Abstract
Nicotine dependence (ND) has a reported heritability of 40–70%. Low-coverage whole-genome sequencing was conducted in 1,889 samples from the UCSF Family study. Linear mixed models were used to conduct genome-wide association tests of ND in this and five cohorts obtained from the database of Genotypes and Phenotypes. Fixed-effect meta-analysis was carried out separately for European (n = 14,713) and African (n = 3,369) participants, and then in a combined analysis of both ancestral groups. The meta-analysis of African participants identified a significant and novel susceptibility signal (rs56247223; P = 4.11 × 10−8). Data from the Genotype-Tissue Expression (GTEx) study suggested the protective allele is associated with reduced mRNA expression of CACNA2D3 in three human brain tissues (P < 4.94 × 10−2). Sequence data from the UCSF Family study suggested that a rare nonsynonymous variant in this gene conferred increased risk for ND (P = 0.01) providing further support for CACNA2D3 involvement in ND. Suggestive associations were observed in six additional regions in both European and merged populations (P < 5.00 × 10−6). The top variants were found to regulate mRNA expression levels of genes in human brains using GTEx data (P < 0.05): HAX1 and CHRNB2 (rs1760803), ADAMTSL1 (rs17198023), PEX2 (rs12680810), GLIS3 (rs12348139), non-coding RNA for LINC00476 (rs10759883), and GABBR1 (rs56020557 and rs62392942). A gene-based association test further supported the relation between GABBR1 and ND (P = 6.36 × 10−7). These findings will inform the biological mechanisms and development of therapeutic targets for ND.
Keywords: Nicotine dependence, genome-wide meta-analysis, susceptibility genes, expression quantitative trait locus, nonsynonymous variants
Introduction
Tobacco usage is the leading cause of preventable mortality worldwide. Vaporized inhaled nicotine from combusted tobacco is an efficient delivery system that frequently leads to nicotine dependence (ND) [Sullivan and Kendler 1999]. The lifetime incidence of ND may be as high as 25% and is similar across diverse populations [Breslau et al., 2001]. ND clusters in families, and large twin studies indicate that ND has a moderate genetic predisposition, with an estimated heritability between 40% and 70% [Kendler et al., 1999; Sullivan and Kendler 1999; Vink et al., 2005].
Over the past few decades, many large-scale genome-wide association (GWA) studies and meta-analyses have identified a number of single nucleotide polymorphisms (SNPs) associated with the risk of ND and smoking-related behaviors [Bierut et al., 2007; David et al., 2012; Gelernter et al., 2015; Gizer and Ehlers 2015; Hancock et al., 2015; Liu et al., 2010; Loukola et al., 2014; Nishizawa et al., 2015; Rice et al., 2012; Thorgeirsson et al., 2008; Thorgeirsson et al., 2010; Tobacco and Genetics Consortium 2010; Uhl et al., 2008; Zuo et al., 2013]. Among them, the α3/α5/β4 cholinergic nicotinic receptor subunit gene cluster on chromosome 15 consistently shows the most significant replicable risk effect on ND, smoking cessation, smoking quantity, and other smoking-related traits [Thorgeirsson et al., 2008; Thorgeirsson et al., 2010]. In 2015, Gelernter et al. conducted a genome-wide meta-analysis on ND, as measured by the Fagerström Test for ND score. They found that more GWA signals were identified in African American than in European American samples, highlighting the benefit of analyzing populations with different genetic backgrounds [Gelernter et al., 2015].
GWA studies can detect common genetic variants (defined as minor allele frequency [MAF] ≥ 1%). These studies have advanced our understanding of the genetics of ND and smoking-related traits, but the identified variants collectively explain only a small fraction of the total heritability of ND. It has been suggested that common variants, which typically exhibit small effect sizes (e.g. R2 < 0.005), are unlikely to be identified using the customary GWA significance threshold and the sample size typical in most studies [Manolio et al., 2009]. Genome-wide meta-analysis, which is used to aggregate individual studies, is expected to improve the statistical power for associations and has been successfully used to identify novel genetic variants not previously discovered by single studies [Cho et al., 2012; David et al., 2012; Liu et al., 2010; Mahajan et al., 2014; Nalls et al., 2014; Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014]. In addition, in the context of mining existing GWA results, the incorporation of functional data in GWA studies and gene-based analysis has the potential to accelerate the discovery of more functionally relevant genes and could also help to elucidate the biological implications of GWA findings [Ioannidis et al., 2009]. Besides common variants, rare and low-frequency variants that have not been fully covered in GWA studies could explain, at least in part, the risk for common diseases/traits [Manolio et al., 2009]. Recently, multiple low-frequency and even rare coding variants have been revealed to be associated with the risk of ND in population-based studies [Doyle et al., 2014; Haller et al., 2012; Olfson et al., 2016; Thorsteinsdottir et al., 2014; Xie et al., 2011; Yang et al., 2015; Zuo et al., 2016], but the role of rare variants in the risk of ND has not been systematically investigated at the genome-wide level.
In the present study, we sought to identify novel common susceptibility SNPs partially responsible for the risk of ND through genome-wide meta-analysis, and explore their possible mechanisms in the pathogenesis of ND. Genome-wide mixed linear model (MLM)-based association analysis on ND was performed for six studies, with subjects stratified into genetically homogeneous European (EUR) and African (AFR) ancestry subgroups. Meta-analyses were carried out in the EUR, AFR, and trans-population merged cohort, which consisted of 18,082 subjects in total. We explored the regulatory effects of the identified SNPs in human brains through expression quantitative trait locus (eQTL) analysis in the Genotype-Tissue Expression (GTEx) data set, and searched for rare nonsynonymous variants in the implicated genes and several previously identified genes in the whole-genome sequencing data in the UCSF Family Alcoholism study.
Materials and Methods
UCSF Family Alcoholism Study Subjects
The UCSF Family Alcoholism study was designed to identify genetic loci that influence susceptibility to alcohol dependence and related phenotypes. It includes small nuclear families and unrelated subjects, and the majority of study subjects are EUR [Vieten et al., 2004]. A total of 2,154 individuals from 970 families from December 1995 through January 2003 were enrolled in this study. The recruitment of subjects has been described previously [Vieten et al., 2004]. Briefly, study probands from the community were recruited through direct mail, press releases, advertisements, and etc. Their relatives were invited by mail to participate. The diagnosis for ND was determined according to the criteria of the Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV). The samples in the UCSF Family Alcoholism study were subjected to low-pass whole-genome sequencing. Genotypes were imputed from the low-pass sequencing data as previously described [Bizon et al., 2014]. Briefly, low-coverage, paired-end read whole-genome sequencing for 1,889 samples was performed on Illumina HiSeq2000 sequencers. Reads from whole-genome sequencing were aligned using BWA version 0.5.8c [Li and Durbin 2010]. Linkage disequilibrium (LD)-aware variant calls were calculated in a multistage process that initially creates genotype-likelihood files for single samples using samtools-hybrid, then builds initial haplotypes using BEAGLE [Browning and Browning 2007], and then runs Thunder [Li et al., 2011] using the BEAGLE haplotypes as input [Bizon et al., 2014]. The mean coverage of the cohort was 4.2X. We applied the following quality control (QC) to the whole-genome sequencing data (MAF ≥ 1%, Hardy-Weinberg equilibrium (HWE) in controls P > 1.00 × 10−6) and included 8,893,347 common SNPs in the single-variant association test. After selection for subjects with diagnostic data and completion of QC steps, 918 subjects were identified as ND cases and 921 were free of ND (Supplementary Table I).
Database of Genotypes and Phenotypes (dbGaP) Subjects
We accessed five GWA data sets (SAGE: Study of Addiction: Genetics and Environment; GASSC: The Genetic Architecture of Smoking and Smoking Cessation; ADGE: Alcohol Dependence GWAS in European and African Americans; ICGHD: International Consortium on the Genetics of Heroin Dependence; OZALC: Alcohol Research using Australian twins and their families) through dbGaP [Bierut et al., 2010; Bierut et al., 2008b; Cornelis et al., 2010; Hinrichs et al., 2006; Shand et al., 2010; Xie et al., 2011]. The ICGHD data set consisted of three subsets (CIDR: Center for Inherited Disease Research; RATRP: Research on Alcohol and Tobacco Related Phenotypes; and ARC: Alcohol Related Conditions) that were genotyped using three different arrays. The sample sizes ranged from 469 to 6,695 individuals. All subjects were recruited for genetic studies of substance use disorders. The DSM-IV criteria for ND were used as a measure of ND for all studies. The procedures for data collection for these cohorts were described in detail in dbGaP (http://www.ncbi.nlm.nih.gov/gap) and in previous literature [Bierut et al., 2008a; Edenberg 2002; Gelernter et al., 2007; Lessov et al., 2004]. The cohorts contained data for small families and unrelated individuals. See Table I for study characteristics and dbGaP accession numbers.
Table I.
Summary statistics for studies
| study | dbGaP accession | array | EUR
|
AFR
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| case | control | No. of SNPs | λgc | case | control | No. of SNPs | λgc | |||
| SAGE | phs000092.v1.p1 | Illumina Human 1M | 1048 | 1268 | 4524998 | 1.009 | 578 | 605 | 5976319 | 1.007 |
| ADGE | phs000425.v1.p1 | Illumina HumanOmni1-Quad | 481 | 181 | 4261404 | 0.995 | 913 | 824 | 8143245 | 1.005 |
| GASSC | phs000404.v1.p1 | Illumina HumanOmni2.5 | 714 | 146 | 5174464 | 1.019 | 378 | 71 | 7132729 | 1.007 |
| ICGHD CIDR | phs000277.v1.p1 | Illumina Human660W-Quad | 1318 | 1079 | 5298585 | 1.031 | 0 | 0 | – | – |
| ICGHD RATRP | phs000277.v1.p1 | Illumina Human610_Quad | 101 | 661 | 4751941 | 1.017 | 0 | 0 | – | – |
| ICGHD ARC | phs000277.v1.p1 | Illumina HumanCNV370-Quad | 306 | 77 | 4720266 | 1.004 | 0 | 0 | – | – |
| OZALC | phs000181.v1.p1 | Illumina HumanCNV370v1 | 3329 | 2165 | 6245839 | 1.039 | 0 | 0 | – | – |
| UCSF | – | Whole-genome sequencing | 918 | 921 | 8393347 | 1.005 | 0 | 0 | – | – |
|
| ||||||||||
| Total | 8215 | 6498 | 3,251,541 | 1.022 | 1869 | 1500 | 5,227,780 | 1.003 | ||
dbGaP: the database of Genotypes and Phenotypes; EUR: European population; AFR: African population; No. of SNPs: number of single nucleotide polymorphisms; SAGE: Study of Addiction: Genetics and Environment; ADGE: Alcohol Dependence GWAS in European- and African Americans; GASSC: The Genetic Architecture of Smoking and Smoking Cessation; ICGHD: International Consortium on the Genetics of Heroin Dependence; OZALC: Alcohol Research using Australian twins and their families. CIDR: Center for Inherited Disease Research), RATRP: Research on Alcohol and Tobacco Related Phenotypes) and ARC: Alcohol Related Conditions.
In order to facilitate the meta-analysis in more specific ancestral populations, we employed principal component analysis (PCA) to identify genetically homogeneous populations [Price et al., 2006]. In each dbGaP data set, all samples were analyzed together with the reference samples from the International HapMap Project [International HapMap Consortium 2003]. Two subsets of genetically homogeneous subjects, described here as EUR and AFR in each study, were selected using the first two principal components. The PCA plots are shown in Supplementary Figure 1.
Genomic Imputation
We applied QC filters on each individual dbGaP GWA data set (sex check, mean call rate ≥ 0.90, MAF ≥ 1%, HWE in controls P > 1.00 × 10−6). Then, whole-genome imputation was performed using the IMPUTE version 2 [Howie et al., 2012] and the integrated haplotypes of the 1000 Genomes Project Phase III reference panel (October 2014 version) [Auton et al., 2015]. Imputation was carried out in each of the seven dbGaP GWA data sets/subsets individually. We applied QC filters on the post-imputed data sets: SNPs with imputation information < 30%, MAF < 1% or HWE P-value in controls < 1.00 × 10−6 were excluded from further analysis. Only bi-allelic autosomal SNPs were analyzed in this study.
Association and meta-analysis
The single-variant association between each common variant and the risk of ND was measured using a MLM or MLM leaving-one-chromosome-out (LOCO)-based association as implemented in GCTA for each dbGaP GWA data set and the UCSF Family Alcoholism study [Yang et al., 2011; Yang et al., 2014]. Based on previous reports, the MLM-based association appropriately corrects for population stratification and sample relatedness [Yang et al., 2014].
We performed an inverse-variance fixed-effects meta-analysis based on the cohort effect sizes and standard errors using METAL, with a Cochran’s Q test to assess between-study heterogeneity [Willer et al., 2010]. Meta-analysis was performed in the 14,713 EUR, 3,369 AFR, and trans-ancestry merged cohorts (Table I). The meta-analysis for the trans-ancestry cohort aggregated ancestry-specific association result from each study. A P-value of < 5.00 × 10−8 was used as the threshold for genome-wide significance [Panagiotou and Ioannidis 2012].
eQTL analysis
We obtained the genotype and transcript expression data for GTEx from the dbGaP by accession number phs000424.v6.p1 [GTEx Consortium 2013]. We included only 114 subjects genotyped by the Illumina HumanOmni2.5 array. The transcript expression level was measured by RNAseq in the unit of Reads Per Kilobase of transcript per Million mapped reads (RPKM) [GTEx Consortium 2013]. Imputation was executed using IMPUTE version 2 and the 1000 Genomes Project Phase III reference panel (October 2014 version) [Auton et al., 2015; Howie et al., 2012]. Analysis of Variance was employed to evaluate the eQTL effect between the SNPs implicated in the meta-analysis and the mRNA expression level of their respective close genes in thirteen brain tissue samples (cortex, nucleus accumbens basal ganglia, amygdala, anterior cingulate cortex, caudate basal ganglia, cerebellum, frontal cortex, cerebellar hemisphere, hippocampus, hypothalamus, substantia nigra, putamen basal ganglia, and spinal cord cervical). The gender, age, and subject race for each tissue donor were included as covariates. The analysis was implemented in R 3.2.2.
Gene-based association test
We implemented a gene-based association analysis through MetaXcan using the summary statistics from the ND meta-analysis in the merged trans-population data set [Barbeira et al., 2016]. Transcript expression-level predictive models in multiple tissues were previously generated in PrediXcan using a subset of genotype and expression data from the GTEx data set. Models for ten brain tissue samples were available when the current study was conducted (cortex, putamen basal ganglia, caudate basal ganglia, frontal cortex, cerebellum, anterior cortex, hypothalamus, cerebellar hemisphere, hippocampus, and nucleus accumbens basal ganglia) [Gamazon et al., 2015]. MetaXcan uses established predictive models and accepted association summary statistics as input; it then imputes the transcript expression levels and tests the association between these imputed transcript expression levels and the phenotype [Barbeira et al., 2016]. The mathematics for the gene-based test has been described previously [Barbeira et al., 2016; Gamazon et al., 2015]. Between 8,530 and 13,570 gene transcripts were predicted with high confidence in the ten predictive models. We used as inputs for MetaXcan the ND meta-analysis association result in the trans-population merged cohort for 12,768,612 common SNPs that appeared in at least one single data set. The gene association significance threshold was set at 3.68 × 10−6 after Bonferroni correction.
Rare-variant association and gene-based test
We looked for an increased frequency of rare nonsynonymous variants in those genes implicated by the present meta-analysis (CACNA2D3, GABBR1, CHRNB2, PEX2, ADAMTSL1, and GLIS3) and several previously well-known ND susceptibility genes (CHRNA3, CHRNA4, CHRNA5, CHRNB4, DISC1, DLC1, KLHL28, and C14ORF28) [Gelernter et al., 2015; Haller et al., 2012; Hancock et al., 2015; Olfson et al., 2016; Rice et al., 2012; Thorgeirsson et al., 2010; Thorsteinsdottir et al., 2014; Xie et al., 2011] using low-pass whole-genome sequencing data in the UCSF Family Alcoholism study. A quantitative phenotype was created by regressing out the covariates of age and gender. This pseudo-quantitative trait was used for single-variant association analysis using Efficient Mixed-Model Association eXpedited (EMMAX) in the Efficient and Parallelizable Association Container Toolbox (EPACTS) package version 3.2.6 [Kang et al., 2010]. In addition, we employed the Sequence Kernel Association Test (SKAT) under the framework of the EMMAX model in the EPACTS package to perform the gene-based test [Kang et al., 2010; Wu et al., 2011]. The gene-based test included only nonsynonymous variants (including nonsynonymous, splicing, and stop-gain variants) with allele frequency ≤ 1% and allele count in the cohort of ≥ 2. We used a significance threshold of P < 0.05 for single rare-variant association and gene-based tests of the described rare variants.
Results
We performed a single-variant association analysis of common variation using MLM on each individual dbGaP GWA data set and the UCSF Family Alcoholism study for the EUR and AFR samples. The quantile-quantile (Q-Q) plots for each study show that the MLM method controlled the potential inflation in each of the cohorts (Supplementary Figure 2). Fixed-effect meta-analysis was performed for 2,976,328, 4,975,009, and 2,590,787 common study-shared SNPs in the EUR, AFR, and merged cohorts, respectively. The EUR, AFR and merged cohorts consisted of 14,713, 3,369, and 18,082 samples, respectively. The Q-Q and Manhattan plots indicate that the population substructure was appropriately controlled in meta-analyses (Supplementary Figures 3–4). Although this was not an independent replication effort given the overlap of study samples with the previously published reports[Gelernter et al., 2015; Hancock et al., 2015], this meta-analyses confirmed rs1051730 in the CHRNA5-CHRNA3-CHRNB4 region and nine other recently reported SNPs in five distinct genomic regions associated with ND with nominal association evidence (P < 0.05, Supplementary Table II).
Meta-analysis in the AFR cohort
In the AFR samples, SNP rs56247223 on chromosome 3 achieved a genome-wide significant association with ND (OR = 0.93, P = 4.11 × 10−8, MAF = 32%; Figure 1a and Supplementary Tables III–IV). SNP rs56247223 is located in an intron of the gene CACNA2D3 (Figure 1b). The minor A allele of rs56247223 showed a modest, but consistent protective effect in the three AFR data sets (Figure 1a). The analysis of the GTEx data suggested that the minor protective A allele of SNP rs56247223 was associated with reduced levels of CACNA2D3 mRNA in human brain cortex, frontal cortex, and putamen (P = 5.53 × 10−3, 4.58 × 10−2, 4.94 × 10−2, Figure 1c and Supplementary Figure 7).
Figure 1. Association of SNP rs56247223 with ND and the eQTL effect on the CACNA2D3 gene in human brain.
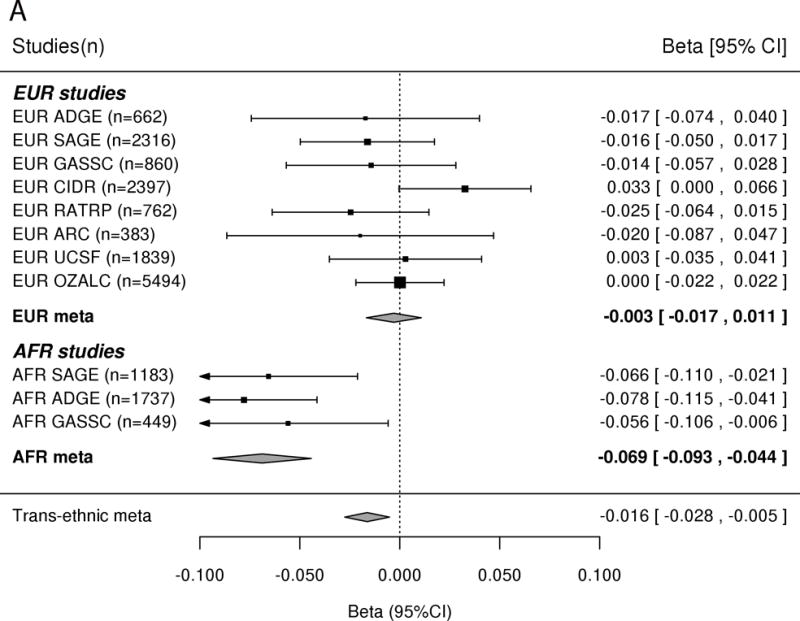
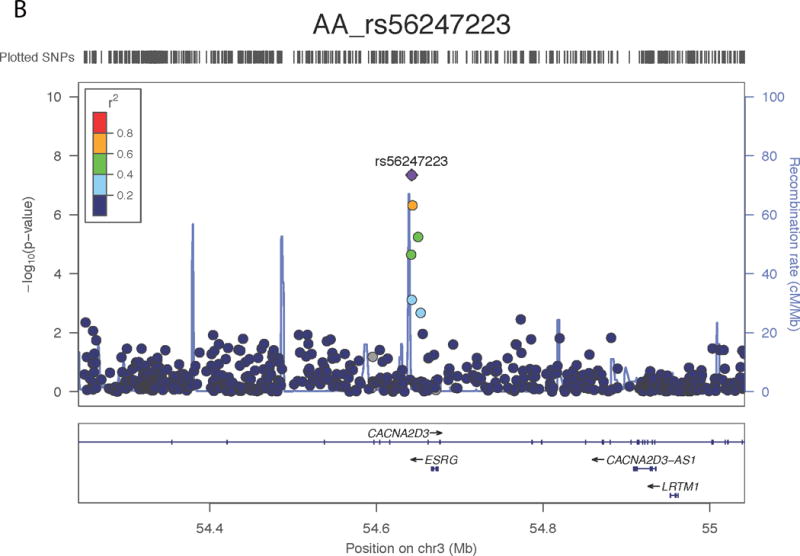
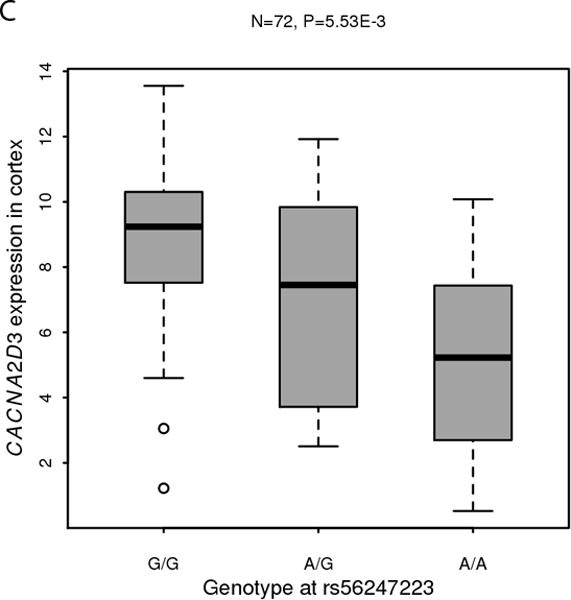
(a) Forest plots of the meta-analysis results. (b) Regional association plots for rs56247223 in AFR samples. The relative location of annotated genes and the direction of transcription are shown in the lower portion of the figure. The chromosomal position is shown on the x-axis. The blue line shows the recombination rate (estimated from HapMap data of AFR sample) across the region (right y-axis), and the left y-axis shows the significance level of the SNP associations (−log10P). The color of the squares and circles indicate the LD status between the SNPs and the leading SNP. (c) The eQTL effect on the CACNA2D3 gene in human brain cortex. The y-axis represents the normalized transcript expression level in the unit of RPKM. SNP: single nucleotide polymorphism; eQTL: expression quantitative trait locus; EUR: European; AFR: African; LD: linkage disequilibrium; RPKM: Reads Per Kilobase of transcript per Million mapped reads. SAGE: Study of Addiction: Genetics and Environment; ADGE: Alcohol Dependence GWAS in European- and African Americans; GASSC: The Genetic Architecture of Smoking and Smoking Cessation; ICGHD: International Consortium on the Genetics of Heroin Dependence; OZALC: Alcohol Research using Australian twins and their families. CIDR: Center for Inherited Disease Research), RATRP: Research on Alcohol and Tobacco Related Phenotypes) and ARC: Alcohol Related Conditions.
Meta-analysis in the EUR cohort
SNP rs56247223 did not show any evidence of association with ND in the EUR cohort (P = 0.68, Figure 1a). No SNPs exceeded the genome-wide significance threshold in the EUR meta-analysis. However, three SNPs, rs1760803, rs17198023, and rs56020557 on chromosomes 1, 6, and 9, respectively, showed moderate and consistent association evidence with ND across samples (P < 5.00 × 10−6, Supplementary Figures 5–6). Among these SNPs, the most significant rs1760803 (OR = 0.97, MAF = 46%, P = 4.26 × 10−6), which resides in the upstream of genes HAX1 and CHRNB2, exhibited a significant eQTL effect on the HAX1 and CHRNB2 genes in human brain cerebellum, hemisphere and hippocampus (P = 3.66 × 10−2, 1.54 × 10−2, and 2.01 × 10−2, respectively, Supplementary Figure 7). The A allele of SNP rs17198023 exhibited considerable association with increased risk for ND in the EUR samples (OR = 1.04, MAF = 13%, P = 4.69 × 10−6), and was correlated with downregulation of the ADAMTSL1 gene mRNA in human brain caudate basal ganglia (P = 3.34 × 10−2, Supplementary Figure 7). Notably, the third SNP, rs56020557 (OR = 1.05, MAF = 6%, P = 4.69 × 10−6), is 5kb upstream of the GABBR1 gene (Supplementary Figures 5–6).
Meta-analysis in the merged cohort
In the merged EUR and AFR meta-analysis, it was noted that three SNPs in high LD (rs12680810, rs56225501, and rs28534373) located on chromosome 8 achieved moderate association evidence (P < 2.00 × 10−6). An examination of the study specific results indicated more consistent results for the EUR than the AFR samples (Supplementary Table V, Supplementary Figures 5–6). Among them, the most significant SNP, rs12680810 (OR = 1.03, MAF = 41% and 22% in the EUR and AFR samples, respectively, P = 9.76×10−7), showed a significant eQTL effect on the PEX2 gene in human brain frontal cortex (P = 1.81 × 10−2, Supplementary Figure 7). Two additional SNPs on chromosome 9 (rs12348139 and rs10759883) were moderately associated with ND in the merged analysis (P < 2.00 × 10−6, Supplementary Table V, Supplementary Figures 5–6); both showed a consistent effect on the risk of ND across all individual studies and achieved at least nominal association evidence in both EUR and AFR ancestry-specific meta-analyses (Supplementary Table V, Supplementary Figures 5–6). rs12348139 is located in an intron of the GLIS3 gene (OR = 1.04, MAF = 7% and 10% in EUR and AFR samples, respectively, P = 1.09 ×10−7); its minor C allele, which was associated with an increased risk for ND, upregulated the expression levels of GLIS3 gene mRNA in human brain putamen (P = 1.98 × 10−2, Supplementary Figure 7). The C allele of rs10759883, located in a long non-coding RNA (LINC00476), was associated with increased risk of ND (OR = 0.98, MAF = 43% and 34% in the EUR and AFR samples, respectively, P = 1.23 × 10−6), and was highly correlated with upregulation of LINC00476 expression levels in nine human brain regions (cortex, frontal cortex, anterior cingulate cortex, caudate basal ganglia, cerebellum, hypothalamus, substantia nigra, putamen and nucleus accumbens basal ganglia, Supplementary Figure 7). The most significant eQTL effect was observed in human brain anterior cingulate cortex (P = 1.95 × 10−5, Supplementary Figure 7). Finally, SNP rs62392942 achieved significant association and showed identical effect direction in the EUR and AFR samples, although it achieved nominal significance only in the EUR meta-analysis (Supplementary Table V and Supplementary Figure 6). SNP rs62392942 resides downstream of the GABBR1 gene on chromosome 6, and is highly correlated with rs56020557 among EUR individuals (LD: r2 = 0.86 and 0.48 in EUR and AFR samples, respectively). Notably, rs56020557 was among the top signals in the EUR meta-analysis (P = 4.50 × 10−6). The C allele of rs62392942, which was associated with increased risk for ND, exhibited an eQTL effect on GABBR1 gene mRNA in human brain putamen (Supplementary Figure 7). Using the merged EUR and AFR meta-analysis summary statistics, the GABBR1 gene was significantly associated with risk for ND in the gene-based MetaXcan analysis (P = 6.36 × 10−7).
Rare variants and gene-based tests
In the UCSF Family Alcoholism study, we performed exploratory single rare-variant association and gene-based tests with the aim of exploring the possible mechanism(s) underlying the associations between the implicated genes, as well as several previously established ND genes. We observed a risk overrepresentation of rare nonsynonymous variants among ND cases relative to controls in the HAX1 gene (gene-based test P = 0.03, Table II). Moreover, a rare nonsynonymous variant in the HAX1 gene conferred a nominally significant protective effect on ND (chr1:154247908:C/T, MAF = 0.14%, OR = 0.31, P = 2.80 × 10−2, Supplementary Table VI). In contrast, the gene-based test of rare nonsynonymous variants in the CACNA2D3 gene was not significant (P = 0.77), though a rare nonsynonymous variant c.C1604T in exon 17 of CACNA2D3 was nominally associated with ND (MAF = 0.16%, OR = 3.66, P = 1.10 × 10−2, Supplementary Table VI). Six heterozygotes subjects were identified with this rare variant, and they were all diagnosed with ND. Similarly, although no aggregate effect of rare nonsynonymous variants was discovered in PEX2 or GLIS3 (Table II), a rare nonsynonymous variant in PEX2 (chr8:77900416:T/C, MAF = 0.16%, OR = 3.33, P = 1.25 × 10−2) and another in GLIS3 (chr9:3898723:C/T, MAF = 0.14%, OR = 0.33, P = 3.90 × 10−2), were nominally associated with risk of ND (Supplementary Table VI). The remaining genes implicated in the present meta-analysis (GABBR1, CHRNB2, ADAMTS1, and LINC00476) were not supported by the rare variant analysis. Among genes previously implicated in the etiology of ND, a rare nonsynonymous variant in DLC1 was nominally associated with ND (Supplementary Table VI), but the gene-based tests did not identify any significant aggregate associations with ND in previously identified susceptibility genes (CHRNA3, CHRNA4, CHRNA5, CHRNB4, DLC1, DISC1, C14ORF28, and KLHL28) (Table II).
Table II.
Summary statistics for the gene-based test of rare variants in the UCSF Family Alcoholism study
| CHR | BEGIN | END | GENE | Variant No. | P |
|---|---|---|---|---|---|
| 1 | 231762651 | 232172555 | DISC1 | 16 | 1.72E-01 |
| 1 | 154245225 | 154247908 | HAX1 | 3 | 2.80E-02 |
| 1 | 154542792 | 154548331 | CHRNB2 | 6 | 1.00E+00 |
| 3 | 54156747 | 55107846 | CACNA2D3 | 14 | 7.71E-01 |
| 6 | 29571342 | 29600126 | GABBR1 | 2 | 1.90E-01 |
| 8 | 12943795 | 13357567 | DLC1 | 29 | 7.29E-01 |
| 8 | 77895667 | 77913120 | PEX2 | 9 | 3.35E-01 |
| 9 | 18533279 | 18906866 | ADAMTSL1 | 20 | 1.00E+00 |
| 9 | 3828355 | 4286526 | GLIS3 | 20 | 3.00E-01 |
| 14 | 45369682 | 45374747 | C14ORF28 | 3 | 5.13E-01 |
| 14 | 45403373 | 45414662 | KLHL28 | 6 | 1.00E+00 |
| 15 | 78873272 | 78885473 | CHRNA5 | 3 | 5.33E-01 |
| 15 | 78885473 | 78911261 | CHRNA3 | 5 | 5.66E-01 |
| 15 | 78917316 | 79012516 | CHRNB4 | 10 | 3.96E-01 |
| 20 | 61978149 | 62005888 | CHRNA4 | 12 | 6.71E-01 |
CHR: chromosome; BEGIN: the start position of gene; END; the end position of gene; Variant No.: the valid available rare nonsynonymous variants. P: the EMMAX SKAT association P-value.
Discussion
ND has a reported heritability of 40–70% [Kendler et al., 1999; Sullivan and Kendler 1999; Vink et al., 2005]. Although GWA studies have previously detected multiple loci associated with ND, even in aggregate they only account for a small fraction of the measured heritability of ND. In the present study, genome-wide meta-analyses were used to identify common SNPs associated with susceptibility for ND. Findings from these meta-analytic efforts were then further explored by using transcriptional expression data in human brains evaluate the possible regulatory effects of the implicated SNPs on gene expression. This study confirmed five previously reported loci (i.e., CHRNA5-A3-B4, KLHL28, C14ORF28, DISC1, and DLC1), identified a novel susceptibility gene CACNA2D3 in the African data set as well as six other potential loci on chromosomes 1, 6, 8, and 9. We provided further evidence suggesting that these implicated SNPs altered mRNA expression levels of genes CACNA2D3, HAX1, CHRNB2, ADAMTSL1, GLS3, LINC00476, and PEX2 through eQTL effects in human brains. Analyses of rare variants conducted in the UCSF Family Alcoholism study provided further evidence for the implicating genes. Specifically, we searched for rare nonsynonymous variants occurring in the implicated genes in low-pass whole-genome sequencing data in the UCSF Family Alcoholism study, and identified four rare nonsynonymous variants in the CACNA2D3, HAX1, PEX2 and GLIS3 genes that conferred risk for ND, as well as an aggregate effect of rare nonsynonymous variants in HAX1 associated with ND diagnostic status. Together, these results demonstrate the utility of meta-analysis, when supplemented with the analysis of expression-level data and sequence-based data, for accelerating the discovery of functionally relevant genes involved in the etiology of ND.
In the present study, a novel genome-wide significant association of rs56247223 on chromosome 3 with risk for ND was identified in the African samples. SNP rs56247223 is located in an intron of CACNA2D3, and was associated with mRNA expression levels of CACNA2D3 in three human brain regions (cortex, frontal cortex, and putamen). In terms of its function, CACNA2D3 encodes a protein member of the voltage-dependent calcium channel complex. Calcium channels mediate the influx of calcium ions into cells and have an important role in neurotransmission [Simms and Zamponi 2014]. Calcium channels have been widely implicated in the pathophysiology of substance dependence, and they are therapeutic targets for addiction medications [Addolorato et al., 2012; Zamponi 2016]. Several different types of calcium channels exist, and each typically consists of four subunits, of which the CACNA2D3 gene encodes the alpha-2/delta subunit [Simms and Zamponi 2014]. Genomic aberrations of the genes encoding the alpha subunit of these channels have been associated with epilepsy and neuropsychiatric disorders such as autism and schizophrenia [Vergult et al., 2015]. This may be important given that smoking behavior is highly prevalent in schizophrenia patients [Dickerson et al., 2013]. Of direct relevance to the present study, the voltage-dependent calcium channel complex alpha-2 and delta subunit family member gene CACNA2D1, was previously associated with risk of ND [Gelernter et al., 2015]. With respect to the association of rs56247223 with ND in present study, the protective A allele of rs56247223 was associated with a decreased expression level of the gene CACNA2D3 in human brain. This finding is consistent with the observation that expression of L-type high voltage-gated calcium channel alpha2/delta subunits increases after chronic nicotine administration in mouse brain [Hayashida et al., 2005]. CACNA2D3 has also been implicated with other smoking phenotypes, including a robust association with smoking cessation, in a convergent analysis of three GWA data sets [Uhl et al., 2008]. Collectively, these results support CACNA2D3 as a susceptibility gene and potential as a therapeutic target for the treatment of ND.
Despite this evidence, it should be noted that rs56247223 was not significantly associated with ND in the European ancestry cohort. Differences in genetic background may explain why an association was detected in the African but not in the European populations. Nonetheless, the detection of a rare nonsynonymous variant in the CACNA2D3 gene confers a large risk (OR > 3) for ND in the UCSF Family Alcoholism study, of which the majority of subjects are of European ancestry, provides additional support that the gene plays a role in ND.
We found an association of the SNP rs1760803 with ND in the European ancestry cohorts. The mRNA expression levels of two nearby genes, HAX1 and CHRNB2, in human brain tissue were found to be associated with this SNP. In the UCSF Family Alcoholism study, one rare nonsynonymous variant in HAX1 was associated with an increased risk of ND, and the presence of rare variants in the HAX1 gene was found to be associated with an increased risk of ND diagnostic status. The HAX1 gene encodes a protein that often binds to several proteins, including cortactin and the product of the polycystic kidney disease 2 gene. Mutations in the HAX1 gene are responsible for autosomal recessive severe congenital neutropenia (SCN), and mutations in the HAX1 protein B isoform are associated with neuropathy in patients with SCN [Boztug et al., 2010]. The nicotine receptor subunit β2 gene CHRNB2, which is also implicated in this region, has been shown to mediate an early physical and psychological response to nicotine [Ehringer et al., 2007]. Therefore more studies are warranted in order to determine which gene in this particular region is actually influencing the ND phenotype.
SNP rs17198023 was found to show consistent association with ND across all European ancestry cohorts. We further found that this SNP modulated mRNA expression level of the ADAMTSL1 gene on chromosome 9 in human brain caudate basal ganglia. The ADAMTS (a disintegrin-like and metalloprotease domain with thrombospondin type-1 repeats) like 1 gene (ADAMTSL1) encodes an ADAMTS family protein. In a previous GWA study, ADAMTSL1 variants were shown to be associated with antidepressant drug response phenotype in depressed patients [Ising et al., 2009]. Additionally, systemic administration of FG7142, an anxiogenic drug and/or a pharmacological stressor that modulates GABA receptors, leads to an increase in mRNA for ADAMTSL1 in the cortex and hippocampus of mice [Kurumaji et al., 2008; Kurumaji and Nishikawa 2012]. More studies will be needed, however exploring how this gene may potentially influence ND.
In the meta-analysis of European and African merged samples, there was a suggestive association found between ND and SNP rs12348139, an intron in gene GLIS3. This result was partially confirmed by the finding of a rare nonsynonymous variant in GLIS3 in the UCSF Family Alcoholism study cohort that conferred a protective effect on ND. The GLIS3 gene is highly expressed in brain [Cruchaga et al., 2013]. SNP rs12348139 was an eQTL site for the GLIS3 gene in human brain. The GLIS3 gene encodes a member of the GLI-similar zinc finger protein family, which is a nuclear protein with five C2H2-type zinc finger domains, and is recognized as a repressor and activator of transcription. The GLIS3 gene has been identified via GWAS with two commonly used biomarkers, cerebrospinal fluid (CSF) tau and tau phosphorylated at threonine 181 (ptau), for Alzheimer’s disease [Cruchaga et al., 2013], and also the risk of type 2 diabetes [Cho et al., 2012]. Notably, chronic nicotine administration exacerbates tau pathology in a transgenic model of Alzheimer’s disease [Oddo et al., 2005], and cigarette smoking is correlated with a higher risk of Alzheimer’s disease and type 2 diabetes [Cataldo et al., 2010; Spijkerman et al., 2014]. Thus, GLIS3 may have a pleiotropic effect on these three disorders that could explain, in part the pathogenesis underlying the clinical link between cigarette smoking, and Alzheimer’s disease, and type 2 diabetes. We also identified SNP rs10759883 in a long non-coding RNA LINC00476 on chromosome 9 that was associated with ND. The minor risk allele C of rs10759883 was found to upregulate the expression level of LINC00476 through eQTL effects in multiple human brain regions. Although the functional relevance of LINC00476 is unclear, recent evidence suggests that non-coding RNAs play an important and dynamic role in transcriptional regulation, epigenetic signaling, stress responses, and plasticity in the nervous system [Sartor et al., 2012]. Therefore, the role of LINC00476 in ND is worthy of further investigations.
Finally, two SNPs in the GABBR1 gene were found to be suggestively associated with the risk of ND in the meta-analyses for both European and African populations. One of these SNPs, rs62392942, was also found to be associated with altered mRNA expression of GABBR1 in human brain putamen. Further, predicted expression levels of the GABBR1 gene in brain, estimated using the recently developed gene-based association method MetaXcan [Barbeira et al., 2016], were shown to be significantly associated with risk for ND. The GABBR1 gene is functionally relevant to ND. More specifically, the gene encodes a receptor for gamma-aminobutyric acid (GABA), which is the main inhibitory neurotransmitter in the mammalian central nervous system [D’Souza and Markou 2013]. Evidence in animal studies suggests that both nicotine intake and nicotine seeking are attenuated when GABA neurotransmission is facilitated [D’Souza and Markou 2013]. Haplotypes of the GABBR1 gene have also been identified with risk of ND in a previous study, and it has been further suggested that the GABBR1 gene contributes to the risk of ND through interactions with GABBR2 [Li et al., 2009]. The current study provides further genetic evidence suggesting a role for GABBR1 variation in risk for ND.
ND is a complex and multi-faceted construct, and as a result it has been operationalized in a number of ways (e.g., DSM-IV diagnosis, Fagerström Test ND score). GWAS of these ND phenotypes have been pursued in multiple independent, large-scale cohorts, but few genomic regions have been consistently and mutually validated. The differences in phenotype definition and phenotypic heterogeneity could be partly responsible for that. In the present study, we used the DSM-IV diagnosis of ND, and thus, it is not surprising that we did not observe significant overlap in association signals previously reported in the GWAS of Fagerström Test ND score [Gelernter et al., 2015]. As noted, phenotypic heterogeneity could have also contributed to the lack of overlapping signals. The presence of co-morbid substance use disorders likely represents one such source of heterogeneity. For example, several of the contributing GWA cohorts were selected on other forms of substance use disorder (e.g., alcohol, cocaine dependence). It is likely that the etiologic factors, including genetic influences, that contribute to the development of ND in individuals with multiple substance use disorders differ, at least in part, from those factors that contribute to the development of ND in individuals with just ND. Unfortunately, data on co-occurring substance dependence diagnoses were not available across all study cohorts, and thus, could not be studied as covariates or as moderators in the association analysis. Nonetheless, the potential impact of polysubstance use on the observed associations, and results from GWAS of substance use disorders more broadly, is an important focus of study that we hope future studies will be able to address.
The findings of this study should be interpreted in light of several limitations. One limitation is that several findings did not exceed the traditional genome-wide significance threshold, however, follow-up analyses such as the eQTL analysis provided some additional support for these findings. The eQTL analyses did not employ genome-wide corrections for multiple testing because they were conducted to address the specific hypothesis that variants in a candidate gene are associated with expression levels when there is prior evidence of association. The second limitation is the lack of an “independent” validation sample. Recent large-scale genome-wide meta-analyses have implemented broadly a two-stage study design in which they perform the initial meta-analysis, pick a set of variants to carry forward, then pursue replication attempts in sets of independent studies, and finally conduct a combined analysis of all available data. Because it has been argued that the validation effort in the meta-analysis of a two-stage design is not a true independent replication [Thomas et al., 2009] and considering the relatively small sample size in the present study, we pursued a single-stage rather than two-stage design with the aim at gathering exhaustively all available data to improve power for discovery..
In summary, we carried out a large-scale GWA meta-analysis on ND, and identified seven susceptibility signals for ND. Notably, our study provides several lines of evidence supporting key role for genes CACNA2D3 and GABBR1 in ND. The respective proteins of these genes are important for calcium channel and GABA neurotransmitter signaling, both of which are currently the main therapeutic targets for the treatment of ND. These findings not only elucidate the pathophysiology of ND, but also highlight potential therapeutic targets.
Supplementary Material
Acknowledgments
We would like to thank all samples participating in each study. This study was supported by National Institutes of Health (NIH) Grant 1-R01-DA030976-01. We acknowledged the support of the database of Genotypes and Phenotypes (dbGaP) to facilitate the access of five genome-wide association studies. The detailed acknowledgements for the dbGaP studies are included in the supplementary materials.
References
- Addolorato G, Leggio L, Hopf FW, Diana M, Bonci A. Novel therapeutic strategies for alcohol and drug addiction: focus on GABA, ion channels and transcranial magnetic stimulation. Neuropsychopharmacology. 2012;37(1):163–177. doi: 10.1038/npp.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira A, Shah KP, Torres JM, Wheeler HE, Torstenson ES, Edwards T, Garcia T, Bell GI, Nicolae D, Cox NJ, Im HK. MetaXcan: Summary Statistics Based Gene-Level Association Method Infers Accurate PrediXcan Results. BioR. 2016;xiv:1–16. [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, Horton WJ, Breslau N, Budde J, Cloninger CR, Dick DM, Foroud T, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Kuperman S, Madden PA, Mayo K, Nurnberger J, Jr, Pomerleau O, Porjesz B, Reyes O, Schuckit M, Swan G, Tischfield JA, Edenberg HJ, Rice JP, Goate AM. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008a;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008b;95(1–2):14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizon C, Spiegel M, Chasse SA, Gizer IR, Li Y, Malc EP, Mieczkowski PA, Sailsbery JK, Wang X, Ehlers CL, Wilhelmsen KC. Variant calling in low-coverage whole genome sequencing of a Native American population sample. BMC Genomics. 2014;15:85. doi: 10.1186/1471-2164-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug K, Ding XQ, Hartmann H, Ziesenitz L, Schaffer AA, Diestelhorst J, Pfeifer D, Appaswamy G, Kehbel S, Simon T, Al Jefri A, Lanfermann H, Klein C. HAX1 mutations causing severe congenital neuropenia and neurological disease lead to cerebral microstructural abnormalities documented by quantitative MRI. Am J Med Genet A. 2010;152A(12):3157–3163. doi: 10.1002/ajmg.a.33748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslau N, Johnson EO, Hiripi E, Kessler R. Nicotine dependence in the United States: prevalence, trends, and smoking persistence. Arch Gen Psychiatry. 2001;58(9):810–816. doi: 10.1001/archpsyc.58.9.810. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo JK, Prochaska JJ, Glantz SA. Cigarette smoking is a risk factor for Alzheimer’s Disease: an analysis controlling for tobacco industry affiliation. J Alzheimers Dis. 2010;19(2):465–480. doi: 10.3233/JAD-2010-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, Takeuchi F, Wu Y, Go MJ, Yamauchi T, Chang YC, Kwak SH, Ma RC, Yamamoto K, Adair LS, Aung T, Cai Q, Chang LC, Chen YT, Gao Y, Hu FB, Kim HL, Kim S, Kim YJ, Lee JJ, Lee NR, Li Y, Liu JJ, Lu W, Nakamura J, Nakashima E, Ng DP, Tay WT, Tsai FJ, Wong TY, Yokota M, Zheng W, Zhang R, Wang C, So WY, Ohnaka K, Ikegami H, Hara K, Cho YM, Cho NH, Chang TJ, Bao Y, Hedman AK, Morris AP, McCarthy MI, Takayanagi R, Park KS, Jia W, Chuang LM, Chan JC, Maeda S, Kadowaki T, Lee JY, Wu JY, Teo YY, Tai ES, Shu XO, Mohlke KL, Kato N, Han BG, Seielstad M. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44(1):67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, Beaty TH, Bennett SN, Bierut LJ, Boerwinkle E, Doheny KF, Feenstra B, Feingold E, Fornage M, Haiman CA, Harris EL, Hayes MG, Heit JA, Hu FB, Kang JH, Laurie CC, Ling H, Manolio TA, Marazita ML, Mathias RA, Mirel DB, Paschall J, Pasquale LR, Pugh EW, Rice JP, Udren J, van Dam RM, Wang X, Wiggs JL, Williams K, Yu K. The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010;34(4):364–372. doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, Bertelsen S, Bailey M, McKean D, Shulman JM, De Jager PL, Chibnik L, Bennett DA, Arnold SE, Harold D, Sims R, Gerrish A, Williams J, Van Deerlin VM, Lee VM, Shaw LM, Trojanowski JQ, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Peskind ER, Galasko D, Fagan AM, Holtzman DM, Morris JC, Goate AM. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78(2):256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza MS, Markou A. The “stop” and “go” of nicotine dependence: role of GABA and glutamate. Cold Spring Harb Perspect Med. 2013;3(6) doi: 10.1101/cshperspect.a012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, Brown WM, Petruzella S, Thacker EL, Kim Y, Nalls MA, Tranah GJ, Sung YJ, Ambrosone CB, Arnett D, Bandera EV, Becker DM, Becker L, Berndt SI, Bernstein L, Blot WJ, Broeckel U, Buxbaum SG, Caporaso N, Casey G, Chanock SJ, Deming SL, Diver WR, Eaton CB, Evans DS, Evans MK, Fornage M, Franceschini N, Harris TB, Henderson BE, Hernandez DG, Hitsman B, Hu JJ, Hunt SC, Ingles SA, John EM, Kittles R, Kolb S, Kolonel LN, Le Marchand L, Liu Y, Lohman KK, McKnight B, Millikan RC, Murphy A, Neslund-Dudas C, Nyante S, Press M, Psaty BM, Rao DC, Redline S, Rodriguez-Gil JL, Rybicki BA, Signorello LB, Singleton AB, Smoller J, Snively B, Spring B, Stanford JL, Strom SS, Swan GE, Taylor KD, Thun MJ, Wilson AF, Witte JS, Yamamura Y, Yanek LR, Yu K, Zheng W, Ziegler RG, Zonderman AB, Jorgenson E, Haiman CA, Furberg H. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson F, Stallings CR, Origoni AE, Vaughan C, Khushalani S, Schroeder J, Yolken RH. Cigarette smoking among persons with schizophrenia or bipolar disorder in routine clinical settings, 1999–2011. Psychiatr Serv. 2013;64(1):44–50. doi: 10.1176/appi.ps.201200143. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Chou AD, Saung WT, Lai AT, Lohoff FW, Berrettini WH. Identification of CHRNA5 rare variants in African-American heavy smokers. Psychiatr Genet. 2014;24(3):102–109. doi: 10.1097/YPG.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health. 2002;26(3):214–218. [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, Schlaepfer I, Smolen A, Stallings MC, Young SE, Zeiger JS. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(5):596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, Cox NJ, Im HK. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, Zhao H, Farrer LA. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77(5):493–503. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, Farrer L, Kranzler HR. Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biol Psychiatry. 2007;61(1):119–126. doi: 10.1016/j.biopsych.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Gizer IR, Ehlers CL. Genome-wide association studies of substance use: considerations regarding populations and phenotypes. Biol Psychiatry. 2015;77(5):423–424. doi: 10.1016/j.biopsych.2014.11.013. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller G, Druley T, Vallania FL, Mitra RD, Li P, Akk G, Steinbach JH, Breslau N, Johnson E, Hatsukami D, Stitzel J, Bierut LJ, Goate AM. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum Mol Genet. 2012;21(3):647–655. doi: 10.1093/hmg/ddr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Reginsson GW, Gaddis NC, Chen X, Saccone NL, Lutz SM, Qaiser B, Sherva R, Steinberg S, Zink F, Stacey SN, Glasheen C, Chen J, Gu F, Frederiksen BN, Loukola A, Gudbjartsson DF, Bruske I, Landi MT, Bickeboller H, Madden P, Farrer L, Kaprio J, Kranzler HR, Gelernter J, Baker TB, Kraft P, Amos CI, Caporaso NE, Hokanson JE, Bierut LJ, Thorgeirsson TE, Johnson EO, Stefansson K. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. doi: 10.1038/tp.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida S, Katsura M, Torigoe F, Tsujimura A, Ohkuma S. Increased expression of L-type high voltage-gated calcium channel alpha1 and alpha2/delta subunits in mouse brain after chronic nicotine administration. Brain Res Mol Brain Res. 2005;135(1–2):280–284. doi: 10.1016/j.molbrainres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, Foroud T, Nurnberger J, Tischfield JA, Kuperman S, Crowe R, Hesselbrock V, Schuckit M, Almasy L, Porjesz B, Edenberg HJ, Begleiter H, Meyerhof W, Bierut LJ, Goate AM. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78(1):103–111. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10(5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, Kohli MA, Hennings JM, Horstmann S, Kloiber S, Menke A, Bondy B, Rupprecht R, Domschke K, Baune BT, Arolt V, Rush AJ, Holsboer F, Muller-Myhsok B. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009;66(9):966–975. doi: 10.1001/archgenpsychiatry.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29(2):299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- Kurumaji A, Ito T, Ishii S, Nishikawa T. Effects of FG7142 and immobilization stress on the gene expression in the neocortex of mice. Neurosci Res. 2008;62(3):155–159. doi: 10.1016/j.neures.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Kurumaji A, Nishikawa T. An anxiogenic drug, FG 7142, induced an increase in mRNA of Btg2 and Adamts1 in the hippocampus of adult mice. Behav Brain Funct. 2012;8:43. doi: 10.1186/1744-9081-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessov CN, Martin NG, Statham DJ, Todorov AA, Slutske WS, Bucholz KK, Heath AC, Madden PA. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34(5):865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Mangold JE, Seneviratne C, Chen GB, Ma JZ, Lou XY, Payne TJ. Association and interaction analyses of GABBR1 and GABBR2 with nicotine dependence in European- and African-American populations. PLoS One. 2009;4(9):e7055. doi: 10.1371/journal.pone.0007055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21(6):940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schafer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Volzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A, Mooser V, Francks C, Marchini J. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukola A, Wedenoja J, Keskitalo-Vuokko K, Broms U, Korhonen T, Ripatti S, Sarin AP, Pitkaniemi J, He L, Happola A, Heikkila K, Chou YL, Pergadia ML, Heath AC, Montgomery GW, Martin NG, Madden PA, Kaprio J. Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol Psychiatry. 2014;19(5):615–624. doi: 10.1038/mp.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jockel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyovalti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Magi R, Mannisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Muhleisen TW, Muller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njolstad I, Nothen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segre AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS, Shu XO, Shuldiner AR, Sigurdsson G, Sijbrands E, Silveira A, Sim X, Sivapalaratnam S, Small KS, So WY, Stancakova A, Stefansson K, Steinbach G, Steinthorsdottir V, Stirrups K, Strawbridge RJ, Stringham HM, Sun Q, Suo C, Syvanen AC, Takayanagi R, Takeuchi F, Tay WT, Teslovich TM, Thorand B, Thorleifsson G, Thorsteinsdottir U, Tikkanen E, Trakalo J, Tremoli E, Trip MD, Tsai FJ, Tuomi T, Tuomilehto J, Uitterlinden AG, Valladares-Salgado A, Vedantam S, Veglia F, Voight BF, Wang C, Wareham NJ, Wennauer R, Wickremasinghe AR, Wilsgaard T, Wilson JF, Wiltshire S, Winckler W, Wong TY, Wood AR, Wu JY, Wu Y, Yamamoto K, Yamauchi T, Yang M, Yengo L, Yokota M, Young R, Zabaneh D, Zhang F, Zhang R, Zheng W, Zimmet PZ, Altshuler D, Bowden DW, Cho YS, Cox NJ, Cruz M, Hanis CL, Kooner J, Lee JY, Seielstad M, Teo YY, Boehnke M, Parra EJ, Chambers JC, Tai ES, McCarthy MI, Morris AP. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R, Ikram MA, Ioannidis JP, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefansson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T, Singleton AB. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa D, Kasai S, Hasegawa J, Sato N, Yamada H, Tanioka F, Nagashima M, Katoh R, Satoh Y, Tagami M, Ujike H, Ozaki N, Inada T, Iwata N, Sora I, Iyo M, Yamada M, Kondo N, Won MJ, Naruse N, Uehara-Aoyama K, Itokawa M, Ohi K, Hashimoto R, Tanisawa K, Arai T, Mori S, Sawabe M, Naka-Mieno M, Yamada Y, Yamada M, Sato N, Muramatsu M, Tanaka M, Irukayama-Tomobe Y, Saito YC, Sakurai T, Hayashida M, Sugimura H, Ikeda K. Associations between the orexin (hypocretin) receptor 2 gene polymorphism Val308Ile and nicotine dependence in genome-wide and subsequent association studies. Mol Brain. 2015;8:50. doi: 10.1186/s13041-015-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Green KN, Liang K, Tran L, Chen Y, Leslie FM, LaFerla FM. Chronic nicotine administration exacerbates tau pathology in a transgenic model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2005;102(8):3046–3051. doi: 10.1073/pnas.0408500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olfson E, Saccone NL, Johnson EO, Chen LS, Culverhouse R, Doheny K, Foltz SM, Fox L, Gogarten SM, Hartz S, Hetrick K, Laurie CC, Marosy B, Amin N, Arnett D, Barr RG, Bartz TM, Bertelsen S, Borecki IB, Brown MR, Chasman DI, van Duijn CM, Feitosa MF, Fox ER, Franceschini N, Franco OH, Grove ML, Guo X, Hofman A, Kardia SL, Morrison AC, Musani SK, Psaty BM, Rao DC, Reiner AP, Rice K, Ridker PM, Rose LM, Schick UM, Schwander K, Uitterlinden AG, Vojinovic D, Wang JC, Ware EB, Wilson G, Yao J, Zhao W, Breslau N, Hatsukami D, Stitzel JA, Rice J, Goate A, Bierut LJ. Rare, low frequency and common coding variants in CHRNA5 and their contribution to nicotine dependence in European and African Americans. Mol Psychiatry. 2016;21(5):601–607. doi: 10.1038/mp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou OA, Ioannidis JP. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41(1):273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rice JP, Hartz SM, Agrawal A, Almasy L, Bennett S, Breslau N, Bucholz KK, Doheny KF, Edenberg HJ, Goate AM, Hesselbrock V, Howells WB, Johnson EO, Kramer J, Krueger RF, Kuperman S, Laurie C, Manolio TA, Neuman RJ, Nurnberger JI, Porjesz B, Pugh E, Ramos EM, Saccone N, Saccone S, Schuckit M, Bierut LJ. CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, St Laurent G, 3rd, Wahlestedt C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Front Genet. 2012;3:106. doi: 10.3389/fgene.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shand FL, Degenhardt L, Nelson EC, Mattick RP. Predictors of social anxiety in an opioid dependent sample and a control sample. J Anxiety Disord. 2010;24(1):49–54. doi: 10.1016/j.janxdis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms BA, Zamponi GW. Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron. 2014;82(1):24–45. doi: 10.1016/j.neuron.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Spijkerman AM, van der AD, Nilsson PM, Ardanaz E, Gavrila D, Agudo A, Arriola L, Balkau B, Beulens JW, Boeing H, de Lauzon-Guillain B, Fagherazzi G, Feskens EJ, Franks PW, Grioni S, Huerta JM, Kaaks R, Key TJ, Overvad K, Palli D, Panico S, Redondo ML, Rolandsson O, Roswall N, Sacerdote C, Sanchez MJ, Schulze MB, Slimani N, Teucher B, Tjonneland A, Tumino R, van der Schouw YT, Langenberg C, Sharp SJ, Forouhi NG, Riboli E, Wareham NJ. Smoking and long-term risk of type 2 diabetes: the EPIC-InterAct study in European populations. Diabetes Care. 2014;37(12):3164–3171. doi: 10.2337/dc14-1020. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res. 1999;2(1 Suppl):S51–57. doi: 10.1080/14622299050011811. discussion S69–70. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Casey G, Conti DV, Haile RW, Lewinger JP, Stram DO. Methodological Issues in Multistage Genome-wide Association Studies. Stat Sci. 2009;24(4):414–429. doi: 10.1214/09-sts288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Gudbjartsson DF, Stefansson K, Doyle GA, Chou AD, Saung WT, Lai AT, Lohoff FW, Berrettini WH. Identification of CHRNA5 rare variants in African-American heavy smokers. Mol Psychiatry. 2014;24(3):102–109. doi: 10.1097/YPG.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65(6):683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergult S, Dheedene A, Meurs A, Faes F, Isidor B, Janssens S, Gautier A, Le Caignec C, Menten B. Genomic aberrations of the CACNA2D1 gene in three patients with epilepsy and intellectual disability. Eur J Hum Genet. 2015;23(5):628–632. doi: 10.1038/ejhg.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten C, Seaton KL, Feiler HS, Wilhelmsen KC. The University of California, San Francisco Family Alcoholism Study. I. Design, methods, and demographics. Alcohol Clin Exp Res. 2004;28(10):1509–1516. doi: 10.1097/01.alc.0000142261.32980.64. [DOI] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35(4):397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Krauthammer M, Cosgrove KP, Oslin D, Anton RF, Farrer LA, Picciotto MR, Krystal JH, Zhao H, Gelernter J. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biol Psychiatry. 2011;70(6):528–536. doi: 10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang S, Yang Z, Hodgkinson CA, Iarikova P, Ma JZ, Payne TJ, Goldman D, Li MD. The contribution of rare and common variants in 30 genes to risk nicotine dependence. Mol Psychiatry. 2015;20(11):1467–1478. doi: 10.1038/mp.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46(2):100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamponi GW. Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat Rev Drug Discov. 2016;15(1):19–34. doi: 10.1038/nrd.2015.5. [DOI] [PubMed] [Google Scholar]
- Zuo L, Tan Y, Li CR, Wang Z, Wang K, Zhang X, Lin X, Chen X, Zhong C, Wang X, Wang J, Lu L, Luo X. Associations of rare nicotinic cholinergic receptor gene variants to nicotine and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2016;171(8):5. doi: 10.1002/ajmg.b.32476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res. 2013;37(5):730–739. doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


