Abstract
Despite its established inter-individual variability, sildenafil has been the subject of only a few pharmacogenetic investigations, with limited data regarding the genetic modulators of its pharmacokinetics. We conducted a pharmacogenetic substudy of patients randomized to sildenafil (n = 85) in the RELAX trial, which investigated the impact of high-dose sildenafil in patients with heart failure with preserved left ventricular ejection fraction (HFpEF). In the overall population, the CYP3A4 inferred phenotype appeared associated with the dose-adjusted peak concentrations of sildenafil at week 12 and week 24 (adjusted P=0.045 for repeated measures analysis), although this P value did not meet our corrected significance threshold of 0.0167. In the more homogeneous Caucasian subgroup, this association was significant (adjusted P=0.0165 for repeated measures). Hence, CYP3A4 inferred phenotype is associated with peak sildenafil dose-adjusted concentrations in patients with HFpEF receiving high doses of sildenafil. The clinical impact of this association requires further investigation.
Keywords: Sildenafil, pharmacokinetics, heart failure, pharmacogenetics
INTRODUCTION
The phosphodiesterase 5 inhibitor (PDE5) sildenafil (Viagra®) has been one of the most widely prescribed drugs in the world for nearly two decades. Despite the established inter-individual variability of its pharmacokinetics and efficacy,1–3 sildenafil has been the subject of only a few pharmacogenomic (PGx) investigations. Limited and inconsistent data from small to moderate size studies have suggested that genetic variants related to a variety of biological pathways could influence sildenafil’s hemodynamic effects in patients with pulmonary hypertension or its efficacy in treating erectile dysfunction.4–9
Considering that a majority of the markers judged to be clinically useful by the National Institutes of Health’s Clinical PGx Implementation Consortium are absorption, distribution, metabolism and elimination (ADME) genes,10 it is surprising that few studies have focused on variants of genes which could modulate sildenafil concentrations and dosing requirements. In particular, given that sildenafil is extensively metabolized in the liver by the cytochrome P450 3A (CYP3A) isoenzymes (major metabolizing route; 79%) and CYP2C9 (20%),11–13 genetic variants coding for these isoenzymes would biologically appear to be likely genetic modulators of the effects of sildenafil. Although small studies (n < 25) have investigated whether CYP3A5 and CYP2C9 were associated with sildenafil concentrations and pharmacokinetics,2,14 the small sample size from these studies limits their statistical power to identify significant associations.
The RELAX (Phosphodiesterase-5 Inhibition to Improve Clinical Status and Exercise Capacity in Heart Failure with Preserved Ejection Fraction) trial, which investigated the impact of high-dose sildenafil on the exercise capacity and clinical status of patients with heart failure with preserved left ventricular ejection fraction (HFpEF), represents a unique opportunity to further explore the genetic determinants of serum concentrations of sildenafil in a larger population than previous studies. Indeed, as part of RELAX, peak sildenafil concentrations were measured after twelve and twenty-four weeks of treatment.15 Because no beneficial hemodynamic or remodelling effects were observed in RELAX, the aim of this pharmacogenetic sub-study was to identify predictors of sildenafil peak concentrations. Based on existing evidence,2,14,16 the primary goal of this ancillary study was to investigate the impact of variants in the CYP2C9, CYP3A4 and CYP3A5 genes on dose-adjusted peak concentrations of sildenafil measured after 12 and 24 weeks of treatment. We hypothesized that patients carrying genotypes associated with a greater metabolizing capacity for these isoenzymes, such as extensive metabolizers (EM), would present lower dose-adjusted peak concentrations than carriers of genetic variations associated with a lower metabolizing capacity, such as intermediate metabolizers (IM) or poor metabolizers (PM).
METHODS
Overview of study design
The methods and results of the RELAX trial (clinicaltrials.gov Identifier: NCT00763867) have been reported previously.15,17 Briefly, RELAX was a multicenter randomized placebo-controlled trial which investigated the impact of high-dose sildenafil on exercise tolerance in patients with HFpEF (LVEF ≥ 50% in the last 12 months presenting New York Heart Association functional class II through IV) whose symptoms were stable while receiving medical therapy.15 The use of significant CYP3A4 inhibitors (e.g. ketoconazole, erythromycin), as well as a current or anticipated future need for nitrate therapy, were exclusion criteria.
Sildenafil was administered orally at 20 mg three times a day for 12 weeks. After 12 weeks, study endpoints were measured as previously described15,17 including peak sildenafil concentrations, which were obtained through phlebotomy 45 to 120 minutes after the scheduled dose. Following this, if the 20 mg three times a day dose was well tolerated, it was then increased to 60 mg three times a day for 12 weeks; otherwise the dose was maintained at 20 mg three times a day. Study endpoint measurements were repeated at week 24, including peak sildenafil concentrations. Sildenafil concentrations were measured by liquid chromatography tandem mass spectrometry (LC-MS/MS) as previously reported.15 Plasma cyclic GMP (cGMP) levels were measured as previously reported at week 24.15 For the current sub–study, we limited our investigations to the 85 patients in the sildenafil group who provided informed consent to participate in the genomic/pharmacogenomic sub-study and who provided a genetic sample.
Genetic analyses
The methods of the Heart Failure Network (HFN) genomics/pharmacogenomics sub-studies, including DNA extraction, genotyping and quality control have been previously reported.18 The genotyping strategy included multiple commercial and custom platforms. Among the commercial platforms, we used Sequenom’s iPLEX® ADME PGx Panel (Sequenom [now Agena Bioscience], San Diego, CA, USA) to genotype functional SNPs related to the absorption, distribution, metabolism and excretion (ADME). For the current report, we limited our investigations to the variants included on Sequenom’s iPLEX® ADME PGx Panel which contains 192 genetic variants, including 183 SNPs which passed genotyping quality control. Following additional data clean-up specific to the group of participants and SNPs of the current analysis (see supplementary information), 75 SNPs with a minor allele frequency (MAF) of ≥0.01 (from 32 genes; see supplementary information) were included.
Metabolizer status inference
Genetic variants genotyped on the Sequenom ADME panel were used to infer the metabolizer status of the primary genes of interest (CYP2C9, CYP3A4, CYP3A5) according to published and recognized scientific evidence in the field (see supplementary information). This approach has the advantage of combining the information of multiple SNPs from a given gene into one variable, thus reducing the number of statistical tests and the severity adjustment made to control for multiple comparisons. This approach was also used for other selected secondary genes (CYP2D6, CYP2C19, CYP2E1, CYP2B6, CYP2A6, CYP2C8, DPYD, SLCO1B1, UGT1A1, SUL1A1, NAT1; see supplementary information), although all secondary genes were also tested as individual SNPs, given that, with the exception of CYP2D6 and CYP2C19, more limited consensus data is available regarding the functional consequences of variants in these genes. Based on previous evidence, and depending on the gene, the metabolizer status was defined, for example, as poor metabolizer (PM), intermediate metabolizer (IM), extensive metabolizer (EM), or ultra metabolizer (UM).
Study objectives and endpoints
The primary objective of this RELAX pharmacogenetic sub-study was to test the association between the predicted phenotype for CYP2C9, CYP3A4 and CYP3A5 and the concentration:dose ratio of sildenafil measured at 12 and 24 weeks. Similar ratios have been previously used to take into account the impact of different doses on concentrations measured.19–22 Another important factor that motivated our selection of this phenotype as a primary outcome is that it was measured on 2 separate occasions in RELAX. Indeed, our group has used such repeated measures for analyses in previous pharmacogenomic studies21,23 to improve statistical power.
The main secondary objective was to evaluate the association between these three genes and concentrations of cGMP at week 24 in patients still receiving sildenafil. As part of the primary RELAX trial, sildenafil was shown to significantly increase cGMP concentrations compared to baseline, although this difference was not statistically significant compared with placebo.
Other secondary objectives included the evaluation of the association between these endpoints and other ADME genes included on the Sequenom iPlex ADME panel. Finally, as an exploratory analysis, we planned to investigate the relationship of variants significantly associated with the concentration: dose ratio of sildenafil with the occurrence of hypotension and flushing, which were more frequent in the sildenafil group than the placebo group in RELAX.
Statistical analysis
For continuous variables, descriptive statistics are presented as mean ± standard deviation, while categorical variables are presented as counts and percentages of each category. Prior to the statistical analyses, the normality test was performed on each endpoint using Shapiro-Wilk test at the 0.01 significance level. As this test appeared to be significant, the normality assumption was rejected and the data was transformed using the log10, square root, inverse and the square functions. The square root of sildenafil peak concentration, the log10 of concentration/dose ratio of sildenafil, and the log10 of cGMP were selected to be the best functions to be statistically analyzed. Data are presented as non-transformed.
Genetic association analyses
The concentration: dose ratio was statistically analyzed using the MIXED procedure of SAS, first including only non-genetic covariates into the analysis model. The repeating factor was the study visit. For cGMP, we used a GLM regression model that, again, was first fitted including non–genetic covariates into the analysis. For the risk of hypotension or flushing, a similar approach was utilized using a logistic regression. For all models, the non-genetic variables investigated were age, sex, body mass index, estimated glomerular filtration rate (eGFR) and amiodarone use. A univariate step selection was conducted between each endpoint and each covariate at 0.05 significance level. When more than one covariate was found to be significantly associated to the endpoint, a stepwise selection method was then performed between each endpoint and these clinical covariates with a P-value of less than 0.1 (entry P-value=0.1, stay P–value≤0.15).
The most important principal components (PC) of the population structure (components 1–3) were included in all genetic analyses to control for genetic ancestry and to avoid confounding by population structure,24 whether or not they were significant at the stepwise level. Thus, all the multivariate genetic models for each of the endpoints were adjusted for the non-genetic covariate meeting the aforementioned criteria and genetic ancestry (PC).
For genes, the inferred metabolizer status was defined, depending on the gene, as poor metabolizer (PM), intermediate metabolizer (IM), extensive metabolizer (EM), or ultra metabolizer (UM) and coded respectively as 0, 1, 2, and 3. For the individual SNP analyses, genotype was coded as 0, 1, or 2 according to the number of copies of minor alleles.
As sensitivity analyses, primary genes and any other genes or SNPs meeting our pre–defined statistical threshold were then analyzed in Caucasians only. Statistical analyses were performed using SAS, PLINK, and existing scripts.
Significance threshold
Statistical tests performed were two-sided and adjusted to account for the multiple testing of genes. For the three genes for the primary analysis (CYP2C9, CYP3A4 and CYP3A5) we used a Bonferroni-adjusted p value threshold of 0.05/3=0.0167. For all other variants, the significance threshold was adjusted using a Bonferroni correction based on the effective number of independent tests (Meff) of all candidate genes, including those of objective 1. The Meff was computed using the method of Gao et al25 and was set at 49. Then, the adjusted significance threshold was calculated as 0.05/49 = 0.001.
Ethical consideration
The RELAX study was approved by all local Institutional Review Boards (IRBs) and all patients provided a written consent to participate in the study. Moreover, in centers electing to participate in the genomics/pharmacogenomics sub-study of the HFN clinical trials, the sub-study was approved by the IRB at each side. Patients taking part in the RELAX trial in these participating centers were offered the possibility to participate in the genetics sub-study. All participants willing to participate in the study provided written informed consent.
RESULTS
A total of 85 patients were included as part of this sub-study (Table 1). Study subjects were more likely to be males (54%), Caucasian (92%) and were 67.9±10.5 years old. The predicted phenotypes and related genotypes of the study participants for CYP2C9, CYP3A4, CYP3A5 are detailed in Table 2.
Table 1.
Baseline characteristics of the study population
| Characteristic | Patients treated with sildenafil (n = 85) |
|---|---|
| Female, n (%) | 39 (45.9) |
| Age, years | 67.9 ± 10.5 |
| Race, n (%) | |
| White | 78 (91.8) |
| Black | 4 (4.7) |
| Other | 3 (3.5) |
| Diabetes, n (%) | 32 (37.6) |
| Blood pressure, mmHg | 126.7 ± 16.6/70.9 ± 10.0 |
| Heart rate, bpm | 69.9 ± 12.2 |
| Baseline LVEF, % | 61.6 ± 6.2 |
| NYHA class, II/III | 44 (51.8)/ 41 (48.2) |
| BMI, kg/m2 | 33.9 ± 7.3 |
| Creatinine, mg/dL | 1.2 ± 0.5 |
| eGFR, ml/min/1.73 m2 | 67.3 ± 23.1 |
| NT proBNP, pg/mL | 1012.5 ± 1212.4 |
| cGMP, pmol/mL | 82.5 ± 36.0 |
| Treatment | |
| ACE inhibitor, n (%) | 34 (40.0) |
| Angiotensin receptor blocker, n (%) | 19 (22.4) |
| Beta blocker use, n (%) | 62 (72.9) |
| Aldosterone antagonist, n (%) | 11 (12.9) |
| Hydrochlorothiazide, n (%) | 11(12.9) |
| Metolazone, n (%) | 2 (2.4) |
| Loop diuretics, n (%) | |
| Furosemide | 52 (61.2) |
| Torsemide | 7 (8.2) |
| Bumetanide | 4 (4.7) |
| Amiodarone, n (%) | 7 (8.2) |
| Calcium channel blocker, n (%) | 20 (23.5) |
Abbreviations: ACE, angiotensin-converting enzyme inhibitors; BMI, Body Mass Index; cGMP, Cyclic guanosine monophosphate; eGFR, estimated glomerular filtration rate; LVEF, left ventricular ejection fraction; NT-proBNP, N-terminal pro-B-type natriuretic peptide.
Table 2.
Inferred phenotypes and related genotypes in the overall population and in the Caucasian subgroup
| Gene | Phenotype | Genotype | Frequency, n (%) |
|---|---|---|---|
|
All patients
| |||
| CYP2C9 | EM | *1/*1 | 52 (62.7) |
| *1/*9 | 2 (2.4) | ||
| IM | *1/*2 | 18 (21.7) | |
| *1/*3 | 7 (8.4) | ||
| PM | *2/*2 | 1 (1.2) | |
| *2/*3 | 3 (3.6) | ||
| CYP3A4 | EM | *1/*1 | 72 (85.7) |
| IM | *1/*22 | 12 (14.3) | |
| CYP3A5 | EM | *1/*1 | 1 (1.2) |
| IM | *1/*3 | 7 (8.2) | |
| PM | *3/*3 | 74 (87.1) | |
| *3/*6 | 2 (2.4) | ||
| *3/*7 | 1 (1.2) | ||
| Caucasians | |||
| CYP2C9 | EM | *1/*1 | 48 (63.2) |
| IM | *1/*2 | 18 (23.7) | |
| *1/*3 | 6 (7.9) | ||
| PM | *2/*2 | 1 (1.3) | |
| *2/*3 | 3 (3.9) | ||
| CYP3A4 | EM | *1/*1 | 66 (85.7) |
| IM | *1/*22 | 11 (14.3) | |
| CYP3A5 | IM | *1/*3 | 6 (7.7) |
| PM | *3/*3 | 72 (92.3) | |
Abbreviations: EM, extensive metabolisers; IM, intermediate metabolizers; PM, poor metabolizers. There were 2 failed samples for CYP2C9, 1 for CYP3A4.
Predictors of dose-concentration ratio of sildenafil
Of the 85 patients enrolled in this sub-study, 77 individuals had at least one sildenafil concentration measured (week 12: n = 71; week 24: n = 66). In univariate analysis, both age and BMI were significantly associated with concentration: dose ratios of sildenafil (all P < 0.0001).
In regards to the primary genes of interest, in the overall population, consistent with their predicted lower metabolising capacity, the IM for the CYP3A4 inferred phenotype presented a higher ratio of sildenafil than EM at both week 12 and week 24 (Figure 1, P = 0.045 for repeated measures analysis, after adjusting for age, BMI and three PCs for genetic ancestry). All IM were CYP3A4*22 carriers. Although this association p value was below the nominal threshold of 0.05, it did not meet our corrected significance threshold of 0.0167. Nevertheless, in the more homogeneous Caucasian subgroup, this association was found to be significant after adjusting for multiple comparisons (P = 0.0165 for repeated measures analysis, after adjusting for age, BMI and three PCs for genetic ancestry [Figure 1]).
Figure 1. Concentration:dose ratios of sildenafil according to CYP3A4 inferred phenotype in (A) all patients and (B) Caucasians.
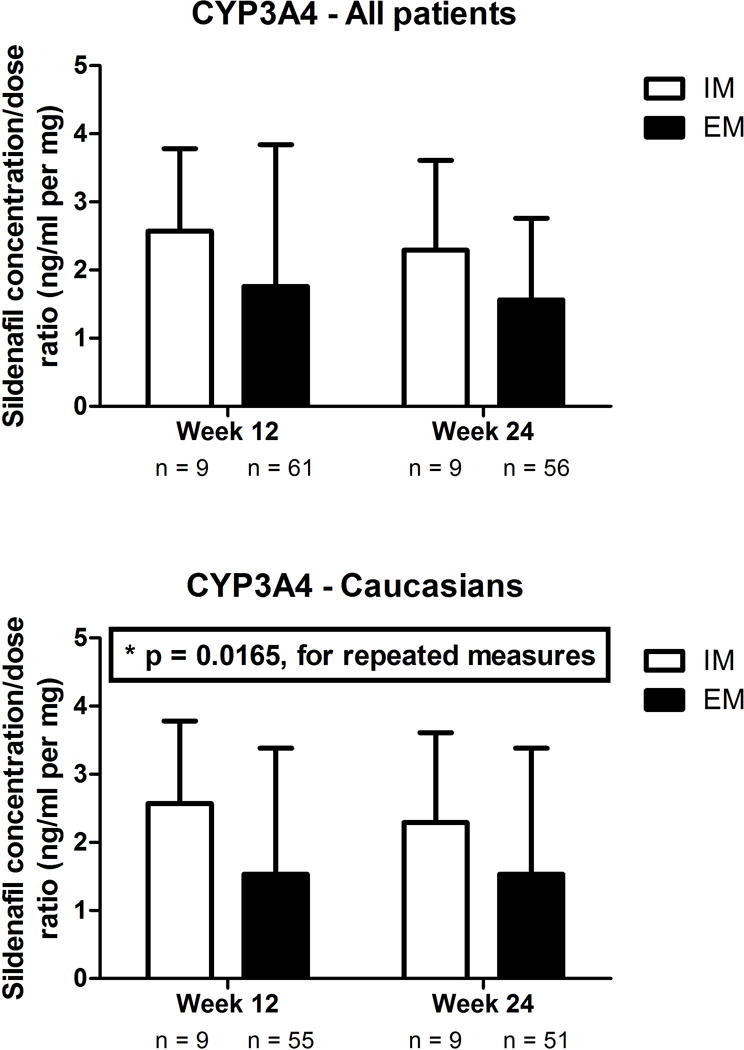
After adjusting for age, body mass index and three principal components for genetic ancestry, CYP3A4 inferred phenotype was significantly associated with the sildenafil concentration:dose ratio in Caucasians (P = 0.0165 for repeated measures analysis). EM, extensive metabolisers; IM, intermediate metabolizers.
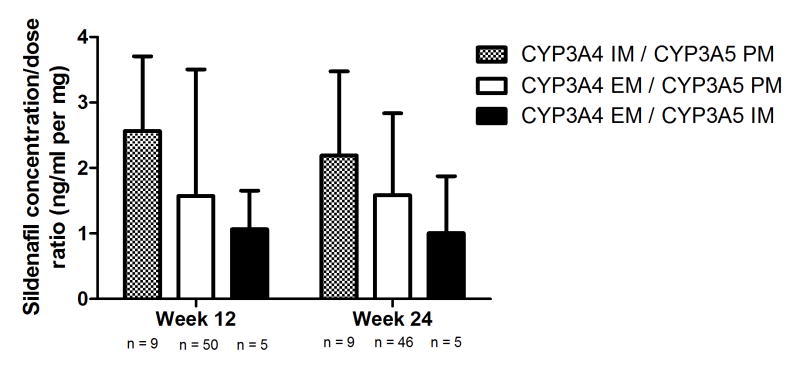
No association was observed between the CYP2C9 inferred phenotype and the sildenafil concentration:dose ratio (Figure 2). In regards to the CYP3A5 inferred phenotype, although PM tended to have higher concentrations, particularly in the Caucasian subgroup, P values did not reach our significance corrected threshold in the overall population nor in the specific Caucasian subgroup (all P values > 0.14, Figure 3). In an exploratory analysis, we grouped Caucasian patients based on their inferred CYP3A4 and CYP3A5 phenotypes. We observed that increasing levels of predicted metabolizing capacity appeared to be associated with decreasing levels of dose-adjusted sildenafil concentrations (F test P = 0.0238; Figure 4). None of the pairwise comparisons between specific groups reached the pre-established threshold.
Figure 2. Concentration:dose ratios of sildenafil according to CYP2C9 inferred phenotype in (A) all patients and (B) Caucasians.
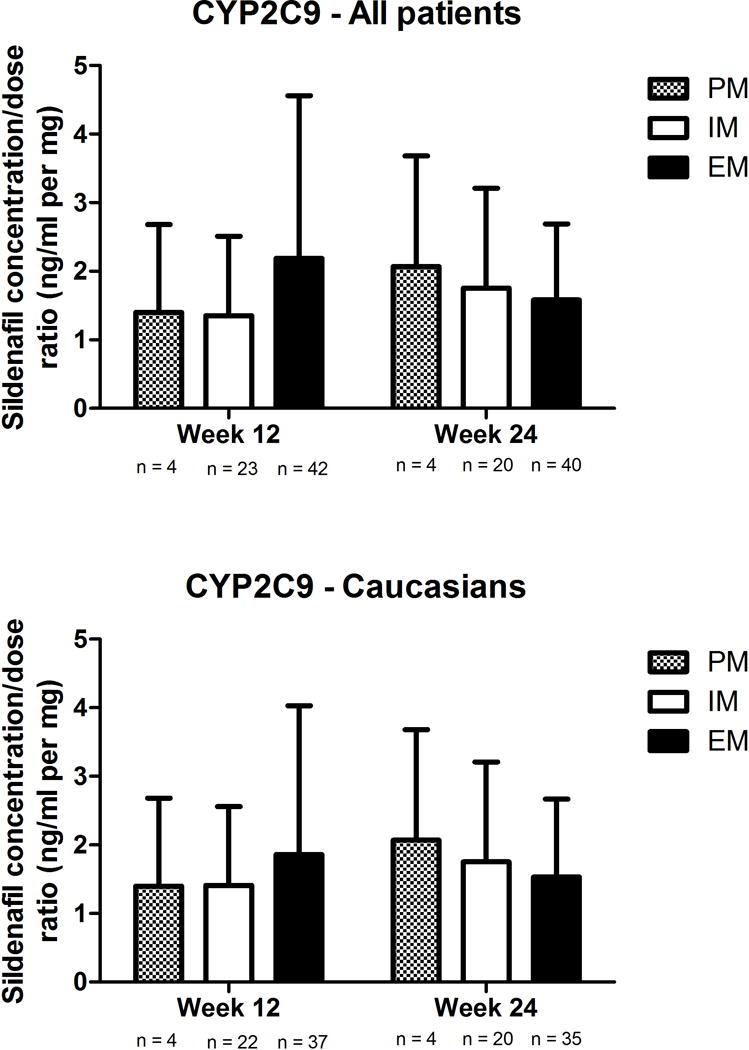
EM, extensive metabolisers; IM, intermediate metabolizers. Data presented as mean ± standard deviation.
Figure 3. Concentration:dose ratios of sildenafil according to CYP3A5 inferred phenotype in (A) all patients and (B) Caucasians.
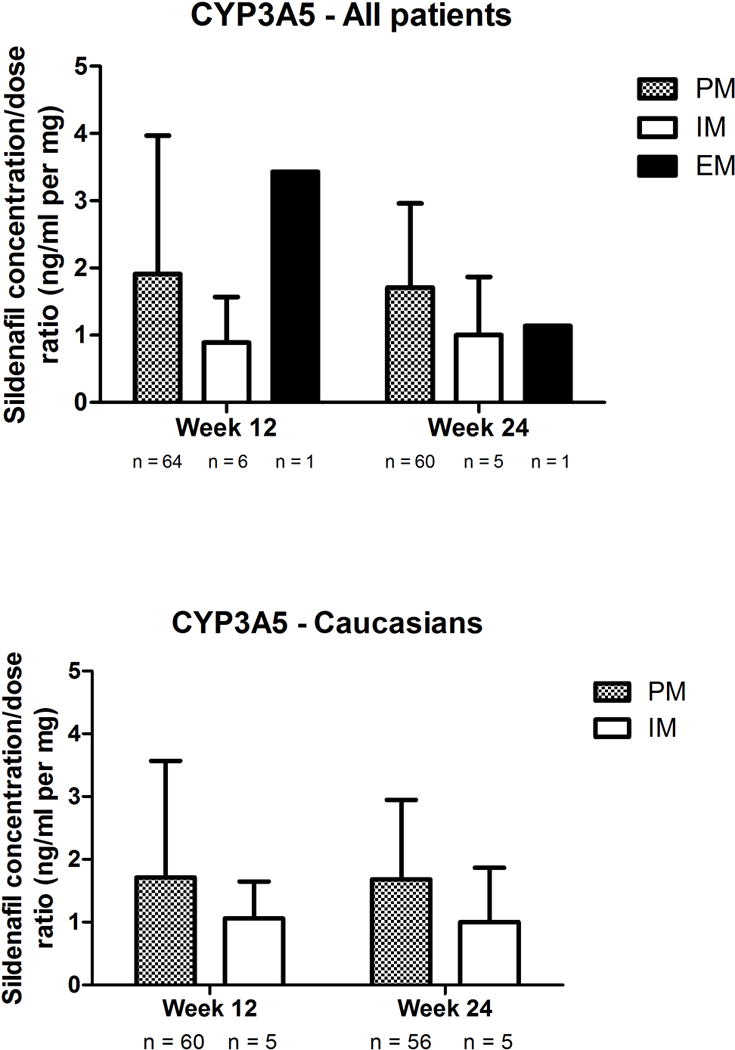
EM, extensive metabolisers; IM, intermediate metabolizers; PM, poor metabolizers. Data presented as mean ± standard deviation.
Figure 4. Concentration:dose ratios of sildenafil according to CYP3A4 and CYP3A5 inferred phenotype in Caucasians.
Increasing levels of predicted metabolizing capacity showed a trend with decreasing levels of dose-adjusted sildenafil concentrations (F test P = 0.0238). EM, extensive metabolisers; IM, intermediate metabolizers; PM, poor metabolizers. Data presented as mean ± standard deviation.
Cyclic GMP
After adjusting for age, BMI, baseline cGMP and three PC of genetic ancestry, none of the primary genes were associated with cGMP at week 24.
Secondary genes
None of the secondary genetic variants or inferred phenotypes reached our pre-established threshold after multiple comparisons (P = 0.001).
Vascular adverse effects
Given the association between the CYP3A4 inferred phenotype and the dose-adjusted concentrations of sildenafil, we explored the association of this genotype with the risk of hypotension and flushing. We observed no significant association, neither in the overall population (IM: 25% vs EM: 17%; P = 0.15), nor with the Caucasian subgroup (IM: 27% vs EM: 18%; P = 0.82).
DISCUSSION
In the current study, we investigated the association between three candidate drug metabolism genes and peak dose-adjusted concentrations of sildenafil in HFpEF patients receiving sildenafil. To the best of our knowledge, this pharmacogenetic sub-study is the largest to have investigated the association between candidate genes and concentrations of sildenafil. Furthermore, we believe we are reporting for the first time that the inferred CYP3A4 phenotype, which in this population corresponded to carriers of the CYP3A4*22 variant, may be associated with dose–adjusted peak concentrations of sildenafil. While this association was only numerically apparent in our entire population, it reached our predetermined significance threshold in the Caucasian subgroup. Although we saw numerically higher dose-adjusted concentrations in CYP3A5 PM, particularly in Caucasians, our results did not reach statistical significance. Moreover we found no indication that genetic variations in the CYP2C9-coding gene (CYP2C9), a minor elimination pathway of sildenafil, influenced sildenafil peak concentrations.
CYP3A4 is the most abundant CYP in the liver.26 It contributes to the metabolism of approximately 30% of all drugs, including sildenafil,13,26,27 through oxidative biotransformation.26 Other substrates of CYP3A4 include the immunosuppressant drug tacrolimus, the lipid-lowering drug atorvastatin and endoxifen, the principal active metabolite of the anti-neoplastic agent tamoxifen.27,28 Existing data suggest that sildenafil undergoes significant first-pass metabolism in the liver and that CYP3A4 is the isoenzyme responsible for a majority of this metabolism,12,13 although CYP3A5 may also contribute to the metabolism of sildenafil by the CYP3A subfamily.16 CYP3A4 is also expressed in the gut wall and this pre-systemic metabolism is also thought to limit the bioavailability (40%) of sildenafil.13
The CYP3A4-coding gene, CYP3A4, is located in a cluster on chromosome 7 along with CYP3A5, CYP3A7, and CYP3A4326,27 Few genetic variants in CYP3A4 have been shown to modulate CYP3A4 activity. The intronic variant rs35599367 (C>T), which is located in intron 6 and which was allocated the *22 allele name by the Human Cytochrome P450 (CYP) Allele Nomenclature Database, is, to our knowledge, the only “common” (MAF > 0.01) SNP convincingly shown to influence CYP3A4 expression.27,29 Existing data suggest that the hepatic CYP3A4 mRNA level and enzyme activity are respectively 1.7- and 2.5-fold greater in individuals with the CC genotype than in carriers of the CT and TT carriers.29 CYP3A4*22’s MAF is reported to be of approximately 4% in Asians and African-Americans and 8% in Caucasians.27 The frequency in our Caucasian subgroup (7%) is thus consistent with existing data. Our observation that CYP3A4*22 carriers present high dose-adjusted concentration of sildenafil is consistent with other reports showing that CYP3A4*22 carriers present higher concentrations of CYP3A4 substrates, including atorvastatin and tacrolimus.30–32
Despite the fact that we observed a significant association between CYP3A4 and sildenafil concentrations, this did not translate into an association with its downstream biomarker cGMP. The hemodynamic effect of sildenafil and other PDE5 inhibitors in the treatment of erectile dysfunction and pulmonary arterial hypertension is secondary to the inhibition of cGMP degradation by PDE5, which leads to smooth muscle relaxation.11,13 Although this lack of association may appear surprising, it is consistent with the findings from the RELAX trial where sildenafil increased cGMP when compared to baseline but was not statistically significant compared to placebo.15 Moreover, the correlation between sildenafil peak concentrations and cGMP was weak in RELAX, perhaps reflecting that multiple other PDE are implicated in the metabolism of cGMP.15 Nevertheless, this lack of association underlines the uncertainty as to whether the observations in the current and other reports30–32 with CYPA4*22 can be extended to the clinical response to drugs.
Given the high degree of similarity in DNA sequence between CYP3A4 and CYP3A5, the known substrate overlap which exist between these isoenzymes,26 and existing data regarding the implication of CYP3A5 in the metabolism of sildenafil,16 we hypothesised that CYP3A5 could also influence the dose-adjusted peak sildenafil concentrations. In a similar fashion to a previous report in 21 healthy Korean males, we observed higher peak dose-adjusted concentrations in Caucasians participants who were CYP3A5 PM in the present study.2 Nevertheless, as with that report, these trends were not statistically significant, which could be the result of our small sample size (see supplementary material).28 Whether combining information from CYP3A4 and CYP3A5 further allows do decipher inter-individual differences in sildenafil’s pharmacokinetics requires additional investigation. If our preliminary observations are confirmed, given the apparent 2.5 fold difference in dose-adjusted concentrations between patients presenting with a CYP3A4 IM / CYP3A5 PM profile and those with a CYP3A4 EM / CYP3A5 IM profile, genotyping could be useful to personalise sildenafil dosing in the clinic.
We did not observe any association between sildenafil concentrations and the CYP2C9 inferred phenotype, a minor route of sildenafil metabolism (20%).11,12 This is consistent with a prior study of 23 healthy males.14 Given the limited metabolism of sildenafil by CYP2C9 but the know functional impact of CYP2C9 variants on the pharmacokinetics of multiple drugs, including warfarin,33 we cannot exclude the possibility that CYP2C9 could exert a modest impact on the pharmacokinetics of sildenafil. Demonstrating such an association would likely necessitate a much larger sample size than the one of the current investigation.
Limitations
Our results should be interpreted while taking into consideration some inherent study limitations. First, our results are issued from the RELAX clinical trial which relied on three times per day dosing of sildenafil. The generalizability of our results to dosing schemes commonly used to treat erectile dysfunction requires further investigation.13 Nevertheless, the RELAX dosing regimen is similar to those investigated in pulmonary artery hypertension11 and thus, the potential clinical consequences of our findings may be of particular interest to investigate in these patients. Second, there was no measure or control for the types of food that may have been taken with sildenafil. Ingesting sildenafil with a high-fat meal is known to reduce its absorption rate and peak concentration,12 while foods that can modify CYP3A4 activity such as grapefruit or pomelo juice could increase concentrations.11,12 Despite this, and the inherent heterogeneity in multicenter studies, we were able to identify a novel genetic association between the predicted CYP3A4 phenotype, ultimately CYP3A4*22, and sildenafil dose-adjusted peak concentrations, the association would be expected to be even stronger in a study controlling for nutritional modifiers of the effect. Third, one could argue that because CYP2C9 inhibitors were allowed as part of the study, this could potentially have contributed to our inability to detect genetic association with CYP2C9 variants. As part of RELAX, restriction on CYP2C9 inhibitors were not required given evidence suggesting that CYP2C9 inhibitors exert no clinically significant impact on the pharmacokinetics of sildenafil.11,12 Consistent with this premise, the use of amiodarone, a known CYP2C9 inhibitor,26 was not associated with our primary endpoint. A fourth limitation stems from the single concentration measure available at each visit, which limited our ability to conduct a more detailed pharmacokinetic study of sildenafil to assess other pharmacokinetic parameters of interest such as bioavailability, the elimination half-life or the area under the curve are influenced by CYP3A4. Fifth, despite constituting the largest pharmacogenetic study investigating the pharmacokinetics of sildenafil, our sample size was not sufficiently large to identify variants or inferred phenotypes with a more modest effect. Finally, it should be noted that there is a possibility that some patients may have carried rarer genetic variants that were not tested as part of our assay which could have contributed to misclassification of their metabolizer status. This may even be more likely for non-Caucasian patients.
In conclusion, we observed that the CYP3A4 inferred phenotype, which reflected the CYP3A4*22 carrier status, was associated with peak sildenafil dose adjusted concentrations in patients with HFpEF receiving high doses of sildenafil. Whether these results can be replicated in other populations for which sildenafil is currently indicated and influence its clinical response requires further investigation. Should this observation be validated, genotype-guided prescription of sildenafil in clinical practice or clinical trials could lead to a more predictable clinical response from this agent.
Supplementary Material
Acknowledgments
Funding: Supported by grant HL084904 from the National Heart, Lung, and Blood Institute.
Footnotes
Conflict of Interest
Simon de Denus has received research grants or been a coinvestigator on grants supported by AstraZeneca, Novartis, Roche and Pfizer. He has received speaker fees from Pfizer and consulting fees from Servier and Novartis. Jean Lucien Rouleau is a consultant for Novartis. Marie-Pierre Dubé has been a coinvestigator on grants supported by AstraZeneca and Pfizer, has received honoraria and research contracts from Roche and is a member of the executive committee of the Dal-GenE trial sponsored by DalCor. A patent was submitted on pharmacogenomic determinants of responses to cardiovascular drugs and Dr Dubé is listed as inventor.
Author’s Contributions
S de Denus designed the substudy, interpreted the results and wrote the first draft of the manuscript.
MP Dubé designed the substudy, analyzed the data and wrote of parts the manuscript
J Rouleau, DL. Mann, GS. Huggins, TP. Cappola, S Shah, NL Pereira designed the substudy and contributed to critical review and writing of the manuscript.
R Fouodjio analyzed the data and wrote of parts the manuscript.
I Mongrain generated genetic data and wrote parts of the manuscripts.
Supplementary information is available at The Pharmacogenomics Journal’s website.
References
- 1.Kanjanawart S, Gaysonsiri D, Tangsucharit P, Vannaprasaht S, Phunikhom K, Kaewkamson T, et al. Comparative bioavailability of two sildenafil tablet formulations after single-dose administration in healthy Thai male volunteers. Int J Clin Pharmacol Ther. 2011;49:525–530. doi: 10.5414/cp201496. [DOI] [PubMed] [Google Scholar]
- 2.Shon JH, Ku HY, Bae SY, Oh MK, Yeo CW, Bae SK, et al. The disposition of three phosphodiesterase type 5 inhibitors, vardenafil, sildenafil, and udenafil, is differently influenced by the CYP3A5 genotype. Pharmacogenet Genomics. 2011;21:820–828. doi: 10.1097/FPC.0b013e32834b79e6. [DOI] [PubMed] [Google Scholar]
- 3.Benard F, Carrier S, Lee JC, Talwar V, Defoy I. Men with mild erectile dysfunction benefit from sildenafil treatment. J Sex Med. 2010;7:3725–3735. doi: 10.1111/j.1743-6109.2010.02015.x. [DOI] [PubMed] [Google Scholar]
- 4.Muniz JJ, Lacchini R, Rinaldi TO, Nobre YT, Cologna AJ, Martins AC, et al. Endothelial nitric oxide synthase genotypes and haplotypes modify the responses to sildenafil in patients with erectile dysfunction. Pharmacogenomics J. 2013;13:189–196. doi: 10.1038/tpj.2011.49. [DOI] [PubMed] [Google Scholar]
- 5.Eisenhardt A, Sperling H, Hauck E, Porst H, Stief C, Rubben H, et al. ACE gene I/D and NOS3 G894T polymorphisms and response to sildenafil in men with erectile dysfunction. Urology. 2003;62:152–157. doi: 10.1016/s0090-4295(03)00137-7. [DOI] [PubMed] [Google Scholar]
- 6.Peskircioglu L, Atac FB, Erdem SR, Deveci S, Verdi H, Ozkardes H. The association between intron 4 VNTR, E298A and IVF 23+10 G/T polymorphisms of ecNOS gene and sildenafil responsiveness in patients with erectile dysfunction. Int J Impot Res. 2007;19:149–153. doi: 10.1038/sj.ijir.3901501. [DOI] [PubMed] [Google Scholar]
- 7.Lacchini R, Muniz JJ, Nobre YT, Cologna AJ, Martins AC, Tanus-Santos JE. VEGF genetic polymorphisms affect the responsiveness to sildenafil in clinical and postoperative erectile dysfunction. Pharmacogenomics J. 2013;13:437–442. doi: 10.1038/tpj.2012.39. [DOI] [PubMed] [Google Scholar]
- 8.Sekine A, Tanabe N, Sugiura T, Shigeta A, Jujo T, Nishimura R, et al. Polymorphism of the G Protein beta3 Subunit Gene Influences the Efficacy of Sildenafil in Patients with Pulmonary Hypertension. Int Med (Tokyo, Japan) 2014;53:291–297. doi: 10.2169/internalmedicine.53.0658. [DOI] [PubMed] [Google Scholar]
- 9.Sperling H, Eisenhardt A, Virchow S, Hauck E, Lenk S, Porst H, et al. Sildenafil response is influenced by the G protein beta 3 subunit GNB3 C825T polymorphism: a pilot study. J Urol. 2003;169:1048–1051. doi: 10.1097/01.ju.0000058369.72348.ba. [DOI] [PubMed] [Google Scholar]
- 10.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaumais MC, Perrin S, Sitbon O, Simonneau G, Humbert M, Montani D. Pharmacokinetic evaluation of sildenafil as a pulmonary hypertension treatment. Expert Opinion Drug Metabolism Toxicol. 2013;9:1193–1205. doi: 10.1517/17425255.2013.804063. [DOI] [PubMed] [Google Scholar]
- 12.Gupta M, Kovar A, Meibohm B. The clinical pharmacokinetics of phosphodiesterase-5 inhibitors for erectile dysfunction. J Clin Pharmacol. 2005;45:987–1003. doi: 10.1177/0091270005276847. [DOI] [PubMed] [Google Scholar]
- 13.Dorsey P, Keel C, Klavens M, Hellstrom WJ. Phosphodiesterase type 5 (PDE5) inhibitors for the treatment of erectile dysfunction. Expert Opinion Pharmacotherapy. 2010;11:1109–1122. doi: 10.1517/14656561003698131. [DOI] [PubMed] [Google Scholar]
- 14.Jetter A, Lazar A, Schomig E, Fuhr U, Kinzig-Schippers M, Sorgel F. The CYP2C9 genotype does not influence sildenafil pharmacokinetics in healthy volunteers. Clin Pharmacol Ther. 2005;78:441–443. doi: 10.1016/j.clpt.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Redfield MM, Chen HH, Borlaug BA, Semigran MJ, Lee KL, Lewis G, et al. Effect of phosphodiesterase-5 inhibition on exercise capacity and clinical status in heart failure with preserved ejection fraction: a randomized clinical trial. JAMA. 2013;309:1268–1277. doi: 10.1001/jama.2013.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ku HY, Ahn HJ, Seo KA, Kim H, Oh M, Bae SK, et al. The contributions of cytochromes P450 3A4 and 3A5 to the metabolism of the phosphodiesterase type 5 inhibitors sildenafil, udenafil, and vardenafil. Drug Metabolism Disposition: Biological Fate Chemicals. 2008;36:986–990. doi: 10.1124/dmd.107.020099. [DOI] [PubMed] [Google Scholar]
- 17.Redfield MM, Borlaug BA, Lewis GD, Mohammed SF, Semigran MJ, Lewinter MM, et al. PhosphdiesteRasE-5 Inhibition to Improve CLinical Status and EXercise Capacity in Diastolic Heart Failure (RELAX) trial: rationale and design. Circ Heart Fail. 2012;5:653–659. doi: 10.1161/CIRCHEARTFAILURE.112.969071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Denus S, Rouleau JL, Mann DL, Huggins GS, Cappola TP, Shah SH, et al. A pharmacogenetic investigation of intravenous furosemide in decompensated heart failure: a meta-analysis of three clinical trials. Pharmacogenomics J. 2016 Mar 1; doi: 10.1038/tpj.2016.4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Zeevi A, Schuetz E, Lamba J, McCurry K, Griffith BP, et al. Tacrolimus dosing in adult lung transplant patients is related to cytochrome P4503A5 gene polymorphism. J Clin Pharmacol. 2004;44:135–140. doi: 10.1177/0091270003262108. [DOI] [PubMed] [Google Scholar]
- 20.Wehland M, Bauer S, Brakemeier S, Burgwinkel P, Glander P, Kreutz R, et al. Differential impact of the CYP3A5*1 and CYP3A5*3 alleles on pre-dose concentrations of two tacrolimus formulations. Pharmacogenet Genomics. 2011;21:179–184. doi: 10.1097/FPC.0b013e32833ea085. [DOI] [PubMed] [Google Scholar]
- 21.de Denus S, Zakrzewski M, Barhdadi A, Leblanc MH, Racine N, Belanger F, et al. Association between renal function and CYP3A5 genotype in heart transplant recipients treated with calcineurin inhibitors. J Heart Lung Transplant. 2011;30:326–331. doi: 10.1016/j.healun.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 22.de Denus S, Zakrzewski M, Barhdadi A, Leblanc MH, Racine N, Belanger F, et al. CYP3A5*1/*3 genetic polymorphism is associated with post cardiac transplant renal dysfunction in patients treated with calcineurin inhibitors. J Cardiac Fail. 2008;14:S3–S3. [Google Scholar]
- 23.Lachance K, Barhdadi A, Mongrain I, Normand V, Zakrzewski M, Leblanc MH, et al. PRKCB is associated with calcineurin inhibitor-induced renal dysfunction in heart transplant recipients. Pharmacogenet Genomics. 2012;5:336–343. doi: 10.1097/FPC.0b013e3283510a35. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Gen. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 26.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Werk AN, Cascorbi I. Functional Gene Variants of CYP3A4. Clin Pharmacol Ther. 2014;96:340–348. doi: 10.1038/clpt.2014.129. [DOI] [PubMed] [Google Scholar]
- 28.Binkhorst L, Mathijssen RHJ, Jager A, van Gelder T. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treatment Rev. 2015;41:289–299. doi: 10.1016/j.ctrv.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11:274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein K, Thomas M, Winter S, Nussler AK, Niemi M, Schwab M, et al. PPARA: a novel genetic determinant of CYP3A4 in vitro and in vivo. Clin Pharmacol Ther. 2012;91:1044–1052. doi: 10.1038/clpt.2011.336. [DOI] [PubMed] [Google Scholar]
- 31.Pallet N, Jannot AS, El Bahri M, Etienne I, Buchler M, de Ligny BH, et al. Kidney transplant recipients carrying the CYP3A4*22 allelic variant have reduced tacrolimus clearance and often reach supratherapeutic tacrolimus concentrations. Am J Transplant. 2015;15:800–805. doi: 10.1111/ajt.13059. [DOI] [PubMed] [Google Scholar]
- 32.Elens L, Capron A, van Schaik RH, De Meyer M, De Pauw L, Eddour DC, et al. Impact of CYP3A4*22 allele on tacrolimus pharmacokinetics in early period after renal transplantation: toward updated genotype-based dosage guidelines. Ther Drug Monit. 2013;35:608–616. doi: 10.1097/FTD.0b013e318296045b. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.