Abstract
The family Circoviridae comprises viruses with small, circular, single-stranded DNA (ssDNA) genomes, including the smallest known animal viruses. Members of this family are classified into two genera, Circovirus and Cyclovirus, which are distinguished by the position of the origin of replication relative to the coding regions and the length of the intergenic regions. Within each genus, the species demarcation threshold is 80 % genome-wide nucleotide sequence identity. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the Circoviridae, which is available at www.ictv.global/report/circoviridae.
Keywords: Circoviridae, Circovirus, Cyclovirus, ICTV report, taxonomy
Abbreviations
ORF, open reading frame; Rep, replication associated protein; Cp, capsid protein; ori, origin of replication; RCR, rolling circle replication.
Virion
Virions, which have only been visualized for a few members of the genus Circovirus, are non-enveloped and have an icosahedral T=1 symmetry with a diameter of 15–25 nm [1–3] (Table 1, Fig. 1). Members of the genus Cyclovirus have only been described through sequence-based analyses and no structural data are available.
Table 1. Characteristics of the family Circoviridae.
| Typical member: | porcine circovirus 1 (AF071879), species Porcine circovirus 1, genus Circovirus |
|---|---|
| Virion | Non-enveloped, icosahedral T=1 symmetry, 15–25 nm diameter |
| Genome | Monopartite, circular, single-stranded DNA of 1.7–2.1 kb |
| Replication | Rolling circle replication |
| Translation | From at least two mRNAs encoding the replication-associated and capsid proteins |
| Host Range | Circovirus: mammals, birds and fish; Cyclovirus: unconfirmed for most species |
| Taxonomy | More than 70 species in the genera Circovirus and Cyclovirus |
Fig. 1.
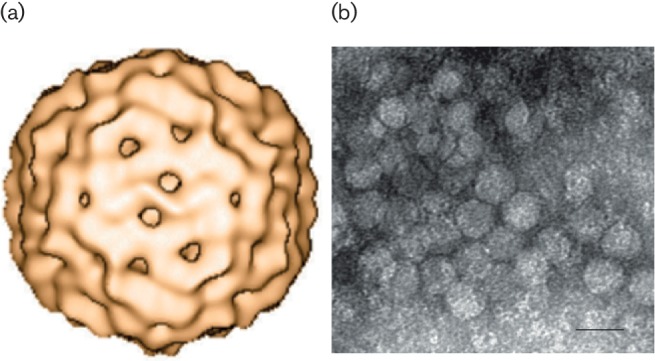
(a) 3D reconstruction of porcine circovirus 2 using cryo-electron microscopy. A structural model comprising 60 subunits (T=1) arranged in 12 pentameric morphological units has been proposed [1]. (b) Negative-stained transmission electron micrograph of porcine circovirus 2 (provided by Carolina Rodríguez-Cariño and Joaquim Segalés, CReSA, Spain). Scale bar=20 nm.
Genome
Both genera include viruses with small, covalently closed, circular ssDNA genomes. Their genomes range in size from 1.7 to 2.1 kb and contain two major (>600 nt) open reading frames (ORFs), which encode the replication-associated (Rep) and capsid (Cp) proteins. Members of the genera Circovirus and Cyclovirus are distinguished by the location of the origin of replication (ori) relative to the coding regions, and the length of the intergenic regions (Fig. 2) [4]. Members of the genus Circovirus have the ori on the same strand as the rep ORF, whereas members of the genus Cyclovirus have the putative ori on the same strand as the cp ORF [5]. Circovirus genomes are characterized by two intergenic regions between the major ORFs; however, the intergenic region between the 3′ ends of the major ORFs in cyclovirus genomes is either absent or consistently smaller [6]. In addition, introns have been identified within the ORFs of several cyclovirus genomes, while none have been observed for members of the genus Circovirus.
Fig. 2.
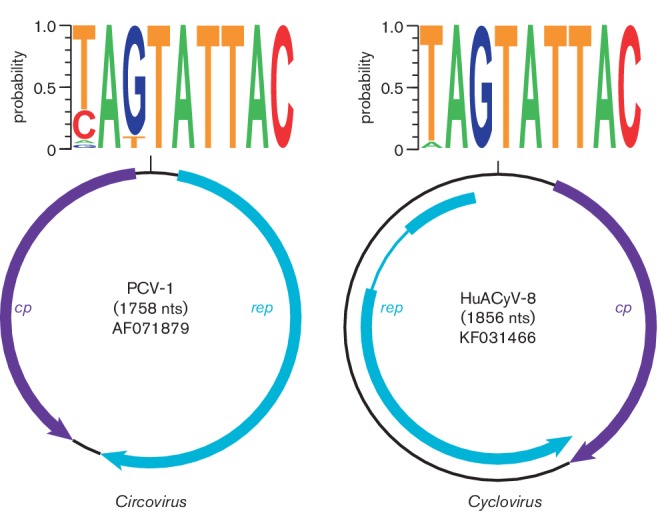
Genome schematics illustrating the major open reading frames (ORFs) characteristic of members of the family Circoviridae. Members of the family have two major ORFs encoding replication-associated (Rep) and capsid (Cp) proteins, as well as a conserved nonanucleotide motif marking the origin of replication. The nonanucleotide motif sequence is depicted through sequence probability logos generated in Weblogo 3. The rep gene of human-associated cyclovirus 8, a representative of the Cyclovirus type species, is interrupted by an intron.
Replication
The ori is characterized by a conserved nonanucleotide motif [(T/n)A(G/t)TATTAC] (Fig. 2) at the apex of a stem–loop structure located between the 5′ ends of Rep- and Cp-encoding ORFs [4, 7]. In characterized members of the genus Circovirus, the Rep protein is thought to initiate replication through the rolling circle replication (RCR) mechanism by nicking the virion-sense strand between positions 7 and 8 of the nonanucleotide motif [8]. RCR involves the production of a dsDNA replicative form by host DNA polymerases and the generation of viral ssDNA from the replicative form template. Both circovirus and cyclovirus Rep proteins contain conserved domains that are important for RCR. Putative Rep-binding domains characterized by iterative sequences near the ori have been identified for members of both genera [9, 10].
Taxonomy
The family Circoviridae includes two genera, Circovirus and Cyclovirus [4]. Members of the genus Circovirus have only been identified in vertebrates, whereas members of the genus Cyclovirus have been identified in both vertebrates and invertebrates [5]. The type species for the genus Circovirus is Porcine circovirus 1 and the type species for the genus Cyclovirus is Human-associated cyclovirus 8. The species demarcation threshold for viruses of the family Circoviridae is 80 % genome-wide nucleotide sequence identity.
Resources
Full ICTV Online (10th) Report: www.ictv.global/report/circoviridae.
Funding information
M. B. and K. R. are supported by grant DEB-1239976 from the Assembling the Tree of Life Program of the US National Science Foundation. Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418A1A).
Acknowledgements
The members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Michael J. Adams, Donald B. Smith, Richard J. Orton and Balázs Harrach.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Crowther RA, Berriman JA, Curran WL, Allan GM, Todd D. Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J Virol. 2003;77:13036–13041. doi: 10.1128/JVI.77.24.13036-13041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ritchie BW, Niagro FD, Latimer KS, Lukert PD, Steffens WL 3rd, et al. Ultrastructural, protein composition, and antigenic comparison of psittacine beak and feather disease virus purified from four genera of psittacine birds. J Wildl Dis. 1990;26:196–203. doi: 10.7589/0090-3558-26.2.196. [DOI] [PubMed] [Google Scholar]
- 3.Todd D, Niagro FD, Ritchie BW, Curran W, Allan GM, et al. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch Virol. 1991;117:129–135. doi: 10.1007/BF01310498. [DOI] [PubMed] [Google Scholar]
- 4.Rosario K, Breitbart M, Harrach B, Segalés J, Delwart E, et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 5.Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol. 2012;157:1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, et al. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk HJ. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol. 1997;71:2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinfeldt T, Finsterbusch T, Mankertz A. Demonstration of nicking/joining activity at the origin of DNA replication associated with the rep and rep' proteins of porcine circovirus type 1. J Virol. 2006;80:6225–6234. doi: 10.1128/JVI.02506-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dayaram A, Potter KA, Moline AB, Rosenstein DD, Marinov M, et al. High global diversity of cycloviruses amongst dragonflies. J Gen Virol. 2013;94:1827–1840. doi: 10.1099/vir.0.052654-0. [DOI] [PubMed] [Google Scholar]
- 10.Steinfeldt T, Finsterbusch T, Mankertz A. Rep and Rep' protein of porcine circovirus type 1 bind to the origin of replication in vitro. Virology. 2001;291:152–160. doi: 10.1006/viro.2001.1203. [DOI] [PubMed] [Google Scholar]


