Abstract
The Bunyaviridae family comprises viruses causing diseases of public and veterinary health importance, including viral haemorrhagic and arboviral fevers. We report the isolation, identification and genome characterization of a novel orthobunyavirus, named Wolkberg virus (WBV), from wingless bat fly ectoparasites (Eucampsipoda africana) of Egyptian fruit bats (Rousettus aegyptiacus) in South Africa. Complete genome sequence data of WBV suggests it is most closely related to two bat viruses (Mojuí dos Campos and Kaeng Khoi viruses) and an arbovirus (Nyando virus) previously shown to infect humans. WBV replicates to high titres in VeroE6 and C6-36 cells, characteristic of mosquito-borne arboviruses. These findings expand our knowledge of the diversity of orthobunyaviruses and their insect vector host range.
Keywords: arbovirus, Rousettus aegyptiacus, Egyptian fruit bat, Bunyaviridae, orthobunyavirus, bat flies, Eucampsipoda africana, pathogen discovery
Abbreviations
CPE, cytopathic effect; EMEM, Eagle’s minimum essential medium; KKV, Kaeng Khoi virus; L, large; M, medium; MDCV, Mojuí dos Campos virus; NP, nucleocapsid protein; RdRp, RNA-dependent RNA-polymerase; RER, rough endoplasmic reticulum; S, small; TCMV, Tacaiuma virus; WBV, Wolkberg virus.
Introduction
The Bunyaviridae family contains over 530 members and has an extensive host range, including arthropods, rodents, large mammals and plants. The typical bunyavirus genome comprises single-stranded RNA separated into small (S), medium (M) and large (L) segments with complementary terminal sequences unique to specific viral genera. The majority of bunyavirus genomes are negative sense, but some viruses use ambisense strategies to express genes from the S segment. The S segment encodes the nucleoprotein, the M segment encodes the glycoprotein precursor and the L segment encodes the RNA-dependent RNA polymerase [1, 2].
The orthobunyavirus genus comprises more than 170 known viruses assembled into 48 species and 19 serogroups [2]. Viruses in the genus, such as La Crosse, Ngari, Oropouche and Nyando viruses, are known to cause disease in humans, ranging from mild febrile illness to more severe complications including encephalitis, haemorrhagic fever and death [3–11]. Viruses of veterinary importance include Schmallenberg, Akabane and Shuni viruses [12–14]. Orthobunyaviruses are primarily transmitted by arthropods. Mojuí dos Campos (MDCV) and Kaeng Khoi (KKV) viruses have been isolated from bats in South America and East Asia, respectively, while KKV was also isolated from bedbugs [15–17]. Some serological evidence suggests that KKV might be of public health importance [18].
Several orthobunyaviruses have been detected in South Africa (Shuni, Pongola and Bunyamwera viruses amongst others) [4, 5, 13], but none are known to be associated with bats or their ectoparasites. We describe the discovery of the first orthobunyavirus to be isolated from bat ectoparasites (Eucampsipoda africana), which were collected from wild-caught Egyptian fruit bats (Rousettus aegyptiacus) in South Africa. The virus has been named Wolkberg virus (WMV), after the location of the cave harbouring the Egyptian fruit bats from which these ectoparasites were collected.
Results and discussion
Virus isolates and growth
From a total of 273 bat ectoparasite pools tested, 11 caused a non-characteristic cytopathic effect (CPE) by day 5–7 post inoculation (from the third passage onwards) in the form of small, rounded, refractory detached cells in the presence of a mostly intact Vero monolayer. All ectoparasite pools originated from apparently healthy Egyptian fruit bats. The 11 isolates replicated to titres in excess of 1×105.0 TCID50 ml−1 in Vero cells by the third passage. A standard curve (R2=0.999) generated by triplicate testing of a dilution series of stock virus enabled us to establish TCID50 equivalents relative to RT-PCR C t values (Table 1). This enabled us to determine TCID50 equivalent values for RT-PCR results obtained in further testing. WBV displayed similar growth curve characteristics in Vero and C6-36 cells over a period of 2 weeks (Fig. 1). However, in HEK293 cells the virus replicated to lower levels relative to Vero and C6-36 cells at the same days post inoculation with the same concentration of stock virus. The difference in yield (TCID50 equivalents) between HEK293 and the other cells lines ranged from 1 log to 3 logs in some cases, especially with the lowest virus input. The ectoparasites from which the viruses were isolated were morphologically identified as nycteribiid bat flies, Eucampsipoda africana Theodor (Diptera: Nycteribiidae) [19, 20].
Table 1. Correlation of real-time RT-PCR C t values to TCID50 .
| Real-time RT-PCR C t value | Corresponding TCID50 equivalent value |
|---|---|
| <16 | >1×106.75 |
| 16–19 | 1×105.75 to 1×106.75 |
| 19–23 | 1×104.75 to 1×105.75 |
| 23–27 | 1×103.75 to 1×104.75 |
| 27–30 | 1×102.75 to 1×103.75 |
| 30–34 | 1×101.75 to 1×102.75 |
| 34–38 | 1×100.75 to 1×101.75 |
| 38–40 | 1×100.25 to 1×100.75 |
| 40–42 | 1×10−0.25 to 1×100.25 |
| 42–45 | 1×10−0.75 to 1×10−0.25 |
Fig. 1.
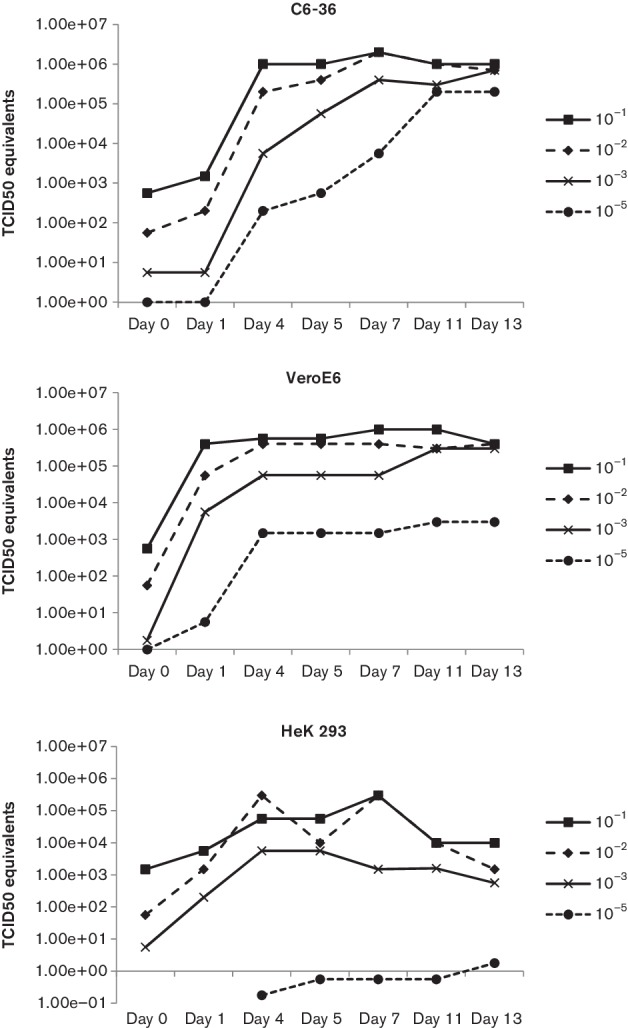
Growth curve of WBV in C6-36, VeroE6 and HEK293 cells. Note: virus present on day 0 is due to inoculum.
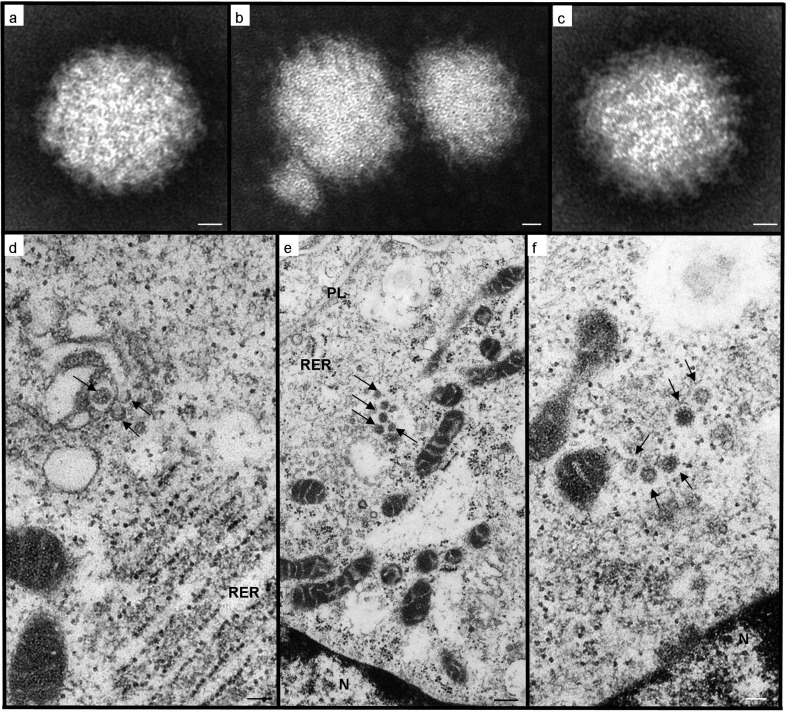
Identification by transmission electron microscopy and next-generation-sequencing negatively-stained preparations of culture supernatants revealed numerous enveloped, roughly spherical virions, heterogeneous in size (between 78 and 111 nm), but with an average diameter of 87 nm (n=100). The envelope appearance varied from indistinct and fuzzy, to fairly regularly spiked, in which projections measured up to 8 nm (Fig. 2a–c). These features conform to those described for viruses of the Bunyaviridae family [2], specifically of the genus Orthobunyavirus [21, 22].
Fig. 2.
Transmission electron microscopy of WBV negatively stained particles (a, b, c) and infected Vero cells (d, e, f). (a) Virion of icosahedral-spherical shape, with an apparently amorphous envelope; (b) heterogeneity in particle size; (c) virus particle in which regularly spaced, envelope projections (right side) contrast with the fuzzy appearance of the envelope (left side); (d) transverse sections through characteristic tubular elements (arrows) forming within Golgi cisternal stacks, which are in close proximity to mitochondrial profiles and distended rough endoplasmic reticulum (RER); (e) developing virions (arrows) within a Golgi-derived vesicle, approaching the plasmalemma (PL) for exocytosis. Note the organelle arrangement with the nucleus (N) and numerous mitochondrial profiles in juxtaposition to the disrupted Golgi body; (f) mature virions (arrows) with glycoprotein envelope spikes, still within an endomembrane that appears continuous with those of the tangentially sectioned mitochondrial profiles. Scale bars: (a, b, c) 12 nm; (d, f) 100 nm; (e) 250 nm.
This taxonomic assignation was supported by ultrastructural details of infected cells (Fig. 2d–f). Virus particles develop within the Golgi body, initially from round and tubular structures, which to date are unique to Orthobunyavirus infections [23]. The Golgi body is located close to one side of the nucleus (frequently within an indentation), and the tubular viral factories are connected to both rough endoplasmic reticulum and numerous mitochondrial profiles, as in Bunyamwera virus morphogenesis [24]. The Golgi body becomes increasingly disrupted, and enveloped particles can be seen within Golgi-derived vesicles being trafficked towards the cell membrane for exocytosis, as described for other orthobunyaviruses [16, 25].
Identification as an orthobunyavirus was confirmed by using an unbiased next-generation sequencing approach that resolves the 5′ and 3′ termini (sequence-independent single-primer amplification combined with rapid amplification of cDNA ends, SISPA-RACE). Assembled contigs were aligned to the nt sequence database (blast, GenBank). All 11 isolates contained contigs that had matches for an S, M and L segment of an orthobunyavirus.
Genome and phylogenetic analysis
Maximum-likelihood trees, constructed with nucleic acid sequence data for the L, M and S segments of representative viruses from the different genera within the Bunyaviridae family (see Figs S1–S3, available in the online Supplementary Material), show the placement of the 11 WBV isolates amongst other orthobunyaviruses in the family. Table 2 provides information on the sequence data obtained from the 11 WBV isolates. All WBV genome segments for which complete sequences were obtained, or at least complete sequences at either terminal end, had identical terminal sequences that were complementary to each other at the 5′ and 3′ ends. These 5′ and 3′ terminal end sequences are 5′-AGTAGTGT and ACACTACT-3′ respectively, in the antigenomic (positive) sense. These terminal complimentary sequences are characteristic of viruses in the Orthobunyavirus genus [2].
Table 2. Sequence information for the 11 WBV isolates.
Complete and coding complete (CC) descriptions were defined previously [44]. If genome is coding complete but does have one of the ends complete, 5C or 3C denotes a complete sequence at the 5′ or 3′ end of the segment sequence, respectively.
| WBV isolate number | Genome segment | Genome segment length (nucleic acid) |
Sequence completeness | ORF length (nucleic acid) |
GenBank accession number |
|---|---|---|---|---|---|
| 2562 | L | 6873 | Complete | 6759 | KX470551 |
| M | 4461 | Complete | 4266 | KX470552 | |
| S | 978 | Complete | 702 | KX470553 | |
| 2761 | L | 6873 | Complete | 6759 | KX470554 |
| M | 4471 | Complete | 4266 | KX470555 | |
| S | 978 | Complete | 702 | KX470556 | |
| 2763 | L | 6800 | CC | 6759 | KX470557 |
| M | 4465 | Complete | 4263 | KX470559 | |
| S | 967 | CC (5C) | 702 | KX470558 | |
| 2795 | L | 6873 | Complete | 6759 | KX470560 |
| M | 4461 | Complete | 4266 | KX470561 | |
| S | 978 | Complete | 702 | KX470562 | |
| 2812 | L | 6873 | Complete | 6759 | KX470563 |
| M | 4461 | Complete | 4266 | KX470564 | |
| S | 978 | Complete | 702 | KX470565 | |
| 2813 | L | 6873 | Complete | 6759 | KX470566 |
| M | 4468 | Complete | 4266 | KX470567 | |
| S | 978 | Complete | 702 | KX470568 | |
| 2818 | L | 6873 | Complete | 6759 | KX470569 |
| M | 4461 | Complete | 4266 | KX470570 | |
| S | 978 | Complete | 702 | KX470571 | |
| 2824 | L | 6873 | Complete | 6759 | KX470572 |
| M | 4465 | Complete | 4263 | KX470573 | |
| S | 979 | Complete | 702 | KX470574 | |
| 3011 | L | 6841 | CC (3C) | 6759 | KX470575 |
| M | 4425 | CC (5C) | 4266 | KX470576 | |
| S | 818 | CC | 702 | KX470577 | |
| 3264 | L | 6873 | Complete | 6759 | KX470578 |
| M | 4461 | Complete | 4266 | KX470579 | |
| S | 978 | Complete | 702 | KX470580 | |
| SM910 | L | 6873 | Complete | 6759 | KX470581 |
| M | 4461 | Complete | 4266 | KX470582 | |
| S | 978 | Complete | 702 | KX470583 |
Maximum-likelihood trees were prepared using the WBV deduced amino acid sequences from the ORFs of the three segments and those of other viruses in the Orthobunyavirus genus (Figs 3–5). Only the ORF encoding the nucleocapsid protein was used from the S segment since WBV does not have a second ORF in the S segment. The ORF encoding the Gn-Gc-NSm polyprotein was used from the M segment.
Fig. 3.
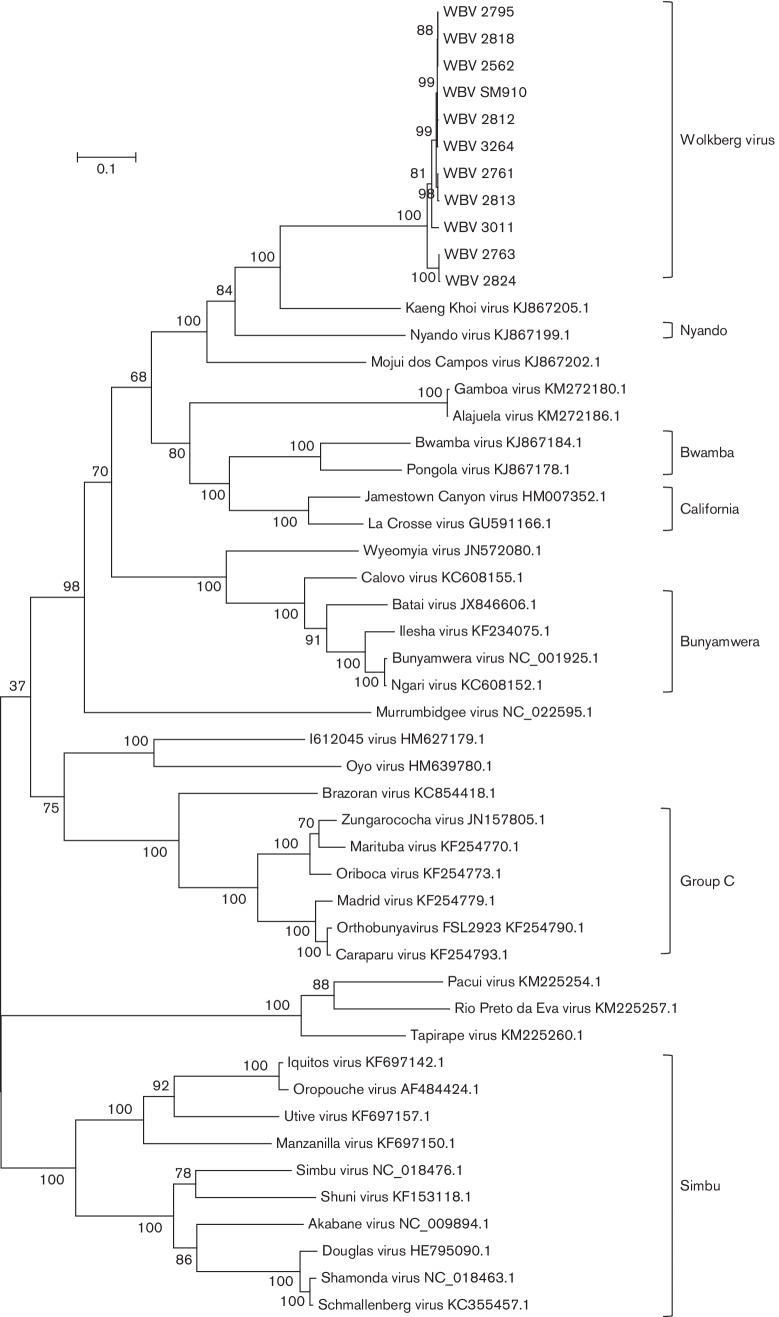
Molecular phylogenetic analysis by the maximum-likelihood method showing representative viruses from different viruses in the Orthobunyavirus genus using L-segment ORF amino acid sequences. GenBank accession numbers are indicated next to virus names (excluding WBV for which accession numbers are provided in Table 2). Medically important serogroups, and WBV isolates, are indicated by brackets.
Fig. 4.
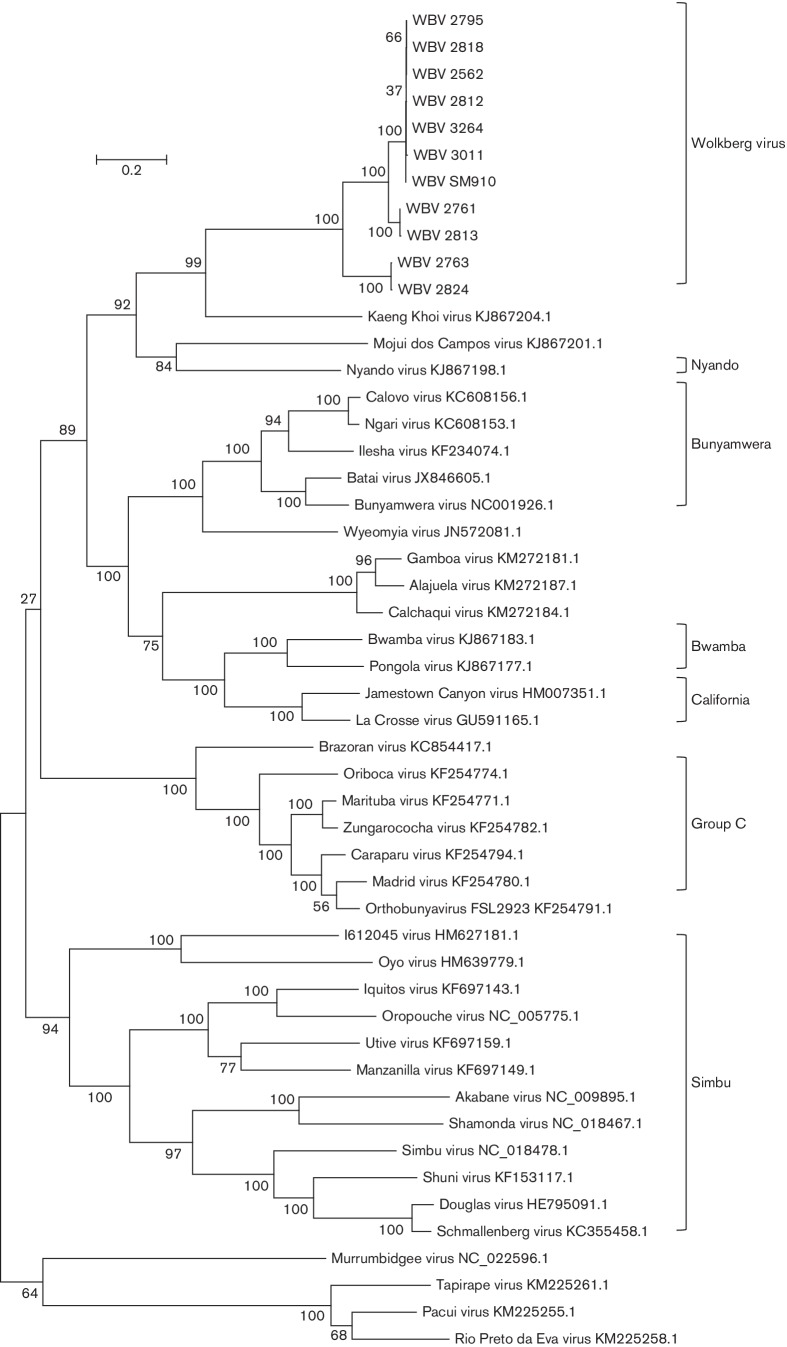
Molecular phylogenetic analysis by the maximum-likelihood method showing representative viruses from different viruses in the Orthobunyavirus genus using M-segment ORF amino acid sequences. GenBank accession numbers are indicated next to virus names (excluding WBV for which accession numbers are provided in Table 2). Medically important serogroups, and WBV isolates, are indicated by brackets.
Fig. 5.
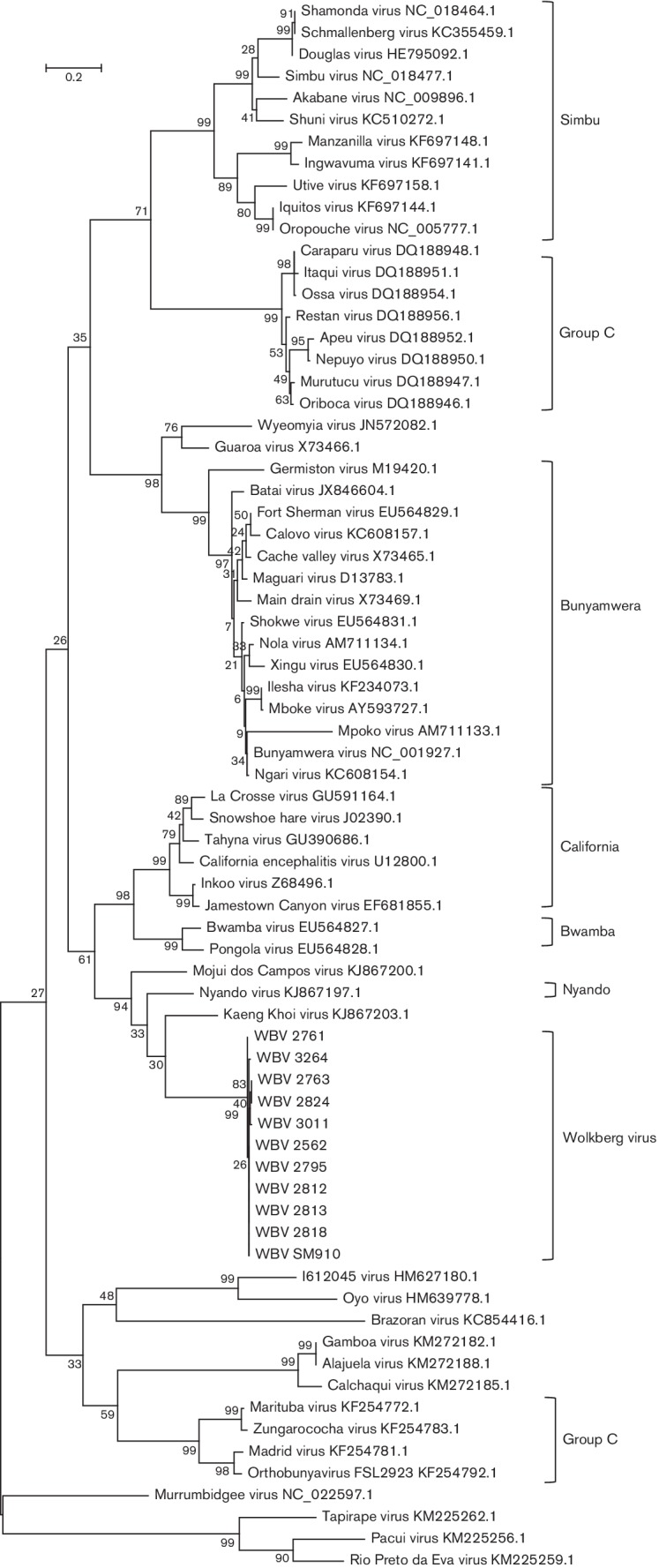
Molecular phylogenetic analysis by the maximum-likelihood method showing representative viruses from different viruses in the Orthobunyavirus genus using the S-segment ORF amino acid sequences. GenBank accession numbers are indicated next to virus names (excluding WBV for which accession numbers are provided in Table 2). Medically important serogroups, and WBV isolates, are indicated by brackets.
A distinct clade is formed by the 11 WBV isolates, KKV, MDCV and Nyando virus (Figs 3–5) within the Orthobunyavirus genus. WBV is most closely related to KKV based on L-, M- and S-segment nucleic acid identity (67.6 %, 56.5 %, 65.5 %) and ORF-deduced amino acid sequences of the L and M segments (66.3 %, 46.4 %), but closest to MDCV based on an S-segment ORF amino acid sequence (65.6 %). The ICTV species demarcation in the Orthobunyavirus genus is complicated by the lack of biochemical characterization of named viruses, and species are thus demarcated based on serological cross-reaction criteria [2]. In the absence of these data, another possible criterion is nucleocapsid protein amino acid difference, where differences of more than 10 % represent different species. Based on the fact that the closest-related virus to WBV based on the nucleocapsid protein amino acid sequence differs by 34.4 % (MDCV, based on currently available sequence data), WBV is likely a new species in the Orthobunyavirus genus. However, this needs to be further investigated by evaluating serological cross-reactivity to closely related viruses. The sequences of the 11 WBV isolates are not identical. There appear to be three sub-lineages of the virus based on the ORF-deduced amino acid sequences of all three segments (Figs 3–5), more noticeably based on the M-segment ORF amino acid sequence.
Percentage pairwise differences in sequences between the 11 isolates range from being identical (all segments) to 4.1 % (L-segment ORF), 28.8 % (M-segment ORF) or 1.3 % (S-segment ORF). Based on the M-segment ORF, the WBV isolates 2763 and 2824 seem to be closely related (0.4 % p-distance) but quite distinct from the other nine isolates (between 27.9 and 28.8 % difference). The close-relatedness between all 11 isolates is based on L- and S-segment ORFs, but clear divergence in two isolates in the M-segment might represent evidence of re-assortment with another variant of WBV that is yet to be isolated, and thus possibly evidence of higher undiscovered divergence of the new virus. Based on limited data which suggest that re-assortment can only occur between viruses in the same species [2] and the fact that the M-segment ORFs of isolates 2763 and 2824 are no more than 46.5 % similar to any other currently known orthobunyavirus (for which sequence data are available), it is likely that re-assortment occurred with another, yet undiscovered, member of the proposed WBV species in the Orthobunyavirus genus.
RT-PCR detection of WBV in bat fly homogenates and bat serum
The original 11 homogenates from which the strains of WBV virus were isolated were subsequently tested by RT-PCR to determine viral loads. Similarly, bat serum pools from around the period of virus isolation were also tested by RT-PCR. The results are summarised in Table 3. WBV was readily detected, at relatively high concentrations, in bat fly homogenates (9/11). However, WBV was only detected in one serum pool from Egyptian fruit bats (1/85 pools of five bat sera), and at relatively low concentration. Considering the transient viraemia of most arboviral infections, this is not surprising. Detection of the virus in the serum of one bat demonstrates that it is likely that Egyptian fruit bats can be infected by WBV, thereby probably playing a role in its maintenance.
Table 3. Detection of WBV in bat fly homogenates and Egyptian fruit bat serum.
| Bat ID number | Real-time RT-PCR C t value | TCID50 equivalent ml−1 |
|---|---|---|
| 2824 | 26.37 | 1×103.75 |
| 2763 | 34.29 | 1×101.75 |
| 2761 | 25.43 | 1×104.2 |
| 2818 | 31.7 | 1×102.3 |
| 2795 | Neg | N/A |
| 2812 | 22.81 | 1×104.75 |
| 2813 | 23.1 | 1×104.75 |
| 2562 | Neg | N/A |
| 3011 | 33.54 | 1×101.9 |
| SM910 | 26.42 | 1×103.75 |
| 3264 | 23.36 | 1×104.8 |
| 5× bat sera/pool (n=85) | 1 positive (C t 38.79) | 1×100.75 |
ORFs
RNA-dependent RNA-polymerase (RdRp)
The deduced 2253 aa size of the WBV RdRp (262 kDa, pI 6.88) corresponds to the size of the same protein of other members of the Orthobunyavirus genus (Bunyamwera virus RdRp 2238 aa) [26].
Gn-Gc-NSm polyprotein
The 1422 aa WBV polyprotein (162 kDa, pI 8.33) is likely co-translationally cleaved into the 951 aa G1 (108.8 kDa, pI 6.56), 285 aa G2 (32 kDa, pI 8.83) and 166 aa NSm (19 kDa, 9.34). Cleavage of the signal peptide is predicted at position 18 relative to the first methionine (SignalP 3.0). The G2 contains the conserved arginine at position 303, which likely represents the cleavage site from the downstream NSm [27]. SignalP predicts the cleavage site between NSm and the downstream G1 at ILI470-EA, which corresponds to other orthobunyaviruses [28]. Six potential glycosylation sites were identified using NetNGlyc 1.0 (www.cbs.dtu.dk/services), of which one is in the G2, three in the NSm and two in the G1 protein.
Nucleocapsid protein
The WBV-deduced nucleocapsid protein (NP) is 234 aa in length (26.5 kDa, pI 9.44) and corresponds to other orthobunyaviruses [28]. It satisfies the ICTV criterion of >10 % divergence from other viruses in the Orthobunyavirus genus to regard it as a separate, new species [2]. The four residues involved in ribonucleoprotein complex formation (P125, G131, Y158 and I231) are all conserved in WBV [29]. Contrary to most orthobunyaviruses, WBV does not encode an NSs protein from the S segment. The NSs protein in most orthobunyaviruses is a non-essential protein during mammalian infection that plays a role in pathogenesis by shutting down host protein synthesis leading to cell death, and counteracting host antiviral responses [30–32]. The protein seems to be important for replication of Bunyamwera virus in mosquito cells [33].
Viruses from the Tete, Anopheles A and Anopheles B serogroups also do not express an NSs protein [34]. It has been proposed that these viruses lack an NSs protein, by assumption being unable to counteract mammalian host innate responses and thus being less virulent, and might be maintained in nature by an insect-only cycle through vertical (transovarial) and/or horizontal (venereal) transmission. If this is the case, these viruses bypass the need for generating the high viraemia in vertebrate hosts needed for transmission of arboviruses. It has been suggested that the NSs protein is a major factor in the zoonotic capacity of orthobunyaviruses, by allowing viruses expressing this protein to efficiently infect and replicate in vertebrates with a functioning innate type I interferon response [35]. However, Tacaiuma virus (TCMV) from the Anopheles A serogroup, has been shown to block interferon beta mRNA production despite lacking an ORF for NSs, suggesting that this virus has developed an alternative mechanism for counteracting the mammalian host innate response. Also, TCMV, which was originally isolated from a monkey and, subsequently, an Anopheles mosquito in South America, has been shown to cause febrile illness in humans and has been associated with other vertebrate hosts (bat, primate, bird and horse) [34, 36, 37]. The ability of WBV to counteract vertebrate host innate responses and cause high viraemia, despite the lack of NSs, needs future investigation through in vitro and in vivo studies. The decreased replication of WBV in HEK293 cells versus Vero cells might indicate that, without the expression of NSs, the virus is not able to replicate as efficiently in the presence of an interferon response, but this requires further in-depth investigation in in vivo interferon-competent versus interferon-deficient mouse models.
Conclusion
Based on sequence divergence of the NP from other known orthobunyaviruses, it is likely that WBV represents a new species in the genus. Evidence of re-assortment of the M segment in two of the 11 isolates described here, which are quite divergent from the other nine isolates, likely indicates that the diversity of viruses in this proposed new species is higher than that indicated by the data. The high percentage of isolates from a rather low number of ectoparasite pools (11 isolates out of 273, amounting to 4 %) might indicate a very high prevalence in this specific bat colony, or may indicate that the timing of collection represents a period of high transmission rates. All isolates were made from ectoparasites collected during the dry winter period in South Africa when fruit is scarce, and also overlaps with the period when young adult bats have lost their maternal immunity. Aspects which need further investigation include whether WBV infects bats and what is the seroprevalence of bats collected from Mahlapitsi cave. Whether this new virus has any public or veterinary health importance is yet to be determined. Antibodies to the virus most closely related to WBV, KKV, have been detected in humans who suffered a mild disease [18]. The discovery of WBV expands the range of currently known orthobunyaviruses and also the arthropod host range. Its global distribution needs further investigation, considering the wide distribution of Egyptian fruit bats, the close association of bat flies with specific bat hosts, and the close association of arboviruses with their arthropod hosts. The ability of WBV to replicate in vitro in a mosquito cell line might indicate its ability to be disseminated by mosquitoes, but this needs further investigation through vector competence studies.
Methods
Virus source and isolation
Egyptian fruit bats (Rousettus aegyptiacus) were sampled between March 2013 and March 2014 at Mahlapitsi cave in the Mahlapitsi Valley, Limpopo province, South Africa, as described before [20]. Catch and release sampling included the collection and pooling of arthropods from parasitized bats into Eagle’s minimum essential medium (EMEM, Lonza). Pools of parasites were homogenized (30 Hz for 8 min by using a Tissuelyzer II and 5 mm stainless steel beads, Qiagen) and clarified supernatants used to inoculate Vero cell cultures. The wells of 24-well tissue culture plates (Nunc) were seeded with Vero E6 cells and grown to 80–90 % confluence in EMEM supplemented with antibiotics (100 U penicillin/ml 100 µg streptomycin/ml 250 ng amphotericinB/ml,Lonza) and 10 % fetal calf serum, at 37 °C and 5 % CO2. Culture medium was removed and the monolayers in individual wells inoculated with 200 µl of ectoparasite pool homogenates (one pool representing parasites from one bat). After 1 h adsorption at 37 °C, the inoculum was removed and fresh EMEM containing antibiotics and 2 % fetal calf serum added. Parasites were morphologically identified, and confirmed by amplification and amplicon sequencing of a region of the cytochrome oxidase 1 subunit gene, as belonging to the Nycteribiidae family, genus Eucampsipoda [20]. Inoculated cultures were monitored for development of CPEs for three blind passages of 14 days each. Cultures displaying CPEs were subjected to two or three more passages in 75 cm2 tissue culture flasks to prepare stocks. Stock virus titres were determined by standard TCID50 titrations on 96-well microtitre plates.
Bat fly homogenate pools and bat serum pools were subsequently tested by RT-PCR for the presence of WBV.
Transmission electron microscopy
Processing of infected cell cultures for transmission electron microscopy was performed as described previously [20]. Briefly, culture supernatants from six of the 11 isolates were concentrated, adsorbed onto coated grids, negatively stained and viewed. Infected monolayers were routinely processed for ultramicrotomy (fixation, postfixation, ethanol dehydration, resin embedding, ultramicrotomy with double staining of 70 nm sections).
Complete genome sequencing
Stock virus culture supernatant was added to Trizol-LS (Life Technologies) at a ratio of 100 µl supernatant to 300 µl Trizol-LS. RNA was extracted using a column based kit (Direct-Zol RNA kit, Zymo Research). To increase sensitivity, rRNA was depleted using the method described by Morlan et al. [38]. Samples were prepared for sequencing using the SISPA-RACE technique described previously [20]. Sequencing was performed either on an Illumina MiSeq or NextSeq 500 using either a 2×150 or 2×250 version2 kit. Illumina and SISPA adapter sequences were trimmed from the sequencing reads using Cutadapt-1.2.1 [39], quality filtering was conducted with Prinseq-lite [40] and reads were assembled into contigs using Ray Meta with kmer length=25 [41]. Resultant contigs were aligned to the NCBI sequence database using blast (www.ncbi.nlm.nih.gov/BLAST/).
Phylogenetic and sequence analysis
The mega (version 6) program was used to prepare alignments (clustal w) of nucleic acid segment sequences, deduced amino acid sequences, phylogenetic trees and pairwise distance calculations [42]. The publicly available bunyavirus sequences used in the analysis were obtained from NCBI-Nucleotide (GenBank). Nucleotide sequences from a small number of viruses from each genus in the Bunyaviridae family were used to prepare a maximum-likelihood tree showing the placement of WBV in the family based on complete sequences of all three segments (L, M and S). Maximum-likelihood trees were prepared using amino acid sequences of all ORFs from all segments, showing the placement of WBV in the Orthobunyavirus genus relative to other viruses in this genus for which sequence is available on GenBank. Virus sequence accession numbers are summarised in Table 2. The evolutionary histories were inferred by using the maximum-likelihood method based on the Tamura–Nei model [43]. The trees with the highest log likelihood are shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbour-joining method to a matrix of pairwise distances estimated using the maximum-composite-likelihood approach. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in mega6 [42]. ORFs were located and deduced protein amino acid sequences prepared by using the CLC Genomics Workbench (Qiagen).
Virus growth
In vitro replication of WBV was evaluated in three cell lines: Vero E6 (source: African green monkey kidney), HEK293 (source: human embryonic kidney) and C6-36 (source: Aedes albopictus mosquitoes). Cells were grown to 50–70 % confluence in 25 cm2 flasks, supernatant removed and respective flasks inoculated with 1 ml of 10−1, 10−2, 10−3 and 10−5 dilutions from stock virus (1×106.75 TCID50 ml1 determined by standard 50 % tissue culture infectious doses titration) in EMEM. After 1 h adsorption at 37 °C (Vero E6 and HEK293) or 28 °C (C6-36), the inoculum was removed, cells washed with 5 ml PBS and fresh EMEM, antibiotics and 2 % fetal calf serum (Hyclone) added. Cultures were incubated for 13 days at 37 °C (Vero E6 and HEK293) or 28 °C (C6-36) while 0.5 ml aliquots of supernatant were collected from each flask directly after inoculation and addition of fresh medium (day 0), followed by days 1, 4, 5, 7, 11 and 13. RNA was extracted from 140 µl of the supernatant collections from respective days (QIamp viral RNA kit, Qiagen) and subjected to TaqMan real-time RT-PCR. A TaqMan real-time RT-PCR was developed to target the S segment of WBV. Primers and probe sequences are: forward MAOBV_783F (5′-TTGGCTTTCTTTGCATTCAG-3′), reverse MAOBV_869R (5′-ATGGTTTCAACCCTGAGGAA-3′) and probe MAOBV_831P (FAM-TCTTCACAAGTGGCAATGC-BHQ), with the number in the oligonucleotide name indicating the nucleic acid position in the S segment. Real-time RT-PCR was performed on the extracted RNA using the Qiagen One-step RT-PCR kit (Qiagen) on a SmartCycler (Cepheid) with the following parameters: reverse transcription (50 °C for 30 min), hot-start Taq activation (95 °C for 15 min) and 50 cycles of amplification (95 °C for 15 s; 52 °C for 25 s plus signal acquisition; 72 °C for 20 s). RNA extracted from diluted stock WBV (final 1×104.75 TCID50 ml1) was used as a qualitative positive control in each run. A standard curve was prepared by testing a dilution series of stock virus (1×106.75 TCID50 ml1) in triplicate and correlating C t values to TCID50 equivalents.
Funding information
The project is jointly funded by the following grants awarded to: Janusz T. Paweska (CDC Global Disease Detection program, GDD 5U19 GH000571-05/96667), Wanda Markotter (South African National Research Foundation, Grant UID 91496, 92524 and 98339; Poliomyelitis Research Foundation, PRF Grant number 12/14) and Petrus Jansen van Vuren (South African National Research Foundation, Incentive Funding for Rated Researchers, Grant UID 85544). The grant holders acknowledge that opinions, findings and conclusions or recommendations expressed in any publication generated by GDD and NRF-supported research are those of the authors and that the GDD and NRF accept no liability whatsoever in this regard.
Acknowledgements
The authors would like to thank the following individuals for their contributions towards fieldwork and technical assistance: Busi Mogodi, Justice Kgatitsoe, Stewart McCulloch, Terence Scott, Joe Kgaladi, Marinda Mortlock, Marike Geldenhuys, Jessica Coertse and Andre Coetzer.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was carried out according to the recommendations of the South African National Standards for the Care and Use of Animals for Scientific Purposes (SANS 10386 : 2008). The field sampling protocols and transport of Rousettus aegyptiacus and samples collected from this species are approved by the National Health Laboratory Service Animal Ethics Committee (AEC 137/12), University of Pretoria Animal Ethics Committee (EC054-14), Department of Economic Development, Environment and Tourism: Limpopo Province Directorate: Wildlife Trade and Regulation Permit (CPM 006806) and the South African Department of Agriculture, Forestry and Fisheries (Section 20 approval 12/11/1/1/8).
Supplementary Data
References
- 1.Elliott RM. Emerging viruses: the Bunyaviridae. Mol Med. 1997;3:572–577. [PMC free article] [PubMed] [Google Scholar]
- 2.Plyusnin A, Beaty BJ, Elliot RM, Goldbach R, Kormelink R, et al. Family Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier; 2012. pp. 725–741. (editors) [Google Scholar]
- 3.Digoutte JP, Gagnard VJ, Bres P, Pajot FX. Nyando virus infection in man. Bull Soc Pathol Exot Filiales. 1972;65:751–758. [PubMed] [Google Scholar]
- 4.Kalunda M, Lwanga-Ssozi C, Lule M, Mukuye A. Isolation of Chikungunya and Pongola viruses from patients in Uganda. Trans R Soc Trop Med Hyg. 1985;79:567. doi: 10.1016/0035-9203(85)90105-1. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez JP, Georges AJ. Bunyaviral fevers: Bunyamwera, Ilesha, Germiston, Bwamba and Tataguine. In: Monath T, editor. The Arboviruses: Epidemiology and Ecology. Boca Raton, FL: CRC Press; 1988. pp. 87–98. (editor) [Google Scholar]
- 6.Lutwama JJ, Rwaguma EB, Nawanga PL, Mukuye A. Isolations of Bwamba virus from south central Uganda and north eastern Tanzania. Afr Health Sci. 2002;2:24–28. [PMC free article] [PubMed] [Google Scholar]
- 7.Gerrard SR, Li L, Barrett AD, Nichol ST. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J Virol. 2004;78:8922–8926. doi: 10.1128/JVI.78.16.8922-8926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briese T, Bird B, Kapoor V, Nichol ST, Lipkin WI. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol. 2006;80:5627–5630. doi: 10.1128/JVI.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastos MS, Figueiredo LT, Naveca FG, Monte RL, Lessa N, et al. Identification of oropouche orthobunyavirus in the cerebrospinal fluid of three patients in the Amazonas, Brazil. Am J Trop Med Hyg. 2012;86:732–735. doi: 10.4269/ajtmh.2012.11-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd BD. La Crosse encephalitis: a persistent arboviral threat in North Carolina. N C Med J. 2016;77:330–333. doi: 10.18043/ncm.77.5.330. [DOI] [PubMed] [Google Scholar]
- 11.Pastula DM, Smith DE, Beckham JD, Tyler KL. Four emerging arboviral diseases in North America: Jamestown Canyon, Powassan, Chikungunya, and Zika virus diseases. J Neurovirol. 2016;22:257–260. doi: 10.1007/s13365-016-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles JA. Akabane virus. Vet Clin North Am Food Anim Pract. 1994;10:525–546. doi: 10.1016/S0749-0720(15)30537-5. [DOI] [PubMed] [Google Scholar]
- 13.van Eeden C, Williams JH, Gerdes TG, van Wilpe E, Viljoen A, et al. Shuni virus as cause of neurologic disease in horses. Emerg Infect Dis. 2012;18:318–321. doi: 10.3201/eid1802.111403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beer M, Conraths FJ, van der Poel WH. 'Schmallenberg virus: a novel orthobunyavirus emerging in Europe. Epidemiol Infect. 2013;141:1–8. doi: 10.1017/S0950268812002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams JE, Imlarp S, Top FH, Cavanaugh DC, Russell PK. Kaeng Khoi virus from naturally infected bedbugs (Cimicidae) and immature free-tailed bats. Bull World Health Organ. 1976;53:365–369. [PMC free article] [PubMed] [Google Scholar]
- 16.Wanzeller AL, Diniz JA, Gomes ML, Cruz AC, Soares MC, et al. Ultrastructural, antigenic and physicochemical characterization of the Mojuí dos Campos (Bunyavirus) isolated from bat in the Brazilian Amazon region. Mem Inst Oswaldo Cruz. 2002;97:307–311. doi: 10.1590/S0074-02762002000300005. [DOI] [PubMed] [Google Scholar]
- 17.Groseth A, Mampilli V, Weisend C, Dahlstrom E, Porcella SF, et al. Molecular characterization of human pathogenic bunyaviruses of the Nyando and Bwamba/Pongola virus groups leads to the genetic identification of Mojuí dos Campos and Kaeng Khoi virus. PLoS Negl Trop Dis. 2014;8:e3147. doi: 10.1371/journal.pntd.0003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne JC, Rupprecht CE, Olson JG, Ksiazek TG, Rollin PE, et al. Isolation of Kaeng Khoi virus from dead Chaerephon plicata bats in Cambodia. J Gen Virol. 2003;84:2685–2689. doi: 10.1099/vir.0.19294-0. [DOI] [PubMed] [Google Scholar]
- 19.Theodor O. An Illustrated Catalogue of the Rothschild Collection of Nycteribiidae (Diptera) in the British Museum (Natural History), with Keys and Short Descriptions for the Identification of Subfamilies, Genera, Species and Subspecies. London, UK: British Museum (Natural History); 1967. [Google Scholar]
- 20.Jansen van Vuren P, Wiley M, Palacios G, Storm N, Mcculloch S, et al. Isolation of a novel fusogenic Orthoreovirus from Eucampsipoda africana bat flies in South Africa. Viruses. 2016;8:65. doi: 10.3390/v8030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden TA, Bitto D, Mclees A, Yeromonahos C, Elliott RM, et al. Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. PLoS Pathog. 2013;9:e1003374. doi: 10.1371/journal.ppat.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albornoz A, Hoffmann A, Lozach P-Y, Tischler N. Early Bunyavirus-host cell interactions. Viruses. 2016;8:143. doi: 10.3390/v8050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontana J, López-Montero N, Elliott RM, Fernández JJ, Risco C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell Microbiol. 2008;10:2012–2028. doi: 10.1111/j.1462-5822.2008.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. J Gen Virol. 2011;92:2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- 25.Goldsmith C. Morphologic differentiation of viruses beyond the family level. Viruses. 2014;6:4902–4913. doi: 10.3390/v6124902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott RM. Nucleotide sequence analysis of the large (L) genomic RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. Virology. 1989;173:426–436. doi: 10.1016/0042-6822(89)90555-2. [DOI] [PubMed] [Google Scholar]
- 27.Fazakerley J, Gonzalezscarano F, Strickler J, Dietzschold B, Karush F, et al. Organization of the middle RNA segment of snowshoe hare bunyavirus. Virology. 1988;167:422–432. doi: 10.1016/S0042-6822(88)90104-3. [DOI] [PubMed] [Google Scholar]
- 28.Savji N, Palacios G, Travassos da Rosa A, Hutchison S, Celone C, et al. Genomic and phylogenetic characterization of leanyer virus, a novel orthobunyavirus isolated in northern Australia. J Gen Virol. 2011;92:1676–1687. doi: 10.1099/vir.0.028308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eifan SA, Elliott RM. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J Virol. 2009;83:11307–11317. doi: 10.1128/JVI.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridgen A, Weber F, Fazakerley JK, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci USA. 2001;98:664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Kessler N, et al. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol. 2002;76:7949–7955. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohl A, Clayton RF, Weber F, Bridgen A, Randall RE, et al. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J Virol. 2003;77:7999–8008. doi: 10.1128/JVI.77.14.7999-8008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szemiel AM, Failloux A-B, Elliott RM. Role of Bunyamwera orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl Trop Dis. 2012;6:e1823. doi: 10.1371/journal.pntd.0001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohamed M, Mclees A, Elliott RM. Viruses in the Anopheles A, Anopheles B, and Tete serogroups in the Orthobunyavirus genus (family Bunyaviridae) do not encode an NSs protein. J Virol. 2009;83:7612–7618. doi: 10.1128/JVI.02080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart TJ, Kohl A, Elliott RM. Role of the NSs protein in the zoonotic capacity of orthobunyaviruses. Zoonoses Public Health. 2008;56:285–296. doi: 10.1111/j.1863-2378.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 36.Causey OR, Causey CE, Maroja OM, Macedo DG. The isolation of arthropod-borne viruses, including members of two hitherto undescribed serological groups, in the Amazon region of Brazil. Am J Trop Med Hyg. 1961;10:227–249. doi: 10.4269/ajtmh.1961.10.227. [DOI] [PubMed] [Google Scholar]
- 37.Iversson LB, Silva RA, da Rosa AP, Barros VL. Circulation of eastern equine encephalitis, western equine encephalitis, Ilhéus, Maguari and Tacaiuma viruses in equines of the Brazilian Pantanal, South America. Rev Inst Med Trop Sao Paulo. 1993;35:355–359. doi: 10.1590/S0036-46651993000400009. [DOI] [PubMed] [Google Scholar]
- 38.Morlan JD, Qu K, Sinicropi DV. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS One. 2012;7:e42882. doi: 10.1371/journal.pone.0042882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 40.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boisvert S, Raymond F, Godzaridis Élénie, Laviolette F, Corbeil J. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;1993:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 44.Ladner JT, Beitzel B, Chain PSG, Davenport MG, Donaldson E, et al. Standards for sequencing viral genomes in the era of high-throughput sequencing. MBio. 2014;5:e01360-14. doi: 10.1128/mBio.01360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.