Abstract
Objectives
To estimate prevalence, examine time trends, and test for clinical correlates and outcomes associated with HIV-1 intersubtype recombination under a full-genome sequencing context in a rural community in Mbarara, Uganda, where HIV-1 subtypes A1 and D co-circulate.
Methods
Near-full-genome HIV-1 Sanger sequence data was collected from plasma samples of 504 treatment-naïve individuals, who then received PI or NNRTI-containing regimens and were monitored for up to 7.5 years. Subtypes were inferred by Los Alamos RIP 3.0 and compared with Sanger/REGA and MiSeq/RIP. “Non-recombinants” and “recombinants” infections were compared in terms of pre-therapy viral load, CD4 count, post-therapy time to virologic suppression, virologic rebound, first CD4 rise above baseline and sustained CD4 recovery.
Results
Prevalence of intersubtype recombinants varied depending on the genomic region examined: gag (15%), prrt (11%), int (8%), vif (10%), vpr (2%), vpu (9%), GP120 (8%), GP41 (18%), and nef (4%). Of the 200 patients with near-full-genome data, prevalence of intersubtype recombination was 46%; the most frequently observed recombinant was A1-D (25%). Sanger/REGA and MiSeq/RIP yielded generally consistent results. Phylogenetic tree revealed most recombinants did not share common ancestors. No temporal trend was observed (all p>0.1). Subsequent subtype switches were detected in 27 of 143 (19%) subjects with follow-up sequences. Non-recombinant versus recombinants infections were not significantly different in any pre- nor post-therapy clinical correlates examined (all p>0.2).
Conclusion
Intersubtype recombination was highly prevalent (46%) in Uganda if the entire HIV genome was considered, but was not associated with clinical correlates nor therapy outcomes.
Keywords: Uganda, Africa, recombinants, non-B subtypes, full-genome sequencing, virologic outcomes, consequence, HIV-1, clinical outcomes, deep sequencing
Introduction
HIV-1 Group M, which currently dominates the glob al epidemic, is classified into subtypes (or clades) A1, A2, A3, A4, B, C, D, F1, F2, G, H, J, K and various circulating recombinant forms (CRFs) such as AB, AE, and AG [1]. Because the virus’ life cycle involves packing two full-length RNA genomes during viral particles assembly, it provides chances for an event called “template switching” during reverse transcription, leading to the generation of recombinant daughter genomes that contains portions of the parental templates [2].
It has been estimated that HIV-1 CRFs and unique recombinant forms (URFs) are currently responsible for 18–20% of the infections worldwide, and are especially prominent in African, Asian and South American countries where multiple subtypes cocirculate [3]. However, most studies that report the prevalence of intersubtype recombinant viruses examine only a specific part of the HIV-1 genome, mainly from pol due to its usage in drug resistance testing and its availability in public databases.
Relevant to this study, prevalence of A1-D recombinants in rural Uganda was estimated to be 6–19% based on pol and/or GP41 sequences conglomerated from multiple cohorts and sequence database in multiple studies [4–6]. A small scale study examined near-full-genome data from 46 patients in Rakai, Uganda and reported 30% recombinants [7]. Another study reported a 30% prevalence of AD recombinants by pol and its enrichment in severely septic patients [8]. Little is known about pre- and post-treatment virologic and immunologic impacts associated with infections by these recombinant viruses.
The Uganda AIDS Rural Treatment Outcomes (UARTO) cohort consists of over 500 HIV-infected patients in Mbarara, Uganda, where HIV-1 subtypes A1 and D co-circulate [9–11]. Plasma samples were available before and after treatment initiation, and virologic and immunologic outcome data were collected for over seven years post-therapy. As such, this cohort provides an excellent opportunity to observe the natural prevalence and clinical impact of HIV recombinants.
The objective of this study is to estimate prevalence, examine time trends, and test for clinical correlates and outcomes associated with infections by HIV-1 intersubtype recombinants. We hypothesize that prevalence of recombinants is higher than previously reported if examined under a near-full-genome-sequencing context, and that infection with HIV-1 recombinants, compared to non-recombinants, is associated with negative pre-therapy clinical correlates and inferior post-therapy virologic and immunologic responses.
Methods
Ethics Statement
The study was approved by the Mbarara University of Science and Technology Human Subjects Committee and Partners Healthcare Human Subjects Committee, the Uganda Council of Science and Technology, the University of British Columbia/Providence Health Care Research Ethics Board (H11-01642) and the University of California Human Research Subjects Committee. All participants provided written informed consent.
Cohort Description
The Uganda AIDS Rural Treatment Outcomes (UARTO) [12–14] is a cross-sectional cohort of 504 initially treatment-naïve HIV-1 infected subjects. They were followed primarily at the Immune Suppression Syndrome (ISS) Clinic in Mbarara, Uganda, a rural community 4.5 hours by automobile from the capital city of Kampala. Subjects were enrolled just before the start of antiretroviral regimen from June 27, 2005 to April 8, 2010, and were longitudinally followed every three to six months to receive viral load and CD4 count monitoring for up to 7.5 years until January 11, 2013 or until lost to follow-up. Among the 504 participants, 296 post-therapy follow up samples were available for HIV-1 RNA sequencing from 143 unique individuals.
HIV-1 Near- full-genome PCR amplification
Total nucleic acid was extracted from 500 μL of plasma samples using NucliSENS easyMag (bioMérieux). All PCR and sequencing primer sequences, and thermocycler methods are listed in Supplementary Tables 1–3. Briefly, reverse transcription and nested-PCR reactions were performed using a three-reaction five-amplicon approach with near-full-genome coverage. The five overlapping amplicons covered gag to protease(pr) (HXB2 coordinate 680–2724), pr to reverse transcriptase(rt) (2011–3798), rt to vpu (3626–5980), vpr to GP120 (5549–7760) and GP41 to nef (7652–9610).
Sanger sequencing
Bulk sequencing was performed on ABI 3730 DNA Sequencer using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Chromatograms were aligned against HXB2 references sequences by our in-house automated alignment and base-calling program RECall [15], resulting in seven sequence databases for each of gag, prrt, int, vif, vpr, vpu, partial GP120, GP41 and nef, which were concatenated to create near-full-genome sequences. For more details on alignment, concatenation and quality control, refer to Supplementary Material and Methods. Los Alamos PhyML 3.0 was used to construct phylogenetic trees from these concatenated near-full-genome sequences along with relevant Los Alamos 2010 HIV-1 subtype references (Figure 1).
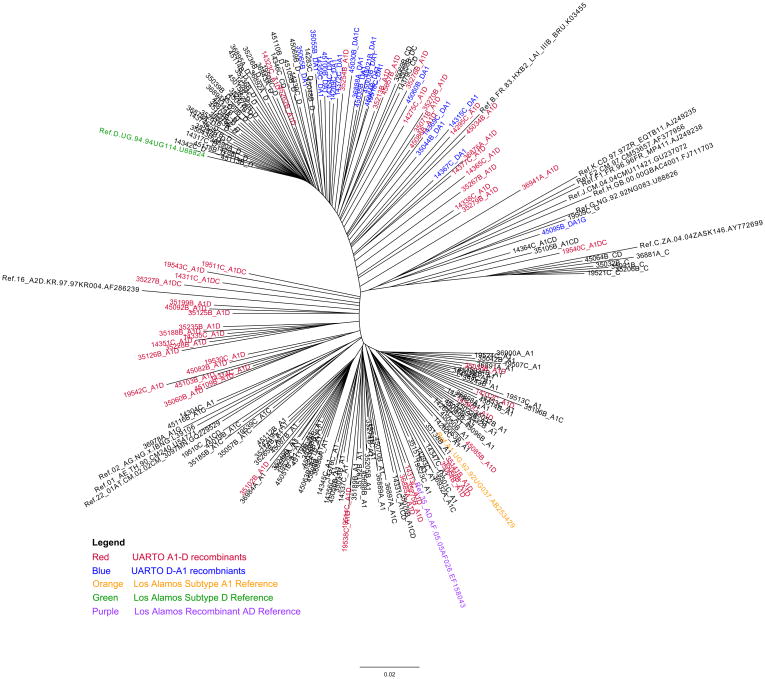
Figure 1.
Phylogenetic analysis by maximum-likelihood tree (PhyML) of the 200 near-full-genome sequences showed that recombinants did not share a common ancestor, suggesting multiple recombination events.
Subtyping and recombinants inferences
Los Alamos RIP 3.0 was used in our primary analysis. In our gene-by-gene examination, RIP window size 400 with confidence interval 95% was arbitrary selected to infer subtype for sequences with HXB2 reference sequence >600 nucleotides in length (gag, prrt, int, GP41, nef), otherwise, RIP window size 100 with confidence interval 90% was used (vif, vpr, vpu, partial GP120). In our near-full-genome examination, RIP window size 400 with confidence interval 95% was used. A sample was called a “non-recombinant” when RIP returned a single subtype inference (eg. A1) and a “recombinant” when RIP detected multiple subtypes within a fragment (eg. A1-D). We defined a “subtype switch” event as having any discordant subtyping results in a series of longitudinal samples from a single patient (eg. switching from A1-D recombinant to D). For comparison, we repeated subtyping and recombinants inferences for prrt using another algorithm REGA 2.0 (BIOAFRICA) with default settings.
Definitions of therapy outcomes
Virologic suppression was defined as <400 copies HIV RNA/mL to reflect the viral load detection limit during the initial years of the follow-up period. Four therapy outcomes were examined: (i) time to virologic suppression (first of two consecutive viral load <400 copies HIV RNA/mL), (ii) time to post-therapy virologic rebound (first of two consecutive viral load ≥400 copies HIV RNA/mL post-suppression, defined as the number of day since the first of two consecutive virologic suppression event), (iii) time to first CD4 rise (any post-therapy CD4 count above baseline), and (iv) time to sustained CD4 recovery (first of two consecutive post-therapy CD4 count increase of >200 cells/μL from pre-therapy count, or first of two consecutive post-therapy CD4 count >350 cells/μL). “Lost to follow-up” was defined as the lack of an event until study cutoff and having the last clinic visit >18 months (548 days) before study cutoff. A sample R script on the definitions and data extraction for virologic outcome is available in Gist (https://gist.github.com/guineverelee/public). Transmitted drug resistance data for this cohort was defined by the WHO list [16] and was previously published by our group [17]. Post-treatment drug resistance was defined using Stanford HIVdb [18].
Statistical Methods
All statistical analyses were performed using R, SAS and/or GraphPad Prism 5.0. Two-tailed Mann-Whitney tests were used to compare age, pre-therapy viral load, pre-therapy CD4 count, and follow up duration. Log-rank tests were used to compare Kaplan-Meier curves of time to virologic suppression, virologic rebound, CD4 rise and CD4 recovery.
Two-tailed Fisher’s exact test was used to compare gender distribution and the extent of lost to follow up. Multivariate Cox Proportional Hazard confounder models were used for time to virologic suppression and time to post-suppression virologic rebound. The initial list of variables included in the full Cox models were “recombinants or non-recombinants,” “age at enrollment,” “gender,” “baseline viral load,” “baseline CD4 count,” “number of follow up visits,” “mean visit interval in days (per 10 increase),” “year of therapy start (dichotomized into >2007 versus ≤2007)” and “type of first regimen (nevirapine-based versus others).” Then, variables were dropped one-at-a-time using the lowest relative change in the coefficient for the variable related to the outcome as a criterion, until the maximum change from the full model exceeded 5% [19]. Statistical significance was defined as p<0.05 in all analyses. Note, Bonferroni correction for multiple comparisons could arguably be used instead (p = 0.05/26 = 0.002); this value is provided here for benchmarking purpose.
MiSeq (Illumina) Deep Sequencing
For each sample, the five nested second-round PCR amplicons from three reactions were pooled (5μL each) and purified using AMPure Beads (Agilent). Library was prepared with Nextera XT kits according to manufacturer’s protocol and sequenced with MiSeq Reagent Kit V2 (500-cycles) with a target coverage depth at 8000. FASTQ outputs were processed by our in-house bioinformatics pipeline (version 6.8). Quality cutoff q15 was chosen. Each genomic region (gag, pr, rt, int, vif, vpr, vpu, env and nef) was aligned using bowtie separately. Briefly, shortgun reads were initially aligned to its corresponding HXB2 reference sequences followed by a reiterative process to obtain sample-specific reference sequences. Then, one consensus sequence per sample per genomic region was created at a 20% nucleotide mixtures cutoff. Sequences with an average coverage depth <10 were excluded from subsequent analyses. “Concordance between Sanger and MiSeq” was defined as “either having completely identical subtype inference results (eg. Sanger A1 versus MiSeq A1) or having partially concordant results (eg. Sanger A1 versus MiSeq A1-D recombinant).” The following sequences were excluded from our analyses for quality control: (1) sequences with an average coverage depth <10 across the target genomic region, (2) sequences that were shorter than 400 or 100 nucleotides in length for RIP window size 400 and 100 analyses respectively, and (3) sequences which RIP failed to yield a subtype inference and returned as “None Significant.” A secondary analysis specifically examined samples with coverage depth between 10 and 100.
Results
Pre-therapy Baseline Characteristics (n=504)
Cross-sectional pre-therapy baseline HIV+ plasma samples were collected from 2005 to 2010 from 504 HIV-infected patients immediately before initiation of antiretroviral therapy. At baseline, 69% were female, median age was 35 (Q1–Q3 29– 39), median baseline viral load and CD4 count were 1×105 copies HIV RNA/mL (Q1–Q3 4×104–4×105) and 132 cells/μL (Q1–Q3 75–200). Initial regimens were primarily NVP- (86%) and EFV-based (12%) in combination with lamivudine (3TC) and zidovudine (AZT).
Prevalence of HIV-1 Intersubtype Recombination
When each genomic region was individually examined, intersubtype recombinants were detected at the following frequencies in pre-treatment samples: gag (15%), prrt (11%), int (8%), vif (10%), vpr (2%), vpu (9%), GP120 (8%), GP41 (18%), and nef (4%) as shown in Table 1 column gag to nef. Of the 200 patients who had sequence data available across all genomic regions, nucleotides from all genomic regions were concatenated to produce near-full-genome HIV data to be re-analyzed by RIP. Prevalence of intersubtype recombination detected anywhere along the genome was 46% (Table 1, column “Near-Full-Genome”). The most frequently detected recombinant was A1-D (25%). Stratification by year revealed no temporal trend in the prevalence of recombinants (all p>0.1, Supplementary Table 4). Phylogenetic analysis by maximum-likelihood tree of these 200 near-full-genome sequences showed that most A1-D and D-A1 recombinants did not cluster into monophyletic groups and did not share common ancestor(s), suggesting multiple recombination events (Figure 1). Recombinant breakpoints scattered across the genome (Figure 2). To assess the consistency of RIP’s subtype inferences compared to other subtyping algorithms, we submitted all prrt sequences to REGA 2.0 (BIOAFRICA) for a representative comparison. Overall, REGA predicted a higher prevalence of recombinants in prrt (20%) than RIP (11%) and their results were 89% concordant.
Table 1.
High prevalence of HIV-1 intersubtype recombinants in Mbarara, Uganda revealed by a HIV-1 near-full-genome Sanger sequencing approach
| INTRA-GENE | INTRA- GENOME Near-Full- Genome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gag | prrt | int | vif | vpr | vpu | GP120 | GP41 | nef | ||
| HXB2 coordinates | 790–2289 | 2253–3749 | 4230–5093 | 5041–5616 | 5559–5847 | 6062–6307 | 6315–7112 | 7758–8792 | 8797–9414 | 790–9414 |
| Sequence length (bp) | 1500 | 1497* | 864 | 576 | 288* | 246 | 797* | 1035 | 618 | 8625 |
| RIP (Window Size, Confidence Interval) | 400, 95% | 400, 95% | 400, 95% | 100, 90% | 100, 90% | 100, 90% | 100, 90% | 400, 95% | 400, 95% | 400, 95% |
| Unique individuals sampled (n) | 479 | 486 | 464 | 458 | 387 | 456 | 277 | 485 | 476 | 200 |
|
| ||||||||||
| Any recombinants (%) | 15% | 11% | 8% | 10% | 2% | 9% | 8% | 18% | 4% | 46% |
|
| ||||||||||
| Breakdown of “non-recombinants” | ||||||||||
|
| ||||||||||
| %A1 | 49.7% | 47.3% | 48.1% | 47.4% | 49.1% | 43.0% | 55.2% | 44.1% | 53.4% | 32.0% |
| %B | None | None | None | None | None | 0.4% | None | None | None | None |
| %C | 4.6% | 4.5% | 4.7% | 7.6% | 10.1% | 9.4% | 3.6% | 5.2% | 5.7% | 2.5% |
| %D | 30.3% | 37.2% | 39.4% | 35.4% | 39.3% | 37.9% | 32.5% | 32.2% | 36.8% | 19.0% |
| %G | 0.4% | 0.4% | 0.2% | None | None | 0.7% | 0.7% | 0.4% | 0.4% | 0.5% |
|
| ||||||||||
| Breakdown of “any recombinants” | ||||||||||
|
| ||||||||||
| %A1-C | 2.5% | 0.8% | 1.3% | 0.4% | 0.5% | 0.2% | 0.7% | 1.0% | None | 4.0% |
| %A1-AE | None | None | None | None | None | None | 0.4% | None | None | None |
| %A1-B | None | None | None | 0.2% | None | None | None | None | None | None |
| %A1-C-D | None | 0.2% | None | None | None | None | 0.4% | None | None | 2.5% |
| %A1-D | 10.2% | 6.0% | 5.2% | 7.0% | 0.3% | 7.0% | 4.7% | 15.1% | 2.7% | 24.5% |
| %A1-D-C | None | None | None | None | None | None | None | None | None | 2.0% |
| %A1-D-G | None | None | None | None | None | None | None | None | None | None |
| %A1-G | 0.4% | None | None | None | None | None | None | 0.2% | None | 0.5% |
| %AE-A1 | None | None | None | None | 0.3% | 0.2% | None | None | None | None |
| %B-D | None | None | None | None | None | 0.2% | None | None | None | None |
| %C-A1 | 0.2% | None | None | 0.2% | None | None | None | None | None | None |
| %C-D | 0.6% | 1.2% | 0.9% | 0.9% | 0.3% | 0.7% | 0.7% | 1.9% | 1.1% | 2.0% |
| %C-D-J | None | None | None | 0.2% | None | None | None | None | None | None |
| %D-A1 | 1.0% | 1.4% | 0.2% | 0.4% | None | None | 0.4% | None | None | 9.0% |
| %D-A1-C | None | 0.2% | None | None | None | None | None | None | None | 0.5% |
| %D-A1-G | None | None | None | None | None | None | None | None | None | 0.5% |
| %D-C | None | 0.4% | None | 0.2% | None | None | 0.4% | None | None | 0.5% |
| %D-K | None | None | None | None | None | None | 0.4% | None | None | None |
| %D-G | None | 0.2% | None | None | 0.3% | None | None | None | None | None |
| %D-H | None | None | None | None | None | 0.2% | None | None | None | None |
Partial prrt was used in this analysis to reflect the actual region used in clinical drug resistance monitoring; *HXB2 vpr had a single thymine (T) out-of-frame insertion, and was manually removed from the reference sequence for alignment purposes; *Partial GP120 was used in this analysis because quasispecies diversity in env relative to other regions resulted in extremely low Sanger sequence quality; *None means zero observation.
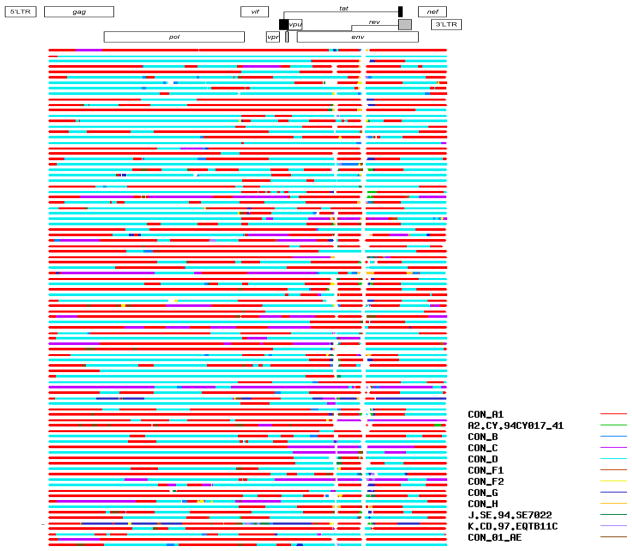
Figure 2.
Recombination mapping by RIP shows that breakpoints were scattered across the HIV genome (n=92 full-genome recombinants; RIP window size 400, confidence threshold 95%).
Longitudinal switches in HIV-1 subtypes
Since none of the 143 individuals whom we had longitudinal sequences for were among the 200 whom we had near-complete full-genome data, we proceeded to estimate prevalence of switches in each genomic region. Longitudinal sequences were available from 77 subjects (gag), 90 (prrt), 67 (int), 63 (vif), 59 (vpr), 77 (vpu), 31 (GP120), 80 (GP41) and 86 (nef). Longitudinal subtype switches were observed in 5% of the subjects (gag), 9% (prrt), 6% (int), 2% (vif), 3% (vpr), 3% (vpu), 13% (GP120), 8% (GP41) and 7% (nef). Details of these 37 cases of subtype switches in 27/143 subjects (19%) are listed in Supplementary Tables 5a–i. To explore potential reasons for switches, we examined phylogenetic trees, and found that one discordant sample from patient MBA1100 was an immediate neighbor of MBA1101 in a phylogenetic tree of all available vif sequences, suggesting potential labeling error (Supplementary Table 5b), whereas in another case nef sequences of patient MBA1435 switched from A1 to G, but all sequences formed a tight monophyletic group on a phylogenetic tree of the entire cohort’s nef sequences, suggesting potential RIP artifacts (Supplementary Table 5i). No potential explanation for other switch cases was found.
Comparison with Illumina (MiSeq) near-full-genome deep sequencing data
To examine whether another sequencing method would yield consistent subtyping results, we performed near-full-genome Nextera XT MiSeq deep-sequencing, consensus nucleotide sequence generation and RIP subtyping on 23 randomly selected samples. Of these, 20 were pre-therapy baseline samples from 20 unique individuals, and three were post-therapy samples from three unique individuals. Paired Sanger and MiSeq data were successfully obtained for 22 (gag), 22 (prrt), 11 (int), 19 (vif), 19 (vpr), 22 (vpu), 19 (partial GP120), 16 (GP41), 20 (nef) and 19 (near-full-genome). RIP subtype inferences were always concordant between Sanger sequences and MiSeq-derived consensus in all of gag, prrt, int, vpr, vpu, GP120, GP41, nef and full-genome data; the two discordant cases were observed in vif (first case Sanger “D” but MiSeq “C”; second case Sanger “D” but MiSeq “B”). Among the 19 pre-therapy samples with near-full-genome MiSeq data, 11 (58%) were inferred by RIP as recombinants, closely resembling the value estimate by Sanger near-full-genome data (46%, Table 1).
Furthermore, among all the above paired results, we observed 30 cases in which the average MiSeq coverage depths were extremely low between 10 and 100, mainly in int (9/30, 30%) and vif (12/30, 40%). Interestingly, in 29/30 cases, RIP subtype inferences at this low coverage depth remained completely concordant between paired Sanger and MiSeq-derived sequences.
The remaining 1/30 case was a GP120 sequence; RIP failed to associate both the Sanger and MiSeq-derived sequences with any known subtype or recombinants. Also of note was that of the 19 paired GP120 Sanger and MiSeq sequences that covered the first 160 nucleotides of GP120, MiSeq successfully yielded full-length GP120 consensus sequences that included all the variable loops in 16/19 (84%) cases. In contrast, we were unable to obtain clean Sanger sequence data beyond approximately the 160th nucleotide in GP120 due to the high sequence diversity in env resulting in base-calling ambiguities.
Baseline Clinical Correlates and Therapy Outcomes (n=200)
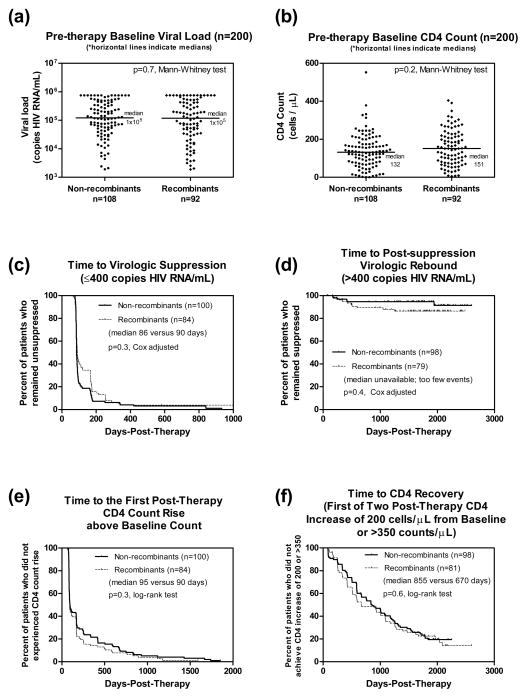
We dichotomized the 200 patients with near-full-genome data into “non-recombinants” (54%) and “recombinants” (46%) HIV-1 infections, and compared their pre- and post-therapy-initiation clinical correlates. At pre-therapy baseline, the two groups were not significantly different in gender (67% versus 70% female, p=0.8 Fisher two-tailed), baseline viral load (Figure 3a, p=0.7 Mann-Whitney), and baseline CD4 count (Figure 3b, p=0.2 Mann-Whitney). Subjects infected with recombinants were slightly younger (median age 35 versus 34, p=0.04 Mann-Whitney). At post-therapy, univariate tests showed marginally significant differences in “time to virologic suppression” (Figure 3c non-adjusted, p=0.03 Log-rank), but were not significantly different in “time to post-suppression virologic rebound” (Figure 3d non-adjusted, p=0.1 Log-rank), “time to first CD4 rise above baseline” (Figure 3e, p=0.3 Log-rank) and “time to sustained CD4 recovery” (Figure 3f, p=0.6 Log-rank). Neither the “proportion of subjects lost to follow up” nor “duration of follow up” were significantly different between groups (all p>0.2). Next, we further explored the marginal differences observed in virologic outcomes using multivariate Cox Proportional Hazard confounder models: After adjustment, both “time to virologic suppression” and “time to post-therapy virologic rebound” were not significantly different between groups (Figure 3c adjusted p=0.3, hazard ratio recombinants/non-recombinants 0.8, 95% confidence interval 0.6–1.1, controlling for visit interval, year of therapy start and type of first regimen; Figure 3d adjusted p=0.4, hazard ratio 1.6, 95% confidence interval 0.6–4.5, controlling for age, gender, baseline viral load and visit interval). Finally, we compared the prevalence of transmitted drug resistance and the prevalence of any major drug resistance mutations in recombinants versus non-recombinant groups and did not find significant difference between groups (p=0.7 and 1.0 Fisher Exact 2-tail).
Figure 3.
Non-recombinant versus recombinants infections were not significantly different in any pre- nor post-therapy clinical correlates examined.
Discussion
In summary, our near-full-genome sequencing approach revealed a high prevalence of infections by intersubtype HIV-1 recombinants (46%, without time trend) in a rural African community where multiple HIV subtypes cocirculate. We also provided evidence that most of these recombinants arose from independent recombination events, and found evidence of longitudinal subtype switches. Importantly, our study provided evidence that infections with recombinant HIV was not associated with any negative pre- nor post-therapy virologic nor immunologic clinical correlates.
Our reported prevalence of Uganda’s HIV-1 recombinants in prrt (11%) fell within the range previously reported (6–19%) [4–6]. Importantly, we showed that the prevalence of intersubtype recombinants is a lot higher under a full-genome context (Table 1, column gag to nef). This finding has several implications. First, it points out that studies that used only part of the genome to estimate recombination prevalence may result in underestimation. Second, it suggests that HIV subtype studies on disease progression and clinical impacts could be biased depending on the genomic region used for subtyping. For instance, numerous studies reported that subtype D infections (by pol) as being more aggressive with a faster disease progression [20–22]. However, in our current report, multiple patients had subtype D in pol, but A1 in GP41. This may also have an implication on subtype-specific vaccine development strategies. Third, our observation that these recombinants did not tend to share common ancestors in a phylogenetic tree suggests that recombination is very frequent in HIV natural biology, and may even contribute to the change of subtype over time that we have observed (on top of other explanations such as undetected sample-mixed-up and/or superinfections). This knowledge of recombination frequency may help further our understanding about the natural “template switch” frequency during the reverse transcription step in HIV’s life cycle.
Our conclusion on the prevalence of recombinants is limited by our choice of subtyping algorithm and its settings. We have arbitrary chosen RIP with a window size 400 and confidence interval of 95% for genes that are >600 nucleotides in length; otherwise window size 100 and confidence interval of 90% is chosen. It should be noted that a lower “input sequence length to RIP window size” ratio corresponds to a decreased sensitivity for recombinants detection. Since these ratios differ in each genomic region examined in our gene-by-gene analyses (Table 1, column gag to nef), comparison of percentage prevalence of recombinants between genomic regions would not be appropriate. A fair estimation of the prevalence of recombinants (46%) was obtained in our near-full-genome concatenation approach: All sequences were 8625 nucleotides in length, and a constant RIP window size of 400 with 95% confidence interval was used. In addition, we also showed that a different subtyping algorithm, REGA, called more recombinants in prrt than RIP did, which highlighted the variability introduced by different algorithms and settings.
Another technical limitation relates to primer mismatch bias, which could potentially lead to a preferential amplification of one of the amplicons over another in our duplex A or dulplex B reactions, and/or preferential amplification of one subtype over another. To address this concern, we reviewed our dataset to compare the number of successful amplifications and the distribution of subtypes across amplicons (Table 1). First, we observed that the number of successful amplifications of each region were comparable, ranging from 387 to 486 successes (excluding GP120, of which Sanger sequence data was uncallable over the variant stretch of the genome, likely due to excessive indels). Since all 504 samples were subjected to the same reactions A1/A2, B1/B2 and C, this comparable success rate implies that there was at least a low degree of bias in amplification efficiency between amplicons. Next, Table 1 shows that the percentage distribution of subtypes between genomic regions were also comparable across amplicons (duplex A1 contained gag; A2 vpu; duplex B1 prrt; B2 GP41, nef; reaction C: int, vif, vpr). Cross these amplicons, %Subtype-A1 ranged from 43.0% to 53.4%, %C ranged from 3.6% to 10.1%, and %D ranged from 30.3% to 39.4%. In other words, we did not observe one subtype being over-represented in one amplicon compared to another. Although we cannot rule out the absence/presence of primer amplification bias due to primer mismatches, these observations suggest comparable amplification efficiency across genome and across subtypes.
Our conclusion on the clinical impacts of recombinant HIV-1 is limited by sample size and sampling intervals. We only had 200 patients who had near-full-genome sequencing data available and were thus included in the clinical outcome analyses. Much larger study with more patients will be need to increase statistical power. Furthermore, these patients were scheduled to receive virologic and immunologic monitoring every three to six months, which resulted in the “staircase-like” shapes observed in the Kaplan-Meier curves (instead of smooth curves) in Figure 3c–f. This factor, compounded with the relatively low number of subjects in both groups (n≤100), might have compromised the statistical comparisons.
Finally, our conclusion that Sanger and MiSeq produced highly concordant subtyping results is limited by the lower PCR/sequencing success rate in int and vif. This was potentially due to our “equal volume pooling” approach (instead of an “equal mole pooling” approach) during the MiSeq library preparation step, which reflects different PCR amplification efficiency between amplicons for this particular set of samples. However, our results also suggest that an average MiSeq coverage depth of 10–100 can still very accurately predict Sanger subtyping results across all genomic regions in these Ugandan non-subtype B HIV samples. This agrees with previous observations that MiSeq can accurately predict Sanger-derived rt sequences at coverage depth below 100 [23]. Furthermore, we showed that MiSeq was more successful in yielding full-length GP120 sequences compared to Sanger sequencing, supporting the move to deep sequencing in HIV env genetic studies.
In conclusion, this study revealed a high prevalence of HIV-1 infections by intersubtype recombinants in a rural African community where subtype A1 and D cocirculate, but showed that infections by recombinants did not impact pre- nor post- therapy virologic nor immunologic clinical correlates and therapy outcomes. Future studies should continue to monitor intersubtype recombinants in this and other similar communities under a full-genome sequencing context to keep check of their spread, evolution and clinical impacts.
Supplementary Material
Acknowledgments
Source of Funding: This work was funded by the Canadian Institutes of Health Research (CIHR), National Institutes of Health (NIH) Centers for AIDS Research (CFAR) Program, National Institute of Mental Health and National Institute on Alcohol Abuse and Alcoholism (R01 MH054907, P30 AI027763, U01 CA066529, UM1 CA181255, R21 AA014784).
We thank all patients participating in the UARTO cohort whom have made this study possible; we also thank Mr. Conan Woods for sequence database management and quality control assistance, and Ms. Benita Yip for advices on data analysis.
Footnotes
Presented in part at: The Annual Canadian Conference on HIV/AIDS Research (CAHR 2015; abstract BS20) and the 8th IAS Conference on HIV Pathogenesis, Treatment & Prevention, Vancouver, BC (IAS 2015; abstract MOPEA040).
Conflicts of Interest: PRH has received consulting fees from ViiV/Pfizer and Quest; holds stock in Merck, Gilead and Illumina. For the remaining authors no conflicts of interest were declared.
Authors contribution: The work presented here was carried out in collaboration between all authors. The study was conceptualized and designed by GQL and PRH. Plasma samples, baseline and follow-up data were collected and managed by ARM, DRB, JNM, PWH, SHK and YB. HIV-1 genotyping laboratory work was done by CL, GQL and TM. Results were analyzed by CJB, GQL, WZ and VDL. GQL wrote the manuscript; all authors contributed to, seen, and approved the manuscript.
References
- 1.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, et al. HIV-1 nomenclature proposal. Science. 2000;288:55–6. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 2.Kuzembayeva M, Dilley K, Sardo L, Hu WS. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454–455:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau Ka, Wong JJL. Current trends of HIV recombination worldwide. Infect Dis Rep. 2013;5:15–20. doi: 10.4081/idr.2013.s1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yebra G, Ragonnet-Cronin M, Ssemwanga D, Parry CM, Logue CH, Cane PA, et al. Analysis of the History and Spread of HIV-1 in Uganda using Phylodynamics. J Gen Virol. doi: 10.1099/vir.0.000107. Published Online First: 27 February 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ssemwanga D, Ndembi N, Lyagoba F, Bukenya J, Seeley J, Vandepitte J, et al. HIV type 1 subtype distribution, multiple infections, sexual networks, and partnership histories in female sex workers in Kampala, Uganda. AIDS Res Hum Retroviruses. 2012;28:357–65. doi: 10.1089/aid.2011.0024. [DOI] [PubMed] [Google Scholar]
- 6.Gale CV, Yirrell DL, Campbell E, Van der Paal L, Grosskurth H, Kaleebu P. Genotypic variation in the pol gene of HIV type 1 in an antiretroviral treatment-naive population in rural southwestern Uganda. AIDS Res Hum Retroviruses. 2006;22:985–92. doi: 10.1089/aid.2006.22.985. [DOI] [PubMed] [Google Scholar]
- 7.Harris ME, Serwadda D, Sewankambo N, Kim B, Kigozi G, Kiwanuka N, et al. Among 46 near full length HIV type 1 genome sequences from Rakai District, Uganda, subtype D and AD recombinants predominate. AIDS Res Hum Retroviruses. 2002;18:1281–90. doi: 10.1089/088922202320886325. [DOI] [PubMed] [Google Scholar]
- 8.Doka NI, Jacob ST, Banura P, Moore CC, Meya D, Mayanja-Kizza H, et al. Enrichment of HIV-1 subtype AD recombinants in a Ugandan cohort of severely septic patients. PLoS One. 2012;7:e48356. doi: 10.1371/journal.pone.0048356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee GQ, Bangsberg DR, Muzoora C, Boum Y, Oyugi JH, Emenyonu N, et al. Prevalence and Virologic Consequences of Transmitted HIV-1 Drug Resistance in Uganda. AIDS Res Hum Retroviruses. 2014;30:896–906. doi: 10.1089/aid.2014.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee GQ. Standard and Enhanced Genotypic Assessment of HIV Drug Resistance and Tropism in Subtype-B and Non-subtype-B HIV-1 Infections. 2014 [Google Scholar]
- 11.Lee GQ, Lachowski C, Cai E, Lima VD, Boum Y, Mocello AR, et al. Non-R5-tropic HIV-1 in Subtype A and D Infections Were Associated with Lower Pre-therapy CD4 Count but not with Therapy Outcomes. In. 54th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); Washington, DC, USA. 2014. p. Abstract #H-1633. [Google Scholar]
- 12.Kaida A, Matthews LT, Kanters S, Kabakyenga J, Muzoora C, Mocello aR, et al. Incidence and Predictors of Pregnancy among a Cohort of HIV-Positive Women Initiating Antiretroviral Therapy in Mbarara, Uganda. PLoS One. 2013;8:e63411. doi: 10.1371/journal.pone.0063411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, et al. Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS. 2011;25:2123–31. doi: 10.1097/QAD.0b013e32834c4ac1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byakwaga H, Boum Y, Huang Y, Muzoora C, Kembabazi A, Weiser SD, et al. The Kynurenine Pathway of Tryptophan Catabolism, CD4+ T-Cell Recovery, and Mortality Among HIV-Infected Ugandans Initiating Antiretroviral Therapy. J Infect Dis. 2014;210:383–91. doi: 10.1093/infdis/jiu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woods CK, Brumme CJ, Liu TF, Chui CKS, Chu AL, Wynhoven B, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936–42. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee GQ, Bangsberg DR, Muzoora C, Boum Y, Oyugi JH, Emenyonu N, et al. Prevalence and virologic consequences of transmitted HIV-1 drug resistance in Uganda. AIDS Res Hum Retroviruses. 2014;30:896–906. doi: 10.1089/aid.2014.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang MW, Liu TF, Shafer RW. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology. 2012;55:98–101. doi: 10.1159/000331998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima VD, Geller J, Bangsberg DR, Patterson TL, Daniel M, Kerr T, et al. The effect of adherence on the association between depressive symptoms and mortality among HIV-infected individuals first initiating HAART. AIDS. 2007;21:1175–1183. doi: 10.1097/QAD.0b013e32811ebf57. [DOI] [PubMed] [Google Scholar]
- 20.Eller MA, Opollo MS, Liu M, Redd AD, Eller LA, Kityo C, et al. HIV Type 1 Disease Progression to AIDS and Death in a Rural Ugandan Cohort Is Primarily Dependent on Viral Load Despite Variable Subtype and T-Cell Immune Activation Levels. J Infect Dis. 2015;211:1574–84. doi: 10.1093/infdis/jiu646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoro MM, Perno CF. HIV-1 Genetic Variability and Clinical Implications. ISRN Microbiol. 2013;2013:481314. doi: 10.1155/2013/481314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pant Pai N, Shivkumar S, Cajas JM. Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996–2010. J Acquir Immune Defic Syndr. 2012;59:382–8. doi: 10.1097/QAI.0b013e31824a0628. [DOI] [PubMed] [Google Scholar]
- 23.Lapointe H, Dong W, Lee GQ, Bangsberg DR, Karakas A, Kirkby D, et al. HIV Drug Resistance Testing by High-Multiplex “Wide” Sequencing on the Illumina MiSeq. XXIV International HIV Drug Resistance Workshop; Seattle, Washington. 2015. p. Abstract #38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.