Abstract
Cytosolic DNA species derived from invading microbes or leaked from the nuclear or mitochondrial compartments of the cell can trigger the induction of host defense genes by activating the endoplasmic reticulum (ER)-associated protein STING. Using a mass spectrometry based approach, we show that after association with cyclic dinucleotides (CDN’s), delivery of Tank binding kinase 1 (TBK1) to interferon regulatory factors (IRF) such as IRF3 relies on K63-linked ubiquitination of K224 on STING. Blocking K224 ubiquitination specifically prevented IRF3 but not NF-κB activation, additionally indicating that STING trafficking is not required to stimulate the latter signaling pathway. By carrying out a limited siRNA screen we have identified mitochondrial E3 ubiquitin protein ligase 1 (MUL1) as an E3 ligase that catalyzes the ubiquitination of STING on K224. These data demonstrate the critical role of K224 ubiquitination in STING function and provide molecular insight into the mechanisms governing host defense responses.
Introduction
Host cells have evolved a variety of germline-encoded pattern recognition receptors (PRRs) to form the first line of defense against microbial invasion (1, 2). Based on structural homology, these PRRs are approximately divided into four families: the Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), and C-type lectin receptors (CLRs) (3, 4). PRRs recognizes structures conserved among microbial species such as viral RNAs, CpG DNA, and lipopolysaccharides to trigger a cascade of signal transduction and the activation of interferon regulatory factor 3 (IRF3) and/or NF-κB, which eventually induce type I interferons(IFNs) and proinflammatory cytokines production to establish an antiviral immune response (2, 3, 5).
In addition, an endoplasmic reticulum (ER)-resident protein referred as stimulator of interferon genes (STING, also known as TMEM173) has been identified as a critical regulator of innate immune signaling triggered by the presence of cytosolic DNA species, such as self-DNA leaked from the nucleus or mitochondria or introduced by microbes harboring DNA genomes such as HSV-1 (6, 7). Sting-deficient cells are dramatically defective in producing type I IFNs and proinflammatory cytokines in response to cytosolic DNA and are more susceptible to HSV-1 or Listeria monocytogenes infection (8). Studies have shown that STING is a sensor activated by binding with cyclic dinucleotides (cyclic di-GMP or cyclic GMP-AMP (cGAMP)) generated directly by invading bacteria or via a DNA binding synthase cGAS (cGAMP synthase, also known as MB21D1) (9–11). Activated STING traffics from the ER to endosomal/lysosomal regions to activate IRF3 and NF-κB (8, 12, 13). Phosphorylated IRF3 and NF-κB then translocate into the nucleus to initiate the transcription of type I IFNs and proinflammatory cytokine genes.
Besides playing an instrumental role in protecting the host against microbial infection, the activation of STING in phagocytes via the DNA of engulfed dying cancer cells, is essential for the generation of cytokines required for the efficient production of anti-tumor adaptive immunity (14). However, while transient STING function is essential for triggering the induction of host defense genes, chronic STING activation has been shown to be associated with lethal inflammatory disease such as Aicardi-Goutieres Syndrome (AGS) and severe systemic lupus erythematosus (SLE) (15–18). The activity of STING therefore requires tight control to prevent the sustained production of cytokines which are responsible for harmful autoinflammatory disease (19). Thus, following transient activation and the initiation of cytokine production, STING activity is negatively controlled by phosphorylation events and rapidly undergoes degradation (20).
In addition to phosphorylation, the control of STING function has also been reported to involve the palmitoylation and ubiquitination of STING, although conflicting reports indicate that the mechanisms of ubiquitination remain to be fully clarified (21–27). For example, the E3 ligases TRIM56, TRIM32 and AMFR have been reported to catalyze K63 or K27-linked polyubiquitination of a number of lysine residues in STING (21, 23, 24). Alternatively, the ubiquitination of STING has been reported to implicate K48-linked ubiquitination processes, an event which reportedly promotes proteasome-mediated degradation (22, 26). To extend and clarify these studies, we adopted a proteomic approach and report that the ubiquitination of STING on lysine K224 is essential for efficient cytosolic DNA-mediated signaling. We further report that the mitochondrial E3 ubiquitin protein ligase 1 (MUL1, also known as GIDE/MAPL/MULAN/RNF218) ubiquitinates STING on K224 via K63-linked polyubiquitination, which facilitates optimal STING trafficking and the transcription of host defense genes.
Results
STING is ubiquitinated on lysine 224 with K63-linked polyubiquitin chains
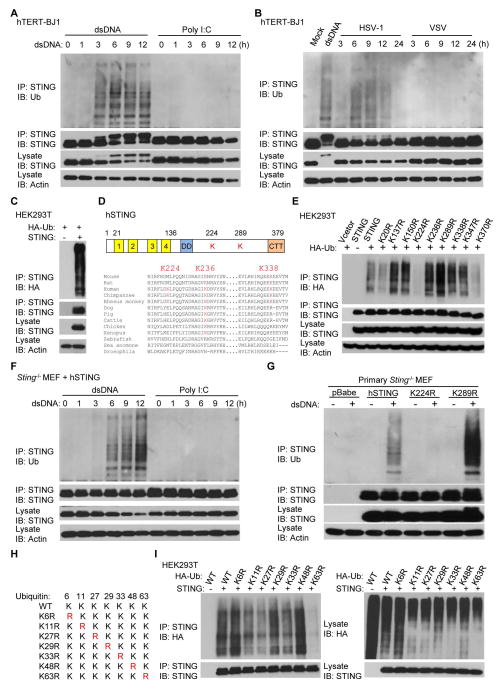
We and others have previously observed that post-translational modification of STING plays an important role in controlling STING-dependent cytokine production (7, 20, 28). To extend these studies we investigated ubiquitination processes in human fibroblasts and confirmed that in the presence of cytosolic dsDNA, but not poly I:C, STING is robustly ubiquitinated within 3 hours (Fig. 1A). The importance of cytosolic dsDNA in mediating these events was confirmed by observing that STING ubiquitination only occurred in cells infected with the dsDNA pathogen herpes simplex virus 1 (HSV-1), but not following exposure to the RNA virus, vesicular stomatitis virus (VSV) (Fig. 1B). To identify which lysine residues in STING are ubiquitinated, we co-transfected HEK293T cells that lack STING, with human STING (hSTING) and hemagglutinin (HA)-tagged ubiquitin. We then purified the ubiquitinated STING from cells using antibodies specific to HA or STING via tandem affinity precipitation (Fig. 1C and fig. S1A). Purified STING protein was analyzed by mass spectrometry which indicated that STING was predominantly ubiquitinated on three lysine residues (K224, K236 and K338) (Fig. 1D and fig. S1B). Of the three sites, K236 appears to be highly conserved in mammals, while K224 is only found to be conserved in human and non-human primates (Fig. 1D). Mass spectrometry analysis identified both K48-linked and K63-linked polyubiquitin chains on STING as have been previously reported (Fig. S1C) (21–23).
Fig. 1. STING is ubiquitinated on lysine 224 with K63-linked polyubiquitin chains.
(A) hTERT-BJ1 cells were transfected with dsDNA (4 μg/ml) or poly I:C (4 μg/ml) for the indicated time periods. Cell lysates were immunoprecipitated with anti-STING antibody and immunoblotted with the indicated antibodies. (B) hTERT-BJ1 cells were transfected with dsDNA (6 hr) or infected with HSV-1 (MOI=10) or VSV (MOI=10) for the indicated time periods. Cell lysates were immunoprecipitated with anti-STING antibody and immunoblotted with the indicated antibodies. (C) HEK293T cells were transfected with hSTING and HA-tagged ubiquitin (HA-Ub) for 30 hr. Cell lysates were immunoprecipitated with anti-STING antibody and immunoblotted with the indicated antibodies. (D) Alignment of STING amino acid sequences. Highlighted amino acids indicate ubiquitinated lysine residues of hSTING detected by mass spectrometry. (E) hSTING or its variants were individually transfected into HEK293T cells along with HA-Ub for 30 hr. Cell lysates were immunoprecipitated with anti-STING antibody and immunoblotted with the indicated antibodies. (F) Primary Sting−/− MEFs were reconstituted with hSTING using retroviruses. Cells were transfected with dsDNA (4 μg/ml) or poly I:C (4 μg/ml) for the indicated time periods, immunoprecipitated with anti-STING antibody and immunoblotted with indicated antibodies. (G) Primary Sting−/− MEFs reconstituted with hSTING or its variants were transfected with dsDNA (4 μg/ml) for 6 hr. Cell lysates were immunoprecipitated with anti-STING antibody and immunoblotted with the indicated antibodies. (H) Schematic presentation of ubiquitin and its variants. (I) HEK293T cells were transfected with hSTING along with HA-Ub or its variants for 30 hr. Cell lysates were immunoprecipitated with anti-STING antibody and then immunoblotted with the indicated antibodies. Each panel of data is a representative of at least two independent experiments which had the same outcome.
To validate our mass spectrometry data, all nine lysine residues that reside in hSTING, including K224, K236 and K338, were individually mutated to arginine and their ability to be ubiquitinated was analyzed following transfection into HEK293T cells. This approach indicated that only substitution of K224, identified by mass spectrometry and previously unreported, was able to significantly reduce the ubiquitination of STING (Fig. 1E). To further confirm this finding, we retrovirally reconstituted Sting−/− murine embryonic fibroblasts (MEFs) with wild type hSTING or STING variants, and confirmed that hSTING was similarly able to undergo ubiquitination in reconstituted MEFs (Fig. 1F). We again observed that substitution of K224, but not the other lysine residues, significantly reduced dsDNA-induced STING ubiquitination events (Fig. 1G and fig. S1D). Through these studies, we also noted that substitution of K289 to K289R greatly increased dsDNA-induced STING ubiquitination, for reasons that remain to be determined (Fig. 1G and S1D).
Previous studies have reported that STING is ubiquitinated via K48-linked polyubiquitin chains following sendai virus infection, an RNA virus, an event which reportedly leads to STING’s proteasomal degradation (22). However, the proteasome inhibitors MG132 or lactacystin did not appear to significantly enrich ubiquitinated STING in hTERT-BJ1 cells, suggesting that STING might not be modified via K48-linked polyubiquitination, at least in the presence of dsDNA (fig. S1, E and F). To further investigate this, we analyzed the ubiquitination profile of STING using ubiquitin harboring various lysine substitutions (Fig. 1H and fig. S1G). We noted that STING ubiquitination was dramatically reduced only when using an ubiquitin harboring a K63R substitution (Fig. 1I and fig. S1H). These data indicate that K63-linked polyubiquitination on K224 is a predominant ubiquitination type required for STING activity although other types of STING ubiquitination may also exist to control STING, such as to facilitate degradation.
Ubiquitination on K224 of STING is required for IRF3 activation, but not NF-κB activation
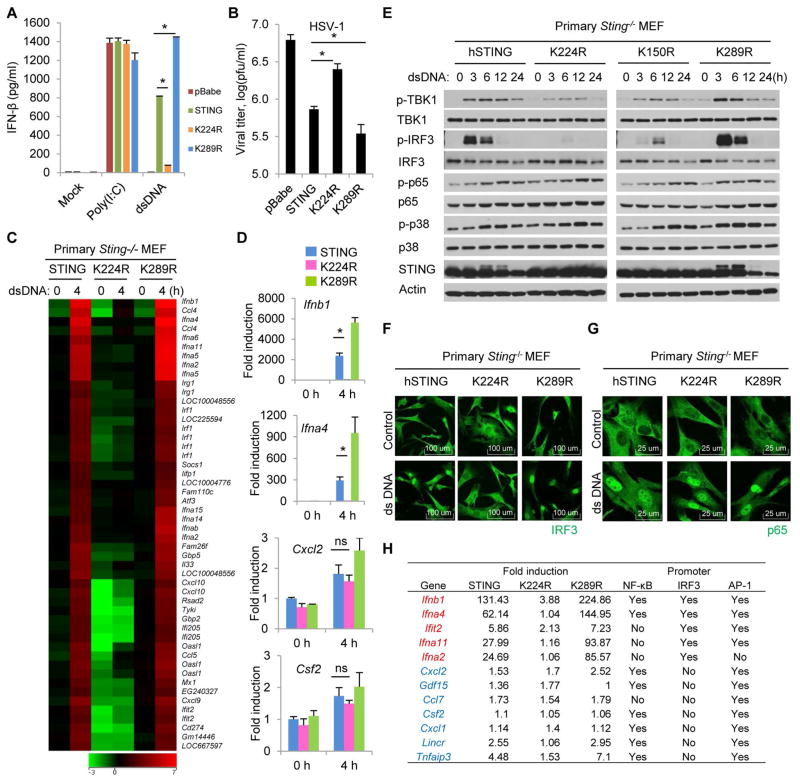
To further investigate the role of K224 ubiquitination in regulating STING function, we stably reconstituted primary Sting−/− MEFs with wild-type hSTING or selected variants (K224R or K289R) and treated the cells with poly I:C or STING-activating dsDNA. ELISA analysis indicated that dsDNA-mediated IFNβ production was inhibited in cells reconstituted with the K224R variant. In contrast, cells containing the K289R variant were noted to exhibit slightly enhanced IFNβ production (Fig. 2A and fig. S2A). Accordingly, cells reconstituted with K224R were seen to enable enhanced HSV-1 replication, probably due to the reduced type I IFN production (Fig. 2B and fig. S2B). To further validate these findings, we treated the reconstituted Sting−/− MEFs with dsDNA and carried out a microarray analysis. This study confirmed that expression of type I IFNs as well as other cytosolic DNA-mediated, STING-inducible genes, including members of the IFIT family, were significantly suppressed in cells reconstituted with the K224R variant (Fig. 2C). Expression profiles were further validated by quantitative real-time PCR, which confirmed that loss of ubiquitination on K224 resulted in a dramatic decrease in STING-inducible gene induction while again, substitution of K289 to arginine may actually enhance cytokine production (Fig. 2D). However, it was noted that the expression of a number of genes such as Cxcl2 and Csf2 remained somewhat unaffected by loss of K224 ubiquitination and appeared readily inducible in the presence of cytosolic DNA (Fig. 2D and fig. S2C). Since type I IFN production is regulated by the coordinated activation of a number of transcription factors, such as IRF3/7, NF-κB and AP-1 (29), we therefore evaluated whether ubiquitination on K224 is required for STING-dependent activation of both IRF3/7 and NF-kB signaling pathways. Immunoblot analysis indicated that dsDNA-induced phosphorylation of TBK1 and IRF3 was significantly inhibited in cells reconstituted with K224R (Fig. 2E and fig. S2D). However, phosphorylation of p65 or p38 was not affected in these cells following stimulation with cytosolic dsDNA (Fig. 2E). A p65 ELISA further confirmed that cells reconstituted with K224R could activate p65 phosphorylation after dsDNA stimulation similar to wild type hSTING (fig. S2, E and F). To extend these studies, immunofluorescence microscopy was carried out, which confirmed that K224R expressing cells did not exhibit IRF3 translocation into the nucleus following treatment with cytosolic DNA (Fig. 2F and fig. S2G). However, the p65 subunit of NF-κB translocated into the nucleus normally in similarly treated K224R expressing cells (Fig. 2G and fig. S2H). Thus, ubiquitination of STING on K224 appears to principally affect IRF3 but not NF-κB signaling. These results may help explain why the transcriptional activation some genes such as Cxcl2 and Csf2 were unaffected by loss of K224 ubiquitination, since they only predominantly require NF-κB signaling for transcriptional activity, unlike type I IFN which requires NF-κB, AP-1 as well as IRF3/7 (Fig. 2H). Taken together, our data confirm that ubiquitination on K224 of STING is essential for dsDNA-mediated activation of IRF3 pathway, but may not affect NF-κB or AP-1 pathways.
Fig. 2. Ubiquitination on lysine 224 of STING is required for IRF3 activity, but not NF-κB activity.
(A) Primary Sting−/− MEFs reconstituted with hSTING or its variants were transfected with dsDNA (4 μg/ml) or poly I:C (4 μg/ml) for 16 hr, and IFNβ production was measured by ELISA. (B) Reconstituted Sting−/− MEFs were infected with HSV-1 (MOI = 0.1) for 24 hr, and viral titer were measured by plaque assay. (C) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for 4 hr. Total RNA was purified and examined for gene expression with Illumina Sentrix BeadChip Array (Mouse WG6 version2). (D) Real-time PCR was carried out with the indicated probes to confirm gene array analysis shown in Figure 2C. (E) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for the indicated time periods, and cell lysates were immunoblotted with the indicated antibodies. (F and G) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for 6 hr, stained with anti-IRF3 (F) or anti-p65 (G) antibodies and imaged by confocal microscopy. (H) Fold induction of selected genes in dsDNA treated Sting−/− MEF reconstituted with hSTING, K224R or K289R (data from gene array analysis shown in Figure 2C). Each panel of data is a representative of at least two independent experiments which had the same outcome. Data in (A) and (D) were presented as average ± SD of duplicated and (B) of triplicated samples from each group. P value was determined by Student’s t test and a p<0.05 was considered statistically significant difference between two groups as indicated by asterisks. ns, not significant.
Loss of ubiquitination inhibits the translocation, phosphorylation and degradation of STING
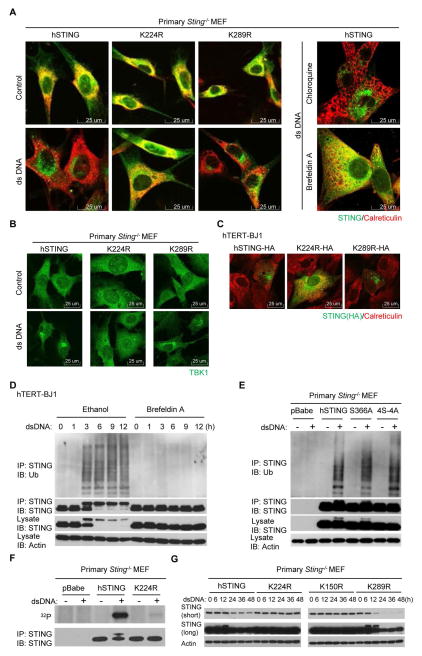
In the presence of cytosolic DNA, STING translocates with TBK1 from the ER to perinuclear vesicles containing transcription factors. STING is subsequently degraded in the lysosomal compartments to avoid chronic cytokine production (8, 20). Blocking STING trafficking from the ER to the Golgi using brefeldin A (BFA) was noted to prevent STING phosphorylation, suggesting that such post-translational events occur after trafficking has commenced (8, 20). Since loss of ubiquitination on K224R renders STING inactive, we similarly investigated at what stage STING function was blocked. Following the reconstitution of STING-deficient MEFs, we observed that out of the nine K to R STING variants examined, only the ubiquitination-defective mutant K224R lost the ability to translocate in response to dsDNA stimulation (Fig. 3A, fig. S3, A and B). This suggests that ubiquitination of STING is required for trafficking to commence from the ER to the Golgi apparatus, or occurs on route between these cellular compartments. Chloroquine, a lysosomal inhibitor which reportedly prevents STING degradation (20), did not block STING trafficking, confirming that STING degradation occurs after translocation (Fig. 3A). Cytosolic DNA-induced TBK1 translocation was also prevented in MEF cells reconstituted with K224R (Fig. 3B), although K224R was observed to associate with TBK1 (fig. S3C). MEF cells reconstituted with K224R also exhibited significantly decreased cytosolic DNA-induced LC3 conversion (fig. S3D). The HA-tagged K224R variant was also observed not to efficiently traffic when expressed in human hTERT-BJ1 cells that express endogenous STING (Fig. 3C). These data suggest that ubiquitination on K224 is essential for promoting dsDNA-mediated STING trafficking in both mouse and human cells. Additional studies indicated that the K to R substitution on 224 residue did not appear to affect STING’s ability to bind to CDN’s or subsequently to dimerize, suggesting that ubiquitination occurred after association with STING activators (Fig. S4). To further evaluate at what stage ubiquitination of STING occurs to facilitate function, we treated cytosolic DNA transfected hTERT-BJ1 cells with BFA and noted that this treatment similarly prevented STING ubiquitination (Fig. 3D). Considering that BFA inhibits protein transport from the ER to the Golgi apparatus, this would suggest that STING ubiquitination probably occurs after activation with CDN’s and during the ER to the Golgi transportation process. This post-translational modification appears important for the regulating STING/TBK1 trafficking and rendezvousing with IRF3.
Fig. 3. Loss of ubiquitination inhibits STING translocation, phosphorylation and degradation.
(A) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for 9 hr, stained with indicated antibodies and imaged by confocal microscopy. Cells treated with chloroquine (50 μM) or brefeldin A (0.05 μg/ml) 1 hour before dsDNA stimulation were included as controls. (B) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for 3 hr, stained with anti-TBK1 antibody and imaged by confocal microscopy. (C) hTERT-BJ1 cells were transfected with HA-tagged hSTING or mutants for 36 hr and immunostained with the indicated antibodies. (D) hTERT-BJ1 cells were incubated with ethanol or brefeldin A (0.05 μg/ml) for 1 hr and then transfected with dsDNA (4 μg/ml) for the indicated time periods. Cell lysates were immunoprecipitated with anti-STING antibody and then immunoblotted with the indicated antibodies. (E) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for 6 hr. Cell lysates were immunoprecipitated with anti-STING antibody and then immunoblotted with the indicated antibodies. 4S-4A stands for S345/358/366/379A. (F) Reconstituted Sting−/− MEFs were incubated with 32P-labeled phosphate for 30 min and then transfected with dsDNA (4 μg/ml) for 3 hr. Cell lysates were immunoprecipitated with anti-STING antibody and then analyzed by autoradiography or immunoblotted with anti-STING antibody. (G) Reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for the indicated time periods, and cell lysates were immunoblotted with the indicated antibodies. Each panel of data is a representative of at least two independent experiments which had the same outcome.
We have previously demonstrated that in the presence of cytosolic DNA, STING is phosphorylated on four major serine residues (20). Phosphorylation of STING on S366 was found to negatively regulate activity. We therefore investigated the relationship between STING ubiquitination and phosphorylation. We noted that the inactive STING S366A variant or a STING variant containing all four serine substitutions could readily undergo ubiquitination following cytosolic DNA stimulation (Fig. 3E). Further, phosphorylation-defective STING variants appeared able to traffic normally in response to dsDNA (fig. S3E). These data indicate that STING ubiquitination and trafficking does not require phosphorylation. Conversely, phosphorylation of the ubiquitination-defective mutant K224R was noted to be significantly impeded, indicating that ubiquitination of STING occurs before phosphorylation (Fig. 3F). In addition, the K224R STING variant did not appear to undergo significant degradation after dsDNA treatment, presumably since subsequent phosphorylation events are hindered and trafficking to lysosomal compartments for degradation are prevented (Fig. 3G). We also noted through these studies that the hyper-active mutant K289R degraded faster compared to the wild-type hSTING (Fig. 2B and 3G), presumably because it is hyper ubiquitinated for reasons that presently remain unclear (Fig. 1G and fig. S1D). Taken together, our data indicate that ubiquitination on K224 is critical for the efficient trafficking of activated STING from the ER to the Golgi, and for further post-translational events, such as phosphorylation, to occur.
We had also noted that the K289R mutation significantly increased dsDNA-induced STING ubiquitination as well as type I IFN production (Fig. 1G, fig. S1D and S2A). The K289R mutant was also more readily phosphorylated and rapidly degraded than wild-type STING in response to dsDNA (Fig. 3G). To investigate the cause of this hyper-activity, we further substituted K224 or the previously reported K150 with arginine within the K289R mutant. Ubiquitination assays demonstrated that the additional substitution of K224, but not K150, abolished the observed increased ubiquitination on K289R (fig. S5A). Accordingly, dsDNA-induced IFNβ production was reduced in cells reconstituted with the K224/289R variant compared to those expressing the K289Rvariant alone (fig. S5, Band C). Collectively, these data suggest that the hyper-activity of K289R could plausibly be due to an increase inK224 ubiquitination.
To confirm that ubiquitination on K224 influences STING function in human cells, we retrovirally reconstituted wild-type hSTING, K150R, K224R or K289R into a human lung cancer cell line CRL-5800, which does not express endogenous STING. Similar to our results using reconstituted Sting−/− MEFs, we observed that ubiquitination of K224R in CRL-5800 was significantly reduced in the presence of cytosolic DNA (fig. S6A). Immunoblot assay indicated reduced phosphorylation of TBK1 as well as delayed phosphorylation of IRF3 in cells reconstituted with the K224R variant following treatment with dsDNA (fig. S6B). In addition, dsDNA-induced STING phosphorylation and degradation was also inhibited with K224R (fig. S6B). Further analysis verified that dsDNA-induced STING trafficking and phosphorylation as well as IRF3 nuclear translocation was suppressed in cells reconstituted with K224R (fig. S6, C and D). Accordingly, CRL-5800 cells expressing K224R exhibited dramatically reduced dsDNA-induced cytokine production (fig. S6, E and F). These data confirm that ubiquitination on K224 is essential for STING trafficking as well as TBK1-mediated IRF3 activation in the presence of cytosolic DNA in human cells.
Identification of MUL1 as an essential activator of dsDNA-mediated STING-dependent pathway
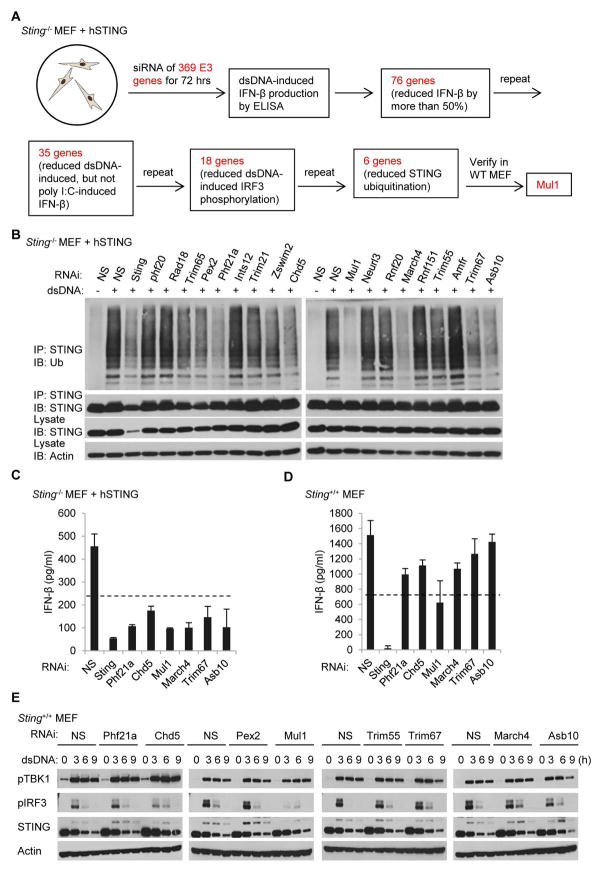
We have demonstrated that ubiquitination of STING was required for dsDNA-induced STING trafficking and activation. To identify the ubiquitin ligase which may facilitate STING ubiquitination, we performed a siRNA-based screening using the primary Sting−/− MEFs reconstituted with hSTING (Fig. 4A). Briefly, siRNA of 369 ubiquitin E3 ligase genes were individually transfected into cells prior to treatment with dsDNA or poly I:C. After 3 days, medium was retrieved and ELISA analysis was used to identify approximately 35 E3 ligases, the silencing of which was seen to significantly inhibit dsDNA-induced, but not poly I:C-mediated IFNβ production. Among them, 6 genes were found to possibly be involved in regulating STING ubiquitination, since RNAi knock down greatly reduced this process (Fig. 4B). Additional ELISA analysis further confirmed that a number of the E3 ligases could influence dsDNA-induced IFNβ production in Sting−/− MEFs reconstituted with hSTING (Fig. 4C). However, only the silencing of Mul1 dramatically reduced dsDNA-induced IFNβ production in wild-type MEFs (Fig. 4D). Further analysis indicates that suppressing Mul1 expression significantly inhibited dsDNA-induced phosphorylation of IRF3 in wild-type MEFs (Fig. 4E). These data indicate that MUL1 might be required for the efficient ubiquitination of STING.
Fig. 4. Identification of MUL1 as a potential STING ubiquitin E3 ligase.
(A) Schematic chart flow of screening procedure for identifying STING ubiquitin ligase. (B) Reconstituted Sting−/− MEFs were transfected with the indicated siRNAs for 72 hr followed with dsDNA (4 μg/ml) treatment for 6 hr. Cell lysates were precipitated with anti-STING antibody and then immunobloted with the indicated antibodies. (C and D) Reconstituted Sting−/− MEF (C) or Wild-type MEF (D) were transfected with the indicated siRNAs for 72 hr followed with dsDNA (4 μg/ml) treatment for 16 hr, and IFNβ production was measured by ELISA. (E) Wild-type MEFs were transfected with the indicated siRNAs for 72 hr followed with dsDNA (4 μg/ml) treatment for the indicated time periods. Cell lysates were immunoblotted with the indicated antibodies. Each panel of data is a representative of at least two independent experiments which had the same outcome. Data in (C) and (D) were presented as average ± SD of duplicated samples from each group.
MUL1 interacts with STING and ubiquitinates STING on K224
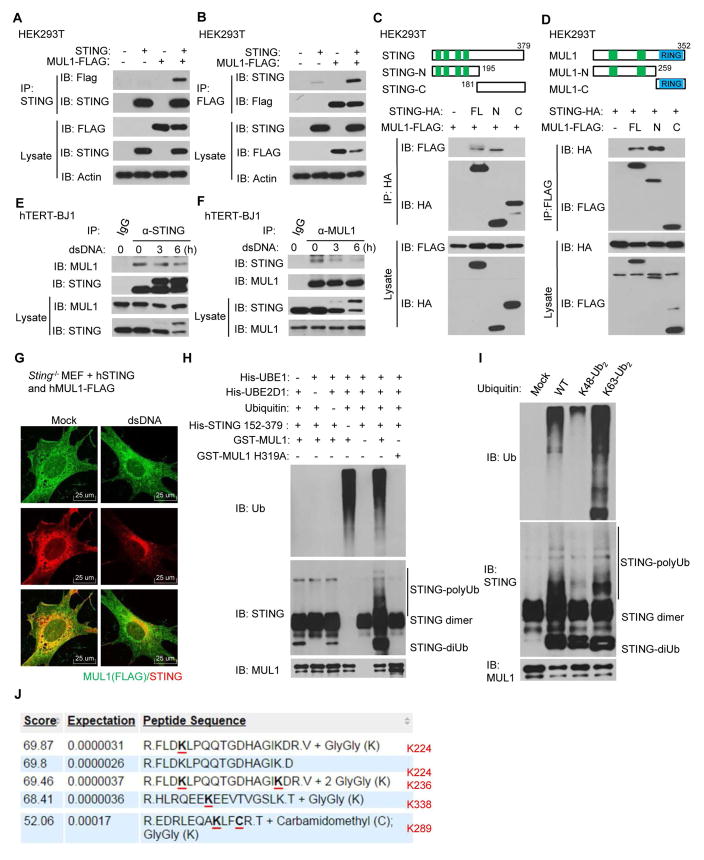
To further confirm the role of MUL1 in STING ubiquitination, we transfected FLAG-tagged MUL1 and STING individually or together into HEK293T cells. Immunoprecipitation assays confirmed that STING could interact with FLAG-tagged MUL1 (Fig. 5, A and B). By using HA-tagged STING truncated variants (Fig. 5C, upper panel), we determined that the amino-terminal region of STING mediated association with MUL1 (Fig. 5C, lower panel). Likewise, the transmembrane region of MUL1 was mapped as facilitating association with STING (Fig. 5D). We further confirmed that MUL1 interacted with STING endogenously in resting hTERT-BJ1 cells using similar immunoprecipitation assays (Fig. 5, E and F). Notably, the endogenous association between MUL1 and STING were substantially reduced after dsDNA stimulation (Fig. 5, E and F). Immunofluorescence microscopy analysis demonstrated that MUL1 colocalized with STING in resting reconstituted MEF cells (Fig. 5G). However, MUL1 did not co-translocate with STING to the peri-nuclear region after dsDNA treatment (Fig. 5G). Thus, MUL1 may associate with STING under non-stimulated conditions, in the vicinity of the ER.
Fig. 5. MUL1 interacts with STING and ubiquitinates STING on K224.
(A and B) STING and FLAG-tagged MUL1 were transfected individually or together into HEK293T cells for 30 hr. Cell lysates were immunoprecipitated with anti-STING (A) or anti-FLAG (B) antibodies and then immunoblotted with the indicated antibodies. (C) HA-tagged STING or its mutants were individually transfected into HEK293T cells along with FLAG-tagged MUL1. Cell lysates were immunoprecipitated with anti-HA antibody and then immunoblotted with the indicated antibodies. (D) FLAG-tagged MUL1 or its mutants were individually transfected into HEK293T cells along with HA-tagged STING. Cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with the indicated antibodies. (E and F) hTERT-BJ1 cells were transfected with dsDNA (4 μg/ml) for the indicated time periods. Cell lysates were immunoprecipitated with rabbit IgG or anti-STING (E) or anti-MUL1 (F) antibodies and then immunoblotted with the indicated antibodies. (G) Primary Sting−/− MEFs were reconstituted with hSTING and FLAG-tagged hMUL1 using retrovirus. The cells were transfected with dsDNA (4 μg/ml) for 9 hr, and then stained with anti-STING and anti-FLAG antibodies. (H) Purified E1, E2, ubiquitin, STING (152–379 aa), GST-MUL1 or GST-MUL1 H319A proteins were mixed together as indicated and incubated at 37°C for 2 hr. The mixture was then analyzed by immunoblot with the indicated antibodies. (I) Purified E1, E2, STING (152–379 aa) and GST-MUL1 proteins were mixed together with ubiquitin, K48-linked diUb or K63-linked diUb proteins and incubated at 37°C for 2 hr. The mixture was then analyzed by immunoblot with indicated antibodies. (J) In vitro ubiquitinated STING by GST-MUL1 as described in Figure 5H was analyzed by mass spectrometry. Highlighted lysine residues were identified as the ubiquitination sites. Each panel of data is a representative of at least two independent experiments which had the same outcome.
To evaluate whether MUL1 directly ubiquitinates STING, we purified glutathione S-transferases (GST)-tagged MUL1 protein as well as a previously-reported MUL1 mutant H319A, which eliminates its ubiquitin ligase activity (30) (fig. S7A). In vitro ubiquitination assays confirmed that MUL1 catalyzed the formation of polyubiquitin chains on STING, whereas the MUL1 H319A could not (Fig. 5H and fig. S7B). As a control, MUL1 was not found to ubiquitinate GFP in vitro (fig. S7C). In vitro ubiquitination assays using pre-linked K48-Ub2 or K63-Ub2 suggested that MUL1 preferably catalyzed K63-linked, but not K48-linked polyubiquitin chains on STING (Fig. 5I). Importantly, mass spectrometric analysis of ubiquitinated STING proteins generated from in vitro ubiquitination assays indicated that MUL1 could catalyze ubiquitination on four lysine residues (K224, K236, K289 and K338) (Fig. 5J). Since we noted that STING variants that do not bind cGAMP were not ubiquitinated after cytosolic dsDNA stimulation (fig. S7, D to F), it is thus plausible that conformational rearrangement of STING dimers after binding with CDNs might enable MUL1 to ubiquitinate STING. Taken together, our data demonstrates that MUL1 directly interacts with STING and can catalyze K63-linked polyubiquitination of STING on lysine 224.
MUL1 regulates dsDNA-induced STING-dependent innate immune response
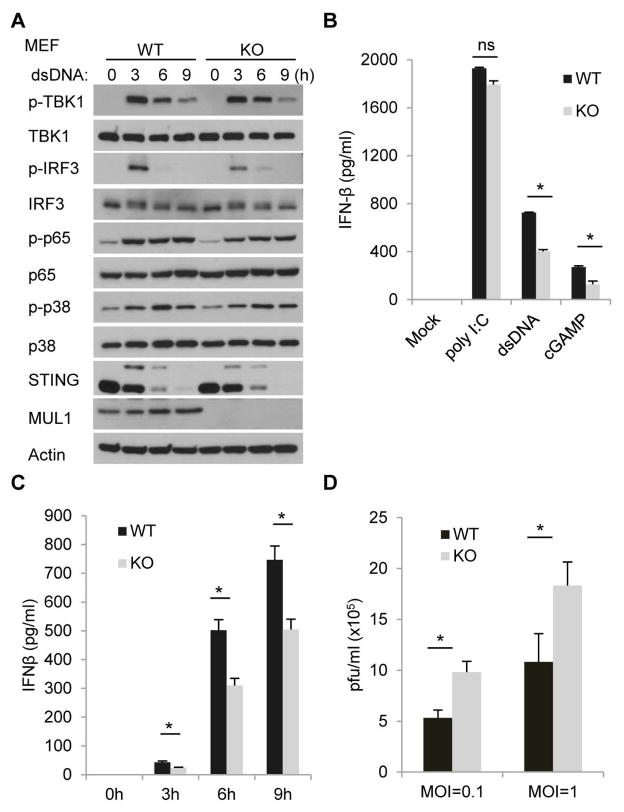
To further explore the function of MUL1 in STING-dependent innate immunity, we suppressed MUL1 expression using siRNA in Sting−/− MEFs reconstituted with hSTING. We observed that silencing of Mul1inhibited dsDNA-induced phosphorylation of TBK1 and IRF3 (Fig. 6A and fig. S8A). However, phosphorylation of p65 or p38 was largely not affected (Fig. 6A). Notably, STING phosphorylation and degradation were also suppressed in Mul1-silenced cells following activation with cytosolic DNA (Fig. 6A). A similar effect was observed in wild-type MEFs treated with RNAi to Mul1 (Fig. 6B and fig. S8B). Significantly, dsDNA-induced STING ubiquitination was greatly ablated when MUL1 expression was suppressed (Fig. 6C). Accordingly, silencing of Mul1 reduced dsDNA-induced, but not poly I:C or 5′ppp dsRNA mediated IFNβ production in reconstituted Sting−/− MEFs (Fig. 6D, 6E and fig. S8C) as well as in wild type MEFs (Fig. 6F, 6G and fig. S8D). We also observed a similar effect in RAW 264.7 cells, an Abelson murine leukemia virus transformed macrophage cells line (fig. S8, E and F). It was noted that suppression of MUL1 did not completely eliminate cytosolic DNA triggered STING activity perhaps due to inefficiency of knockdown or more likely to the existence of alternate ubiquitin ligases (21, 23). Nevertheless, our studies also indicated the suppression of STING trafficking as well as IRF3 but not p65 translocation into the nucleus of Mul1-silenced cells in response to cytosolic DNA (Fig. 6, H to J). However, silencing of Mul1 did not affect translocation of IRF3 or p65 into the nucleus following treatment with poly I:C, confirming that MUL1 largely mediates cytosolic DNA triggered STING function (Fig. 6,H and I, fig. S8, G and H). To extend this analysis further, we infected Mul1 siRNA-treated cells or control cells with γ34.5-deleted HSV-1 (HSV-1 γ34.5), which robustly triggers type I IFN production. Immunoblot analysis indicated that viral-mediated activation of the TBK1-IRF3 axis was also inhibited in Mul1-silenced, reconstituted Sting−/− MEFs as well as in wild-type MEFs (Fig. 6, K and L). Accordingly, loss of MUL1 expression was noted to reduce viral induction of IFNβ mRNA and facilitate HSV-1 γ34.5 replication (Fig. 6, M and N). These findings were complemented in human cells following suppression of MUL1 (fig. S9). To further substantiate the function of MUL1 in cytosolic DNA-mediated signaling pathway, we obtained Mul1−/− MEFs and observed that MUL1 deficiency attenuated dsDNA-induced phosphorylation of IRF3 (Fig. 7A). However, phosphorylation of p65 or p38 was less affected (Fig. 7A). Accordingly, dsDNA or cGAMP-induced, but not poly I:C-mediated IFNβ production was reduced in Mul1−/− MEFs (Fig. 7, B and C). Moreover, HSV-1 replication was seen to be facilitated in Mul1−/− MEFs (Fig. 7D). However, similar to RNAi analysis, deficiency of MUL1 did not completely abrogate dsDNA-induced IRF3 phosphorylation or type I IFN production, indicating that other E3 ligases likely play a role in regulating STING activity. Nevertheless, taken together, our data demonstrate that K224 is a key site for ubiquitination-mediated control of STING function.
Fig. 6. MUL1 regulates dsDNA-induced STING-dependent innate immune response.
(A and B) siRNA-treated reconstituted Sting−/− MEFs (A) or wild-type MEFs (B) were transfected with dsDNA (4 μg/ml) for the indicated time periods, and cell lysates were immunoblotted with indicated antibodies. NS, nonspecific siRNA. (C) siRNA-treated reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for the indicated time periods. Cell lysates were immunoprecipitated with anti-STING antibody and then immunoblotted with the indicated antibodies. (D and F) siRNA-treated reconstituted Sting−/− MEFs (D) or wild-type MEFs(F) were transfected with poly I:C (1 μg/ml), 5′ppp dsRNA (1 μg/ml), or dsDNA (4 μg/ml) for 16 hr, and IFNβ production was measured by ELISA. (E and G) siRNA-treated reconstituted Sting−/− MEFs (E) or wild-type MEFs (G) were transfected with dsDNA (4 μg/ml) for the indicated time periods, and IFNβ production was measured by ELISA. (H and I) siRNA-treated reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) or poly I:C (4 μg/ml) for 6 hr, and then stained with anti-IRF3 (H) or anti-p65 (I) antibodies. (J) siRNA-treated reconstituted Sting−/− MEFs were transfected with dsDNA (4 μg/ml) for the indicated time periods, and then stained with the indicated antibodies. (K and L) siRNA-treated reconstituted Sting−/− MEFs (K) or wild-type MEFs (L) were infected with HSV-1 γ34.5 (MOI=10) for the indicated time periods, and cell lysates were immunoblotted with the indicated antibodies. (M) siRNA-treated reconstituted Sting−/− MEFs were infected with HSV-1 γ34.5 (MOI=10) for 6 hr, and induction of IFNβ mRNAs was measured by real-time PCR. (N) siRNA-treated reconstituted Sting−/− MEFs were infected with HSV-1 γ34.5 (MOI=2 or 10) for 24 hr, and viral titer was measure by plague assay. Each panel of data is a representative of at least two independent experiments which had the same outcome. Data in (D) - (G), (M) were presented as average ± SD of duplicated and (N) of triplicated samples from each group. P value was determined by Student’s t test and a p<0.05 was considered statistically significant difference between two groups as indicated by asterisks. ns, not significant.
Fig. 7. MUL1 deficiency attenuates cytosolic DNA-mediated innate immune response.
(A) Wild type (WT) or Mul1 knockout (KO) MEFs were transfected with dsDNA (4 μg/ml) for the indicated time periods, and cell lysates were immunoblotted with indicated antibodies. (B) WT or Mul1 KO MEFs were transfected with poly I:C (4 μg/ml), dsDNA (4 μg/ml) or cGAMP (8 μg/ml) for 16 hr, and IFNβ production was measured by ELISA. (C) WT or Mul1 KO MEFs were transfected with dsDNA (4 μg/ml) for the indicated time periods, and IFNβ production was measured by ELISA. (D) WT or Mul1 KO MEFs were infected with HSV-1 (MOI=0.1 or 1) for 24 hr, and viral titer was measure by plague assay. Each panel of data is a representative of at least two independent experiments which had the same outcome. Data in (B) and (C) were presented as average ± SD of duplicated and (D) of triplicated samples from each group. P value was determined by Student’s t test and a p<0.05 was considered statistically significant difference between two groups as indicated by asterisks. ns, not significant.
Discussion
The presence of dsDNA species in the cytoplasm of the cell triggers the production of host defense proteins including pro-inflammatory cytokines and is a signaling event that is controlled by the cellular ER resident protein STING (7). STING trafficking is critical for the activation of TBK1 and IRF3 as well as NF-κBrequired for the robust activation of type I IFN (8, 31). Several post-translational events, including phosphorylation and ubiquitination, have been reported to control STING activity. However, the data frequently appears conflicting and has predominantly involved a mutagenic approach without the use of mass spectrometry. By adopting this method, we identified three ubiquitination sites (K224, K236 and K338) on STING using a mass spectrometry approach following expression of STING in a 293T system which was necessary to obtain sufficiently modified STING for ubiquitination analysis. Our results indicated that only substitution of K224, but not other lysine residues, almost completely abolished STING ubiquitination in the presence of cytosolic DNA. Although substitution of other lysines, such as K150 and K236, partially inhibited dsDNA-induced IFNβ production, they did not appear to significantly influence STING ubiquitination in our model. Our data also indicate that STING is predominantly ubiquitinated on K224 with K63-linked polyubiquitin chains. We further found that ubiquitination of STING on K224R was required for STING/TBK1 trafficking and the activation of IRF3. Of note, K224R substitution did not appear to affect STING’s ability to associate with CDN’s or to affect dimerization. These new observations were underscored by demonstrating that STING K224R was able to trigger NF-κB signaling in the presence of cytosolic DNA. Thus, K224 ubiquitination probably is not required for regulating the NF-κB pathway. Since STING K224R did not appear to efficiently traffic from the ER, our data would indicate that STING’s ability to regulate NF-κB occurs prior to this event, likely in the ER or Golgi following association with CDN’s.
Our previous data have also demonstrated that STING is phosphorylated predominantly on four serine residues including S366 after trafficking through the Golgi. Since we show that STING ubiquitination is required for trafficking, it is not surprising to observe that STING K224R is unable to be phosphorylated after activation with cytosolic DNA species. In contrast, phosphorylation-deficient STING variants were readily able to be ubiquitinated, confirming that phosphorylation is not required for STING ubiquitination, but rather the opposite. Thus, ubiquitination and phosphorylation events may have evolved to control STINGs ability to regulate the TBK1/IRF3 axis and not the more evolutionarily conserved NF-kB pathway (32).
By using an ubiquitin E3 ligase siRNA library, we further identified mitochondrial E3 ubiquitin protein ligase 1 (MUL1) as a putative STING ubiquitin E3 ligase. MUL1 is a transmembrane protein located in the mitochondrial outer membrane as well as in the ER/mitochondria region of contact(33, 34). MUL1 has been implicated to play extensive rolesin various processes including mitochondrial dynamics, cell growth, apoptosis, and mitophagy, through its ubiquitin E3 ligase activity or SUMO E3 ligase activity (30, 33–36). Our data here indicate that MUL1 can interact with and ubiquitinate STING on K224, preferentially via K63-linked polyubiquitin chains. Suppression of MUL1 expression by siRNA inhibited dsDNA-induced STING ubiquitination and the production of type I IFN. MUL1 may possibly interact with STING in resting cells, although STING does not appear to be ubiquitinated until after association with CDN’s. How MUL1 ubiquitinates STING in the presence of cytosolic DNA has yet to be determined, although a conformational change following association with CDN’s may facilitate this process. Suppression of MUL1 or MUL1 deficiency did not completely abrogate STING trafficking and function however, confirming that other E3 ligases may also play a role in this process, as the literatures describe.
In summary, our data indicate that ubiquitination on K224 of STING is required to initiate cytosolic DNA-mediated STING trafficking, which is essential to activate the production of antimicrobial cytokines and chemokines. Our study provides new mechanistic on the regulation of cytosolic dsDNA-mediated signaling and might assist the development of novel therapeutics designed to prevent a variety of autoinflammatory disorders and cancers manifested by chronic inflammatory signaling(15, 16, 37).
Materials and Methods
Mice
Mul1 ES cells were purchased from KOMP. Mul1 chimera mice were generated by Transgenic and Gene Targeting Mouse Model Core Facility at University of Miami on a C57BL/6 background. Mul1 chimera mice were further crossed to C57BL/6 mice to generate Mul1−/− mice. Mice were genotyped by standard PCR. All experiments were performed with Institutional Animal Care and Use Committee (IACUC) approval and in compliance with IACUC guidelines.
Mass spectrometric analysis
HEK293T cells were co-transfected with 5 μg pcDNA-hSTING and 10 μg pcDNA-HA-ubiquitin plasmids per 10cm dish for 30 h. Cell lysates were first immunoprecipitated with HA affinity beads (Covance, AFC-101P) and eluted with 100 μg/ml HA peptide (Sigma). The whole elute was then subjected to a second immunoprecipitation using rabbit IgG or STING affinity beads respectively. Ubiquitinated STING proteins were eluted with 100mM triethylamine (pH 11.5) and neutralized by 0.1M glycine (pH 2.7). The elute was digested with trypsin and peptides were separated by a Waters nanoACQUITY with MS analysis on a AB Sciex 5600 Triple Toff mass spectrometer at Yale University MS & Proteomics W.M. Keck Foundation Biotechnology Resource Laboratory.
RNA microarray analysis and real-time PCR
Reconstituted Sting−/− MEF cells were transfected with 5 μg/m dsDNA for 4 h and total RNA was then isolated by the RNeasy RNA extraction kit (QIAGEN). Preparation of cDNA and microarray analysis was performed at the Oncogenomics Core Facility, University of Miami. The Illumina Sentrix BeadChip Array (Mouse WG6 version 2; Affymetrix) was used for the analysis. Data analysis was performed at the Center of Computational Science, University of Miami. Promoter sequence of listed genes (−1,000 to +200) in Figure 2H were obtained through DBTSS (http://dbtss.hgc.jp) and analyzed by TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) at threshold score 85.
Ubiquitin E3 ligase screening
A library containing siRNA to 369 genes (329 genes were from Mouse ON-TARGETplus siRNA Library Ubiquitin Conjugation Subset 3, 40 genes were from customized siRNA library, Dharmacon) were transfected into Sting−/− MEFs reconstituted with hSTING for 72 hr and then transfected with 4 μg/ml dsDNA for 16 hr. IFN-β production was measured by ELISA and compared with cells transfected with non-targeting siRNA. Genes which reduce IFN-β production by more than 50% were selected and further analyzed by immunoblot and ubiquitination assay to identify the E3s, suppression of which significantly reduced dsDNA-induced IRF3 phosphorylation and STING ubiquitination.
In vitro ubiquitination assay
Purified His-STING (152–379 aa, 500 nM) was mixed with GST-MUL1 (300 nM), UBE1 (100 nM), UBE2D1 (1 μM), ubiquitin (50 μM) (UBPBio) in a reaction buffer containing 20 mM Tris–HCl, pH 7.6, 50mM NaCl, 5 mM MgCl2, 2mM ATP, 1 mM β-ME and 5% glycerol. The reaction was carried out at 37°C for 2 hr and then subjected to SDS-PAGE followed by immunoblot with indicated antibodies.
Statistical analysis
Statistical significance of differences in cytokine levels, mRNA expression, and viral titers were determined using Student’s t-test (two-tailed). For all tests, a p value of < 0.05 was considered statistically significant.
Supplementary Material
Fig. S1. STING is ubiquitinated on lysine 224 with K63-linked polyubiquitin chains.
Fig. S2. Ubiquitination on lysine 224 is essential for STING activity.
Fig. S3. Ubiquitination on lysine 224 is required for STING translocation.
Fig. S4. K224R mutation does not affect STING dimer formation, nor its interaction with CDNs.
Fig. S5. Hyper-activity of STING K289R is caused by increased ubiquitination on K224.
Fig. S6. Ubiquitination on K224 is essential for STING activity in human cells.
Fig. S7. MUL1 ubiquitinates STING in vitro.
Fig. S8. MUL1 regulates dsDNA-induced STING-dependent innate response.
Fig. S9. MUL1 partially regulates STING activity in human cells.
Fig. S10. Entire Western blots.
Raw data.
siRNA screening data.
Acknowledgments
We thank Delia Gutman for technical assistance, Auristela Rivera for mice breeding, Dr. Biju Issac of the Sylvester Comprehensive Cancer Center Bioinformatics Core Facility for Gene expression array analysis, Dr. Kathy Stone and Dr. TuKiet Lam of Keck Biotechnology Resource Laboratory at Yale University for mass spectrometry analysis.
Funding: This study was supported by NIH grant R01 AI079336.
Footnotes
This manuscript has been accepted for publication in Science Immunology. This version has not undergone final editing. Please refer to the complete version of record at http://immunology.sciencemag.org. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: G.N. designed and performed the experiments, and performed the statistical analysis. H.K. assisted in protein purification, 32P-labeled autoradiography, and performed MEF isolation. G.N.B designed and supervised all the work and wrote the manuscript with contribution from all authors.
Competing interests: The authors declare that they have no competing financial interests.
Data and materials availability: The microarray dataset was deposited to GEO under the accession code GSE94845.
References and Notes
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern - recognition receptors and its relevance in autoimmunity. Immunological reviews. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nature medicine. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunological Reviews. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbs N, et al. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn J, Ruiz P, Barber GN. Intrinsic self-DNA triggers inflammatory disease dependent on STING. The Journal of Immunology. 2014;193:4634–4642. doi: 10.4049/jimmunol.1401337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn J, Barber GN. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuchida T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhong B, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41:919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Qin Y, et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014;10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. TRIM30alpha Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015;11:e1005012. doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukai K, et al. Activation of STING requires palmitoylation at the Golgi. Nature Communications. 2016;7 doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis ME, Gack MU. Ubiquitination in the antiviral immune response. Virology. 2015;479:52–65. doi: 10.1016/j.virol.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–910. doi: 10.1038/cr.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe T, et al. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmore TD, Wolenski FS. NF - κ B: where did it come from and why? Immunological reviews. 2012;246:14–35. doi: 10.1111/j.1600-065X.2012.01096.x. [DOI] [PubMed] [Google Scholar]
- 33.Li W, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prudent J, et al. MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Mol Cell. 2015;59:941–955. doi: 10.1016/j.molcel.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Li J, et al. Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy. Autophagy. 2015;11:1216–1229. doi: 10.1080/15548627.2015.1017180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yun J, et al. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife. 2014;3:e01958. doi: 10.7554/eLife.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn J, et al. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:5166. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. STING is ubiquitinated on lysine 224 with K63-linked polyubiquitin chains.
Fig. S2. Ubiquitination on lysine 224 is essential for STING activity.
Fig. S3. Ubiquitination on lysine 224 is required for STING translocation.
Fig. S4. K224R mutation does not affect STING dimer formation, nor its interaction with CDNs.
Fig. S5. Hyper-activity of STING K289R is caused by increased ubiquitination on K224.
Fig. S6. Ubiquitination on K224 is essential for STING activity in human cells.
Fig. S7. MUL1 ubiquitinates STING in vitro.
Fig. S8. MUL1 regulates dsDNA-induced STING-dependent innate response.
Fig. S9. MUL1 partially regulates STING activity in human cells.
Fig. S10. Entire Western blots.
Raw data.
siRNA screening data.