Abstract
Kalanchoe crenata popularly known as “dog’s liver” is used in most African countries for the treatment of chronic diseases such as diabetes, asthma and HIV/AIDS related infections. The evaluation of K. crenata for herb-drug interactions has not been reported. This study therefore aims to evaluate the risk of K. crenata for herb-drug interaction in vitro. Crude methanol and fractions of K. crenata were incubated and preincubated with recombinant human CYP2C19 and CYP3A4. Comparative studies were conducted in both human liver microsomes and recombinant human CYP to ascertain the inhibition profile of the crude extract and the various fractions. The cocktail approach of recombinant human CYPs was conducted to confirm the inhibition potential of the fractions in the presence of other CYPs. The results showed significant time-dependent inhibition of tested samples on CYP3A4 with crude methanol (39KC), fractions 45A, 45B and 45D given IC50 fold decrease of 3.29, 2.26, 1.91 and 1.49, respective. Time dependent kinetic assessment of 39KC and 45D showed KI and kinact values for 39KC as 1.77µg/mL and 0.091 min-1 while that of 45D were 6.45 µg/mL and 0.024 min-1, respectively. Determination of kinact based on IC50 calculations yielded 0.015 and 0.04 min-1 for 39KC and 45D, respectively. Cocktail approach exhibited fold decreases in IC50 for all test fractions on CYP3A4 within the ranges of 2.10 – 4.10. At least one phytoconstituent in the crude methanol extract of Kalanchoe crenata is a reversible and time-dependent inhibitor of CYP3A4.
Keywords: Human liver microsomes, Kalanchoe crenata, recombinant human CYPs, reversible inhibition, time-dependent inhibition
INTRODUCTION
Kalanchoe c renata is a herbaceous plant belonging to the family of Crassulaceae, also known as the Bryophyllum. Although K. crenata originated from Madagascar, it is now found in many tropical regions of Africa [1]. Traditional health practitioners (THP) extensively use K. crenata for the treatment of several diseases. Juice from fresh leaves is employed for the treatment of asthma, headache, convulsion abdominal pain and smallpox. In the West Province of Cameroon and in other African countries, it is widely used by the THP to treat diabetes mellitus [2]. The adult human doses of K. crenata calculated from different prescriptions are 75 mg/kg and 100 mg/kg when given orally. Anecdotal reports indicate that the use of K. crenata has gained popularity among the geriatric population, and in patients with chronic diseases such as HIV/AIDS, who might be taking orthodox medications for these health conditions.
Consumption of herbal products concurrently with orthodox medications has the potential to endanger the health of an individual through pharmacokinetic and pharmacodynamic herb-drug interactions, interfering with the efficacy or enhancing the toxicity or adverse effects of the orthodox medications.
Cytochrome P450s (CYPs) are enzymes belonging to a large family of heme proteins involved in biotransformation of both endogenous and xenobiotic substances [3]. There are more than 400 known CYP isozymes, with CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 contributing to elimination of about 90% of xenobiotics. CYP3A4 is the most predominant CYP located in the human liver (~40%) responsible for over 50% metabolism of current marketed drugs. Many cancer chemotherapeutic agents and antiretroviral drugs are substrates of CYP3A4. The CYP2C subfamily constitutes the second most abundant CYPs (~20% of total hepatic CYP which accounts for approximately 16–20% of clearance of drugs used clinically. CYP2C19 plays a key role in the metabolism of proton pump inhibitors and the anti-epileptic medications diazepam and mephenytoin. Inter-individual variations in the activity of CYP2C19 have beenreported due to polymorphism which could lead to different degrees of drug interactions [4].
St. John’s Wort (SJW), widely known as a prominent inducer of CYP3A4, is also known to induce other CYPs such as CYP2C19 [5]. An investigation by Wang et al. [5], showed that ingestion of SJW led to significantly increased activity of CYP2C19 in extensive metabolisers carrying the CYP2C19*1/*1 allele. The urinary excretion of 4-hydroxymephenytoin increased by 150%. Thus co-administration of SJW with medications that are substrates of CYP2C19 in such individuals may reduce efficacy and increase levels of metabolites of parent drugs, with possible toxicological consequences. An estimated 11–25% allele frequency of CYP2C19*2 has been found in black Africans. However, such individuals are poor metabolisers of CYP2C19 substrates and may not generate significant interaction with herbal products. Genetic polymorphism is considered as an important factor among different ethnicities due to variations in the expressions of drug-metabolizing enzyme proteins within and/or between populations. African populations are known to have higher levels of genetic diversity than the others [6,7]. The genetic diversity within African populations put them at higher risk for herb-drug interactions due to drug-metabolizing enzyme and/or transporter polymorphisms.
Initial screening of K. crenata on five major CYPs (CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4) using recombinant human enzymes showed that it is a potent inhibitor of CYP3A4 and CYP2C19 (result not reported in this paper). Though a common herbal product in several African countries, no report on the potential of K. crenata to cause herb-drug interactions has been reported. This study therefore aims to evaluate the risk for K. crenata to cause herb-drug interactions (HDI) in both human liver microsomes (HLMs) and recombinant CYPs.
Different in vitro models are employed for the identification of CYP isozymes involved in the metabolism of conventional drugs. Combination of methods such as solid phase extraction and LC/MS/MS or application of cocktail of substrates has emerged as recent techniques to enhance screening of compounds [8]. The microtiter plate (MTP) assay involving the application of recombinant cDNA CYPs expressed in baculovirus-insect cell or an Escherichia coli based expression system has primarily been used in many pharmaceutical industries and research laboratories to evaluate the inhibitory profile of NCE [9]. In recombinant CYPs, only single enzyme is expressed, hence a highly selective substrate is required [10]. The absence of contribution of other metabolic enzyme in recombinant CYP assays, however, diminishes its application as an ideal in vitro model to represent in vivo conditions [11]. The inhibitory effects of a drug candidate in HLMs might be lower than that in recombinant CYP due to the presence of other CYPs in HLMs which could be involved in the metabolism of the candidate drug. A study to determine the IC50 of new drugs on CYP3A4 activity in both HLMs and recombinant CYP showed lower values in the latter compared to that in HLMs [12]. Although the utilization of HLMs is accepted as a standard practice for drug-drug interactions (DDI) and HDI, assays involving HLMs utilize substrates which require analytical methods such as LC/MS/MS, UPLC and HPLC which are time- and labour consuming. Recombinant CYPs are therefore recommended as an HTS method for rapid evaluation and prediction of drugs’ potential to cause interactions in vivo. Most importantly, the use of cocktail recombinant human CYPs and substrates is reported to generate inhibition profiles close to those observed in HLMs [13]. Introduction of such in vitro methodologies will enhance prediction of HDI while reducing the disadvantages associated with the use of conventional HLMs substrate which demands rigorous sample analysis.
For herbal products consisting of multiple phyto-constituents, it is important to screen their potential to cause drug interactions in both HLMs and recombinant human CYPs. Evaluation of herbal products for reversible and time-dependent inhibitions using both HLMs and recombinant human CYPs as a mechanistic approach helps in the categorization of inhibitory potential of such products. The current FDA and the European Medicines Agency (EMA) regulations require in vitro evaluations of the reversible and irreversible inhibitory potential of a new drug candidate on CYPs. The characterization of time-dependent inhibition (TDI) potency of a new drug candidate involves determination of inhibition parameters; maximum rate of inactivation (kinact) and inhibition constant (KI) values from complex in vitro study. It is therefore waste of time, labour and resource to conduct such studies in compounds with weak TDI potential. The preliminary TDI screening processes such as IC50 shift decrease, normalized ratio, and NADPH- and time-dependent inhibition reduce the tendency for conducting TDI kinetics on weak drug candidates, saving time, labour and money. Hence, this study employs the categorization of K. crenata fractions TDI potential using preliminary in vitro HTS methodologies such as the IC50 shift, NADPH, time and concentration dependent and the normalized ratio prior to the conduct of TDI kinetics. Comparative screening assay in HLMs and recombinant human CYP3A4 was also conducted to evaluate the differences in the inhibitory effects of K crenata. In addition, the influence of K. crenata on clearance of testosterone will be evaluated in human cryopreserved hepatocytes.
MATERIALS AND METHODS
Chemicals
3-Cyano-7-ethoxycoumarin (CEC), 7-Benzyloxy-4-trifluoromethylcoumarin (BFC), tranylcypromine, keto-conazole, troleandomycin and 0.5M potassium phosphate buffer (pH 7.4) were from the BD Gentest (Woburn, MA, USA). Troleandomycin was from the Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Testosterone, 6 β-hydroxytestosterone, magnesium chloride, and corticosterone were from the Sigma-Aldrich (St Louis, MO, USA). NADPH regeneration solution A and B were purchased from the Corning Discovery Labware (Woburn, MA, USA). HPLC grade acetonitrile, methanol and formic acid were from Fisher Scientific (Waltham, MA, USA). Solid Phase Extraction (SPE) cartridges (silica gel 10 g/60 mL, code 57134) were purchased from the Supelco (Bellefonte, PA, USA). Organic solvents (chloroform and methanol) were purchased from the Fisher Scientific (Fair Lawn, NJ, USA).
Biologicals
Bactosomes prepared from Escherichia coli cells coexpressing recombinant human NADPH-P450 reductase and individual human P450s (CYP3A4 and CYP2C19), pooled mixed-gender cryopreserved hepatocytes prepared from 20 donors and Hepatozyme media were purchased from BD Gentest (Woburn, MA, USA). Mixed gender pooled human liver microsomes prepared from 25 individual donors with protein concentration (20–26 mg/mL) and total P450 concentration (≥0.320 nmol/mg) were obtained from the BioreclamationIVT (Baltimore, USA).
Plant Material
The mixed parts of K. crenata (whole herb) were obtained from the repository of the National Center for Natural Products Research, The University of Mississippi, USA. A sample specimen (voucher number 2865) is available at the National Center for Natural Product Research, University of Mississippi. The plant material was air-dried at room temperature and ground into a fine powder. Solid Phase Extraction (SPE) cartridges (silica gel 10 g/60 mL, code 57134) were purchased from Supelco (Bellefonte, PA, USA). Organic solvents (chloroform and methanol) were purchased from Fisher Scientific (Fair Lawn, NJ, USA).
Extraction and Fractionation of Plant Material
The dried plant material of K. crenata (10.0 g) was extracted four times with 100 mL of methanol by sonication (30 min) and filtered. The solvent was evaporated under reduced pressure at 37 °C to give a dried methanolic crude extract (882.3 mg). The crude extract (820.0 mg) was then suspended in methanol (7 mL) and adsorbed on celite (1 g). The solvent was evaporated and the residual material was powdered, loaded onto a 10 g SPE silica gel cartridge and fractionated using CHCl3 (100 mL), CHCl3:MeOH 9:1 (100 mL), CHCl3:MeOH:H2O 8:2:0.25 (150 mL), and methanol (150 mL). Fractions of 25 mL were collected and combined according to their TLC profile to give four main fractions: F1 (88.9 mg), F2 (60.7 mg), F3 (147.1 mg) and F4 (409.6 mg). The fractions F1-F2 were re-assigned the following codes: F1 (45A), F2 (45B), F3 (45D) and F4 (45E) which were used throughout. These fractions along with the crude methanolic extract were tested for their potential to inhibit CYP3A4 and CYP2C19 using recombinant human CYP and HLMs.
DETERMINATION OF IC50 FOR REVERSIBLE INHIBITION
All determinations of IC50 values for reversible inhibition were carried out as previously described with slight modification [14]. Preliminary studies showed that 100 µg/mL of K. crenata methanol extract produced the maximum inhibitory effect on CYP2C19 and CYP3A4, hence such concentration was used as the highest throughout the experiment. Three microliters (3 µL) of crude methanol extract and fractions of K. crenata of final concentration of 100 µg/mL in 150 µL reaction mixture consisting of phosphate buffer (0.1M), glucose 6-phosphate dehydrogenase (0.6 units/mL in 5mM sodium citrate buffer 7.5), control protein (0.15 mg/mL) and cofactors (0.14 mM NADP+, 7.2mM MgCl2, 7.2 mM glucose 6-phosphate) were serially diluted (1:3) six consecutive times in Costar 96-well black plates. The plates were pre-warmed for 10 mins. 100 µL of enzyme/substrate reaction mixture of 25pmol CYP3A4/10 µM BFC or 25 pmol CYP2C19/3 µM CEC in phosphate buffer (0.1M) was added to each well except for the blank wells and incubated for 30 mins as recommended in the BD Gentest protocol. The reaction was terminated by the addition of an ice-cold 20% Tris base/80% acetonitrile (ACN). Activity of the enzymes was monitored by measuring the formation of fluorescent metabolite at specific excitation and emission wavelengths of 405/460 for CYP2C19 and 405/535 for CYP3A4, respectively.
Determination of IC50 for Time-Dependent Inhibition
Three microliters (3 µL) of crude methanol extract and fractions of K. crenata of final concentration of 100 µg/mL in 150 µL enzyme/phosphate buffer reaction mixture consisting of phosphate buffer (0.1M), glucose 6-phosphate dehydrogenase (0.6 units/mL in 5mM sodium citrate buffer 7.5), control protein (0.15 mg/mL) and cofactors (0.14 mM NADP+, 7.2mM MgCl2, 7.2 mM glucose 6-phosphate) was serially diluted as described above. The concentrations of CYP3A4 and CYP2C19 were similar to that use in the reversible IC50 assay. The plates were pre-incubated for 30 mins followed by the addition of 100 µL of substrate reaction mixture of 10 µM BFC or 3 µM CEC for CYP3A4 and CYP2C19, respectively, in phosphate buffer (0.1M) to each well except for the blank wells and incubated for additional 30 mins. The reaction was terminated and the activity of the enzymes was monitored by measuring the formation of fluorescent metabolite at excitation/emission wavelengths similar to that of reversible IC50.
Single Point NADPH and Time Dependent Inhibition Screening
Potential TDI fractions selected based on the IC50 shift reduction were preincubated (50 µg/mL) with 250 pmol CYP3A4 or CYP2C19/well reaction mixture in the absence and presence of NADPH for 30 mins at 37oC. The single point NADPH dependent inhibition was repeated for 10 and 20 mins preincubation. For time-dependent inhibition, the primary incubate was preincubated for 0, 5, 10 and 20 mins. At the end of each preincubation time point, 10 µL aliquot of the primary incubates was added to 90 µL of substrate mixture in appropriate wells for respective CYPs and incubated for additional 30 mins. The reaction was terminated and the activity of the enzymes was monitored by measuring the formation of fluorescent metabolite as at excitation/ emission wavelengths described previously. The single point NADPH dependent assay was used to determine the normalized ratio as described in our previous study [14].
Kinetics of CYP3A4 Inactivation
The two-step incubation method was used to characterize the time- and concentration-dependent inhibition of CYP3A4 by the active methanol extract and fractions of K. crenata. In the inactivation assay, varying concentrations of the fractions (ranging between 1 – 100 µg/mL) were incubated with CYP3A4 (250 pmol/well), cofactor, control protein, and 0.1 M phosphate buffer, pH 7.4. At 0, 5, 10, 20 and 30 mins preincubation time points, an aliquot of the preincubation mixture (1:10 dilution) was taken and added to the activity assay (similar to the TDI assay), and a further 30 mins incubation was carried out at 37°C. The reaction was terminated by the addition of ice-cold 20% Tris base/80% ACN solution, and activity was followed by measuring the formation of HFC.
HLMs and Recombinant Human CYP3A4
6β-hydroxytestosterone formation was used as a marker reaction for the activity of CYP3A4. The incubation mixture contained HLMs (0.6 mg/mL protein) or recombinant CYP3A4 (with protein concentration of 0.04 mg/mL leading to activity identical to that of HLMs incubations) in 0.1M potassium phosphate buffer (pH 7.4). The fractions of K. crenata (25 – 100 µg/mL) were added to the enzyme mixture and pre-warmed for 10 mins. NADPH regeneration solution B (1.0 mM) was added to the mixture and incubated for 30 mins at 37oC. The reaction was terminated by the addition of ice-cold ACN containing 5µL corticosterone as an internal standard. The mixture was centrifuged at 10,000 rpm for 5 mins and the supernatant transferred into vials for HPLC analysis.
HPLC Analysis
Testosterone samples were analyzed using a Waters 2695 separation module C18 column (4 µm, 150×3.9 mm), with an isocratic elution and UV detection (254 nm) according to the methods described by Sonderfan et al., [15]. The HPLC mobile phase consisted of 30% water / 69.9% ACN (A) and 99.9% ACN (B) both containing 0.1% formic acid at a flow rate of 1.0 mL/min. Each run was followed by 5 min wash with 100% acetonitrile and equilibration period of 10 min. The peak area ratio between 6β-hydorxytestosterone and the internal standard were used to determine the percent remaining activity.
Assessment of Reversible IC50s using a Cocktail Substrate Approach with Recombinant Human CYPs
As described in the conventional IC50 determination, 3 µL of methanol crude extract and fractions of K. crenata of final concentration 100 µg/mL in 150 µL reaction mixture consisting of phosphate buffer (0.1M), glucose 6-phosphate dehydrogenase (0.6 units/mL in 5mM sodium citrate buffer 7.5), control protein (0.15 mg/mL) and cofactors (0.14 mM NADP+, 7.2mM MgCl2, 7.2 mM glucose 6-phosphate) was serially diluted in Costar 96-well black plates. The plates were pre-warmed for 10 mins. 100 µL of enzymes/substrates reaction mixture of 25pmol CYP3A4/10 µM BFC, 25 pmol CYP2C19/3 µM CEC, 30 pmol CYP2D6/ 15 µM AMMC and 2.5 pmol CYP1A2/ 3 µM CEC mixture in phosphate buffer (0.1M) was added to each well except for the blank wells and incubated for 30 mins at 37oC. The reaction was terminated by addition of an ice-cold 20% Tris base/80% ACN. Activity of the enzymes was monitored by measuring the formation of fluorescent metabolite at specific excitation and emission wavelengths of 405/460 for CYP2C19 and CYP1A2, 390/460 for CYP2D6 and 405/535 for CYP3A4.
Incubation Procedure
Cryopreserved hepatocytes were thawed and the cells were suspended in HepatoZYME buffer (BD Gentest). The cell viability was monitored at the beginning and the end of the incubation in the presence and absence of K. crenata with the use of Bio-Rad TC20 automated cell counter following the instructions of the supplier. The cell density was adjusted with HepatoZYME buffer to 1.0 million viable cell/ml. 100 µM of testosterone (TST) and 100 µg/mL of K. crenata were coincubated with 1 mL hepatocyte mixture at 37oC under humidified atmosphere of 95% air and 5% CO2 in Heraeus incubator. Aliquots of the incubation mixture were taken for HPLC analysis at 0, 0.5, 1, 2, 4, and 6 hours of incubation. The metabolic reaction was stopped by the addition of ACN containing 5 µL of corticosterone as an internal standard. Samples were kept at −80oC for at least 4 hours to break the cell walls, thawed and centrifuged at 13000×g for 5 mins. The supernatant was transferred into HPLC vials for analysis.
PHYTOCHEMICAL INVESTIGATION BY HPLC- UV
The crude extract of K. crenata and its active fractions were investigated using an Agilent 1290 Infinity HPLC-UV system. The separations were achieved using a UPLC BEH C18 column (1.7 µm 2.1× 100 mm, Waters). The mobile phase consisted of water (A) and acetonitrile (B), both containing 0.05% formic acid. A gradient elution was performed as follows: 95% A to 20% A in 15 min, then to 0% in 5 min and held for 5 min. The flow rate was 0.3 mL/min.
DATA ANALYSIS
Prediction of TDI from Fold Decrease IC50
The percentage of residual activity was plotted against the log transformed concentrations of the inhibitors tested. A sigmoid curve was then fitted using non-linear regression, and the IC50 value calculated using GraphPad Prism (GraphPad Software Inc., San Diego, CA). The differences between the IC50 values obtained without preincubation incubation and the IC50 (shifted IC50) values obtained with preincubation were compared, and the fold decrease was determined. The test compound with an IC50 fold-shift decrease ≥ 1.3 is considered as time-dependent inhibitor as recommended by Sekiguchi et al., [26]. However, a more conservative approach was taken here, in that a ratio of 1.5 was used as the cut off for tested fractions to be classified as TDI or non-TDI.
Single Point NADPH and Time Dependent Inhibition Screening
The percentage of residual activity was plotted against the concentrations of the methanol extract and fractions in the absence and presence of NADPH using a bar graph. For TDI, connecting lines were used to join the % residual activity to respective preincubation time. The TDI effect of selected fractions on the activity of CYP3A4 in the absence and presence of NADPH at 10 and 20 mins preincubation were expressed as the normalized ratio as demonstrated in the equation below:
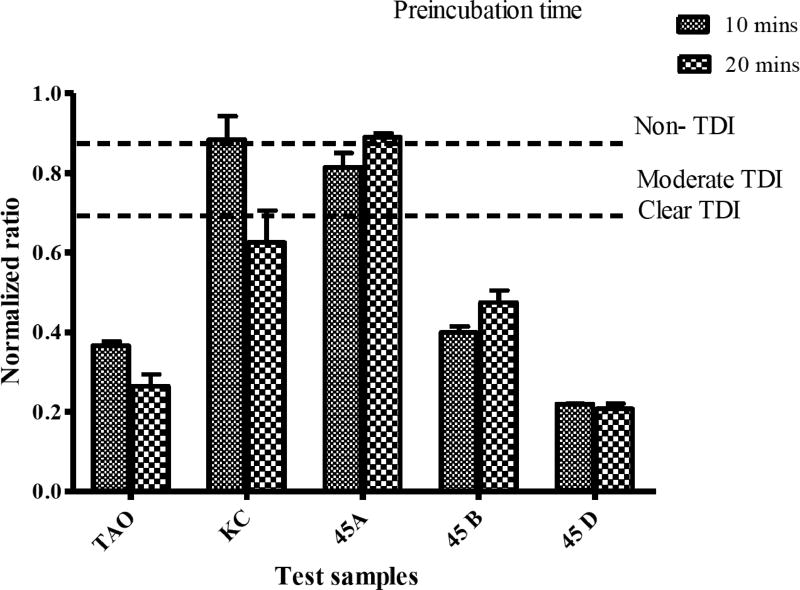
Where (R + 1NADPH ) is the rate of reaction when incubation was performed in the presence of both inhibitor and NADPH; (R -1NADPH ) is the rate of reaction when incubation was performed in the presence of NADPH but in the absence of inhibitor; (R + 1noNADPH ) is the rate of reaction when incubation was done in the presence of inhibitor without NADPH; while (R -1noNADPH ) is the rate of reaction when incubation was performed in the absence of both inhibitor and NADPH. The normalized ratio was used to classify each test sample as follows: below 0.7 (clear TDI), between 0.7 and 0.9 (intermediate zone with undefined TDI), and above 0.9 (non-TDI) as described by Atkinson et al., [16].
Kinetics of CYP3A4 Inactivation
The natural logarithm of the percentage of remaining activity was plotted against the preincubation time at each inhibitor concentration to obtain the kobs (slope). The kobs is the rate constant describing the inactivation at each inhibitor concentration. The values of kinact and KI were determined by the double-reciprocal plotting of the values of kobs and I from the equation below: kobs = kinact x[I] KI + [I] where kinact is the maximal rate of inactivation; KI is the inhibitor concentration required for half-maximal inactivation; and [I] is the preincubation concentration of inhibitor. Th Ie C50 values from reversible and irreversible inhibition were also used to predict the Kinact as recommended by Sekiguchi et al., [26].
Hepatic Clearance Calculations
The half-life (t1/2) and the elimination rate constants (lambda = In2/t1/2) of TST in hepatocytes incubates were determined using log linear regression of TST concentration profile against incubation time. The intrinsic clearance in vitro was calculated from the lambda and the cell density and estimation of intrinsic clearance in vivo was made from in vitro data employing the human liver mass (27.7 g/kg body mass) and hepatocellularity (number of hepatocytes per gram of liver = 99.0 million cells/kg liver):
Based on CLint in vivo and the hepatic blood flow [Q (20.7 ml/min)/kg body mass], the hepatic metabolic clearance (CLh,b) was predicted using the well-stirred model:
The free fractions in vitro or in vivo were assumed to be identical; hence no corrections were made as reported by Davies and Morris [17].
STATISTICAL ANALYSIS
All values were expressed as Mean ± SEM. The percent activity remaining of TST in human liver microsomes and recombinant human CYPs were compared using unpaired t test with P<0.05 considered as significance.
RESULTS
Prediction of TDI from IC50 Fold Decrease
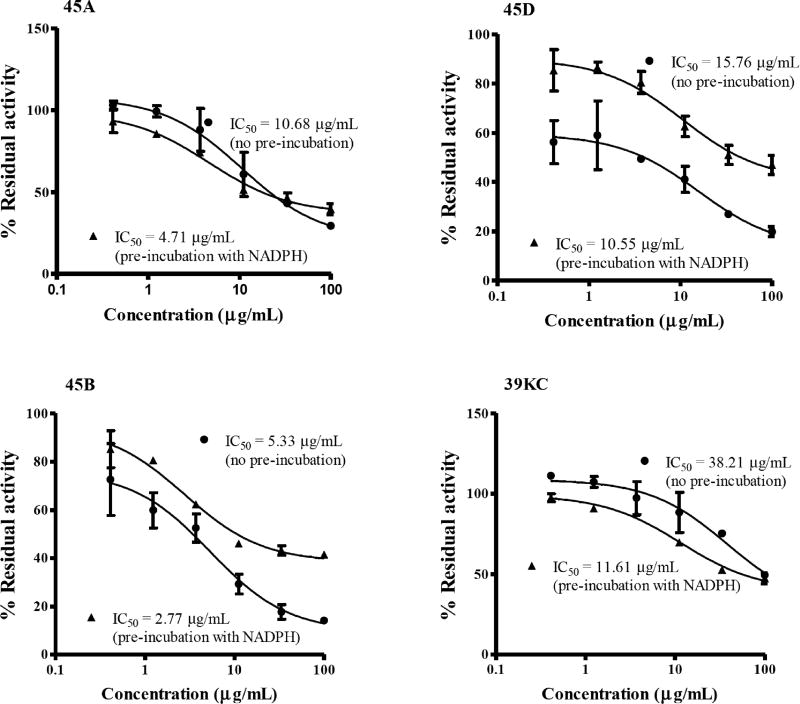
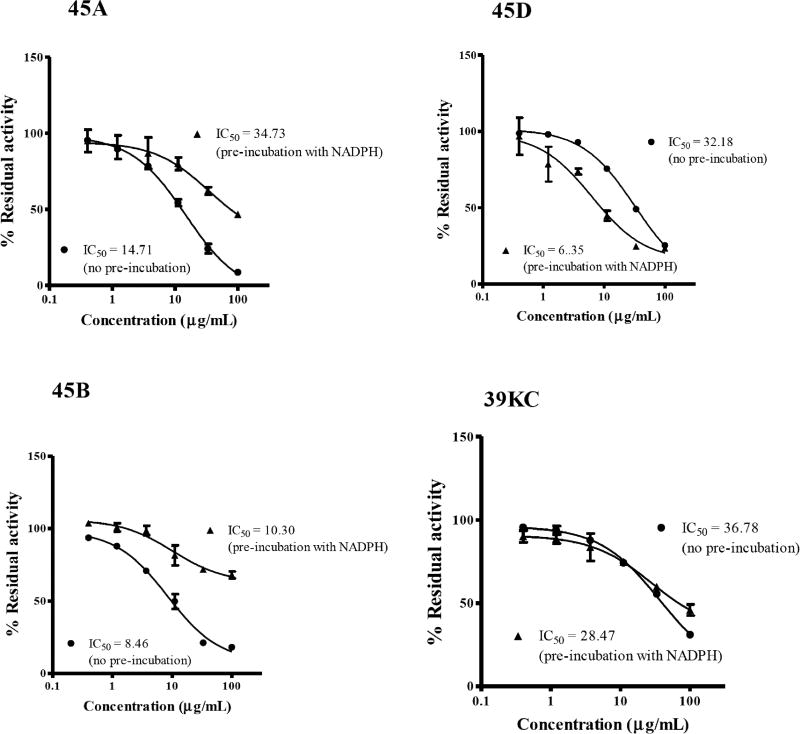
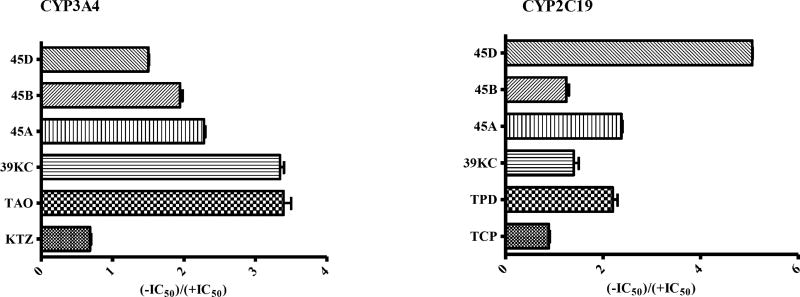
The IC50-shift assay with single substrate for CYP2C19 and CYP3A4 was used to evaluate the TDI potencies of the various fractions. An inhibition curve (shift IC50 value for TDI) obtained in the presence of NADPH was compared with inhibition curve (IC50 for reversible inhibition) to determine the TDI potency of individual fractions as shown in Figs. 1 and 2. Fold decrease in IC50 as a ratio of the reversible and TDI IC50s was employed to determine the potencies of each fraction by categorizing samples as positive TDI or negative TDI (Fig. 3). Test samples with fold decrease in IC50 ≥1.5 were classified as positive TDIs. The fractions 45A and 45D showed TDI potencies with an IC50 fold decrease of 2.26 and 1.50 for CYP3A4 and, 2.36 and 5.07 for CYP2C19, respectively. The crude methanol K. crenata extract (39KC) and the fraction 45B were observed as positive TDIs of CYP3A4 with fold decrease of 3.29 and 1.91, respectively. However, both 39KC and 45B showed a non-TDI effect on CYP2C19 with observed fold decrease of 1.30 and 1.20. Troleandomycin and ketoconazole produced a fold decrease of 3.29 and 0.67, respectively which were in agreement with previous studies [25]. The positive and negative controls of CYP2C19, ticlopidine (TPD) and tranylcypromine (TCP) yielded a fold decrease of 2.10 and 0.55, respectively. The fraction 45E did not show any activity on CYP3A4 and CYP2C19.
Fig. (1).
Inhibition profiles for BFC metabolism at various concentrations of inhibitors in recombinant CYP3A4. Values represent the average of duplicate experiments. Inhibition percentages for no preincubation (closed circles) and 30 min preincubation (close triangle) are shown.
Fig. (2).
Inhibition profiles for CEC metabolism at various concentrations of inhibitors in recombinant CYP2C19. Values represent the average of duplicate experiments. Inhibition percentages for no preincubation (closed circles) and 30 min preincubation (close triangle) are shown
Fig. (3).
Effects of preincubation on inhibitory potential against recombinant CYP3A4 and CYP2C19 activities. Bars represent the ratio of IC50 values from the no preincubation assays to the 30 min preincubation assays for the test samples listed. Data are the mean ± SEM (n = 2).
Single Point NADPH-, Time and Concentration- Dependent Inhibition Screening
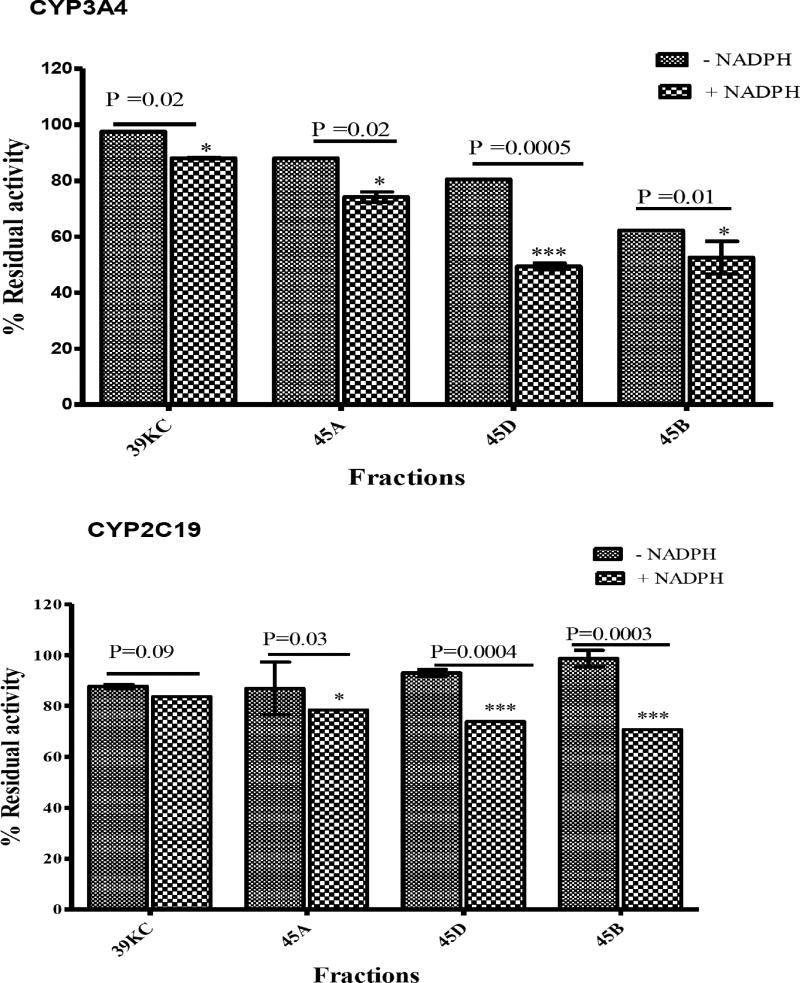
Treatment of all fractions with NADPH before preincubation led to a reduction in the percent of activity remaining on CYP3A4 compared to those without NADPH. The fractions 45A, 45B and 45D showed a significant reduction in percent residual activity after preincubation with NADPH (Fig. 4). Fractions 45A and 45D showed an NADPH-dependent inhibitory effect on both CYP3A4 and CYP2C19. However, 39KC had a non-significant effect after preincubation with NADPH as the inhibition potential was reduced when compared to those without NADPH preincubation with CYP2C19. The categorization of fractions into clear TDI (≤0.70), intermediate TDI (0.7 – 0.9) or non-TDI (>0.9) based on normalized ratio showed 39KC as a non-TDI and an intermediate TDI at 10 mins and 20 mins preincubation times, respectively (Fig. 5). All other fractions exhibited clear TDI regardless of the duration for the preincubation.
Fig. (4).
Inhibitory effects of 50 µg/mL of each fraction on CYP3A4 and CY2C19 preincubated for 30 mins in the presence or absence of NADPH. The percent residual activities were plotted against respective fractions. Two tailed unpaired t-test were used to compare the percent activity remaining for each fraction with P < 0.05 considered significant.
Fig. (5).
Time dependent inhibition (TDI) classification of test compounds based on normalized ratio. Known TDI compound, troleandomycin (TAO, 20 µM) and non-TDI compound, ketoconazole (KTZ, 1 µM) were used as positive and negative controls. Single concentration of each fraction 50 µg/mL was preincubated with CYP3A4 for 10 and 20 mins. The normalized ratio was calculated as described under Materials and Methods.
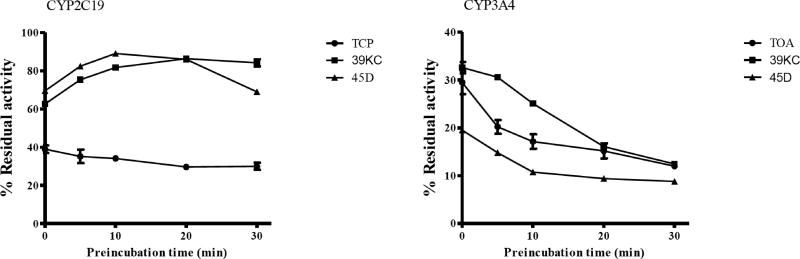
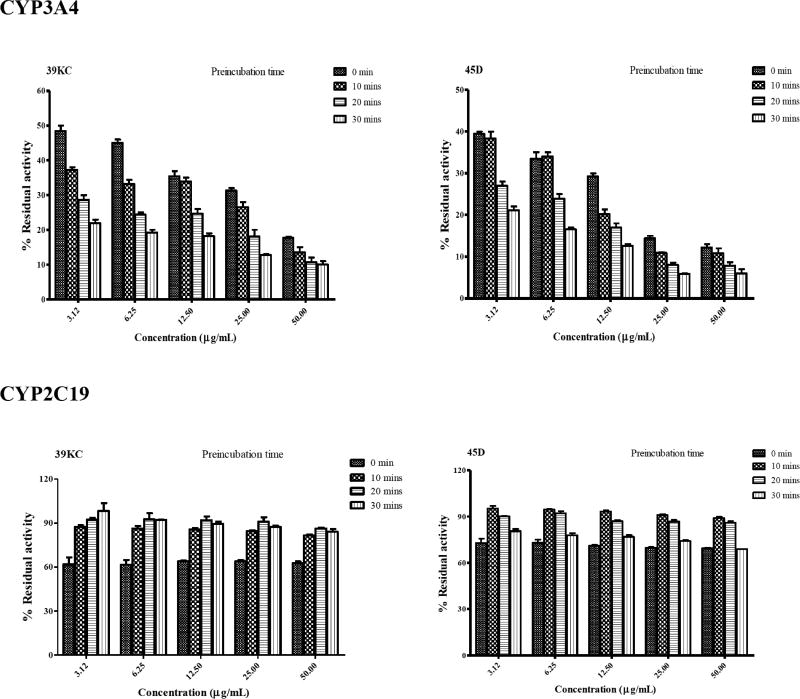
Preincubation of fractions 39KC and 45D for 0, 10, 20 and 30 mins showed time-dependent inactivation of CYP3A4 as the degree of inhibition increased with time. However, 39KC did not indicate TDI following preincubation with CYP2C19. The fraction 45D also exhibited TDI on CYP2C19 as shown in Fig. 6. Inactivation of CYP3A4 by 39KC and 45D followed a time- and concentration-dependent pattern. However, inactivation of CYP2C19 by 39KC and 45D did not indicate any time- and concentration dependency, as the inhibition profile became weaker with increase in both time and concentration of the respective fractions applied to the incubation mixture (Fig. 7).
Fig. (6).
Time-dependent inhibition of CYP3A4 and CYP2C19 by 50 µg/mL of 39KC (closed rectangular box) and 50 µg/mL of 45D (closed triangular box) for 0, 10, 20 and 30 min preincubation time. The percent remaining activity was plotted against respective time point through connecting lines. Each point represents the Mean ± SEM of duplicate determinations.
Fig. (7).
Concentration- and time-dependent inhibition of CYP3A4 and CYP2C19 by 39KC and 45D for 0, 10, 20 and 30 min preincubation time. The percent remaining activity was plotted against respective time point through connecting lines. Each point represents the Mean ± SEM of duplicate determinations.
Kinetics of CYP3A4 Inactivation
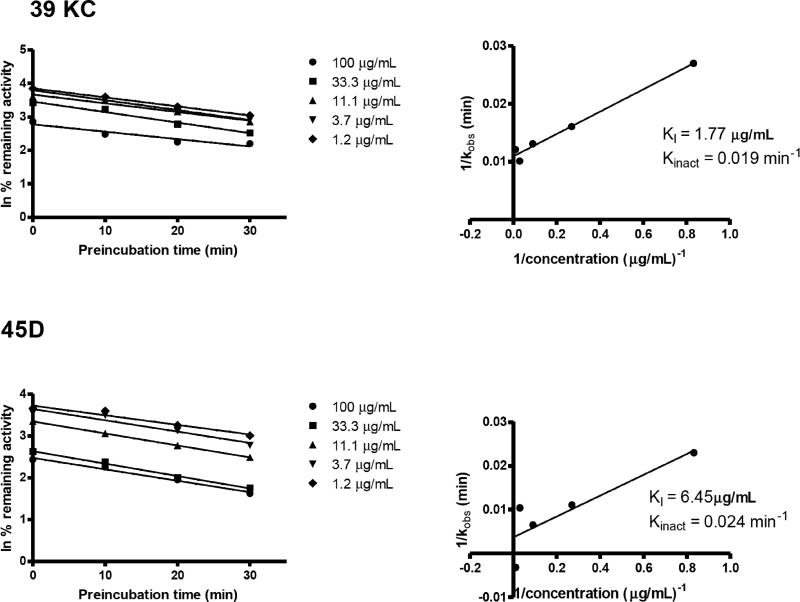
CYP3A4 HFC activity was inactivated by 39KC and 45D in a time- and concentration-dependent manner in the presence of NADPH regeneration system. The loss of activity followed pseudofirst-order kinetics. Linear regression analysis of the time course data were used to determine the kobs values at various concentrations of 39KC and 45D (Fig. 8). The KI and kinact values were determined from double reciprocal plots of kobs values and inactivator concentrations. The KI and kinact values for 39KC were 1.77µg/mL and 0.091 min-1 while that of 45D were 6.45 µg/mL and 0.024 min-1, respectively. Determination of kinact based on IC50 calculations yielded 0.015 and 0.04 min-1 for 39KC and 45D, respectively.
Fig. (8).
Time- and concentration-dependent inhibition of CYP3A4 by 39KC and 45D. Concentrations of fractions are shown on the right of the plots (A). Double-reciprocal plots of kobs and inhibitor concentrations were plotted on linear graph to determine inactivation parameters KI and kinact in plot (B). Each point represents the mean of duplicate determinations.
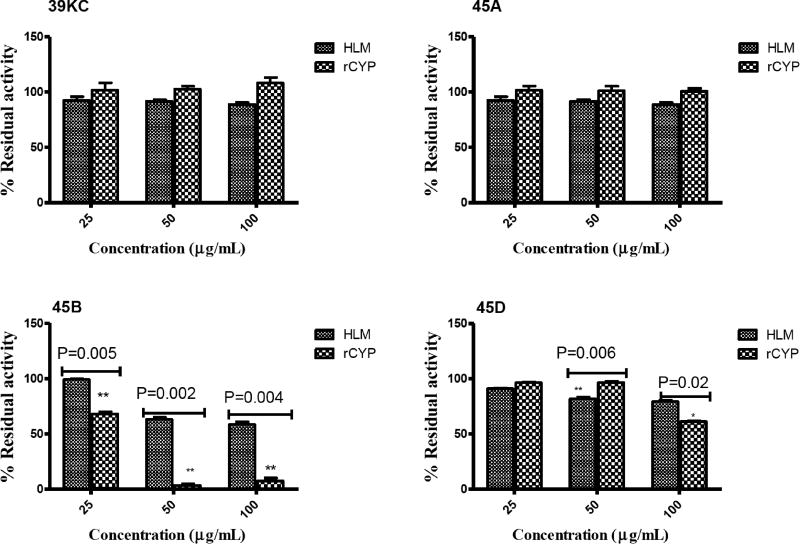
HLMs Versus Recombinant CYP3A4 Activity Comparative evaluation of the inhibitory effects of various fractions of K. crenata on CYP3A4 activity on 6β-hydroxylation of TST in HLMs and recombinant human CYP3A4 was monitored using HPLC. The concentration of TST used in this assay was approximately the Km, as a substrate probe in both HLMs and recombinant human CYP3A4. As shown in Fig. 9, the fraction 45D significantly reduced (P=0.006) the activity of CYP3A4 in HLM compared to that of rCYP at 50 µg/mL. Conversely, the activity of CYP3A4 was reduced significantly (P=0.02) in rCYP compared to that in HLM by fraction 45D at 100 µg/mL. The fraction 45B indicated significant reduction in CYP3A4 activity in rCYP compared to that of HLM at all the three different concentrations tested. Crude methanol extract (39KC) and fraction 45A showed no significant differences in the activity of CYP3A4 when tested in both HLM and rCYP.
Fig. (9).
Inhibitory effect of fractions (25 – 100 µg/mL) on CYP3A4-catalyzed 6β-Hydroxytestosterone formation in HLMs and recombinant human CYP3A4.
A COMPARISON OF IC50 OBTAINED WITH SINGLE- AND COCKTAIL RECOMBINANT HUMAN CYPS-SUBSTRATES APPROACHES
The IC50 values of various fractions were re-evaluated using cocktail enzyme/substrate for recombinant human CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4. A fold decrease in the inhibitory potency of an inhibitor was determined as a ratio of cocktail and single IC50 values for individual fractions as shown in Table 1. Crude methanol of K. crenata, fractions 45B and 45D showed a significant reduction of 3.32, 2.55 and 4.8 inhibition potencies on CYP3A4. The inhibition effect of 39KC and 45B was not detected in CYP2C19 for most fractions. Fraction 45D however showed a decline in inhibition potency of 4.36 on CYP2C19.
Table 1.
Comparison of inhibitory potency of fractions in cocktail and single recombinant human CYPs.
| Test Samples | Conventional IC50 (µg/mL) |
Cocktail IC50 (µg/mL) |
Decrease Inhibitory Potency |
|---|---|---|---|
| CYP3A4 | |||
| 39 KC | 38.21 | 127 | 3.32 |
| 45B | 5.33 | 13.6 | 2.55 |
| 45D | 15.76 | 75.72 | 4.80 |
| CYP2C19 | |||
| 39KC | 36.78 | ND | - |
| 45B | 10.30 | ND | - |
| 45D | 32.18 | 140.4 | 4.36 |
The decrease inhibitory potency = cocktail IC50/conventional IC50.
The viability of hepatocytes (percentage viable cell relative to total cells) reduced from 78.10% at time zero to 52.3% without the test inhibitor and 50.5% with K. crenata. Hence, the cell degradation is not attributable to the presence of K. crenata. TST was also stable in the HepatoZYME buffer. Cellular activity is therefore, a responsibility for the degradation of the hepatocytes.
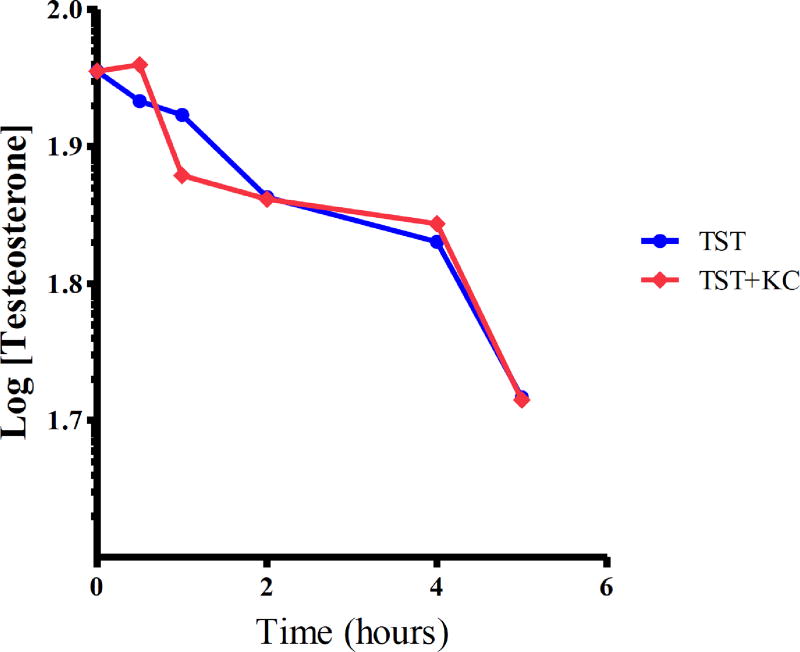
The intrinsic clearance of TST in the presence and absence of K. crenata was monitored in human hepatocytes (Fig. 10). Estimation of hepatic blood clearance was done using the well-stirred liver model. K. crenata reduced the clearance of TST by 9.3% (Table 2)
Fig. (10).
Metabolic clearance of testosterone in hepatocytes in the presence and absence of K. crenata.
Table 2.
The intrinsic clearance of testosterone in human hepatocytes in the absence and presence of K. crenata.
| Parameter | TST Alone | TST + K. crenata |
|---|---|---|
| t½ (min) | 565.3 | 618.1 |
| CLint in vitro ((µL/min)/million cells) | 2.45 | 2.24 |
| CLint in vivo ((mL/min)/kg body mass) | 6.24 | 5.71 |
| CLh,b ((mL/min)/kg body mass) | 4.79 | 4.47 |
| CLh,b (% hepatic blood flow) | 23.16 | 21.61 |
PHYTOCHEMICAL INVESTIGATION BY HPLC- UV
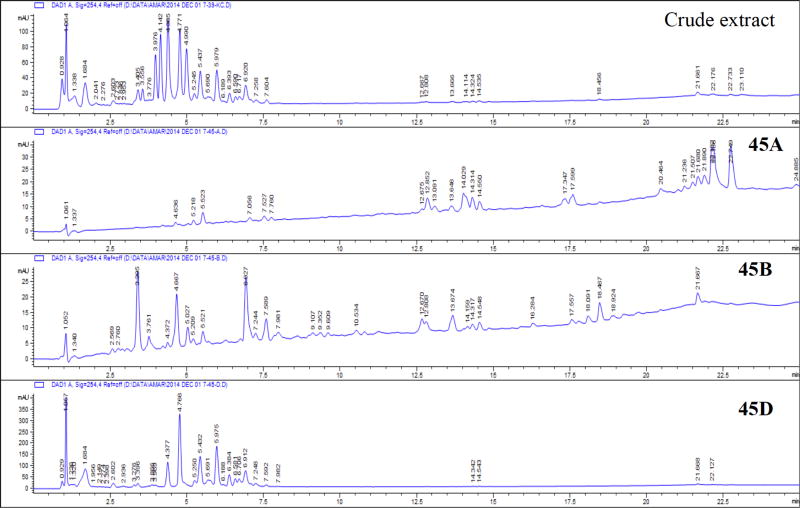
The crude methanolic extract of K. crenata was enriched in polar compounds. The HPLC-UV profile indicated the presence of at least 13 flavonoid glycoside candidates. After fractionation, fraction 45D yielded 18% of the crude extract and contained at least 10 of the 13 components. Fractions 45A and 45B contained minor non-polar and medium polarity constituents, respectively (Fig. 11).
Fig. (11).
UV profile of crude and active fractions of K. crenata.
DISCUSSION
The conduct of drug-drug interactions to screen new chemical entities has now become a routine practice in most pharmaceutical companies. Drug regulators including the US FDA have published guidance for in vitro and in vivo drug interaction studies. With recent popularization of herbal medications globally, most pharmaceutical companies and research institutions have adopted the FDA guidelines for the conduct of DDI to flag herbs with potential risk for herb-drug interactions [18,19]. Such studies have resulted in the revision of labels on products such as indinavir and postmarket withdrawal of mibefradil [20,21]. The FDA guidelines on the conduct of DDI however do not address the specific designs of the studies, and this has resulted in variations in the models and approaches for in vitro evaluations of new chemical entities [22].
Usually, novel compounds are evaluated for their potential to cause CYP-mediated drug interaction based on single concentration screening while allowing for later detailed characterization involving IC50 and Ki determinations for reversible inhibition. Additionally, evaluation of compounds as time-dependent or mechanism-based inhibitors following reversible inhibition has become mandatory recently, as the consequences are more severe. However, due to the complexity of mechanism-based inhibition, pharmaceutical companies have developed approaches to evaluate the potential of time-dependent inactivation, funneling compounds through single concentration, single time point experiments, and determining IC50 shift to avoid waste of time, labour and money on compounds that are weak CYP inactivators [23]. The choice of in vitro models for the conduct of mechanism-based screening of compounds is important, because false positives or negatives can result to inappropriate compound progression or de-selection. HLMs or recombinant human CYPs are routinely applied for the conduct of such studies some of which have generated under- and over-estimation of inhibition profile of compounds on few occasions. As preliminary studies to evaluate the potential of K. crenata to cause herb-drug interactions, a combination of HLMs, recombinant human CYPs and the cocktail approaches was adopted to avoid false positive predictions.
Limited information is currently available regarding the interactions of K. crenata with clinically prescribed medications. This is an issue of potential safety concern as K. crenata is consumed by geriatrics and patients with chronic diseases such as diabetes, HIV/AIDS and asthma. Drugs commonly used by these patients include analgesics (diclofenac, ibuprofen, celecoxib -CYP2C9 substrates), protein pump inhibitors like omeprazole (CYP2C19 substrate), antiasthmatics (theophylline –CYP1A2 substrate) and nevirapine (CYP3A4); some of them have narrow therapeutic indices and high tendencies for DDI. Furthermore, such patients are prone to co-morbidities and infections, which might require the use of other CYP substrate medications like the sulphonylureas (tolbutamide and glipizide), antituberculosis (isoniazid, rifampicin), macrolide antibiotics (e.g., erythromycin, clarithromycin). Hence, there exists a high potential risk for the interaction when any of these medications are consumed concurrently with K. crenata which contains multiple phyto-constituents.
Recently, the Pharmaceutical Research and Manufacturers Association recommended the use of IC50-shift approach as the first step for TDI assays [24]. This method involves the application of HLMs or recombinant human CYPs to evaluate whether the potency of the NCE to inhibit CYP increases when the drug candidate is preincubated in the presence of NADPH. The IC50 shift approach was subsequently used by Obach et al., [23] to evaluate the TDI potential and demonstrated a progressive flow strategy in which NCE are selected for detailed kinetics. Therefore, this study used the IC50 shift approach to evaluate the TDI potency of various fractions of K. crenata. The fractions were preincubated with single recombinant human CYPs in the presence of NADPH as a primary incubate followed by the dilution of the reaction mixture with substrate mixture for additional 30 mins incubation. Such approach showed a leftward shift in the inhibition curves after preincubation compared to the curves without preincubation (Fig. 1). Although many pharmaceutical companies and research institutions are adopting the IC50 shift approach for screening of TDI potential compounds, there is currently no definitive criterion for the classification of compounds as negative TDI or positive TDI. Many pharmaceutical companies have been using the fold-shift criterion of 1.2 to 3-fold whilst Berry et al., [25] also concluded that an IC50-shift decrease of >1.5 indicated significant potency. In our studies, the Berry et al., criterion was adopted whereby compounds with a fold decrease ≥1.5 where tagged as potential TDI candidates. The fractions tested showed TDI potencies of 3.29, 2.26, 1.91 and 1.5 for crude methanol extract (39KC), 45A, 45B and 45D, respectively on CYP3A4. TDI potencies of 3.29 and 0.67 was observed for troleandomycin and ketoconazole, known TDI and non-TDI compounds for CYP3A4 which were in agreement with the result reported by Sekiguchi et al., [26]. The fractions 45A and 45D also exhibited positive TDI on CYP2C19 with fold decrease ratio of 2.36 and 5.07, respectively. However, the crude methanol extract (39KC) and 45B showed negative for TDI, with respective fold decreases of 1.30 and 1.20.
To confirm the positive TDI of fractions evaluated in this assay, the normalized ratio was used as described by Atkinson et al., [16]. The normalized ratio categorizes TDI positive compounds into clear TDI, intermediate TDI and non-TDI based on preincubation with NADPH and without NADPH. The normalized ratio approach showed that 39KC had borderline activity, with non-TDI and intermediate TDI characteristics after 10 and 20 mins preincubations, respectively. The exhibition of intermediate TDI after 20 mins preincubation may be attributable to the adequate generation of reactive metabolite with a potential of inactivating CYP3A4, which is insufficient at 10 mins of preincubation. Other fractions 45A, 45B and 45D showed clear TDI on CYP3A4 regardless of the preincubation duration. The use of normalized ratio as secondary approach to confirm the criterion for the determination of TDI potencies of K. crenata has shown the possibility of overestimation and/or underestimation of compounds or herbal products with only the IC50 shift approach. It is here suggested that the use of NADPH- and time-dependent normalized ratio approach can serve as a confirmatory procedure for the categorization of TDI potencies of NCE and herbal products. Inactivators of CYPs are known to be time- and concentration dependent, hence effects of such parameters on fractions were tested. The results showed that none of the fractions was a time-dependent inactivator of CYP2C19, as the inhibition profile weakened on incubation. Such a phenomenon can be due to recovery of enzyme as activity, since the inhibition potency of the phytoconstituents weakens with time regardless of the concentration applied. However, fractions 39KC and 45D exhibited time- and concentration-dependent inhibition as their inhibition profile strengthened in CYP3A4. Thus, 39KC and 45D were identified as TDIs of CYP3A4 based on the observations that the inhibitory effects of 39KC and 45D on CYP3A4 activity were dependent on: 1) the preincubation, 2) concentration, and 3) NADPH. Next, the inactivation kinetics parameters of 39KC and 45D were determined in recombinant human CYP3A4. The kinetic parameters for the inactivation of CYP3A4 by 39KC and 45D were: KI, 1.77 and 6.45 µg/mL; Kinact, 0.019 and 0.02 min-1, respectively. Prediction of Kinact based on reversible and irreversible inhibition IC50 yielded values of 0.015 and 0.04 min-1 for 39KC and 45D, respectively which were closer to the results obtained from the dilution method. Thus, shift IC50 values can be used to predict inactivation constant of time-dependent inhibition of herbal medicines as recommended earlier by Sekiguchi et al., [26].
Recombinant human CYPs from insects contain a single enzyme, and thus lack the potential effect of other CYP isozymes which might influence the true inhibitory activity of a drug candidate likely to be observed in vivo. Although the recombinant human CYPs are used as an HTS model, the HLMs are preferred as an ideal in vitro tool for evaluation of the inhibitory activity of new drug candidates by employing specific substrates. In comparing the inhibitory effects of various fractions tested, HLMs and recombinant activity were therefore monitored using TST as probe substrate of CYP3A4. The results showed a general weak inhibition profile of the fractions in HLMs. This could be attributable to some phytoconstituents of the fractions 39K and 45D acting as substrates, therefore metabolized via other pathways than CYP3A4. Earlier studies comparing inhibitory potency of known CYP3A4 inhibitors in both HLMs and recombinant observed higher IC50 values in HLMs. Such weak potencies of inhibitors in HLMs were attributed to such compounds acting as substrates for other isozymes, and therefore metabolized to less active forms which may weakly inhibit CYP3A4. However, in a situation where strong metabolites are generated due to the activities of other isozymes in HLMs, lower IC50 values are observed [27, 28]. This is attributed to reactive metabolites produced through the activities of other enzymes in HLMs causing an inactivation of a specific enzyme(s) as a potential irreversible inhibitor thereby reducing the efficiency of the affected enzyme(s). Thus the inhibitory potency of a specific compound depends on the functional group and the type of isozymes available. In the current study, although weaker inhibition activity was observed in HLMs assay, the difference was only marginal when compared with that of recombinant human CYP. These results agree with the findings by Bapiro et al., [9] and Bell et al., [29] showing positive correlation between compounds screened with both HLMs and recombinant human CYP for their inhibition effects. However, strong inhibition was observed in recombinant human CYP compared to HLMs for fractions 45B. Thus, the results indicate the possibility of underestimation of potential inhibitor when HLMs are used as the sole model for the prediction of drug candidates’ potential for DDI or HDI. Conversely, the use of recombinant human CYP might also lead to overestimate of such compounds; hence combination of these models should be used in screening herbs purported to have potential risk for HDI to avoid any missing information. As a method of confirming the inhibition effects of the various fractions in the presence of other isozymes, the cocktail approach was adopted. A general decrease in inhibition profile was observed as the IC50 values for the cocktail were higher compared to the single CYPs. In an attempt to establish differences between potencies of known inhibitors using conventional HLMs or recombinant human CYPs against the cocktail approach, Dierks et al., [30] observed a strong correlation between the two models. For example, identical IC50 values of ketoconazole, quinidine, sulfaphenazole and tranylcypromine were observed for their respective CYPs in both conventional single CYP and the cocktail approach. However, for herbal extracts or fractions with multi-phytochemicals, there is a high probability of at least a constituent acting as a substrate to other isozymes or binding to other CYPs. Such effects might be responsible for the weak inhibitory effects observed in cocktail approach. This confirms the presence of other CYP isozymes in HLMs contributing to the reduction of inhibition activity of fractions tested as opposed to the single CYP.
Compare with recombinant human CYPs and HLMs, in vitro data obtained from the cryopreserved hepatocytes are closer to in vivo conditions due to total expression of cell matrix and presence of other drug metabolizing enzymes. The 9.3% reduction in metabolic clearance of TST by K. crenata shows it is a weak candidate for interaction with CYP3A4 substrates in humans. However, caution must be exercised by patients taking K. crenata due to potential variations in the phytoconstituents attributable to differences in time of collection and dosage which might affect other organs in the future.
Based on chemical characterization of other Kalanchoe species and on HPLC-UV-MS results, the crude methanolic extract of K. crenata may be composed of flavonoid glycosides. The HPLC-UV profile indicated the presence of at least 13 flavonoid glycoside candidates. This is in agreement with the previous literature [31]. The majority of these compounds was present in fractions D and E (data not included), which constitute 18% and 50%, respectively in weight of the crude extract. Considering the widespread occurrence of these compounds in several plant species, it is doubtful that flavonoid glycosides may be related to the drug interaction effects observed with K. crenata. This assumption is supported by the lack of activity for the fraction 45E, which mainly contains flavonoid glycosides. Minor fractions were more active than fraction 45E, it is thus possible that its interaction with drugs may be related to the minor constituents.
CONCLUSION
This study has evaluated the reversible and time-dependent inhibition of K. crenata using HLMs and recombinant human CYPs. Data from this study have shown that the constituents in fraction F3 (45D) of K. crenata reversibly and irreversibly inhibit CYP3A4 in recombinant human CYP. It will therefore be important to further evaluate the inhibition effect of isolated compounds from fraction 45D on CYP3A4 in vitro. The findings of this study have also indicated that combination of HLMs and recombinant human CYPs as in vitro models reduces the tendency for over- and under-estimation of K. crenata potential to cause HDI. K. crenata however has a mild inhibitory effect on clearance of TST in human cryopreserved hepatocytes.
Acknowledgments
This study was funded by the NIH-Fogarty International Center training grant-Brown AIDS International Training and Research Program (Grant# D43TW000237). The United States Department of Agriculture, Agricultural Research Service, Specific Cooperative Agreement No. 58-6408-1-603 is also acknowledged for partial support of this work.
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Adjanohoun JC, Aboubakar N, Dramasse K, Ebot ME, Ekpere JA, Enow-Orock EG, Focho D, Gbile ZO, Kamanyi A, Kamsu Kom P, Jr, Keeta A, Mbenkum T, Mbi CM, Mbielle AL, Mbome IL, Mubiru NK, Namey WL, Nkongmeneck B, Stabie B, Sofowa A, Tanze V, Wirmum CK. CNPMS, Portonovo. Benin; 1996. Traditional Medicine and Pharmacopeia-Contribution to Ethnobotanical and Floristic studies in Cameroon; p. 505. [Google Scholar]
- 2.Kamgang R, Mboumi RY, Fondjo AF, Tagne MA, N'dillé GP, Yonkeu JN. Antihyperglycaemic potential of the waterethanol extract of Kalanchoe crenata (Crassulaceae) J Nat Med. 2008;62(1):34–40. doi: 10.1007/s11418-007-0179-y. [DOI] [PubMed] [Google Scholar]
- 3.Soars MG, McGinnity DF, Grime K, Riley RJ. The pivotal role of hepatocytes in drug discovery. Chem. Biol. Interact. 2007;168:2–15. doi: 10.1016/j.cbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Küpfer A, Preisig R. Pharmacogenetics of mephenytoin: a new drug hydroxylation polymorphism in man. Eur. J. Clin. Pharmacol. 1984;26:753–759. doi: 10.1007/BF00541938. [DOI] [PubMed] [Google Scholar]
- 5.Wang LS, Zhu B, Abd El-Aty AM, Zhou G. The influence of St. John's Wort on CYP2C19 activity with respect to genotype. J. Clin. Pharmacol. 2004;44:577–581. doi: 10.1177/0091270004265642. [DOI] [PubMed] [Google Scholar]
- 6.Dandara C, Masimirembwa CM, Magimba A, Sayi J, Kaaya S, Sommers DK, Snyman JR, Hasler JA. Genetic polymorphism of CYP2D6 and CYP2C19 in east- and southern African populations including psychiatric patients. Eur. J. Clin. Pharmacol. 2001;57(1):11–7. doi: 10.1007/s002280100282. [DOI] [PubMed] [Google Scholar]
- 7.Ioannides C. Pharmacokinetic interactions between herbal remedies and medicinal drugs. Xenobiotica. 2002;32:451–78. doi: 10.1080/00498250210124147. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, Racha J, Li SY, Olejnik N, Satoh H, Moore D. Automated high throughput human CYP isoform activity assay using SPE-LC/MS method: application in CYP inhibition evaluation. Xenobiotica, 2000, 30(2), 141–54. In Vitro Evaluation of Reversible and Time-Dependent Inhibitory Drug Metabolism Letters. 2015;9(115) doi: 10.1080/004982500237749. [DOI] [PubMed] [Google Scholar]
- 9.Bapiro TE, Egnell AC, Hasler JA, Masimirembwa CM. Application of higher throughput screening (HTS) inhibition assays to evaluate the interaction of antiparasitic drugs with cytochrome P450s. Drug Metab. Dispos. 2001;29(1):30–5. [PubMed] [Google Scholar]
- 10.Foti RS, Wienkers LC, Wahlstrom JL. Application of cytochrome P450 drug interaction screening in drug discovery. Comb. Chem. High Throughput Screen. 2010;13:145–158. doi: 10.2174/138620710790596718. [DOI] [PubMed] [Google Scholar]
- 11.Kramer MA, Tracy TS. Studying cytochrome P450 kinetics in drug metabolism. Expert Opin. Drug Metab. Toxicol. 2008;4:591–603. doi: 10.1517/17425255.4.5.591. (2008) [DOI] [PubMed] [Google Scholar]
- 12.Nomeir AA, Ruegg C, Shoemaker M, Favreau LV, Palamanda JR, Silber P, Lin CC. Inhibition of CYP3A4 in a rapid microtiter plate assay using recombinant enzyme and in human liver microsomes using conventional substrates. Drug Metab. Dispos. 2001;29(5):748–53. [PubMed] [Google Scholar]
- 13.Kozakai K, Yamada Y, Oshikata M, Kawase T, Suzuki E, Haramaki Y, Taniguchi H. Cocktail-substrate approach-based high-throughput assay for evaluation of direct and time-dependent inhibition of multiple cytochrome P450 isoforms. Drug Metab Pharmacokinet. 2014;29(2):198–207. doi: 10.2133/dmpk.dmpk-13-rg-093. [DOI] [PubMed] [Google Scholar]
- 14.Awortwe C, Bouic PJ, Masimirembwa CM, Rosenkranz B. Inhibition of Major Drug Metabolizing CYPs by Common Herbal Medicines used by HIV/AIDS Patients in Africa-Implications for Herb- Drug Interactions. Drug Metab. Lett. 2014;7(2):83–95. doi: 10.2174/1872312808666140129123210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonderfan AJ, Arlotto MP, Dutton DR, McMillen SK, Parkinson A. Regulation of testosterone hydroxylation by rat liver microsomal cytochrome P450. Arch. Biochem. Biophys. 1987;255:27–41. doi: 10.1016/0003-9861(87)90291-8. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson A, Kenny JR, Grime K. Automated assessment of time-dependent inhibition of human cytochrome P450 enzymes using liquid chromatography-tandem mass spectrometry analysis. Drug Metab. Dispos. 2005;33(11):1637–47. doi: 10.1124/dmd.105.005579. [DOI] [PubMed] [Google Scholar]
- 17.Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 18.Izzo AA, Ernst E. Interactions between herbal medicines and prescribed drugs: an updated systematic review. Drugs. 2009;69(13):1777–98. doi: 10.2165/11317010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Zhang YD, Zhao P, Huang SM. Predicting drugdrug interactions: an FDA perspective. AAPS J. 2009;11(2):300–6. doi: 10.1208/s12248-009-9106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clauson KA, Santamarina ML, Rutledge JC. Clinically relevant safety issues associated with St. John's wort product labels. BMC Complement Altern. Med. 2008;8:42. doi: 10.1186/1472-6882-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley J, Wilton LV, Shakir SA. A post-marketing observational study to assess the safety of mibefradil in the community in England. Int. J. Clin. Pharmacol. Ther. 2002;40(6):241–8. doi: 10.5414/cpp40241. [DOI] [PubMed] [Google Scholar]
- 22.Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, Kao J, King SP, Miwa G, Ni L, Kumar G, McLeod J, Obach RS, Roberts S, Roe A, Shah A, Snikeris F, Sullivan JT, Tweedie D, Vega JM, Walsh J, Wrighton SA Pharmaceutical Research and Manufacturers of America (PhRMA) Drug Metabolism/Clinical Pharmacology Technical Working Group; FDA Center for Drug Evaluation and Research (CDER) The conduct of in vitro and in vivo drug-drug interaction studies: a Pharmaceutical Research and Manufacturers of America (PhRMA) perspective. Drug Metab. Dispos. 2003;31(7):815–32. doi: 10.1124/dmd.31.7.815. [DOI] [PubMed] [Google Scholar]
- 23.Obach RS, Walsky RL, Venkatakrishnan K. Mechanism-based inactivation of human cytochrome p450 enzymes and the prediction of drug-drug interactions. Drug Metab. Dispos. 2007;35(2):246–55. doi: 10.1124/dmd.106.012633. [DOI] [PubMed] [Google Scholar]
- 24.Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, Kao J, King SP, Miwa G, Ni L, Kumar G, McLeod J, Obach RS, Roberts S, Roe A, Shah A, Snikeris F, Sullivan JT, Tweedie D, Vega JM, Walsh J, Wrighton SA Pharmaceutical Research and Manufacturers of America (PhRMA) Drug Metabolism/Clinical Pharmacology Technical Working Group; FDA Center for Drug Evaluation and Research (CDER) J. Clin. Pharmacol. 2003;43:443–69. [Google Scholar]
- 25.Berry LM, Zhao Z. An examination of IC50 and IC50-shift experiments in assessing time-dependent inhibition of CYP3A4, CYP2D6, and CYP2C9 in human liver microsomes. Drug Metab. Lett. 2008;2:51–59. doi: 10.2174/187231208783478407. [DOI] [PubMed] [Google Scholar]
- 26.Sekiguchi N, Higashida A, Kato M, Nabuchi Y, Mitsui T, Takanashi K, Aso Y, Ishigai M. Prediction of drug-drug interactions based on time-dependent inhibition from high throughput screening of cytochrome P450 3A4 inhibition. Drug Metab. Pharmacokinet. 2009;24(6):500–10. doi: 10.2133/dmpk.24.500. [DOI] [PubMed] [Google Scholar]
- 27.Patki KC, Von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes p450: role of CYP3A4 and CYP3A5. Drug Metab. Dispos. 2003;31(7):938–44. doi: 10.1124/dmd.31.7.938. [DOI] [PubMed] [Google Scholar]
- 28.Nomeir AA, Ruegg C, Shoemaker M, Favreau LV, Palamanda JR, Silber P, Lin CC. Inhibition of CYP3A4 in a rapid microtiter plate assay using recombinant enzyme and in human liver microsomes using conventional substrates. Drug Metab. Dispos. 2001;29(5):748–53. [PubMed] [Google Scholar]
- 29.Bell L, Bickford S, Nguyen PH, Wang J, He T, Zhang B, Friche Y, Zimmerlin A, Urban L, Bojanic D. Evaluation of fluorescence- and mass spectrometry-based CYP inhibition assays for use in drug discovery. J. Biomol. Screen. 2008;13(5):343–53. doi: 10.1177/1087057108317480. [DOI] [PubMed] [Google Scholar]
- 30.Dierks EA, Stams KR, Lim HK, Cornelius G, Zhang H, Ball SE. A method for the simultaneous evaluation of the activities of seven major human drug-metabolizing cytochrome P450s using an in vitro cocktail of probe substrates and fast gradient liquid chromatography tandem mass spectrometry. Drug Metab. Dispos. 2001;29(1):23–9. [PubMed] [Google Scholar]
- 31.Kamgang R, Mboumi RY, Fondjo AF, Tagne MAF, N’dillé GPRM, Yonkeu JN. Antihyperglycaemic potential of the water–ethanol extract of Kalanchoe crenata (Crassulaceae) J. Nat. Med. 2008;62(1):34–40. doi: 10.1007/s11418-007-0179-y. [DOI] [PubMed] [Google Scholar]