Abstract
Atrazine, a herbicide used on agricultural crops is widely applied in the Midwestern United States as well as other areas of the globe. Atrazine frequently contaminates potable water supplies and is a suspected endocrine disrupting chemical. Previous studies have reported morphological, hormonal, and molecular alterations due to developmental and adulthood atrazine exposure; however, studies examining epigenetic alterations are limited. In this study, the effects of atrazine exposure on DNA methyltransferase (DNMT) activity and kinetics were evaluated. Global DNA methylation levels and dnmt expression in zebrafish larvae exposed to 0, 3, or 30 parts per billion (ppb) atrazine throughout embryogenesis was then assessed. Results indicate that atrazine significantly decreased the activity of maintenance DNMTs and that the inhibition mechanism can be described using non-competitive Michaelis-Menten kinetics. Furthermore, results show that an embryonic atrazine exposure decreases global methylation levels and the expression of dnmt4 and dnmt5. These findings indicate that atrazine exposure can decrease the expression and activity of DNMTs, leading to decreased DNA methylation levels.
Keywords: atrazine, dnmt, epigenetics, methylation, zebrafish
Graphical Abstract
1. Introduction
Epigenetic programming plays an important role in an organism’s reaction to environmental stressors during development. Numerous definitions have been proposed to define epigenetics, the most encompassing representation is that epigenetics is heritable, reversible, and self-perpetuating phenotypic alterations elicited by gene expression changes resulting from structural adaptations of chromosomal regions that take place without any alteration of the integral DNA sequence where these modifications occur.1–5 Three types of known epigenetic mechanisms include DNA methylation, histone modifications, and small non-coding RNAs that regulate gene expression.6 Of these, the alteration of DNA methylation patterns and subsequent reprogramming of developmental processes by changing transcriptional gene expression has received most attention, especially as it pertains to the heritability of these epigenetic alterations.7–12
DNA methylation occurs with high frequency at CpG islands, which typically have greater than 50% GC content with the CpG ratio of at least 60%.13–15 DNA methylation is critical for normal cellular development because of its role in regulating gene transcription, maintaining transposon inactivation, X-chromosome inactivation, and genomic imprinting. DNA methylation is mediated by the family of DNA methyltransferases (DNMTs) that catalyze the transfer of a methyl group from S-adenosyl methionine (SAM) to cytosine of DNA. Mammals possess five DNMTs (DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L); however, only DNMT1, DNMT3A, and DNMT3B possess methyltransferase activity. DNMT1 functions to maintain proper methylation maintenance, while DNMT3A and DNMT3B contribute to de novo methylation during development.14
Endocrine disrupting chemicals (EDCs) are exogenous agents that disrupt endogenous hormone signaling pathways. These chemicals are diverse in structure and are found in numerous products such as plasticizers, pharmaceuticals, and pesticides, making human exposure to these chemicals a likely event.16 Studies show that EDCs can alter tissue formation, reproduction, and play a role in the onset of obesity and cancer.16–23 Furthermore, studies implicate that exposure to EDCs can alter different levels of epigenetic processes which can act transgenerationally.9–13, 24–27
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is a pre-emergent herbicide that is applied throughout the Midwestern United States and other parts of the globe. Atrazine is applied on a variety of agricultural crops including corn, sorghum, sugar cane, and wheat.28–30 Due to atrazine’s water solubility, mobility in soil, and long half-life, atrazine frequently contaminates potable water supplies and reaches levels above the maximum contaminant level (MCL) as set by the U. S. Environmental Protection Agency (EPA) of 3 parts per billion (ppb; µg/L).31–32 The greatest risk of drinking water source contamination occurs following rainfall events after agricultural field application in spring and summer with recent monitoring data measuring over 300 ppb in the Midwestern US.31 Widespread water contamination issues led the European Union to ban the use of atrazine in 2003.33–34 Epidemiological studies show several potential adverse health effects associated with maternal atrazine exposure including an increased risk of babies born small for their gestational age (SGA), intrauterine growth retardation (IUGR), and birth defects.35–38 Laboratory studies utilizing various animal models report numerous hormonal and molecular alterations caused by atrazine exposure.20, 39–43 Many of these studies report adverse effects on the neuroendocrine system through the hypothalamus-pituitary-gonadal axis. For example, atrazine exposure disrupted the hypothalamic control of pituitary-ovarian function with additional studies reporting robust alterations in reproductive hormones including gonadotropin-releasing hormone (GnRH), luteinizing hormone (LH), and follicle-stimulating hormone (FSH).20, 39–43 However, understanding the epigenetic alterations contributing to atrazine toxicity is still under investigation.44–47
We have previously reported that an embryonic atrazine exposure [0.3, 3, or 30 ppb throughout embryogenesis, 1–72 hours post fertilization (hpf)] resulted in alterations to the transcriptome of zebrafish larvae in genes associated with reproductive system function and development, cell cycle regulation, and cancer.48 In addition, morphological alterations in head size of the zebrafish larvae exposed to 0.3, 3, or 30 ppb atrazine during embryogenesis was observed.48 Furthermore, a decrease in spawning was observed in adult zebrafish exposed to atrazine only during embryogenesis with persistent morphological alterations in their offspring.49 These adult females also had an increase in ovarian progesterone and follicular atresia, alterations in levels of 5-hydroxyindoleacetic acid (5-HIAA, a serotonin metabolite) and serotonin turnover in brain tissue, and transcriptome changes in brain and ovarian tissue supporting neuroendocrine alterations similar to what has been reported in other animal studies.49–50 In a previous study, we also first began to investigate epigenetic mechanisms behind the observed alterations in larvae and adult zebrafish by evaluating changes in microRNA (miRNA) expression following the same atrazine exposure regimen (i.e., an embryonic atrazine exposure at 0.3, 3, or 30 ppb throughout embryogenesis, 1–72 hpf).47 Results from that study revealed alterations in miRNAs that were associated with angiogenesis, cancer, and neurodevelopment, but there are still questions on additional epigenetic mechanisms of atrazine toxicity including DNA methylation.47
In order to investigate DNA methylation parameters associated with atrazine exposure, in the current study we evaluated the effects of atrazine on DNMT activity and kinetics. Next, methylation levels and expression of dnmts was quantified in zebrafish that were exposed to 0, 3, or 30 ppb atrazine throughout embryogenesis (1–72 hpf) to evaluate the current U.S. EPA MCL and a concentration 10X higher. The zebrafish is a strong complementary vertebrate model when used in investigating epigenetic dysfunction and developmental toxicity due to its ex utero fertilization and development, short developmental periods, relatively short life span, and high genetic homology to humans.51–52 While there are some differences between DNA methylation reprogramming between zebrafish and mammals, all DNA methyl transferases identified in mammals are present in zebrafish.53–58 In addition, in contrast to mammals, which have three active DNMTs (DNMT1, DNMT3A, and DNMT3B); the zebrafish genome contains multiple paralogs.53–58 Zebrafish express dnmt1 (homolog to mammalian DNMT1) and six DNMT3 paralogs, classified as dnmt3–8.53–57 The DNMT3A orthologs of zebrafish are dnmt6 and dnmt8; while DNMT3B orthologs are dnmt3, dnmt4, dnmt5, and dnmt7.58
2. Materials and Methods
2.1 Evaluation of DNMT activity using enzyme-linked immunosorbent assay (ELISA)
An enzyme-linked immunosorbent assay (ELISA) was performed to assess enzyme activity of human methyltransferases (DNMT1, DNMT3A, DNMT3B) and bacterial methyltransferase (M. SssI) in the presence and absence of atrazine. All methyltransferases were obtained commercially (NEB and Epigentek, NY). The reaction was performed using a commercial ELISA kit (DNMT1 and DNMT3A: EpiQuik DNMT activity assay kit (Colorimetric), Epigentek, NY; and DNMT3B: DNMT3B Inhibitor Screening Assay Kit, Abcam, MA). Atrazine concentrations of 0, 3, and 30 parts per billion (µg/L; ppb) were tested (CAS #1912-24-9; Chem Service, 98% purity). The specific enzyme activity (min−1 ng−1) was determined following Eq.1.
| (Eq. 1) |
To evaluate the effect of atrazine on DNMT activity, we reported the relative activity of DNMTs as the ratio of the enzyme activity in presence and absence of atrazine.
2.2 The effect of atrazine on DNMT kinetics
Kinetic assays were conducted using DNA fragments with defined sequence content (Supplementary Table 1) at varying DNA concentrations (0.15 – 0.75 µM). Atrazine was added to a reaction buffer (50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM MgCl2 and 1 mM DTT) at concentrations of 0, 3, and 30 ppb. The reaction was carried out using excessive amounts of Ado-Met (SAM, 160 µM), initiated by the addition of bacterial methyltransferase (M.SssI, ~ 20 nM) and incubated at 37°C. M.SssI resembles maintenance DNA methyltransferase activity and has a much higher biochemical activity. As such, M.SssI was used as model enzyme to determine atrazine effects on maintenance DNMTs. The reaction was stopped at selected time points by heating the reaction mixture at 65°C for 20 minutes to deactivate the methyltransferase. Then, the reacted DNA fragments were digested with a restriction enzyme, HpaII. The endonuclease activity of HpaII is completely blocked by meCG. HpaII is thus typically used in methylation-sensitive restriction enzyme assays to evaluate the methylation level of DNA fragments.59 HpaII digestion products were analyzed using polyacrylamide gel electrophoresis (PAGE). The relative band intensities of unmethylated and methylated DNA (quantified using Image J), which are correlated with the digested (107 bp) and undigested (147 bp) DNA bands, respectively, were used to determine the methylation level of DNA fragments. The meCG concentration was calculated following Eq.2.
| (Eq. 2) |
where, IU and ID are the intensity of the undigested and digested band, respectively, and NCG is the number of CG sites per DNA strand. The characterized [meCG] was then used to determine the initial methyltransferase rate (V) under different reaction conditions. Apparent kinetics parameters, i.e., Km and Vmax, were obtained by fitting the kinetic data using a Michaelis-Menten model (Eq. 3).
| (Eq. 3) |
Control experiments were performed to test if the addition of atrazine or phenol may affect the endonuclease activity of HpaII. Typical PAGE results are shown in Supplementary Figure 1. Under the selected digestion condition, the presence of neither atrazine nor phenol affects the digestion results.
2.3 Zebrafish husbandry and embryonic atrazine exposure
Zebrafish (wild-type AB strain) were housed in a Z-Mod System (Aquatic Habitats, Apopka, FL) on a 14:10 hour light:dark cycle and maintained at 28 ± 1°C with a pH of 7.0–7.2 and conductivity range of 470–520 µS. Adult zebrafish aged 5–7 months were bred in groups in cages and embryos were collected, staged, and rinsed with system water as described previously for experimental use.47–50 Zebrafish embryos were exposed to 0, 3, or 30 ppb atrazine (CAS #1912-24-9; Chem Service, 98% purity) from 1–72 hours post fertilization (hpf) as previously described.47–50 Atrazine water sample concentrations were verified using a U.S. EPA approved immunoassay kit for atrazine (Abraxis Atrazine ELISA kit, Warminster, PA) as previously described.47–50 Similar to our past studies no differences in mortality rates or increases in gross malformations were observed in the atrazine exposed groups compared to the controls with no atrazine treatment.47–50 All animal protocols were approved and performed in accordance with Purdue University’s Institutional Animal Care and Use Committee Guidelines.
2.4 Genomic DNA isolation and measurement of global methylation levels
Following atrazine exposure, zebrafish larvae (72 hpf) were rinsed and genomic DNA was extracted following a standard protocol.60 Briefly, zebrafish embryos were homogenized in 0.5 mL Proteinase K lysis buffer (50 mM Tris, 100 mM EDTA, 100 mM NaCl, 1% SDS, 100 mg/mL Proteinase K) and incubated at 55°C overnight. Following incubation, samples were transferred to Phase Lock Gel (PLG) light 15 mL tubes (5 Prime, Hilden, Germany) and phenol (Phenol-Tris saturated, pH 8) (Roche, Indianapolis, IN) was added. Next, 1 mL chloroform:isoamyl alcohol (24:1) (American Bioanalytical, Natick, MA) was added and samples were centrifuged at room temperature for 5 minutes at 1,500 rcf. The upper aqueous phase was placed into a second PLG tube. The addition of phenol and chloroform:isoamyl alcohol (24:1) and centrifugation was repeated. The aqueous phase was then placed into a 15 mL tube. 0.1× the volume of 3M sodium acetate and 1× isopropanol was added. Samples were inverted until DNA began to aggregate. Next, samples were incubated at room temperature for 20 minutes and then centrifuged at 4°C for 10 minutes at 800 rcf. Excess liquid was removed and the DNA pellet was washed in 70% ethanol and centrifuged at 4°C for 5 minutes at 800 rcf. The DNA pellet was rehydrated in 1× TLE buffer and incubated overnight at 55°C. The DNA concentration was tested on a NanoDrop® ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). The DNA was then sonicated into fragments of 100 – 300 bp in length following an established procedure.61 Fragmented DNA samples were analyzed using a 1% Agarose gel as shown in (Supplementary Figure 2). DNA fragments (~ 500 nM) were then mixed with a fluorescein-labeled DNA methylation probe that was created by our group to determine the methylation level of gDNA following our established approach.62,63 This probe can quantify the total amount of methylated CG dinucleotides above 20 nM independent of DNA sequence contexts.
2.5 Quantitative polymerase chain reaction (qPCR) of dnmt expression in zebrafish larvae
Zebrafish embryos were exposed to 0, 3, or 30 ppb atrazine from 1–72 hpf as described above. Following the exposure period, 50 embryos were pooled per sample (n=9–10), homogenized in Trizol® Reagent (Life Technologies, Carlsbad, CA), and flash frozen in liquid nitrogen. Total RNA was isolated and purified following similar methods described in Freeman and Peterson using Trizol Reagent followed by RNA clean-up with Qiagen RNEasy Mini Kit.64 The RNA samples were quality and quantity checked using a NanoDrop® ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and further converted to cDNA with the Super Script® First-Strand Synthesis System for RT-PCR (Life Technologies, Carlsbad, CA). qPCR was performed on all seven DNMT genes found in zebrafish (dnmt1, dnmt3, dnmt4, dnmt5, dnmt6, dnmt7, dnmt8) using the BioRad iQ SYBR Green Supermix kit according to manufacturer’s recommendations. Primers specific to target genes were designed using the Primer3 website (Table 1). qPCR was performed following a similar method as previously described following MIQE guidelines.47–50, 65 Similar to previous studies in our laboratory, several genes were assessed to determine the best reference gene to be used for this data set (data not shown).47–50 β-actin was found to be the most consistent, not altered by atrazine exposure, and least variable for this analysis. Experimental samples were run in triplicate (technical replicates), quantitative copies calculated using a standard curve, and gene expression normalized to β-actin. Efficiency and specificity were checked with melting and dilution curve analysis and no-template controls.
Table 1.
qPCR primer list for dnmt analysis in zebrafish larvae
| NCBI Accession Number |
Gene Name | Primer Sequence |
|---|---|---|
| NM_131189.2 | dnmt1 | Forward: GAGGAGGATGTGTTGCCAGTTA |
| Reverse: CCTCATTTTCCACACGCACTTT | ||
|
| ||
| NM_131386.1 | dnmt3 | Forward: TAGAGTCATGTTGAACTGGGCC |
| Reverse: TCAGGTCCAGAGATTCAGGGAT | ||
|
| ||
| NM_001025450.1 | dnmt4 | Forward: AAGATTTACCCTGCAGTCCCAG |
| Reverse: CTCGCATACTTCTGACGCAAT G | ||
|
| ||
| NM_001020479.1 | dnmt5 | Forward: TTATCCACCCACTGTTCGAAGG |
| Reverse: ATGACCACACAGAATGACCTCC | ||
|
| ||
| NM_001018140.1 | dnmt6 | Forward: GTGTGGGGAAAGTTACGAGGAT |
| Reverse: TGCTTATTGTAGGTTGGCTGGT | ||
|
| ||
| NM_001020746.1 | dnmt7 | Forward: AGGCAGCTTTTCGGGATTTAGA |
| Reverse: CGATTTCTTGACCATCACGAGC | ||
|
| ||
| NM_001018134.1 | dnmt8 | Forward: CTTTGCCTGTTAATGAAGCCCC |
| Reverse: TGTGAAGTGTCCTGTGGTTGAA | ||
|
| ||
| NM_181601 | β-actin | Forward: CTAAAAACTGGAACGGTGAAGG |
| Reverse: AGGCAAATAAGTTTCGGAACAA | ||
2.6 Statistical analysis
Statistical analysis was completed using OriginPro (Version 2015, Origin Lab Corp, North Hampton, MA) or SAS (Version 9.4) statistical software. ELISA, DNMT kinetics, and global DNA methylation levels were analyzed by a one-way analysis of variance (ANOVA) followed by a Duncan’s multiple range post-hoc test when a significant ANOVA was observed (α < 0.05). qPCR data was analyzed with an ANOVA and a post hoc least significant difference (LSD) test when a significant ANOVA was observed (α < 0.05).
3. Results
3.1 Effects of atrazine on DNMT activity
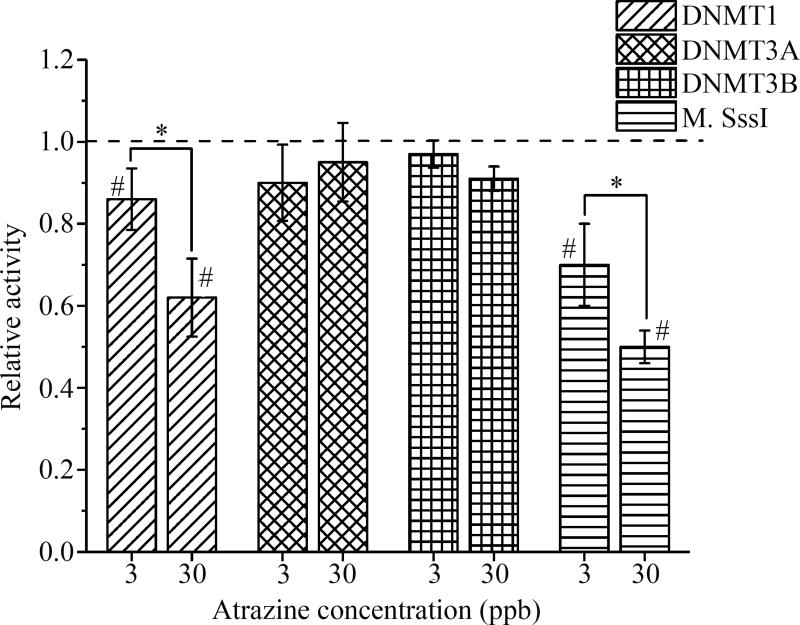
The kinetics of DNMT1 was significantly reduced in a dose-dependent manner by 13% and 38% following an atrazine exposure of 3 and 30 ppb, respectively. No significant alterations in kinetic activity were observed in DNMT3A or DNMT3B. M.Sss1 activity was significantly reduced in the 3 and 30 ppb atrazine treatment groups by approximately 25 and 50%, respectively (p<0.05; Figure 1).
Figure 1. Atrazine altered DNMT1 and M.SssI activity.
Relative activity of DNMT1, DNMT3A, DNMT3B, and M.SssI following an atrazine exposure of 0, 3, or 30 ppb was assessed. The activity of DNMT1 and M.SssI were significantly decreased following atrazine exposure compared to the control (#p<0.05). No significant changes were observed in the activity of DNMT3A or DNMT3B. Data is expressed as mean ± standard deviation (n≥3; *indicates a significant difference (p<0.05) between the 3 and 30 ppb samples)
3.2 Atrazine is a non-competitive inhibitor for DNA methyltransferase reactions
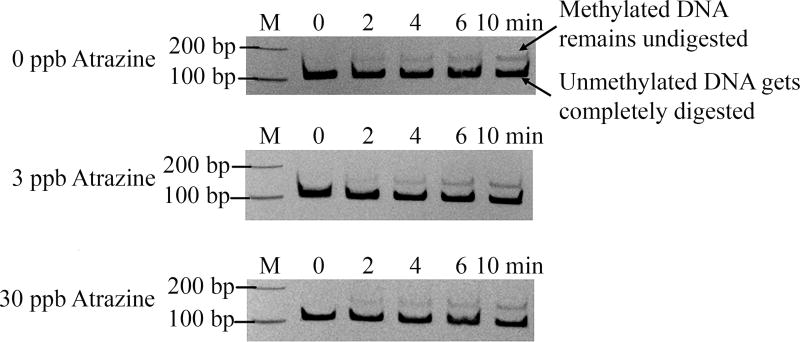
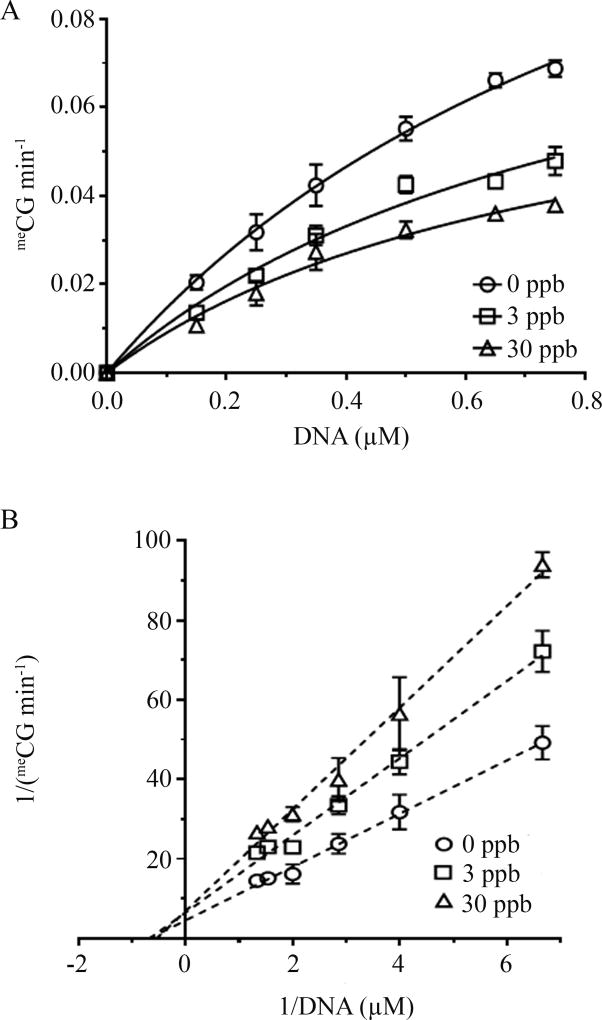
M.SssI was used as a model enzyme to determine atrazine effects on maintenance DNMT. Initial results demonstrated methylation levels at various time points (0, 2, 4, 6, 8, and 10 minutes) at 0, 3, or 30 ppb atrazine examined using a methylation-sensitive restriction enzyme digestion assay (Figure 2). The initial reaction rate was determined in order to calculate the kinetic parameters (Vmax and Km). Results show that an atrazine exposure of 3 or 30 ppb significantly decreases methylation rate (p<0.05; Figure 3A) at various DNA substrate concentrations. The kinetic data were fitted using a Michaelis-Menten model, and the parameters Vmax and Km were summarized (Table 2). The addition of atrazine significantly reduces the Vmax value while no significant difference was observed for Km. To validate the non-competitive inhibition, a Lineweaver-Burke analysis was completed (Figure 3B). The linearized kinetic data exhibit distinctive intercept (Vmax), but similar Km depicted by almost identical abscissa intercepts. Both are characteristic features of non-competitive inhibitors.
Figure 2. Effect of atrazine on the methylation activity of M.SssI.
Changes in methylation levels due to an atrazine exposure of 0, 3, or 30 ppb at 2, 4, 6, 8, and 10 minutes. Reactions were run with 0.15 µM of DNA.
Figure 3. Atrazine inhibited DNA methyltransferase reactions.
Results show that an atrazine exposure of 3 or 30 ppb significantly decreased methylation rate (A). A Lineweaver-Burke analysis of kinetic data obtained under varying atrazine concentrations (B). (n≥3; *p<0.05).
Table 2.
Kinetic parameters of DNA methyltransferase reactions following atrazine exposure catalyzed by M.SssI.
| 0 ppb | 3 ppb | 30 ppb | |
|---|---|---|---|
| Vmax(meCGµM min−1) | 0.170±0.018 | 0.106±0.013* | 0.080±0.010* |
| KM (µM) | 1.06±0.16 | 0.88±0.18 | 0.78±0.17 |
| KCat (min−1) | 7.31±0.75 | 4.56±0.57* | 3.43±0.45* |
p<0.05
3.3 Embryonic atrazine exposure in zebrafish decreased CpG methylation
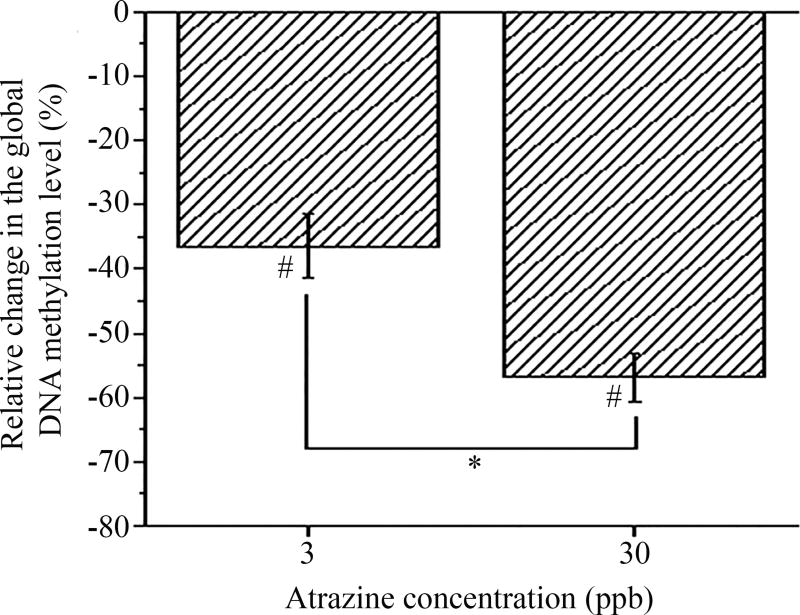
In agreement with our past studies using a similar atrazine exposure regimen, no differences in mortality, hatching rate, or gross malformations were observed following the embryonic exposure. Developmental exposure to atrazine did cause a significant dose-dependent reduction in the average methylation level of the genomic DNA (gDNA). The relative methylation level of gDNA with respect to the control (0 ppb atrazine) was reduced by 36 ± 5% and 56 ± 3% in zebrafish embryos exposed to 3 ppb and 30 ppb atrazine, respectively (p<0.05; Figure 4 and Supplementary Figure 3).
Figure 4. Developmental atrazine exposure decreased CpG methylation.
Zebrafish were treated with 0, 3, or 30 ppb atrazine from 1–72 hpf and gDNA was isolated at 72 hpf. Results indicated a significant reduction in the relative global methylation in both the 3 and 30 ppb atrazine treatments compared with the 0 ppb control (#p<0.05). Data is expressed as mean ± standard deviation (n≥5; *indicates a significant difference (p<0.05) between the 3 and 30 ppb samples)
3.4 Embryonic atrazine exposure in zebrafish altered dnmt expression
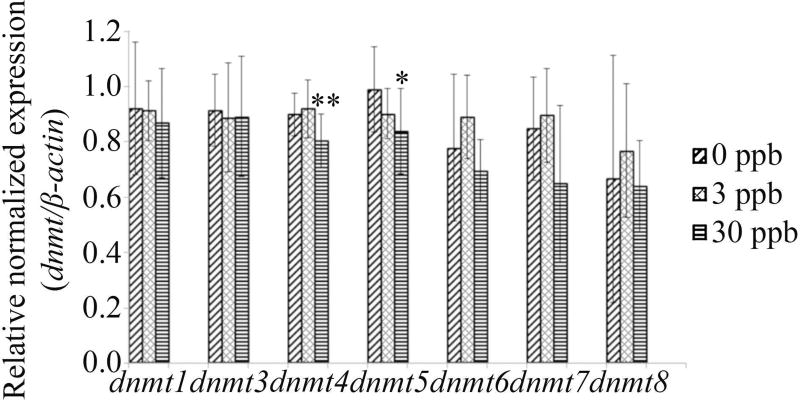
A significant decrease in gene expression in the 30 ppb atrazine treatment was observed for dnmt4 and dnmt5 (p=0.0083 and p=0.0419, respectively). No significant differences in gene expression was observed for any other dnmts tested (dnmt1: p=0.8128; dnmt3: p=0.9055; dnmt6: p=0.0744; dnmt7: p=0.6016; dnmt8: p=0.4802) (Figure 5).
Figure 5. Developmental atrazine exposure alters dnmt expression.
Zebrafish were exposed to 0, 3, or 30 ppb atrazine from 1–72 hpf. Following the exposure period, qPCR was performed to obtain expression levels of dnmts. A significant decrease in the 30 ppb atrazine treatment was observed in dnmt4 and dnmt5. No alterations were reported for the remaining dnmts tested in any of the atrazine concentrations (p>0.05). Data is expressed as mean ± standard deviation (n=9–10; *p<0.05; **p<0.01).
4. Discussion
Studies have begun investigating DNA methylation disruption in response to EDCs in numerous animal models with varying reports of alterations.26,27,66 For example, exposure to bisphenol A in mice or phthalates in rats results in DNA methylation changes, but in contrast a maternal exposure to the anti-androgenic compounds vinclozolin, flutamide, or procymodine in rats had no effects on DNA methylation in male offspring.26,27,66 There is currently very limited knowledge on epigenetic mechanisms of atrazine toxicity with only a couple published studies focused on DNA methylation, miRNA deregulation, or histone modifications.44–47 Based on previous findings from our laboratory identifying immediate and later-in-life neurological and reproductive system impacts of an embryonic atrazine exposure in zebrafish and persistent morphological alterations in their subsequent generation48–50 combined with rodent models also reporting later-in-life alterations from an embryonic atrazine exposure,67–70 it is important to identify the effects of atrazine on DNA methylation parameters as a potential mode of action.
In the current study, the effects of atrazine on DNA methyltransferase activity in vitro was assessed and an observed decrease in the activity of DNMT1 and M.SssI was noted. This similar response can be explained by the conserved sequence motifs in the catalytic and the Ado-met-binding site of both enzymes.71 In addition, M.SssI is the only prokaryotic methyltransferase that can methylate cytosine of a CG similar to DNMT1.72 Meanwhile, the activity of the de novo methyltransferase DNMT3A and DNMT3B remained unaffected by atrazine under the selected concentrations. As our results indicated that atrazine alters maintenance DNMTs in vitro, it was necessary to identify the potential mechanism of inhibition caused by atrazine exposure. To accomplish this task, M.SssI was chosen as the model enzyme due to its large in vitro activities and similarity to DNMT1.72 The decrease in methylation and initial methylation reaction rate following atrazine exposure that were observed suggest that atrazine acts as a noncompetitive inhibitor for M.SssI. To further support this hypothesis, validation through a Lineweaver-Burke analysis showed linearized kinetic data that exhibited a distinctive intercept (Vmax), but similar Km values depicted by almost identical abscissa intercepts, which are characteristic kinetic features of non-competitive inhibitors.73 This noncompetitive inhibition mechanism suggests that atrazine can bind to the enzyme (either in a free or an enzyme-substrate complex form) at a site different from the catalytic center. The binding can result in reduced activities with respect to the DNA substrate.
Exposure to chemicals during critical developmental periods can induce heritable and stable alterations in DNA methylation, therefore potentially altering gene regulation and contributing to disease onset. In this study, the effect of an embryonic atrazine exposure on global methylation in zebrafish larvae was investigated. Assessment during this period is ideal as the establishment of methylation patterns can take up to 96 hpf to complete in the zebrafish;74 therefore, providing a snapshot into the effects of atrazine during primary methylation events. A decrease in global methylation in zebrafish larvae were observed in both the 3 and 30 ppb treatments. To our knowledge, only one other set of studies has evaluated DNA methylation changes following atrazine exposure in vivo. These two recent studies investigated the effects of atrazine on DNA methylation in various tissues of the common carp.44–45 Atrazine concentrations as low as 4.28 ppb were found to cause global hypomethylation patterns in the liver, kidney, gill, gonad, and brain tissue of juvenile carp.44–45 In those studies, reduction in global DNA methylation ranged from 10 to 45% depending on the analyzed tissue. Although these studies were completed with various tissues in juvenile carp, we report a similar global hypomethylation pattern in zebrafish exposed to 3 or 30 ppb atrazine during embryogenesis.
We then assessed expression patterns of zebrafish dnmts following an embryonic atrazine exposure at the same concentrations. Multiple studies have characterized the developmental expression of dnmts in zebrafish embryos, larvae, and adults.57–58,74–76 In addition, studies have also begun investigating EDCs and their effects on global methylation and dnmt expression with the zebrafish. For example, Aluru et al. reported altered dnmt expression in zebrafish with an exposure to dioxin observing upregulation of some dnmts and downregulation of others, but they did not observe global methylation changes, while Fang et al. reported decreased global methylation with exposure to benzo[a]pyrene.77–78 Following an embryonic atrazine exposure, we report a significant decrease at 30 ppb in gene expression of dnmt4 and dnmt5 (both are orthologs of human DNMT3B). Currently, the developmental characterization of dnmt4 and dnmt5 has been identified in the zebrafish. By 72 hpf, dnmt4 is located in the ciliary marginal zone, pharyngeal arches, auditory capsule, pectoral fin buds, intestine, pancreas, and liver. In addition, dnmt4 is also expressed in the tectum, dorsal and ventral thalamus, and in the pallium region.75 dnmt5 expression has also been identified throughout the anterior zebrafish embryo with expression localizing to the ciliary marginal zone of the retina by 72 hpf.76
Our dnmt expression results are similar with the previous studies conducted in juvenile common carp (Cyprinus carpio L.), which reported a consistent reduction in the expression of dnmt1, dnmt3, dnmt4, dnmt5, dnmt6, dnmt7, and dnmt8 following a chronic 40 day atrazine exposure at concentrations ranging from 4.2–42.8 ppb in brain, gonad, liver, kidney, and gill tissue.44–45 Although no significant gene expression alterations were identified in the remaining dnmts (dnmt1, dnmt3, dnmt6, dnmt 7, dnmt8) in our study, a decreasing trend was noted, especially within the 30 ppb atrazine treatment group. The discrepancy between these studies regarding the expression levels of dnmt1, 3, 6, 7, and 8 may be due to the differences between embryonic and juvenile exposure, dosing periods, and atrazine concentrations or are based on the specific function of the genes.
Understanding the toxicological implications of dnmt alterations by EDCs, their impact on later-in-life function, and impacts to future generations requires further investigation. In addition, studies are needed to further compare in vitro and in vivo alterations in maintenance and de novo methyltransferases and gene-specific changes. Further studies are also needed to define the relationship between downregulation of dnmt4 and dnmt5 and adverse effects observed in other studies of atrazine toxicity.47–50
5. Conclusions
Overall in this study, an atrazine exposure was shown to elicit epigenetic dysfunction through decreasing maintenance DNMT activities in vitro and decreased global methylation and dnmt expression in developing zebrafish. The results from this and a previous study conducted from our laboratory support that atrazine elicits developmental toxicity at the epigenetic level therefore, providing a potential mode of action behind the molecular and morphological alterations.47 Further studies are warranted in order to determine specific genes that have altered methylation status. In addition, future studies are needed to determine if epigenetic dysfunction persists into adulthood, and if so, does it contribute to previously identified genomic and functional alterations.49–50 In addition, it will be important to determine if epigenetic alterations are present in future generations.
Supplementary Material
Highlights.
Atrazine exposure reduces activity of maintenance DNMTs
Atrazine inhibition mechanism is described using non-competitive Michaelis-Menten kinetics
Zebrafish embryonic atrazine exposure decreases global methylation
Zebrafish embryonic atrazine exposure decreases dnmt4 and dnmt5 expression
Acknowledgments
This work was supported by the National Institutes of Health, National Institute of Environmental Health Sciences (R15 ES019137 to J.L.F.), a Bilsland Dissertation Fellowship (S.E.W. and J.L.F), the US Army Medical Research (Award Number: W81XWH-14-1-0012 to C.Y.), the School of Chemical Engineering, Purdue University (C.Y.), and the Administrative Department of Science, Technology and Innovation (COLCIENCIAS) from Colombia and Fulbright (Grant 529) (O.F.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: The authors declare that they have no competing interests.
References
- 1.Bernal AJ, Jirtle RL. Epigenomic disruption: the effects of early developmental exposures. Birth. Defects. Res. A. Clin. Mol. Teratol. 2010;88:938–944. doi: 10.1002/bdra.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 3.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes. Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riddihough G, Zahn LM. Epigenetics. What is epigenetics? Introduction. Science. 2010;330:611. doi: 10.1126/science.330.6004.611. [DOI] [PubMed] [Google Scholar]
- 6.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147:S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 8.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013100. Pii, e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inawaka K, Kawabe M, Takahashi S, Doi Y, Tomigahara Y, Tarui H, Abe J, Kawamura S, Shirai T. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol. Appl. Pharmacol. 2009;237:178–187. doi: 10.1016/j.taap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- 12.Vandegehuchte MB, Lemière F, Vanhaecke L, Vanden Berghe W, Janssen CR. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009;151:278–285. doi: 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero-Bosagna Carlos, Luis Valladares. Endocrine-Disrupting Chemicals. Humana Press; 2007. Endocrine disruptors, epigenetically induced changes, and transgenerational transmission of characters and epigenetic states; pp. 175–189. [Google Scholar]
- 14.Portela A, Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 15.Roman T, Aumueller E, Berner C, Haslberger AG. Interaction of Hereditary and Epigenetic Mechanisms in the Regulation of Gene Expression. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2010. [Google Scholar]
- 16.Swedenborg E, Rüegg J, Mäkelä S, Pongratz I. Endocrine disruptive chemicals: mechanisms of action and involvement in metabolic disorders. J. Mol. Endocrinol. 2009;43:1–10. doi: 10.1677/JME-08-0132. [DOI] [PubMed] [Google Scholar]
- 17.Birnbaum LS, Fenton SE. Cancer and developmental exposure to endocrine disruptors. Environ. Health. Perspect. 2003;111:389–394. doi: 10.1289/ehp.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y, Zhang P, Wang F, Yang J, Yang Z, Qin H. The relationship between early embryo development and tumourigenesis. J. Cell. Mol. Med. 2010;14:2697–2701. doi: 10.1111/j.1582-4934.2010.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prins GS, Birch L, Tang W, Ho S. Developmental estrogen exposures predispose to prostate carcinogenesis with aging. Reprod. Toxicol. 2007;23:374–382. doi: 10.1016/j.reprotox.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper RL, Stoker TE, Tyrey L, Goldman JM, McElroy WK. Atrazine disrupts the hypothalamic control of pituitary-ovarian function. Toxicol. Sci. 2000;53:297–307. doi: 10.1093/toxsci/53.2.297. [DOI] [PubMed] [Google Scholar]
- 21.Hatch EE, Troisi R, Wise LA, Titus-Ernstoff L, Hyer M, Palmer JR, Strohsnitter WC, Robboy SJ, Anderson D, Kaufman R, Adam E, Hoover RN. Preterm birth, fetal growth, and age at menarche among women exposed prenatally to diethylstilbestrol (DES) Reprod. Toxicol. 2011;31:151–157. doi: 10.1016/j.reprotox.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy JR, Chakraborty S, Chakraborty TR. Estrogen-like endocrine disrupting chemicals affecting puberty in humans--a review. Med. Sci. Monit. 2009;15:137–145. [PubMed] [Google Scholar]
- 23.Wetzel LT, Luempert LG, 3rd, Breckenridge CB, Tisdel MO, Stevens JT, Thakur AK, Extrom PJ, Eldridge JC. Chronic effects of atrazine on estrus and mammary tumor formation in female Sprague-Dawley and Fischer 344 rats. J. Toxicol. Environ. Health. 1994;43:169–182. doi: 10.1080/15287399409531913. [DOI] [PubMed] [Google Scholar]
- 24.Casati L, Sendra R, Poletti A, Negri-Cesi P, Celotti F. Androgen receptor activation by polychlorinated biphenyls: epigenetic effects mediated by the histone demethylase Jarid1b. Epigenetics. 2013;8:1061–1080. doi: 10.4161/epi.25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casati L, Sendra R, Colciago A, Negri-Cesi P, Berdasco M, Esteller M, et al. Polychlorinated biphenyls affect histone modification pattern in early development of rats: a role for androgen receptor-dependent modulation? Epigenomics. 2012;4:101–112. doi: 10.2217/epi.11.110. [DOI] [PubMed] [Google Scholar]
- 26.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Arguelles DB, Papadopoulos V. Identification of hot spots of DNA methylation in the adult male adrenal in response to in utero exposure to the ubiquitous endocrine disruptor plasticizer di-(2-ethylhexyl) phthalate. Endocrinology. 2015;156:124–33. doi: 10.1210/en.2014-1436. [DOI] [PubMed] [Google Scholar]
- 28.Barr DB, Panuwet P, Nguyen JV, Udunka S, Needham LL. Assessing exposure to atrazine and its metabolites using biomonitoring. Environ. Health. Perspect. 2007;115:1474–1478. doi: 10.1289/ehp.10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eldridge JC, Stevens JT, Breckenridge CB. Atrazine interaction with estrogen expression systems. Rev. Environ. Contam. Toxicol. 2008;196:147–160. doi: 10.1007/978-0-387-78444-1_6. [DOI] [PubMed] [Google Scholar]
- 30.Solomon KR, Carr JA, Du Preez LH, Giesy JP, Kendall RJ, Smith EE, Van Der Kraak GJ. Effects of atrazine on fish, amphibians, and aquatic reptiles: a critical review. Crit. Rev. Toxicol. 2008;38:721–772. doi: 10.1080/10408440802116496. [DOI] [PubMed] [Google Scholar]
- 31.US EPA. EPA-HQ-OPP-2013-0266-0315. Office of Pesticide Programs; Washington, DC: 2016. Refined ecological risk assessment for atrazine. [Google Scholar]
- 32.Rohr JR, McCoy KA. A qualitative meta-analysis reveals consistent effects of atrazine on freshwater fish and amphibians. Environ. Health. Perspect. 2010;118:20–32. doi: 10.1289/ehp.0901164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Commission E. Review report for the active substance atrazine; Finalized in the Standing Committee on the Food Chain and Animal Health at its meeting on 3 October 2003 in support of a decision concerning the non-inclusion of atrazine in Annex I of Directive 91/414/EEC and the withdrawal of authorization for plant protection products containing this active substance: European Commission Health and Consumer Protection Direc-torate-General. 2003 SANCO/10496/2003-final. [Google Scholar]
- 34.Sass JB, Colangelo A. European Union bans atrazine, while the United States negotiates its continued use. Int. J. Occup. Environ. Health. 2006;12:260–267. doi: 10.1179/oeh.2006.12.3.260. [DOI] [PubMed] [Google Scholar]
- 35.Mattix KD, Winchester PD, Scherer LR. Incidence of abdominal wall defects is related to surface water atrazine and nitrate levels. J. Pediatr. Surg. 2007;42:947–949. doi: 10.1016/j.jpedsurg.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 36.Munger R, Isacson P, Hu S, Burns T, Hanson J, Lynch CF, Cherryholms K, Van Drope P, Hausler WJ. Intrauterine growth retardation in Iowa communities with herbicide-contaminated drinking water supplies. Environ. Health. Perspect. 1997;105:308–314. doi: 10.1289/ehp.97105308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochoa-Acuña H, Frankenberger J, Hahn L, Carbajo C. Drinking-water herbicide exposure in Indiana and Prevalence of small-for-gestational-age and preterm delivery. Environ. Health. Perspect. 2009;117:1619–1624. doi: 10.1289/ehp.0900784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winchester PD, Huskins J, Ying J. Agrichemicals in surface water and birth defects in the United States. Acta. Paediatrica. 2009;98:664–669. doi: 10.1111/j.1651-2227.2008.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fa S, Pogrmic-Majkic K, Samardzija D, Glisic B, Kaisarevic S, Kovacevic R, Andric N. Involvement of ERK1/2 signaling pathway in atrazine action on FSH-stimulated LHR and CYP19A1 expression in rat granulosa cells. Toxicol. Appl. Pharmacol. 2013;270:1–8. doi: 10.1016/j.taap.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 40.Foradori CD, Zimmerman AD, Hinds LR, Zuloaga KL, Breckenridge CB, Handa RJ. Atrazine inhibits pulsatile gonadotropin-releasing hormone (GnRH) release without altering GnRH messenger RNA or protein levels in the female rat. Biol. Reprod. 2013;88:1–7. doi: 10.1095/biolreprod.112.102277. [DOI] [PubMed] [Google Scholar]
- 41.Foradori CD, Hinds LR, Hanneman WH, Legare ME, Clay CM, Handa RJ. Atrazine inhibits pulsatile luteinizing hormone release without altering pituitary sensitivity to a gonadotropin-releasing hormone receptor agonist in female Wistar rats. Biol. Reprod. 2009;81:40–45. doi: 10.1095/biolreprod.108.075713. [DOI] [PubMed] [Google Scholar]
- 42.Pogrmic-Majkic K, Samardzija D, Fa S, Hrubik J, Glisic B, Kaisarevic S, Andric N. Atrazine enhances progesterone production through activation of multiple signaling pathways in FSH-stimulated rat granulosa cells: evidence for premature luteinization. Biol. Reprod. 2014;91:1–10. doi: 10.1095/biolreprod.114.122606. [DOI] [PubMed] [Google Scholar]
- 43.Pogrmic-Majkic K, Fa S, Dakic V, Kaisarevic S, Kovacevic R. Upregulation of peripubertal rat Leydig cell steroidogenesis following 24 h in vitro and in vivo exposure to atrazine. Toxicol. Sci. 2010;118:52–60. doi: 10.1093/toxsci/kfq227. [DOI] [PubMed] [Google Scholar]
- 44.Wang C, Zhang Z, Yao H, Zhao F, Wang L, Wang X, Xing H, Xu S. Effects of atrazine and chlorpyrifos on DNA methylation in the liver, kidney and gill of the common carp (Cyprinus carpio L.) Ecotoxicol. Environ. Saf. 2014;108:142–151. doi: 10.1016/j.ecoenv.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Xing H, Wang C, Wu H, Chen D, Li S, Xu S. Effects of atrazine and chlorpyrifos on DNA methylation in the brain and gonad of the common carp. Comp. Biochem. Physiol. Part C. Toxicol. Pharmacol. 2015;168:11–19. doi: 10.1016/j.cbpc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Gely-Pernot A, Hao C, Becker E, Stuparevic I, Kervarrec C, Chalmel F, Primig M, Jégou B, Smagulova F. The epigenetic processes of meiosis in male mice are broadly affected by the widely used herbicide atrazine. BMC. Genomics. 2015 doi: 10.1186/s12864-015-2095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirbisky SE, Weber GJ, Schlotman KE, Sepúlveda MS, Freeman JL. Embryonic atrazine exposure alters zebrafish and human miRNAs associated with angiogenesis, cancer, and neurodevelopment. Food. Chem. Toxicol. 2016b doi: 10.1016/j.fct.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber GJ, Sepúlveda MS, Peterson SM, Lewis SL, Freeman JL. Transcriptome alterations following developmental atrazine exposure in zebrafish are associated with disruption of neuroendocrine and reproductive system function, cell cycle, and carcinogenesis. Toxicol. Sci. 2013;132:458–466. doi: 10.1093/toxsci/kft017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wirbisky SE, Weber GJ, Sepúlveda MS, Lin TL, Jannasch AS, Freeman JL. An embryonic atrazine exposure results in reproductive dysfunction in adult zebrafish and morphological alterations in offspring. Sci. Rep. 2016a doi: 10.1038/srep21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirbisky SE, Weber GJ, Sepúlveda MS, Xiao C, Cannon JR, Freeman JF. Developmental origins of neurotransmitter and transcriptome alterations in adult female zebrafish exposed to atrazine during embryogenesis. Toxicology. 2015;333:156–167. doi: 10.1016/j.tox.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Esch C, Slieker R, Wolterbeek A, Woutersen R, De Groot D. Zebrafish as potential model for developmental neurotoxicity testing: A mini review. Neurotoxicol. Teratol. 2012;34:545–553. doi: 10.1016/j.ntt.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mhanni AA, McGowan RA. Variations in DNA (cytosine-5)-methyltransferase-1 expression during oogenesis and early development of the zebrafish. Dev. Genes Evol. 2002;212:530–533. doi: 10.1007/s00427-002-0275-7. [DOI] [PubMed] [Google Scholar]
- 54.Mudbhary R, Sadler KC. Epigenetics, development, and cancer: zebrafish make their mark. Birth. Defects. Res. C. Embryo. Today. 2011;93:194–203. doi: 10.1002/bdrc.20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seritrakul P, Gross JM. Expression of the De Novo DNA Methyltransferases (dnmt3 – dnmt8) During Zebrafish Lens Development. Dev. Dyn. 2014;243:350–356. doi: 10.1002/dvdy.24077. [DOI] [PubMed] [Google Scholar]
- 56.Smith THL, Dueck CC, Mhanni AA, McGowan RA. Novel splice variants associated with one of the zebrafish dnmt3 genes. BMC. Dev. Biol. 2005;5 doi: 10.1186/1471-213X-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campos C, Valente LMP, Fernandes JMO. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene. 2012;500:93–100. doi: 10.1016/j.gene.2012.03.041. [DOI] [PubMed] [Google Scholar]
- 58.Smith THL, Collins TM, McGowan RA. Expression of the dnmt3 genes in zebrafish development: similarity to DNMT3A and DNMT3B. Dev. Genes. Evol. 2011;220:347–353. doi: 10.1007/s00427-010-0347-z. [DOI] [PubMed] [Google Scholar]
- 59.Oakeley EJ. DNA methylation analysis: a review of current methodologies. Pharmacol. Therapeut. 1999;84:389–400. doi: 10.1016/s0163-7258(99)00043-1. [DOI] [PubMed] [Google Scholar]
- 60.Dobrinski KP, Brown KH, Freeman JL, Lee C. Molecular cytogeneic methodologies and BAC probe banel resource for genomic analysis in the zebrafish. Methods Cell Biol. 2011;104:237–257. doi: 10.1016/B978-0-12-374814-0.00014-8. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J, Russell DW. Fragmentation of DNA by sonication. CSH protocols. 2006;4 doi: 10.1101/pdb.prot4538. [DOI] [PubMed] [Google Scholar]
- 62.Kim SE, Chang M, Yuan CL. One-pot approach for examining the DNA methylation patterns using an engineered methyl-probe. Biosens. Bioelectron. 2014;58:333–337. doi: 10.1016/j.bios.2014.02.064. [DOI] [PubMed] [Google Scholar]
- 63.Jabbari K, Bernardi G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 2004;333:143–149. doi: 10.1016/j.gene.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 64.Peterson SM, Freeman JL. RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. Journal of visualized experiments : JoVE. 2009 doi: 10.3791/1470(30). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 66.Inawaka K, Kawabe M, Takahashi S, Doi Y, Tomigahara Y, Tarui H, Abe J, Kawamura S, Shirai T. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol. Appl. Pharmacol. 2009;237:178–187. doi: 10.1016/j.taap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 67.Rosenberg BG, Chen H, Folmer J, Liu J, Papadopoulos V, Zirkin BR. Gestational exposure to atrazine: effects on the postnatal development of male offspring. J. Androl. 2008;29:304–311. doi: 10.2164/jandrol.107.003020. [DOI] [PubMed] [Google Scholar]
- 68.DeSesso JM, Scialli AR, White TEK, Breckenridge CB. Multigeneration reproduction and male development toxicity studies on atrazine in rats. Birth. Defects. Res. B. Dev. Reprod. Toxicol. 2014;101:237–253. doi: 10.1002/bdrb.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin Z, Roede JR, He C, Jones DP, Filipov N. Short term oral atrazine exposure alters the plasma metabolome of male C57BL/6 mice and disrupts a-linolenate tryptophan, tyrosine and other major metabolic pathways. Toxicology. 2014a;326:130–141. doi: 10.1016/j.tox.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Lin Z, Dodd CA, Xiao S, Krishna S, Ye X, Filipov NM. Gestational and lactational exposure to atrazine via the drinking water causes specific behavioral deficits an selectively alters monoaminergic systems in C57BL/6 mouse dams, juvenile and adult offspring. Toxicol. Sci. 2014b;141:90–102. doi: 10.1093/toxsci/kfu107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Svedružić ŽM. Chapter 6 - Dnmt1: Structure and Function. In: Xiaodong C, Robert MB, editors. Progress in Molecular Biology and Translational Science. Vol. 101. Academic Press; 2011. pp. 221–254. [DOI] [PubMed] [Google Scholar]
- 72.Koudan EV, Bujnicki JM, Gromova ES. Homology Modeling of the CG-specific DNA Methyltransferase SssI and its Complexes with DNA and AdoHcy. J. Biomol. Struct. Dyn. 2004;22:339–345. doi: 10.1080/07391102.2004.10507005. [DOI] [PubMed] [Google Scholar]
- 73.Lineweaver H, Burk D. The determination of enzyme dissociation constants. J. American. Chemical. Society. 1934;56:658–666. [Google Scholar]
- 74.Fang X, Corrales J, Thornton C, Scheffler BE, Willett KL. Global and gene specific DNA methylation changes during zebrafish development. Comp. Biochem. Physiol.B. Biochem. Molec. Biol. 2013a;166:99–108. doi: 10.1016/j.cbpb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takayama K, Shimoda N, Takanaga S, Hozumi S, Kikuchi Y. Expression patters of DNMT3Aa, DNMT3Ab, and dnmt4 during development and fin regeneration in zebrafish. Gene. Expr. Patterns. 2014;14:105–110. doi: 10.1016/j.gep.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 76.Seritrakul J, Gross JM. Expression of De novo DNA methyltransferase (dnmt3-dnmt8) during zebrafish lens development. Dev. Dyn. 2014;243:350–356. doi: 10.1002/dvdy.24077. [DOI] [PubMed] [Google Scholar]
- 77.Aluru N, Kuo E, Helfrich LW, Karchner SI, Linney EA, Pais JE, Franks DG. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio) Toxicol. Appl. Pharmacol. 2015;284:142–151. doi: 10.1016/j.taap.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang X, Thornton C, Scheffler BE, Willett KL. Benzo[a]pyrene decreases global and gene specific DNA methylation during zebrafish development. Environ. Toxicol. Pharmacol. 2013b;36:40–50. doi: 10.1016/j.etap.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.