mol. Phys. 113, 184 (2015). 10.1080/00268976.2014.952696 [DOI] [Google Scholar]
- 77.Mardirossian N. and Head-Gordon M., Phys. Chem. Chem. Phys. 16, 9904 (2014). 10.1039/c3cp54374a [DOI] [PubMed] [Google Scholar]
- 78.Gill P. M., Johnson B. G., and Pople J. A., Chem. Phys. Lett. 209, 506 (1993). 10.1016/0009-2614(93)80125-9 [DOI] [Google Scholar]
- 79.Grimme S., Antony J., Ehrlich S., and Krieg H., J. Chem. Phys. 132, 154104 (2010). 10.1063/1.3382344 [DOI] [PubMed] [Google Scholar]
- 80.Grimme S., Ehrlich S., and Goerigk L., J. Comput. Chem. 32, 1456 (2011). 10.1002/jcc.21759 [DOI] [PubMed] [Google Scholar]
- 81.See https://www.chemie.uni-bonn.de/pctc/mulliken-center/software/dft-d3/dft-d3 for a FORTRAN program implementing the DFT-D3 method and a file with available C6 coefficients, Grimme Research Group.
- 82.Grimme S., Brandenburg J. G., Bannwarth C., and Hansen A., J. Chem. Phys. 143, 054107 (2015). 10.1063/1.4927476 [DOI] [PubMed] [Google Scholar]
- 83.Pople J. A., in Energy, Structure, and Reactivity: Proceedings of the 1972 Boulder Summer Research Conference on Theoretical Chemistry, edited by Smith D. W. and McRae W. B. (Wiley, New York, 1973), p. 51. [Google Scholar]
- 84.Vanommeslaeghe K. and A. D. MacKerell, Jr., J. Chem. Inf. Model. 52, 3144 (2012). 10.1021/ci300363c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vanommeslaeghe K., Raman E. P., and A. D. MacKerell, Jr., J. Chem. Inf. Model. 52, 3155 (2012). 10.1021/ci3003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brooks B. R., Brooks C. L., A. D. MacKerell, Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., and Karplus M., J. Comput. Chem. 30, 1545 (2009). 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., and A. D. MacKerell, Jr., J. Comput. Chem. 31, 671 (2010). 10.1002/jcc.21367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu W., He X., Vanommeslaeghe K., and A. D. MacKerell, Jr., J. Comput. Chem. 33, 2451 (2012). 10.1002/jcc.23067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dewar M. J. S., Zoebisch E. G., Healy E. F., and Stewart J. J. P., J. Am. Chem. Soc. 107, 3902 (1985). 10.1021/ja00299a024 [DOI] [Google Scholar]
- 90.Korth M., Pitoňák M., Řezác J., and Hobza P., J. Chem. Theory Comput. 6, 344 (2010). 10.1021/ct900541n [DOI] [PubMed] [Google Scholar]
- 91.MOPAC2009, J. J. P. Stewart, Stewart Computational Chemistry, Colorado Springs, CO, USA, 2008, http://OpenMOPAC.net.
- 92.Databases UBQ and S22x5 are sSAPT0/jun-cc-pVDZ (a truncated aug-cc-pVDZ basis shown to pair well with SAPT0), BBI and SSI are SAPT2+/aug-cc-pVDZ, and HSG is SAPT2+3(CCD)/aug-cc-pVTZ. All methods are defined in Ref. 61, though that publication employed for SAPT levels >SAPT0 while this work uses .
- 93.Missing GAFF computations are SSI ion: 130ARG-248GLU-1; SSI npl: 219VAL-272ALA-1; SSI TT: 060MET-070GLU-1, 087CYS-119ASP-1, 123MET-140ASN-1, 130ARG-248GLU-1, 219VAL-272ALA-1.
- 94.Missing CGenFF computations are SSI ion: 130ARG-248GLU-1 153ASP-156ARG-1; SSI npl: 219VAL-272ALA-1; SSI TT: 060MET-070GLU-1, 123MET-140ASN-1, 130ARG-248GLU-1, 153ASP-156ARG-1, 219VAL-272ALA-1.
- 95.Occasionally generalized gradient approximation (GGA) functionals will have difficulty converging +/− contacts, so various results among 001ARG-005ASP-1, 010GLU-014LYS-1, 012ASP-024LYS-2, 020ASP-082LYS-1, 023LYS-029GLU-1, 045LYS-084GLU-1, 103GLU-177ASP-1, and 153ASP-156ARG-1 are missing.
- 96.Sinnokrot M. O., Valeev E. F., and Sherrill C. D., J. Am. Chem. Soc. 124, 10887 (2002). 10.1021/ja025896h [DOI] [PubMed] [Google Scholar]
- 97.Burns L. A., Vázquez-Mayagoitia Á., Sumpter B. G., and Sherrill C. D., J. Chem. Phys. 134, 084107 (2011). 10.1063/1.3545971 [DOI] [PubMed] [Google Scholar]
- 98.Goerigk L., Kruse H., and Grimme S., ChemPhysChem 12, 3421 (2011). 10.1002/cphc.201100826 [DOI] [PubMed] [Google Scholar]
- 99.See https://github.com/loriab/qcdb for Python module, databases, and data.
- 100.Pérez F. and Granger B. E., Comput. Sci. Eng. 9, 21 (2007). 10.1109/mcse.2007.53 [DOI] [Google Scholar]
- 101.Burns L. A., Marshall M. S., and Sherrill C. D., J. Chem. Theory Comput. 10, 49 (2014). 10.1021/ct400149j [DOI] [PubMed] [Google Scholar]