Abstract
Early-life stress (ELS) can alter neurodevelopment in variable ways, ranging from producing deleterious outcomes to stress resilience. While most ELS studies focus on its harmful effects, recent work by our lab and others shows that ELS elicits positive effects in certain individuals. We exposed Wistar-Kyoto (WKY) rats, known for a stress reactive, anxiety-/depression-like phenotype, to maternal separation (MS), a model of ELS. MS exposure elicited anxiolytic and antidepressant behavioral effects as well as improved cardiovascular function in adult WKY offspring. The present study interrogates an epigenetic mechanism (DNA methylation) that may confer the adaptive effects of MS in WKY offspring. We quantified global genome methylation levels in limbic brain regions of adult WKYs exposed to daily 180-min MS or neonatal handling from postnatal day 1–14. MS exposure triggered dramatic DNA hypermethylation specifically in the hippocampus. Next-generation sequencing methylome profiling revealed reduced methylation at intragenic sites within two key nodes of insulin signaling pathways: the insulin receptor and one of its major downstream targets, mitogen activated protein kinase kinase kinase 5 (Map3k5). We then tested the hypothesis that enhancing DNA methylation in WKY rats would elicit adaptive changes akin to the effects of MS. Dietary methyl donor supplementation improved WKY rats’ anxiety/depression-like behaviors and also improved cardiovascular measures, similar to previous observations following MS. Overall these data suggest a potential molecular mechanism that mediates a predicted adaptive response whereby ELS induces DNA methylation changes in the brain that may contribute to successful stress coping and adaptive physiological changes in adulthood.
Keywords: Depression, Anxiety, Epigenetics, Folate, Methyl Depletion, Resilience
Introduction
Adverse early-life experiences have long-term effects on the brain and biobehavioral responses to stress in many species (Suomi, 1991; Daniels et al., 2004; Rensel et al., 2010; Thompson et al., 2012). Although most work on ELS highlights its harmful effects such as increasing risk for neuropsychiatric disorders (Hofer et al., 1996; Sanchez et al., 2001; Maccari et al., 2014), smoking, physical inactivity, obesity, and cardiovascular disease in humans (Dong et al., 2004), emerging evidence suggests a more nuanced view. The match/mismatch hypothesis of disease states that ELS can have adaptive value to individuals facing later stressful experiences (Santarelli et al., 2014). For instance, repeated childhood stress exposure confers protective neuroendocrine effects when adult offspring face stress (Lyons & Parker, 2007). This suggests that repeated ELS elicits a predictive adaptive response (PAR) where past stressful experiences augment coping with future stress (Gluckman et al., 2007). PAR is thought to be strongest in stress-susceptible individuals and may be evolutionarily conserved by rapid cross-generational environmental changes (Gluckman et al., 2007; van der Doelen et al., 2013). Our recent work lends further support to this theory. We exposed Wistar-Kyoto (WKY) offspring, a stress susceptible rat strain that exhibits behavioral abnormalities commonly associated with depression and anxiety disorders (Nam et al., 2014), to daily 3-hour maternal separation (MS-180) during the first two weeks of life. While MS-180 has frequently been used to demonstrate deleterious effects on adult rodent emotional behavior [for review see (Sanchez et al., 2001)], we found that MS-180 elicits several adaptive effects in adult WKY offspring, including diminished behavioral despair, increased social exploration (Rana et al., 2015), and improved cardiovascular measures, such as decreased baseline heart rate (HR) and increased HR variability (Rana et al., 2016).
The present study exploited the WKY-MS model to identify neurobiological and epigenetic mechanisms that mediate a PAR to convey adaptive behavioral and physiological changes in adult MS-180-exposed WKY offspring. We hypothesized that MS-180 triggers DNA methylation changes in the brains of WKY offspring, which may contribute to their enhanced stress resilience in adulthood (Rana et al., 2015). To test this, we first examined global DNA methylation (5-methylcytosine) levels in multiple brain regions of adult WKY rats that were exposed to MS-180 (or control condition). Because we found dramatic MS-180-induced hypermethylation in the WKY hippocampus, we then used next-generation sequencing to interrogate gene-specific methylation changes that occurred following MS-180 exposure. Our final experiment tested the hypothesis that enhancing DNA methylation in adult WKYs (by increasing dietary methyl donor content) would elicit adaptive behavioral and cardiovascular changes akin to what occurred following MS-180 exposure. Specifically, we predicted that feeding adult male WKYs a methyl-donor fortified diet would: (a) have anxiolytic and antidepressant effects in tests of anxiety- and depression-like behavior; and (b) elicit positive cardiovascular changes, including reduced baseline HR and greater HR variability, similar to what we previously observed in MS-180-exposed WKY offspring (Rana et al., 2015; Rana et al., 2016).
Materials and Methods
All experiments were approved by the Committee on the Use and Care of Animals at the University of Alabama at Birmingham. This work was performed in accordance with the National Institutes of Health (USA, 2011) and National Research Council (UK, 1996) guidelines on animal research.
Animals
Adult male and female WKY rats (n = 8/sex) were purchased from Charles River Laboratories (Kingston, NY) and housed in a temperature-controlled facility (kept at 21–23 °C, 50–55% humidity) with a 12/12 h light-dark cycle (lights on at 6:00 a.m.). For the first phase of the experiment, male/female pairs were mated for 14 days, and at birth (postnatal day 0) litters were randomly assigned to one of two groups: (a) MS-180 group or (b) neonatal handling (NH) group (n = 4 litters/group/strain). Litters assigned to the MS-180 and NH groups were separated from their dam daily for 180 min and 15 min, respectively, between 8:30 a.m. – 12:00 p.m. from postnatal day (P)1-P14 as previously described (Clinton et al., 2014; Rana et al., 2015). Dams remained in the home cages, while separated litters were transferred to a different room in a small cage placed on a heating pad (~37°C). Littermates remained in close contact throughout the separation period and were returned to home cage after the conclusion of the 15- (NH) or 180- (MS-180) min period. After the final separation on P14, litters remained undisturbed until weaning on P21. Afterwards, male rats of the same early-life experience (NH or MS-180) were housed together three per cage, and a subset of these animals (n = 10 NH, n = 10 MS-180) were utilized in subsequent behavioral and physiological studies that have been reported previously (Rana et al., 2015; Rana et al., 2016). Details of the behavioral and physiological studies can be found in these reports. The animals were euthanized during postnatal weeks 49–50, and their tissue was used for molecular studies reported here.
The second experiment manipulated dietary methyl donor content in a separate group of adult male WKY rats (n = 28) that was purchased from Charles River Laboratories (Kingston, NY). Upon arrival, rats were housed two per cage of the in our animal housing facility. Following a one-week acclimatization period, rats were assigned to either a diet depleted of (n = 14) or supplemented with (n = 14) methyl donors (see below). After four weeks of receiving these modified diets, rats were evaluated on a behavioral test battery and later instrumented with radiotelemetry probes (PA-C40, DSI International; n = 8 per group) for cardiovascular assessments (procedural details in subsequent sections).
Assessing global DNA methylation (5-methylcytosine) levels in MS-exposed WKY brain
We examined global DNA methylation (5-methylcytosine) levels in several brain regions of adult MS-180- and NH- exposed male WKY offspring. Brains were removed, flash frozen in isopentane cooled to −30°C on dry ice, and then stored at −80°C. Brains were sectioned on a cryostat at −10 to −12°C, and alternating sections of 20 and 300 μm were collected. The 20 μm sections were stained with cresyl violet to identify target anatomical regions in the 300 μm sections. Portions of the hippocampus, paraventricular nucleus of the hypothalamus (PVN), amygdala, septum, bed nucleus of the stria terminalis (BNST), and the medial prefrontal cortex (mPFC) were removed from the 300 μm-thick sections using a 0.5 mm tissue punch (Harris Micro-Punch, Ted Pella, Redding, CA). One group of samples was used to isolate DNA (DNeasy, Qiagen, Hilden, Germany; n=10/group), which was later quantified using the Nanodrop ND-1000 (Wilmington, DE), and stored at −20°C. Due to the small volume of tissue collected from PVN, these samples were pooled in pairs yielding an experimental sample size of five per group. We assessed levels of 5-methylcytosine (an indicator of DNA methylation) using the Epigentek MethylFlash Methylated DNA Quantification Kit (Colorimetric) per manufacturer’s instructions as described (Simmons et al., 2013). Briefly, 200 ng of genomic DNA was loaded per well and the samples were run in triplicate. Sample values were compared to a standard curve and to calculate the percent of methylated DNA as described in the manufacturer’s protocol.
Methylated DNA capture coupled with next-generation sequencing (MethylCap-Seq) in hippocampus of MS-exposed WKYs
To obtain a more detailed understanding of DNA methylation differences in the adult hippocampus of MS-180- vs. NH- exposed WKY offspring, we performed a MethylCap-Seq experiment. Methylated DNA was captured using a MethylMiner DNA enrichment kit (Applied Biosystems, Grand Island, NY, ME10025) according to the manufacturer’s recommended protocol. Adult MS-180/NH WKY hippocampus DNA samples (n = 4/group) were sheared by sonication using a Bioruptor Pico device (Diagenode, Denville, NJ, B01060001). The sonication cycle consisted of 15 s on-45 s off for seven total cycles to generate fragments of (on average) 325 base pairs for the methylated DNA enrichment protocol (Brinkman et al., 2010). Methylated DNA fragments were captured with a methyl-binding domain 2 (MBD2) protein coupled to paramagnetic Dynabeads® M-280 Streptavidin per manufacturer instructions and as described (Brinkman et al., 2010). Starting material for the enrichment protocol was 1μg of sonicated DNA, and captured fragments were eluted using 3,500 mM NaCl. Captured material was purified using QIAquick PCR purification spin columns (Qiagen, Valencia, CA); it was then quantified using Quant-it high sensitivity DNA Assay Kit (Invitrogen, Grand Island, NY, Q-33120) and the Agilent 2100 Bioanalyzer high sensitivity chip kit (Agilent Technologies, Santa Clara, CA). Samples were sent to HudsonAlpha Genomic Services Laboratory (Huntsville, AL; http://gsl.hudsonalpha.org) for next-generation sequencing using NEBNext reagents (New England Biolabs, Ipswich, MA) according to manufacturer’s recommendations. Barcoded DNA fragment libraries were created, checked for quality, and quantified with the Kapa Library Quant Kit (Kapa Biosystems, Wilmington, MA). Afterwards, each library was used for high-throughput sequencing on an Illumina HiSeq2000 (Illumina, San Diego, CA) with 25M total 50 base pair single-end reads per sample. Up to six barcoded samples were loaded per lane of a flowcell and sequenced using a paired-end 50-base pair protocol (according to manufacturer’s recommendations). We sequenced four biological replicates per group (with each derived from unique litters) as well as an input (non-captured) control for normalization.
To ensure that MBD2 protein capture resulted in specific enrichment of methylated DNA, we performed control reactions. In these reactions, gDNA was spiked with synthetic methylated and non-methylated DNA fragments (1 pg each, Methyl Miner kit, Invitrogen, Grand Island, NY) prior to immunoprecipitation with recombinant MBD2. We performed PCR with primers for these synthetic fragments within the methylated and non-methylated DNA capture, using both the captured (MBD2-bound) and unbound fractions. Our results demonstrated the presence of methylated DNA fragments in the captured sample, and absence of methylated DNA in the unbound fraction.
After next-generation sequencing was complete, we imported data files into Galaxy (https://usegalaxy.org/), an online data analysis system that facilitates large-scale genome analyses (Giardine et al., 2005; Blankenberg et al., 2010; Goecks et al., 2010). Raw single-end sequenced reads were quality controlled and filtered for read quality (FastQC, Galaxy). Sample reads were mapped onto the rat genome reference sequence (Rn5 assembly) using the high-performance alignment software Bowtie for Illumina (http://bowtie-bio.sourceforge.net/index.shtml). Overall we obtained an average of 27M mapped single-end reads from Methyl-Cap samples. Genome-aligned sequenced reads were examined using SeqMonk (Babraham Institute; www.bioinformatics.babraham.ac.uk/projects/seqmonk) and methylation levels were assessed via built-in analysis pipelines. We used Model-based Analysis of ChIP-Seq (MACS) for methylated peak detection through comparison to input and enriched samples and applying a p-value of 0.05 (Zhang et al., 2008). We then performed read count quantitation with reads per million and probe length corrections. Data were visualized using a Manhattan plot via qqman, an R package (Turner, 2014). Statistical analysis compared intensity differences between WKY-MS-180 and WKY-NH samples (p-value of 0.1 with Benjamini-Hochberg (BH) multiple test corrections) across the genome to evaluated distinct MS-180 vs. NH methylation patterns within several genomic features, including: CpG islands (GC content≥50%, length >200 base pairs); exons and introns within specific genes; and potential gene promoters (2 kb upstream from 5′ transcription start sites (TSS). Where applicable, methylation level is described in normalized read counts, which were normalized by reads per million and by the size of methylated peak (probe length). We performed Gene Ontology (GO) analysis for biological processes through WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/) with genes that have differentially methylated regions within the gene body or within 2 kb upstream of the TSS.
Quantitative real-time PCR confirmation of select genes showing differential methylation in hippocampus of MS-exposed WKY rats
In order to confirm that MS-180-related DNA methylation differences in the hippocampus lead to predicted mRNA expression changes within select genes, we conducted quantitative Real Time PCR (qRT-PCR) using a StepOne Plus (Applied Biosystems, Grand Island, NY, USA) with TaqMan detection chemistry as described by manufacturer. We examined the expression of the following genes: Insr (Rn00690703_m1; RefSeq: NM_017071), Map3k5 (Rn01440422_m1; RefSeq: NM_001277694.1), Igf1r (Rn00583837_m1; RefSeq: NM_052807.2) and Grik4 (Rn00561331_m1; NM_012572) and used Abl1 (Rn01436239_m1; NM_001100850) as a housekeeping gene. RNA was isolated (NucleoSpin RNA II) from hippocampal samples collected from adult WKY rats that were exposed to MS-180 or control condition in early life (n = 5 per condition, with each derived from unique litters). We reverse transcribed 100 ng of RNA to cDNA using Invitrogen SuperScript VILO (Invitrogen, Grand Island, NY). Reactions were carried out in 96-well qPCR plates (Qiagen), with each well loaded with 20 μl of a mix of cDNA, TaqMan qPCR mastermix (ThermoFisher Scientific), RNase-free water, and TaqMan primers. The PCR began with a two-min. hold at 50°C and then Taq Polymerase was activated by heating the plate to 95°C for ten min. Amplifications were performed for 40 cycles, each one consisting of 95°C for 15 s and 60°C for one min. Amplifications of all samples were carried out in triplicate, and the average cycle threshold (CT) was calculated as a geometric mean for each sample. Relative fold changes between WKY-MS-180/NH groups were compared for a given gene at a particular time point were calculated using the ΔΔCT method.
Dietary methyl donor manipulation, behavioral and physiological assessment in adult WKY rats
In order to begin to assess the functional implications of MS-180-induced DNA methylation changes in WKY offspring, we conducted an experiment to test whether inducing DNA hypermethylation in the adult WKY rat brain mimics behavioral and cardiovascular effects akin to what we observed in adult offspring with a history of early-life MS (Rana et al., 2015). To do so, we manipulated methyl donor content in the diet of adult WKY males by feeding them either a diet that was depleted of methyl donors or a diet supplemented with high levels of methyl donors. Adult male WKY rats purchased from Charles River were allowed to acclimate to our housing facilities for one week before being randomly assigned to receive either: (1) chow that was 90% depleted of methyl donors (WKY-DEP); or (2) a methyl supplemented diet (WKY-SUP; n = 14 WKY males per group). The WKY-DEP group received a rodent diet lacking 90% of normal requirements of choline, folate, and methionine (diet no. A04062402, Research Diets Inc, New Brunkswick, NJ), and the WKY-SUP group received chow fortified with increased amounts of cofactors and methyl donors (folic acid, choline, methionine, and Vitamin B12; diet. No. A04062403, Research Diets Inc, New Brunkswick, NJ; Table 1). The diets otherwise were matched for their overall caloric content and their L-amino acid content (except for L-methionine). Animals within both diet groups were matched for their caloric intake (Konycheva et al., 2011; Ishii et al., 2014) to receive 35 g of chow/cage/day during the first three weeks, and then increased to 40 g of chow/cage/day during the remaining 7 weeks of the experiment when behavioral testing and later cardiovascular assessments were conducted. Each cage housed two rats. Animals were weighed each week starting on the first day of diet exposure.
Table 1.
Composition of methyl-donor manipulated diets.
| Dietary component | Methyl-donor supplemented diet (SUP) | Methyl-donor depleted diet (DEP) |
|---|---|---|
| Methionine (g) | 7.50 | 0.60 |
| Choline (g) | 15.00 | 0.08 |
| Folic Acid (mg) | 15.00 | 0.20 |
| Zinc (mg) | 150.51 | 29.12 |
| Vitamin B12 (mg) | 0.01 | 0.01 |
| Betaine (g) | 15.00 | 0.00 |
| Methionine (g) | 7.50 | 0.60 |
| Choline (g) | 15.00 | 0.08 |
| Folic Acid (mg) | 15.00 | 0.20 |
| Zinc (mg) | 150.51 | 29.12 |
WKY rats were randomly assigned to either (1) a methyl donor supplemented (SUP) group receiving a diet enriched with higher than usual amounts of methyl donors and cofactors (folic acid, choline, methionine, and Vitamin B12; diet. No. A04062403, Research Diets Inc, New Brunkswick, NJ; termed methyl donor supplemented group (SUP); or (2) a methyl donor depleted (DEP) group fed chow that was 90% depleted of methyl donors (diet no. A04062402, Research Diets Inc, New Brunkswick, NJ).
Behavioral test battery
After receiving the methyl-depleted or methyl-supplemented diets for four weeks, rats were evaluated in a behavioral test battery comprised of several classic rodent tests of anxiety- and depression-related behavior: a) Open Field Test; b) a Social Interaction test; and c) the Forced Swim Test (FST). All rats were subjected to the full test battery in this test order, with 1–2 days’ rest between tests. Tests were performed between 8:00 a.m. – 12:30 p.m. and conducted under dim lighting (30 lux).
The open field test was conducted in a Plexiglas white box (100×100×50 cm) with a black floor. At the beginning of the test, a rat was placed into the corner of the box and was permitted to explore the apparatus for 5 min. The latency to enter the center of the open field, the amount of time spent and distance traveled in the center, periphery, and corners of the apparatus were quantified utilizing Ethovision® XT 8.0 videotracking software (Noldus, Wageningen, The Netherlands) set up with a digital video camera. A trained observer, who was blinded to experimental groups, manually assessed grooming and rearing behavior using a computerized system provided in the software.
The social interaction test was performed in a black Plexiglas box (30 × 90 × 60 cm) that was divided into three chambers (zones) separated by two black Plexiglas dividers with openings in the center to allow experimental rats to move freely between zones. The test was comprised a single 10-min session where the test rat was placed in the neutral zone (middle chamber), while one of the adjacent chambers contained a novel object (empty cage) and the third chamber contained a novel male stimulus rat within metal cylindrical interaction cage. The metal bars of the interaction cage allowed rats to interact, but prevented any aggressive encounters between animals. Stimulus male rats were age-matched and of the same strain as the test animals, and were previously habituated to interaction cages. Rats’ behavior was videotaped, and Ethovision® XT 8.0 videotracking software (Noldus, Wageningen, The Netherlands) was used to analyze the latency for experimental animals to enter chambers containing social stimulus rats as well as the number of visits and time spent in each chamber.
Porsolt’s FST was performed as we previously described (Nam et al., 2014) with 30-cm deep 25°C water in Plexiglas containers (45 cm high × 20 cm diameter). On FST day 1, rats were placed (1/cylinder) into the water for 15-min; 24 h later, the rats were returned to the water-filled cylinder and tested for another 5 min. Water was changed after every swim session, so that every rat swam in clean water. Rats were videotaped during both test days and immobility was scored using the Ethovision ® XT 8.0 software (Noldus, Wageningen, The Netherlands). We focused on the immobility measure since it is classically considered an indicator of behavioral despair and depressive-like behavior (Porsolt et al., 1977), and it can be clearly defined and easily distinguishable from active coping measures such as swimming and climbing, which are sometimes difficult to reliably distinguish across experimental observers (Cryan et al., 2005).
Radiotelemetry to assess cardiovascular function
In addition to assessing behavioral effects of manipulating dietary methyl donor content in adult male WKY rats, we also used radiotelemetry to monitor baseline and stress-evoked cardiovascular measures (e.g., heart rate [HR], HR variability [HRV], blood pressure) in WKY-DEP and WKY-SUP rats. We hypothesized that WKY-SUP rats would exhibit cardiovascular changes (relative to WKY-DEP) that resembled physiological alterations observed in MS-180-exposed WKY offspring.
At the conclusion of behavior testing, a subset of WKY males from the SUP and DEP groups (n = 7/condition) were randomly selected for radiotelemetry probe implantation for cardiovascular monitoring. Anesthesia was induced with 5% isoflurane and maintained with 2.0–2.5% isoflurane in oxygen delivered at 1 L/min. Pressure transducer at the tip of the catheter was implanted into the abdominal aorta and glued in place with surgical glue (Vetbond Tissue Adhesive; 3M, http://solutions.3m.com/), while the body of the device (PA-C40, Data Sciences International, St. Paul, MN) was sutured into the abdominal wall. Aseptic techniques were used throughout all the surgeries, and the animals were injected with carpofen (5 mg/kg; s.c.) and buprenorphine (0.1 mg/kg; s.c.) for pain control prior to surgery. Rats recovered from anesthesia in a warm, clean cage with water provided in a petri dish. They were then single-housed after the surgery and recovered for one week before the first baseline recordings were collected. Daily routine checks for weight, signs of distress (e.g. spiked coat, lethargy, rings around eyes), food and water intake, and excretory functions were performed throughout the recovery period. Animals showing signs of distress were administered buprenorphine (0.1 mg/kg s.c.). Rats were also treated with a topical antibiotic cream applied to the incision site on a daily basis during recovery.
Following the one week surgical recovery, rats were housed in a recording room equipped with DSI hardware and software (ART 4.3; Data Sciences International). Baseline systolic blood pressure (SBP), mean arterial pressure (MAP), diastolic blood pressure (DBP), HR, and locomotor activity were recorded over a 24-h period during week seven to eight of the study. Continuous blood pressure recordings were acquired with a sampling rate of 500 Hz with data averaged over ten seconds. Rats were kept undisturbed during recording sessions. Data were extracted from the DSI ART 4.3 analysis software as 30-min moving averages. HR, BP and activity recordings were analyzed separately during the 12 h light (inactive) and the 12 h dark (active) phases of the 24 h cycle. HRV was analyzed in the time domain and was calculated from the 24 h continuous blood pressure recordings that were binned into 5-min segments. The segment length was selected as per the recommendation from the Task Force of the European Society of Cardiology the North American Society of Pacing Electrophysiology (1996). HRV was calculated using standard deviation of inter-beat-interval (SDNN) during the 5-min segments. Data was averaged during the 12-h light phase and 12-h dark phase and further analyzed.
One week after the baseline cardiovascular measures were collected, rats were exposed to homecage intruder stress during the midpoint of the light phase of the 24 h cycle to determine stress-evoked cardiovascular responsivity in WKY-SUP versus WKY-DEP rats. Experimental rats (WKY-SUP or WKY-DEP animals) were subjected to homecage intrusion stress when an age-matched socially-housed intruder rats was introduced into their homecage for ten min. We sampled blood pressure and HR at 500 Hz and digitized data as ten second moving averages. Data were then averaged over one-min bins and extracted for offline analyses. Data were acquired for three epochs: (1) an initial baseline period 1–2 h before the intruder was introduced; (2) ten min during the home cage intrusion; and then (3) two h following intruder exposure to assess recovery. Maximal responses for HR and MAP in terms of percent change from baseline were calculated for each animal. We also quantified cardiovascular recovery from maximal responses using curve-fitting analysis. One phase decay for MAP and HR was used as the model to fit the data, using the formula: Y= (Y0 - Plateau)*exp(−K*X) + Plateau, where Y0 is maximal MAP or HR, K is the rate constant/decay constant, and X is time.
Tissue collection and processing
One day after the final cardiovascular assessment, WKY-DEP and WKY-SUP rats were euthanized by rapid decapitation to collect brain tissue, which was removed, flash frozen in isopentane cooled to −30°C on dry ice, and then stored at −80°C. Tissue punches from the hippocampus were collected from each rat as described above. Hippocampal samples were shipped to the Vanderbilt University Neurochemistry Core (https://medschool.vanderbilt.edu/vbi-core-labs/neurochemistry-core; Nashville, TN) for amino acid content quantification.
Statistical analysis
Data analyses for the MethylCap-seq experiment are described above. Data from the global DNA methylation (5-methylcytosine) assessment, qRT-PCR study, and methyl donor diet behavioral and cardiovascular studies were analyzed using GraphPad Prism Software (Version 6.0 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com). All data sets were first verified to be normally distributed using the D’Agostino & Pearson omnibus normality test. In the methyl donor diet manipulation studies, rats’ weight was analyzed by two-way ANOVA, with age and diet condition as independent variables. Data from the qRT-PCR and behavioral studies were analyzed with a two-tailed Student’s t-test, or with the Mann-Whitney U test if not normally distributed. For all analyses, α<0.05. Because some of the samples used in the global DNA methylation assay were derived from animals from the same litter, a covariate analysis was performed using litter as an additional factor. Accordingly, ANCOVA was performed using SPSS (Version 24) with MS/NH treatment as the independent variable, litter as covariate, and 5-methylcytosine as dependent variable. Blood pressure and HR data averaged over 30 min intervals were analyzed using repeated measures two-way ANOVA, with time and diet-manipulation treatment as factors. Significance was set at p < 0.05, and results are presented as mean ± SEM.
Results
Early-life MS elicits global DNA hypermethylation in the hippocampus of WKY offspring
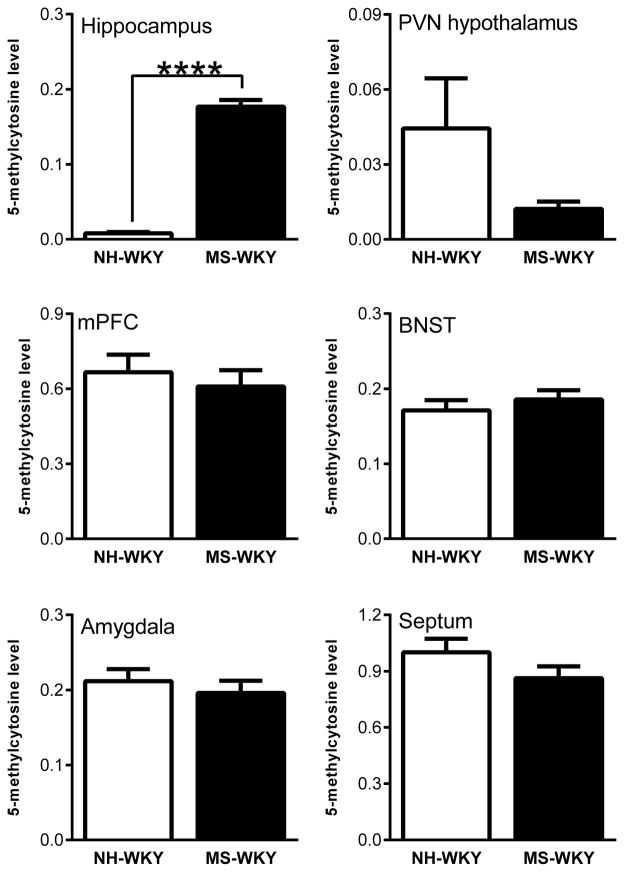
Since early-life experiences are known to shape adult behavior and physiological stress responses, in part, via DNA methylation modifications in brain regions that regulate stress responsivity (Weaver et al., 2002a; Meaney & Szyf, 2005; Szyf et al., 2005; Weaver et al., 2006), we quantified global DNA methylation (5-methylcytosine) levels in several limbic brain regions (hippocampus, PVN of the hypothalamus, amygdala, BNST, septum, and mPFC) in MS-180-exposed vs. control WKY adult males. MS-180-exposure elicited a dramatic increase in DNA methylation specifically in the hippocampus of WKY offspring compared to NH-exposed controls (F=35.257, p< 0.001). Litter was not a significant source of variation (F=1.114, p = 0.31). There was no effect of MS-180 on 5-methylcytosine levels in the PVN (t (8) = 1.59, n.s.), mPFC (t (15) = 0.60, n.s.), amygdala (t (16) = 0.683, n.s.), septum (t (16) = 1.46, n.s.), or BNST (t (8) = 0.78, n.s.; Fig. 1).
Figure 1.
Effects of neonatal maternal separation (MS) on global DNA methylation (5-methylcytosine) in the adult Wistar Kyoto (WKY) rat brain. WKY rats were exposed to daily 180-min MS or 15-min neonatal handling (NH) between postnatal days (P) 1–14. Adult offspring were sacrificed and several brain regions were dissected: the hippocampus, paraventricular nucleus (PVN) of the hypothalamus, amygdala, septum, medial prefrontal cortex (mPFC), and bed nucleus of the stria terminalis (BNST). DNA was extracted and used to measure 5-methylcytosine, an indicator of global DNA methylation. MS exposure dramatically increased 5-methylcytosine levels in the adult WKY hippocampus relative to NH controls, but did not significantly alter DNA methylation levels in the PVN, mPFC, amygdala, septum or BNST Data represent mean ± SEM. **** Statistically significant differences at p < 0.0001.
Next-generation sequencing reveals disparate genome-wide as well as gene loci-specific DNA methylation patterns in the MS- versus NH-exposed WKY hippocampus
To delve more deeply into DNA methylation changes elicited in the hippocampus of MS-180- exposed WKY male offspring, we utilized next-generation methylome sequencing. This technology allowed us to sequence methylated DNA fragments with the resolution of ~300 bp. High-throughput sequencing collected >25M reads per sample, which were mapped onto the rat genome reference sequence (rn5), and analyzed with MACS peak calling to examine total number of sites enriched for DNA methylation. This analysis identified 308,904 methylated genomic sites. These sites were subjected to further analysis to identify the sites that displayed differential methylation between MS-180 and control groups. For analysis of these data, we chose to use two methods: 1) first, we subjected the data to stringent thresholds to obtain only the sites with the high level of differential methylation, 2) we utilized more liberal statistical thresholds to allow identification of possible patterns within genetic elements and to find groups of genes that were enriched in differentially DNA methylation in MS/NH samples.
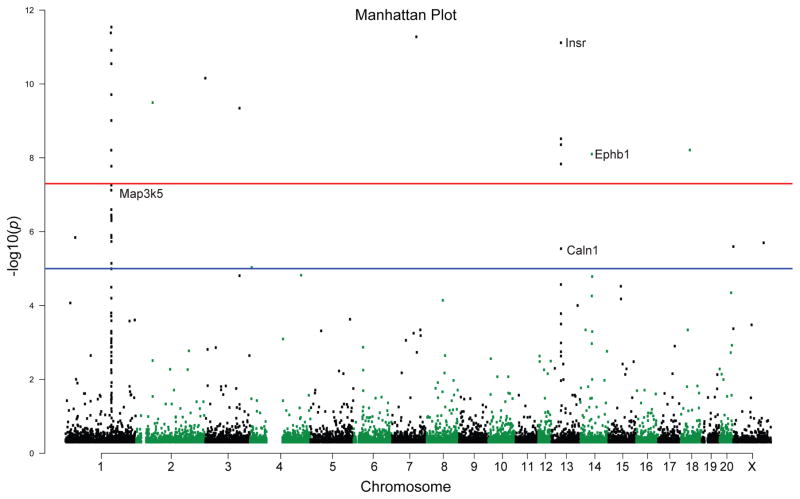
To visualize the occurrence of differentially methylated regions (DMRs) across the genome, we created a Manhattan Plot (Fig. 2). We examined genes that surpassed two thresholds: a) a threshold p-value < 1 × 10−5, the suggested threshold in genome-wide studies (indicated by a blue line in Fig. 2); and then b) a second more stringent threshold p-value < 5 × 10−8 (indicated by a red line in Fig. 2). A group of 56 DMRs, which included 15 gene regulatory sites (a region within the gene body or up to 10 kb upstream of a gene) surpassed the lower (blue line) threshold. A group of 40 DMRs within 12 gene regulatory sites passed the more conservative analytical parameters of the higher (red line) threshold. Some example genes are displayed in the plots in Fig. 2, including: Caln1, Map3k5, Insr and Ephb1.
Figure 2.
Manhattan plot illustrating differentially methylated sites in the adult hippocampus of MS-exposed versus control WKY rats. Differentially methylated regions (DMRs) are shown by chromosome (along the x-axis) and the negative logarithm of the association p-value for each block (−log10 [p-value]) on the y-axis). We examined whether DMRs surpassed two thresholds: a) a threshold p-value less than 1 × 10−5, the suggested threshold in genome-wide studies, indicated by a blue line; and then b) a second more stringent threshold p-value less than 5 × 10−8, indicated by a red line. There were 56 DMRs in the MS versus control adult hippocampus that surpassed the lower (blue line) threshold. This group of DMRs included 15 gene regulatory sites (a region within the gene body or up to 10 kb upstream of a gene). When considering the more stringent parameters of the higher (red line) threshold, we found 40 DMRs within 12 gene regulatory sites that passed that threshold. Some example genes that met these criteria are labelled: ephrin type-B receptor 1 (Ephb1), insulin receptor preprotein (Insr), mitogen-activated protein kinase kinase kinase 5 (Map3k5), calcium-binding protein 8 (Caln1)
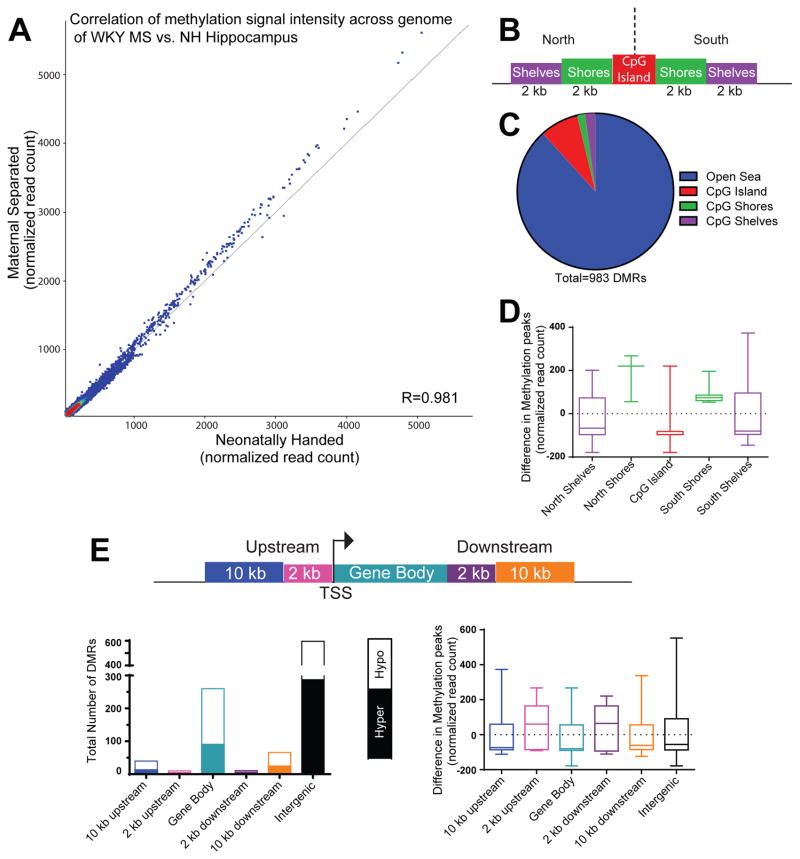
We also visualized MS-180/control hippocampal methylation differences by creating a correlational matrix with intensity levels for methylation differences (normalized reads per kb) across 308,904 genomic sites (MS-180 samples represented along the y-axis and NH samples along the x-axis). There was a strong correlation in signal across these sites (R=0.981; Fig. 3A).
Figure 3.
Early-life maternal separation (MS) exposure elicits lasting DNA methylation changes within several genomic regions of WKY hippocampus. (A) Correlation analysis demonstrated a strong correlation in signal across 308,904 genomic sites that were differentially between samples from MS-exposed and control rats. MS samples represented along the y-axis and neonatal handle control samples along the x-axis. The shift towards increased signal intensity above the diagonal line indicates greater methylation signal in MS versus control hippocampal samples. (B) Graphic depicts an example CpG Island (in red) surrounded by CpG island north and south shores (in green, 0–2 kb from CpG island edges) and CpG island north and south shelves (in purple, 2–4 kb from island edges). Genomic regions >4 kb from any CpG island edge is considered “open sea”. (C) Of the 983 differentially methylated regions (DMRs) between MS and control WKY hippocampal samples, the majority were located in “open sea”, with approximately 12% of the DMRs occurring within CpG islands, shores and shelves. (D) Within the CpG islands and surrounding shores and shelves, sites within the CpG islands, north and south shelves were hypomethylated in MS versus control samples, while sites within north and south shores were hypermethylated between the groups. (E) Methylation data were also classified depending upon where DMRs fell within specific genomic regions, including within: 10 kb or 2 kb upstream of the transcription start site (TSS), the gene body, 2 kb or 10 kb downstream of the gene body. The highest number of DMRs occurred within exons or intergenic regions. On average, sites within regions 2 kb upstream or downstream of a gene body were hypermethylated in MS compared to control samples; DMRs within gene bodies, 10 kb upstream or 10 kb downstream of genes were hypomethylated.
Our next phase of analysis determined the number of MS-180/NH DMRs (p< 0.1 BH correction) within specific genomic features (3B–E), first focusing on CpG islands, defined as >200 base pair stretches of DNA with GC percentage greater than 50% and observed-to-expected CpG ratio greater than 0.6 (Fig. 3B–D). Of the 983 DMRs identified between MS-180 and control hippocampal samples, 7.93% of those sites were within CpG islands; 1.63% of the sites were located within CpG shores (0–2 kb from CpG island edges); and 2.14% of the sites were located within CpG shelves (2–4 kb from island edges). Thus, the majority of DMRs were located in “open sea”, defined as genomic regions >4 kb from any CpG island edge (Fig. 3C). We also separately considered DMRs within regions located either upstream (“north”) or downstream (“south”) of CpG islands, considering relative methylation level (normalized reads per kb) in MS-180 versus control samples. Here, we found that, on average, sites within CpG islands as well as north and south shelves were hypomethylated in MS-180 versus control samples, while sites within north and south shores are hypermethylated in MS-180 versus control (Fig. 3D). We next annotated methylation data based the type of genomic element where DMRs were located, including within: 10 kb or 2 kb upstream of the transcription start site (TSS), the gene body, 2 kb or 10 kb downstream. On average, sites within regions 2 kb upstream or downstream of a gene body were hypermethylated in MS-180 compared to control samples; DMRs located within gene bodies, 10 kb upstream or 10 kb downstream of genes were hypomethylated. While many studies of MS-180 investigate the differentially methylation at gene promoters, our data show that the highest number of DMRs was located within intra- and inter- genic regions (Fig. 3E).
We next performed Gene Ontology (GO) analysis on the 983 DMRs (which corresponded to 46 mapped genes) and identified several biologically relevant ontological terms that were significantly over-represented among the genes differentially methylated in the MS-180-exposed versus control WKY hippocampus. Table 2 shows a list of the top ten GO terms and associated genes, which are involved in a host of neuronal processes including: cell proliferation, axonal guidance, tyrosine kinase signaling, and synaptogenesis and synaptic transmission. Table 3 provides details on the methylation differences detected within these genes. From this gene list we selected four genes – the insulin receptor preproprotein gene (Insr), two molecules downstream of insulin receptor signaling (Map3k5 and Igf1r), and glutamate receptor kainate-4 (Grik4) – for follow-up qRT-PCR experiments to test whether observed MS-180-induced methylation changes within these genes was accompanied by a significant increase in mRNA levels. Our MethylCap-seq revealed hypomethylated DMRs in Insr and Map3k5 and hypermethylated DMRs in Igf1r and Grik4. Our results revealed a 1.60 fold increase in hippocampal Insr mRNA levels in MS-180-exposed WKY hippocampus relative to control (t8= 2.87, p< 0.05). For Map3k5, we found a 1.40 fold change increase in MS-180 vs. NH rats (t8=2.53, p<0.05). For Igf1r, we found a 1.45 fold change increase in MS-180 vs. NH rats (t8=3.37, p<0.01). For Grik4, we found a 1.49 fold change increase in MS-180 vs. NH rats (t8=3.98, p<0.01). As negative controls, we also measured the gene expression in Jmjd8 (t8= 0.9431, n=5, p = 0.37) and Dnaz (t8= 0.02514, p = 0.98). These results are summarized in Table 4.
Table 2.
Biological pathways enriched in genes showing differential methylation in maternal separation versus control WKY rat hippocampal tissue.
| Biological Process Pathways | p-value | enrichment ratio | Genes in Pathway | |
|---|---|---|---|---|
| 1 | positive regulation of cell proliferation | 0.008 | 5.42 | Sphk2, Nf1, Nacc1, Cdh13, Peli1, Efnb1, Insr, St8sia1, Ntn1 |
| 2 | cell proliferation | 0.015 | 3.42 | Sphk2, Nf1, Nacc1, Cdh13, Peli1, Efnb1, Insr, St8sia1, Ntn1, Ephb1, Vpreb1, Fgfrl1 |
| 3 | transmembrane receptor protein tyrosine kinase signaling pathway | 0.023 | 6.62 | Cdh13, Efnb1, Insr, Ephb1, Fgfrl1, Atxn1 |
| 4 | regulation of cell proliferation | 0.023 | 3.53 | Sphk2, Nf1, Nacc1, Cdh13, Peli1, Efnb1, Insr, St8sia1, Ntn1, Fgfrl1 |
| 5 | protein modification process | 0.023 | 3.53 | Eya2, Ephb1, Nf1, Peli1, Hus1, Insr, Lrp8, St8sia1, Fgfrl1, Prdm5, Hs3st5, Hecw2 |
| 6 | macromolecule modification | 0.090 | 2.28 | Eya2, Ephb1, Nf1, Peli1, Hus1, Insr, Lrp8, St8sia1, Fgfrl1, Prdm5, Hs3st5, Hecw2 |
| 7 | locomotion | 0.090 | 2.20 | Ephb1, Nf1, Cdh13, Efnb1, Insr, Lrp8, Hs3st5, Ntn1 |
| 8 | cellular protein modification process | 0.090 | 3.19 | Eya2, Ephb1, Nf1, Peli1, Hus1, Insr, Lrp8, St8sia1, Fgfrl1, Prdm5, Hs3st5, Hecw2 |
| 9 | synaptic transmission | 0.090 | 2.28 | Grik4, Ephb1, Nf1, Atxn1, Lrp8 |
| 10 | regulation of locomotion | 0.090 | 3.98 | Nf1, Hs3st5, Cdh13, Insr, Ntn1 |
This list represents the top ten gene onology (GO) biological processes (p < 0.1) along with their associated p-value for enrichment, ratio of enrichment (calculated based on number of genes observed versus those expected), and the genes that appeared within each pathway. The gene list used for this analysis included those containing differentially methylated regions (DMRs) that occurred within the gene body or within 2 kb upstream of gene (p < 0.1).
Table 3.
Methylation differences in genes enriched in Gene Ontology biological processes
| Gene Symbol | Description | Methylation Difference MS-NH (corr reads per million) | Corr p- value | Distance from TSS (bp) |
|---|---|---|---|---|
| Ephb1 | Ephrin type-B receptor 1 | −18.918 | < 1E-10 | 73,848 |
| Insr | insulin receptor preproprotein | −13.984 | < 1E-10 | 145,195 |
| Peli1 | pellino homolog 1 | −0.731 | 0.00533 | 160 |
| Fgfrl1 | Fibroblast growth factor receptor-like 1 | −0.844 | 0.02583 | 3,050 |
| Nf1 | neurofibromin | 0.587 | 0.04151 | 115,808 |
| Efnb1 | ephrin-B1 precursor | 0.643 | 0.04255 | 8,937 |
| St8sia1 | alpha-N-acetylneuraminide alpha-2,8-sialyltransferase | 0.660 | 0.05003 | 44,729 |
| Ntn1 | netrin-1 precursor | 0.658 | 0.05732 | 55,487 |
| Nacc1 | Nucleus accumbens- associated protein 1 | −0.733 | 0.07153 | 166 |
| Vpreb1 | pre-B lymphocyte 1 precursor | 0.588 | 0.07535 | 39,103 |
| Sphk2 | sphingosine kinase 2 | −0.808 | 0.07799 | 2,666 |
| Hs3st5 | heparan sulfate glucosamine 3-O- sulfotransferase 5 | −7.537 | 0.00029 | 182,073 |
| Grik4 | glutamate receptor, ionotropic kainate 4 precursor | 0.613 | 0.09167 | 250,769 |
| Atxn1 | Ataxin-1 | 0.622 | 0.07918 | 265,815 |
| Lrp8 | LDL Receptor Related Protein 8 | −0.887 | 0.00661 | 41,604 |
| Eya2 | eyes absent homolog 2 | 0.629 | 0.09848 | 35,749 |
| Hus1 | HUS1 checkpoint Clamp component | 0.636 | 0.05732 | 4,889 |
| Prdm5 | PR domain 5 | 0.854 | 0.04823 | 113,670 |
| Hecw2 | E3 ubiquitin-protein ligase | −0.713 | 0.09527 | 219,332 |
| Cdh13 | cadherin-13 precursor | 0.612 | 0.09065 | 506,167 |
The genes listed here were found to be differentially methylated in MS versus NH hippocampus after statistic threshold of p < 0.1 and were a part of the top ten GO biological processes shown in Table 3. For each gene, the methylation difference between MS vs. NH groups are displayed in corrected reads per million. For the differentially methylated region (DMR) within the gene, the corrected p-value and the distance of the DMR from the transcription start site (TSS) are listed.
Table 4.
Genes showing differential methylation and expression in adult WKY rat hippocampus following maternal separation
| Gene Symbol | Description | Difference in methylation MS-NH | Corr P- value | Distance from TSS (kb) | qPCR FC | qPCR p-value |
|---|---|---|---|---|---|---|
| Ephb1 | Ephrin type-B receptor 1 | −18.918 | < 1E-10 | 73.85 | 1.05 | NS |
| Insr | insulin receptor preproprotein | −13.984 | < 1E-10 | 145.20 | 1.60 | p < 0.05 |
| Map3k5 | mitogen-activated protein kinase kinase kinase 5 | −9.045 | 6.60E-07 | 146.41 | 1.40 | p < 0.05 |
| Caln1 | Calcium-binding protein 8 | 0.994 | 2.73E-06 | 336.07 | ||
| Grik4 | glutamate receptor, ionotropic kainate 4 precursor | 0.613 | 0.092 | 250.77 | 1.49 | p < 0.01 |
| Igf1r | Insulin-like growth factor 1 receptor | 0.322 | > 0.1 | 65.27 | 1.45 | p < 0.01 |
This list includes genes that were found to pass the suggested threshold for genome-wide studies and candidate genes (shaded gray). Predicted gene sequences not shown. Igf1r was chosen due to its relationship with Insr. Grik4 was chosen based on previous work showing differentially methylation in other models. The corrected p-value, distance from TSS, and difference in methylation between MS and NH (corrected reads per million) are shown for each gene. Selected targets were also chosen for qPCR and their fold change (FC) and p-value is shown.
Increasing methylation (via dietary methyl donor supplementation) improves WKY rats’ behavior and physiology, closely resembling the effects of MS
To begin to examine the functional implications of MS-180-elicited increases in DNA methylation, we manipulated dietary methyl donor content in diet of adult WKY males (by either depleting methyl donor content or supplementing it). Since our prior studies found that MS-180 exposure produced several adaptive behavioral and physiological changes in WKY offspring together with dramatically increased hippocampal DNA methylation, we predicted that dietary methyl donor supplementation would mimic these adaptive effects. At the conclusion of behavioral and cardiovascular assessments in WKY rats receiving either a diet enriched with (WKY-SUP) or 90% depleted of (WKY-DEP) methyl group donors, brain tissue was collected. To test the efficacy of the diet manipulation, we assessed amino acid content within the hippocampus of WKY-SUP and WKY-DEP rats and found that methionine levels were increased by 31% in WKY-SUP versus WKY-DEP brain tissue (WKY-SUP 2.48 ± 0.11 versus WKY-DEP 1.89± 0.11; t14=3.74, p=0.002).
Effect of methyl donor diet manipulation on body weight
As noted in the Methods section, all groups were food-restricted to ensure equal feeding between groups since previous studies found significantly diminished food intake in rodents receiving a methyl-depleted diet (Konycheva et al., 2011; Ishii et al., 2014). The WKY-SUP and WKY-DEP groups were weighed once per week starting on the first day of diet exposure through the end of behavior testing. All rats gained weight during the diet manipulation period (effect of time [F12, 312=1211, p<0.0001]), although WKY-SUP rats were consistently heavier than WKY-DEP rats (WKY-SUP average weight over the course of the study was 219±2 g versus WKY-DEP average weight of 185±2 g; effect of diet [F1, 26=119, p<0.0001]), which is consistent with previous reports (Konycheva et al., 2011; Ishii et al., 2014).
Behavioral effects of increasing methylation via dietary methyl donor supplementation
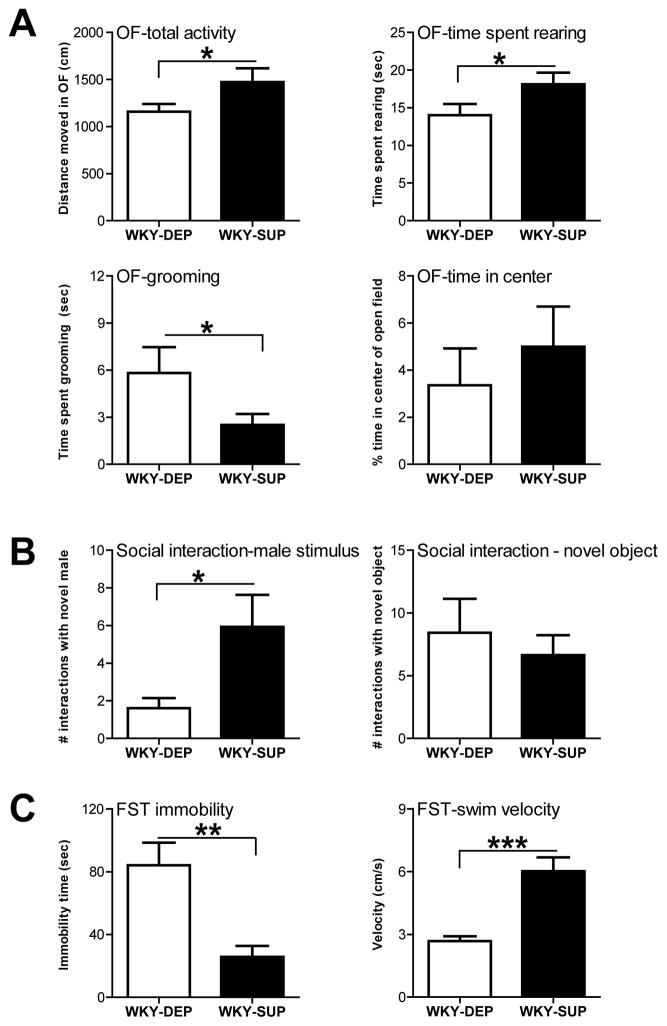
Adult WKY males that were subjected to either the methyl donor supplemented or depleted diets were evaluated in multiple tests of anxiety- and depression-like behavior. WKY rats that received the methyl donor supplemented diet (WKY-SUP) showed greater exploratory behavior in the open field (OF), traversing a greater distance during the test (t (26) = 1.89, p<0.05), showing more rearing (t (26) = 1.93, p<0.05), and decreased grooming behavior (t (26) = 1.87, p<0.05) compared to WKY rats receiving a methyl donor depleted diet (WKY-DEP). Although both groups spent a similar amount of time in the center of the OF (t (26) = 0.71, p=0.24; Fig. 4A). In the social interaction test, WKY-SUP rats made more frequent visits to a male social stimulus rat compared to WKY-DEP rats (t (26) = 2.33, p<0.05); both groups made a similar number of visits to an inanimate novel object (t (26) = 0.57, n.s.; Fig. 4B). In the FST, WKY-SUP rats showed less depression-like behavior compared to WKY-DEP rats, spending significantly less time immobile (t (26) = 3.65, p<0.001) and greater swimming velocity (t (26) = 4.76, p<0.0001; Fig. 4C).
Figure 4.
Dietary methyl donor supplementation in WKY rats has anxiolytic and antidepressant effects. (A) WKY rats were fed either a diet depleted (WKY-DEP) of methyl donors (e.g., folate, choline, etc.) or a diet supplemented (WKY-SUP) with excess methyl donors for 4 weeks before behavioral testing commenced and throughout the remainder of the study. In the Open Field Test, WKY-SUP rats exhibited increased exploration in the novel environment compared to WKY-DEP rats. WKY-SUP rats traversed a greater distance in the open field and spent more time rearing compared to WKY-DEP rats. Although the two groups spent a similar amount of time in the center of the open field, WKY-SUP rats showed less grooming behavior, suggesting a subtle decrease in anxiety-like behavior. (B) In the social interaction test, methyl donor supplementation subtly improved WKYs’ typically low levels of social interaction. WKY-SUP animals made more frequent visits to the male social stimulus rats compared to the WKY-DEP group, but showed similar exploration of a novel object. (C) In the Forced Swim Test (FST), dietary methyl donor supplementation had an antidepressant effect, with WKY-SUP rats showing decreased immobility and greater swimming velocity relative to WKY-DEP rats. Data represent mean ± SEM. *Statistically significant differences at p < 0.05; ** indicates p <0.001; *** indicates p <0.0001.
Baseline cardiovascular effects of increasing methylation via dietary methyl donor supplementation
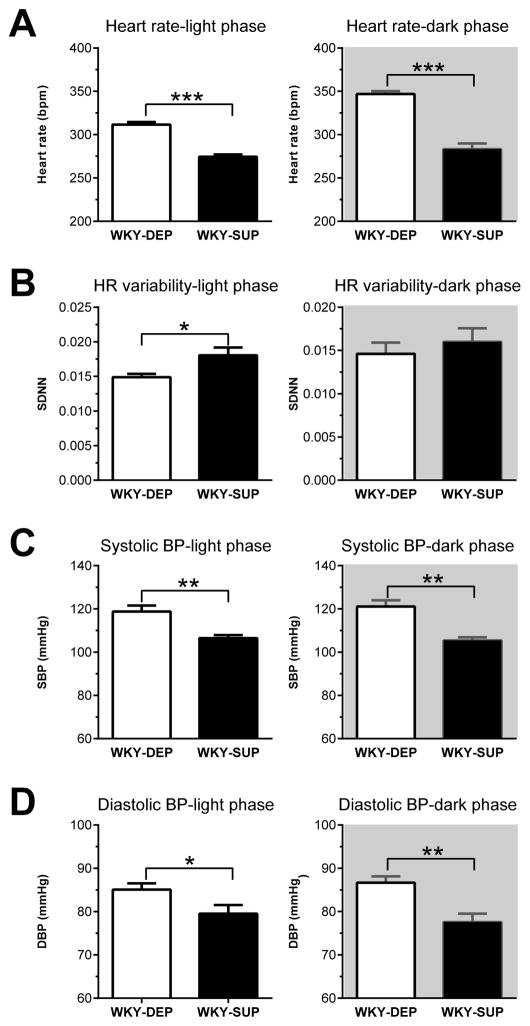
For resting HR, WKY rats that received a methyl-donor supplemented diet (WKY-SUP) showed decreased HR during both the light (t (12) = 8.96, p<0.0001) and dark phase (t (12) = 8.63, p<0.0001) of the 24 h cycle compared to methyl donor depleted rats (WKY-DEP; Fig. 5A). These diet-induced decreases in resting HR were accompanied increased HRV (SDNN) in WKY-SUP versus WKY-DEP rats during the light phase (t (12) = 2.63, p<0.05), but not the dark phase (t (12) = 0.69, n.s.; Fig. 5B). Dietary methyl donor supplementation also decreased resting systolic blood pressure (SBP) and resting diastolic blood pressure (DBP) relative to WKY-DEP rats during both the light phase (SBP: t (12) = 3.94, p<0.001; DBP: t (12) = 2.27, p<0.05) and dark phase (SBP: t (12) = 4.91, p<0.001; DBP: t (12) = 3.71, p<0.01) of the 24 h cycle (Fig. 5C–D). Supplemental Fig. 1 presents HR and BP data in 30-min increments for SUP and DEP rats across the 24-h assessment period.
Figure 5.
Dietary methyl donor supplementation leads to reduced baseline heart rate (HR), blood pressure (BP), and heart rate variability in WKY rats. (A) WKY rats that received a methyl donor-supplemented diet (WKY-SUP) showed substantially lower resting HR compared to those receiving a methyl donor depleted diet (WKY-DEP) during both the light and dark phases of a 24 h period. (B) These diet-induced changes in resting HR were accompanied by increased heart rate variability (standard deviation of inter-beat-interval; SDNN) in WKY-SUP versus WKY-DEP rats during the light phase, but not the dark phase (C–D) WKY-SUP rats also showed lower SBP and DBP compared to WKY-DEP rats during the light and dark phases. Data represent mean ± SEM. *Statistically significant differences at p < 0.05; ** indicates p <0.001; *** indicates p <0.0001.
Stress-Evoked Cardiovascular Responses
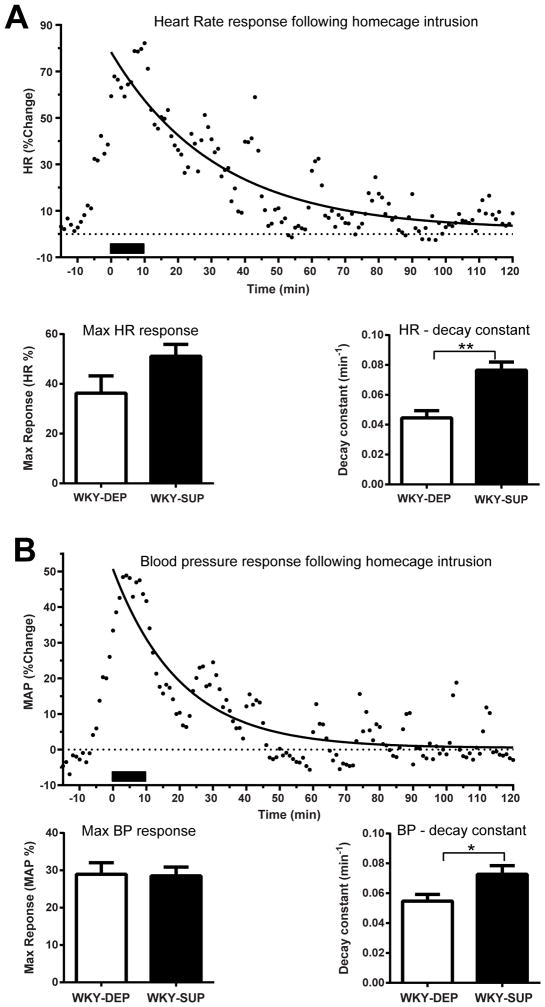
Rats were briefly exposed to 10 min home cage intrusion when a novel age-matched male intruder rat entered their cage. Maximal HR and MAP response was determined by the maximal change in the value from baseline during the test period. In addition, curve-fitting analyses were performed to evaluate potential differences in HR and MAP post-stress recovery. This analysis revealed significantly different curves that best represent HR recovery data between groups (F3, 1688 = 14.28, p < 0.0001). The R2 (i.e. goodness of fit index) for each curve was 0.33 and 0.49 for DEP and SUP groups, respectively, with the following estimated values: Y0 – 45.05 (DEP), 70.86 (SUP); Plateau – −1.94 (DEP), 0.88 (SUP), K – 0.045 (DEP), 0.076 (SUP). Data from individual rats were then curve fitted to estimate group differences in specific parameters of the recovery. Although SUP and DEP groups displayed similar maximal HR responses following home cage intrusion, SUP rats exhibited greater decay constant than the WKY-DEP group (t (12) = 4.37, p < 0.001), indicating faster HR recovery (Fig. 6A). Mean Arterial Pressure (MAP) responses to intruder exposure were also assessed in a similar fashion to HR data. Curve-fitting of grouped data (R2 = 0.45 DEP, R2 = 0.45 SUP) showed a significant difference between groups (F3,1688 = 4.268, p = 0.0052) with the following estimated values: Y0 – 36.72 (DEP), 38.19 (SUP); Plateau – −2.49 (DEP), −0.31 (SUP), K – 0.055 (DEP), 0.073 (SUP). Although SUP and DEP groups showed similar maximal MAP responses to home cage intrusion, SUP rats exhibited a modest, albeit significant increase in the decay constant (t (12) = 2.45, p < 0.05), suggesting faster MAP recovery (Fig. 6B).
Figure 6.
Dietary methyl donor supplementation decreases WKY rats’ cardiovascular response to acute stress of home cage intrusion. (A) A novel age-matched male intruder rat was introduced into the home cage of each methyl donor supplemented (WKY-SUP) or depleted (WKY-DEP) WKY test rat for 10 min. Maximal heart rate (HR) response was determined by the maximal change in the value from baseline (15 min average prior to the beginning of the test) during the test period. Curve-fitting analysis was also performed to determine HR recovery after stress; solid line indicates curve-fitting analysis in a representative animal. WKY-SUP and WKY-DEP groups displayed similar maximal HR responses to the intrusion, although WKY-SUP rats showed greater HR decay constant, indicating steeper rate of recovery. (B) Similarly, the two groups displayed similar maximal mean arterial pressure (MAP) response to home cage intrusion, with the WKY-SUP rats showing increased MAP decay. *Statistically significant differences at p < 0.05; ** indicates p <0.001.
Discussion
Stressful experiences during early life can shape the developing brain and lead to life-long behavioral and physiological effects (Heim et al., 2002; Apter-Levy et al., 2013). Although a preponderance of studies focuses on the harmful effects of early-life stress, accumulating evidence suggests that such experiences can lead to positive effects in certain individuals (Gluckman et al., 2007; Santarelli et al., 2014; Rana et al., 2015). WKY rats are known to be a highly stress-susceptible strain, exhibiting abnormalities within behavioral domains commonly associated with depression and anxiety disorders, high levels of hypothalamic pituitary adrenal axis (HPA) stress reactivity, as well as proclivity to stress-induced ulcers (Pare & Redei, 1993; Malkesman et al., 2006; Overstreet, 2012; Nam et al., 2014). Although WKY rats are vulnerable to stress exposure during adulthood, our recent study showed that early-life MS-180 elicits protective effects in WKYs, leading to increased exploratory behavior, enhanced sociability, decreased anxiety-like behavior, diminished helplessness in the FST (Rana et al., 2015), while producing the opposite effects in the stress resilient Wistar rats (Rana et al., 2015). These positive behavioral changes are accompanied by adaptive cardiovascular alterations (i.e. decreased baseline HR and increased HRV) (Rana et al., 2016). The present experiments aimed to identify neural and molecular mechanisms that may confer these adaptive behavioral and physiological changes in MS-180-exposed WKY offspring. We found that MS-180 elicits a dramatic increase in global DNA methylation in the hippocampus, but not other brain regions studied, suggesting that this epigenetic process contributes to the effects of MS-180 in WKY rats. Next-generation sequencing identified a number of gene-specific methylation differences in the MS-180-exposed WKY hippocampus, including altered methylation of genes encoding the insulin receptor and its downstream targets. Our final experiment manipulated DNA methylation in WKY rats by depleting or supplementing dietary methyl donor content to test the hypothesis that enhancing DNA methylation in brains of adult WKY rats (via increased dietary methyl donor content) would recapitulate behavioral and cardiovascular changes akin to the effects of MS-180. As predicted, dietary methyl donor supplementation improved WKY’s anxiety- and depression-like behavior and improved cardiovascular parameters relative to dietary methyl donor-depleted WKY rats. Overall these data suggest a potential molecular mechanism that mediates a PAR whereby early-life stress induces DNA methylation changes in the brain that may contribute to successful stress coping in adulthood.
MS-exposure specifically affected WKY hippocampus, with no significant methylation changes in other limbic brain regions
Early-life MS-180 has been shown to elicit a number of structural and molecular changes in the developing and adult brain, particularly in the hippocampus, prefrontal cortex, and hypothalamus. For example, MS-180 was found to alter hippocampal neurogenesis (Lajud et al., 2012), cell proliferation (Hulshof et al., 2011), dendritic length (Monroy et al., 2010), dendritic spine density (Monroy et al., 2010), and mossy fiber density within the hippocampal subfields (Huot et al., 2002). These structural changes appear to be accompanied by a host of molecular alterations, including altered expression of glucocorticoid receptors (Meaney et al., 1996; Ladd et al., 2004), glutamate receptor subunits (Pickering et al., 2006), histone deacetylases (Suri et al., 2014), and histone methyltransferases (Suri et al., 2014) along with increased histone methylation (Suri et al., 2013) and hypermethylation of the glucocorticoid receptor gene (Kundakovic et al., 2013). MS-180 exposure has also been shown to trigger structural and molecular changes in the prefrontal cortex, including decreased dendritic length and spine density (Monroy et al., 2010), atrophy of basal dendritic tree and reduced spine density apical/basal dendrites in layer II/III pyramidal neurons, impaired long-term potentiation, and increased expression of AMPA glutamate receptor subunits (GLUR1 and GLUR2), PSD95 and CamKII (Chocyk et al., 2013). In the hypothalamus, aside from MS-180-induced changes in corticotrophin releasing hormone (CRH) and arginine vasopressin (AVP) production (Plotsky & Meaney, 1993; Ladd et al., 2005; Plotsky et al., 2005), early-life MS-180 also leads to greater overall volume of the PVN and supraoptic nucleus as well as decreased serotonergic innervation of the hypothalamus (Lukas et al., 2010). Based on these previous findings of MS-180 impacting multiple limbic regions, our first experiment examined possible MS-180-induced global DNA methylation differences in several brain areas (the hippocampus, mPFC, PVN of the hypothalamus as well as the amygdala, septum, and BNST) of MS-180-exposed and control WKY adult males. MS-180 exposure triggered a robust increase in global methylation selectively in the hippocampus of WKY rats, with no significant changes occurring in the other areas.
Our finding of increased DNA methylation levels specifically in the hippocampus of adult MS-180-exposed WKYs suggests that the adaptive behavioral and cardiovascular effects of MS-180 within WKY offspring may be mediated by changes in hippocampal function. Extensive evidence implicates a role for the hippocampus in regulating each of the behavioral domains that were affected in MS-180-exposed WKY rats, including exploratory behavior, sociability, and FST immobility (Kimble, 1968; Altman et al., 1973; Wallace et al., 1977; Sherman & Petty, 1980; Sams-Dodd et al., 1997; Becker et al., 1999; Joca et al., 2003; Hajszan et al., 2009; Shishkina et al., 2010; Telensky et al., 2011). These behaviors are mediated by multiple hippocampal outflow circuits, including a projection that reaches PVN via a synaptic relay within BNST (Herman et al., 1989; Herman et al., 1996; Herman & Mueller, 2006). Because the PVN also contributes to regulation of cardiovascular function via neurons with direct projections to autonomic regulatory regions (Porter & Brody, 1985; Kerman, 2008; Michelini & Stern, 2009), the cardiovascular effects of MS-180 in WKY rats may be mediated via a hippocampal projection to PVN.
Impact of MS on DNA methylation in brain
Environmental factors and various life experiences modify DNA methylation and other epigenetic processes in a variety of tissues, including the brain (McGowan et al., 2008; McGowan et al., 2009; Murgatroyd et al., 2009; Roth et al., 2009; LaSalle, 2011; Franklin et al., 2012; Naumova et al., 2012; Klengel et al., 2014). Most studies to-date that examine neural DNA methylation changes following early-life adversity focus on genes related to the HPA stress axis, such as the glucocorticoid receptor (GR), CRH, and AVP (Murgatroyd, 2014; Bockmuhl et al., 2015; Palma-Gudiel et al., 2015; van der Doelen et al., 2015; Turecki & Meaney, 2016). Fewer studies have reported methylation changes on a genome-wide scale. Work in humans has largely been restricted to examining genome-wide methylation patterns in peripheral samples (i.e. whole blood, buccal cells) collected from children exposed to stress or abuse (Klengel et al., 2014; Lutz & Turecki, 2014). Rodent studies have begun to evaluate global methylation changes in the brains of rats or mice exposed to early-life stress, although results vary across studies based on the specific stress paradigm used, strain, sex, or age of offspring studied (Kember et al., 2012; Anier et al., 2014; Doherty et al., 2016). For example, one study found that exposing Wistar rats to MS-180 in early life led to decreased global DNA methylation levels in the nucleus accumbens of adult offspring (Anier et al., 2014). Another study utilized a different early-life adversity model (transiently exposing Long-Evans rat pups to an abusive caregiver) and showed that it increased global DNA methylation levels in the hippocampus of adolescent male offspring (Doherty et al., 2016). The current study revealed a robust increase in global DNA methylation in the hippocampus of adult WKY male offspring that were exposed to daily 3-h MS from P1-14. Our next-generation sequencing results corroborated this MS-180-induced increase in methylation and identified 983 genomic regions that were differentially methylated in the MS-180 versus control WKY hippocampus. The majority of DMRs occurred within exons or intergenic regions, with approximately 10% of the DMRs localized in or around CpG islands. Our analysis identified a number of differentially methylated genes, including molecules involved in cell proliferation, axonal guidance, tyrosine kinase signaling, and synaptogenesis and synaptic transmission.
Although our analysis identified numerous genes that were differentially methylated in the hippocampus of MS-exposed offspring compared to controls, it was interesting to find that certain genes previously shown to be sensitive to early life stress and maternal care (such as the glucocorticoid receptor gene, Nr3c1) were not affected in our study. Other groups found that Nr3c1 methylation is regulated by naturally-occurring differences in maternal care (Weaver et al., 2002b), although there are conflicting reports of the effect of early life stress on Nr3c1 methylation. Two such studies reported MS-induced increase in NR2c1 methylation in DBA/6J and C57BL/6J males (Kember et al., 2012; Kundakovic et al., 2013), but Kember et al. found no change in C57BL/6J mice and Kundakovic found no differences in Balb/cJ mice. These disparate results (as well as the lack of NR2c1 methylation changes in our present experiment) are likely related to different mouse or rat strains used across studies as well as technical differences in the maternal separation protocols strain (Kember et al., 2012; Kundakovic et al., 2013).
One of the top molecular pathways that emerged from our methylome analysis of the MS-180-exposed WKY hippocampus involved genes related to insulin receptor signaling. Our analysis revealed large and significant (p < 10−6) MS-180-induced methylation decreases at intragenic sites within two key nodes of insulin signaling pathway: a) the insulin receptor (Insr) and b) mitogen activated protein kinase kinase kinase 5 (Map3k5), a major downstream target of Insr. It may be surprising that we found MS-associated hypomethylation at sites related to these genes when the global methylation assay (data presented in Fig. 1) pointed to dramatic MS-induced hypermethylation. Methylation patterns can vary dramatically across the genome and environmental factors such as MS exposure likely elicit disparate effects across the genome, with the potential for increasing methylation at some points, but decreasing it elsewhere. There are host of molecular processes and structural aspects of certain genomic regions that regulate methylation, which may provide protection of select genes (such as insulin-related genes) against MS-related hypermethylation. For example, the presence and functionality of distinct DNA methyltransferase enzymes, transcription factors, and methyl binding proteins at distinct gene sites may lead to differential regulation of methylation status and later transcription. To the best of our knowledge, these factors are not well understood for insulin-related genes identified in our analyses, although we are certainly interested to pursue such topics in our ongoing work related to this project.
Our qRT-PCR experiment indicated that hypomethylation of sites within the insulin receptor gene was associated with altered gene expression, with a 1.62-fold increase in Insr mRNA levels in the hippocampus of MS-180 versus control groups. Insulin signaling in the brain has been implicated in diverse functions, including metabolism, reproduction, memory, and neuronal survival (Plum et al., 2005). Brain-specific deletion of insulin receptor precipitates depressive- and anxiety- like behaviors in mice, which are reversible with antidepressant drug treatment (Kleinridders et al., 2015). Insulin signaling has also been implicated in hippocampal function, with insulin administration in humans leading to improved hippocampal-dependent verbal memory and delayed word recall (Benedict et al., 2004; Reger et al., 2006; Benedict et al., 2007; Hallschmid et al., 2008; Reger et al., 2008) as well as improved mood (Benedict et al., 2004; Hallschmid et al., 2008). Results from rodent studies are consistent with these findings, showing that injecting insulin into the brain improves performance on hippocampal-dependent tasks, including memory consolidation and spatial memory (Park et al., 2000; Moosavi et al., 2006; Moosavi et al., 2007; Haj-ali et al., 2009; McNay et al., 2010), and memory in a passive-avoidance task (Babri et al., 2007). Future experiments in our laboratory plan to explore the role of insulin receptor signaling in mediating the adaptive effects of MS-180 on WKY behavior.
Increasing dietary methyl donor content leads to improved anxiety- and depressive-like behavior as well as positive cardiovascular effects in WKY rats
Diet represents one of several environmental factors that can potently influence DNA methylation since dietary folate, choline, and methionine act as methyl donors for one-carbon transfer reactions like DNA methylation. DNA methyltransferases (DNMTs) transfer methyl groups from S-adenosylmethionine (SAM) to cytosine (Jones & Takai, 2001), so diets lacking folate or other methyl donors can impede SAM synthesis, thereby leading to DNA hypomethylation (Ghoshal et al., 2006; Pogribny et al., 2008; Chen et al., 2010). Depleting dietary methyl donor content has been shown to decrease DNA methylation markers in the brain (Pogribny et al., 2008), and impede fear memory (Ishii et al., 2014; Tomizawa et al., 2015). On the other hand, boosting levels of methyl donors (or SAM itself) increases DNA methylation levels in brain (Paternain et al., 2016), and elicits antidepressant effects in rats and mice, such as decreased immobility in the FST and improved stress-induced anhedonia (Czyrak et al., 1992; Benelli et al., 1999; Brocardo et al., 2008; Molina-Hernandez et al., 2011; Paternain et al., 2016). The present study showed that increasing WKY male rats’ dietary methyl donor content elicited a series of positive behavioral effects including: increased exploratory behavior, increased social interaction, and decreased behavioral despair (immobility) in the FST. These data together with results from other rodent studies are generally consistent with findings in humans showing that treatment with DNA methylation-promoting agents (i.e. SAM (Delle Chiaie et al., 2002; Mischoulon & Fava, 2002; Hardy et al., 2003) or L-methylfolate (Papakostas et al., 2012; Roberts & Tranter, 2013; Shelton et al., 2013)) improves depressive symptoms in some patients.
Increased methyl donor content also leads to multiple cardiovascular changes in WKY rats, including decreased resting HR, increased HRV, decreased resting blood, as well as improved cardiovascular recovery to social stress (increased decay constants for HR and MAP following intruder exposure). These findings are similar to a study in Wistar-Hanover rats where a methionine-enriched diet led to decreased MAP (Mariotti et al., 2006), although other studies have reported that a methionine-enriched diet increases SBP in other rat strains (Robin et al., 2003; Robin et al., 2004). Taken together these observations suggest that the effects of methyl donor supplementation in rats may depend on the strain and on baseline blood pressure levels. Interestingly, in clinical populations folate supplementation leads to a decrease in stroke incidence in hypertensive individuals (Huo et al., 2015). It is thought that these effects are mediated by changes in circulating homocysteine levels and their effects of vascular function and plaque formation (Santilli et al., 2016). Our present results raise the possibility that such protective effects may be mediated by DNA methylation changes in the brain. Future studies in which we alter methyl donor availability only in the brain will be required to address this issue. Additional studies should also determine whether the methyl donor supplemented diet changes hippocampal gene expression within genes that were found to be altered by MS-180 exposure; because MS-180 exposure and the diet manipulation induce similar behavioral and cardiovascular effects, it would be interesting to determine whether they potentially act through similar molecular mechanisms in the hippocampus.
Technical considerations
An important limitation of the present study is the fact that our experiments used only male WKY offspring. There is a relative lack of information about epigenetic differences in the developing and adult brains of males versus females (Menger et al., 2010; McCarthy & Nugent, 2015) and how such differences may contribute to sexually dimorphic risk for emotional disorders (Uddin et al., 2013). Thus, it will be important for studies like the current one to determine whether early-life stress elicits distinct epigenetic changes in female versus male offspring and how those differences may impact behavioral outcomes. Future experiments in our laboratory aim to determine whether MS-180 elicits similar neural, behavioral and physiological effects in WKY females, and whether methyl donor depletion and/or supplementation influences WKY females as it does in males.
The repertoire of techniques for interrogating genome-wide patterns of epigenomic marks is rapidly developing. Each technique has strengths and weaknesses, and each offers distinct types of information that contribute to our understanding of epigenome structure and function (Harris et al., 2010). We chose MethylCap-Seq because it offers a balance between cost and genome coverage. A limitation of MethylCap-Seq is that it does not offer single-base resolution and shows preference for CpG dinucleotides, whereas bisulfite-based methods can offer this resolution without the preference. On the other hand, it is noteworthy that an advantage of MethylCap-Seq over bisulfite sequencing is that it offers a clean 5-methylcytosine signal whereas bisulfite sequencing cannot resolve 5-methylcytosine and 5-hydroxymethylcytosine without further oxidative steps. Another caveat of our approach is that our tissue punch samples contain a heterogeneous collection of cells (i.e. neurons and glia) that exhibit distinct epigenetic patterns (Lister et al., 2013). Because MethylCap-Seq requires a large amount of genomic DNA, tissue punches are preferable to methods such as cell-sorting techniques or laser-capture microdissection of distinct cell populations. As the techniques improve, we hope to be able to sequence from these specific cell populations. A future goal will be to identify how epigenetic and transcriptional sites differ within cell populations in these brain regions.
One limitation of the current study is that we attempted to manipulate brain DNA methylation by exposing rats to a diet that is either supplemented with or depleted of methyl group donors. Our observation of the significantly greater methionine levels in the hippocampi of the methyl donor supplemented rats suggests increased DNA methylation in the brains of these animals (Anderson et al., 2012). However, it is also likely that this diet-based manipulation also impacted DNA methylation levels throughout the body in a variety of tissues. Two prior studies reported that prenatal exposure to a methyl donor supplemented diet increased the occurrence of colitis coupled with microbiome and epigenetic changes in colonic mucosal tissue of adult offspring (Schaible et al., 2011; Mir et al., 2013). However, these studies did not assess behavioral outcomes, and we are not aware of such studies using the adult diet manipulation akin to our present experiment. It is also feasible that such effects may not have been restricted to the DNA. For example, availability of methyl groups has been suggested to alter circulating catecholamine levels, because methyl group availability may impact the function of methyltransferases that regulate catecholamine synthesis and breakdown (Zhou et al., 2011). Future studies that utilize manipulations to specifically alter DNA methylation levels in the brain will help address this issue.
Conclusions
In summary, our recent work found that exposing stress-reactive WKY rats to early-life MS-180 stress elicited paradoxically positive behavioral and physiological effects, including reduced levels of anxiety- and depressive-like behavior (Rana et al., 2015). Results of the present studies suggest that the adaptive effects of MS-180 in WKY offspring may be mediated, at least in part, through MS-180-induced DNA methylation changes in the hippocampus. MS-180 exposure led to significantly elevated total DNA methylation specifically in the hippocampus of WKY male offspring; moreover, enhancing DNA methylation levels in adult WKY rats (via dietary methyl donor supplementation), elicited positive behavioral and cardiovascular effects that resembled the effects of MS-180. Next-generation sequencing analysis identified a number of molecular pathways with genes that were differentially methylated following MS-180 exposure, including molecules important in insulin receptor signaling. Future experiments will continue to interrogate molecular processes, including DNA methylation as well as insulin receptor signaling, that may contribute to the adaptive effects of MS-180 in WKY offspring and increased stress resilience.
Supplementary Material
Acknowledgments
The authors would like to thank Nateka Jackson for excellent technical assistance with the behavioral studies. The authors would like to thank Dr. David Pollock and Dr. Jennifer Pollock for their help with radiotelemetry studies. The study was funded by: NIMH R00 MH081927-04 (IAK); American Heart Association Predoctoral Fellowship 13PRE16940050 (SR); NIH 4R00MH085859-02 (SMC), NIH R01MH105447-01 (SMC), P30 NS047466PI (JMW), and 5T90DE022736-04 (covering CRM). The authors declare no competing financial interests.
References
- Heart rate variability: standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation. 1996;93:1043–1065. [PubMed] [Google Scholar]
- Altman J, Brunner RL, Bayer SA. The hippocampus and behavioral maturation. Behav Biol. 1973;8:557–596. doi: 10.1016/s0091-6773(73)80144-0. [DOI] [PubMed] [Google Scholar]
- Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23:853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Pruus K, Aonurm-Helm A, Zharkovsky A, Kalda A. Maternal separation is associated with DNA methylation and behavioural changes in adult rats. Eur Neuropsychopharmacol. 2014;24:459–468. doi: 10.1016/j.euroneuro.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Apter-Levy Y, Feldman M, Vakart A, Ebstein RP, Feldman R. Impact of maternal depression across the first 6 years of life on the child’s mental health, social engagement, and empathy: The moderating role of oxytocin. The American journal of psychiatry. 2013;170:1161–1168. doi: 10.1176/appi.ajp.2013.12121597. [DOI] [PubMed] [Google Scholar]
- Babri S, Badie HG, Khamenei S, Seyedlar MO. Intrahippocampal insulin improves memory in a passive-avoidance task in male wistar rats. Brain Cogn. 2007;64:86–91. doi: 10.1016/j.bandc.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Becker A, Grecksch G, Bernstein HG, Hollt V, Bogerts B. Social behaviour in rats lesioned with ibotenic acid in the hippocampus: quantitative and qualitative analysis. Psychopharmacology (Berl) 1999;144:333–338. doi: 10.1007/s002130051015. [DOI] [PubMed] [Google Scholar]
- Benedict C, Hallschmid M, Hatke A, Schultes B, Fehm HL, Born J, Kern W. Intranasal insulin improves memory in humans. Psychoneuroendocrinology. 2004;29:1326–1334. doi: 10.1016/j.psyneuen.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Benedict C, Hallschmid M, Schmitz K, Schultes B, Ratter F, Fehm HL, Born J, Kern W. Intranasal insulin improves memory in humans: superiority of insulin aspart. Neuropsychopharmacology. 2007;32:239–243. doi: 10.1038/sj.npp.1301193. [DOI] [PubMed] [Google Scholar]
- Benelli A, Filaferro M, Bertolini A, Genedani S. Influence of S-adenosyl-L-methionine on chronic mild stress-induced anhedonia in castrated rats. British journal of pharmacology. 1999;127:645–654. doi: 10.1038/sj.bjp.0702589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Chapter 19(Unit 19):10, 11–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl Y, Patchev AV, Madejska A, Hoffmann A, Sousa JC, Sousa N, Holsboer F, Almeida OF, Spengler D. Methylation at the CpG island shore region upregulates Nr3c1 promoter activity after early-life stress. Epigenetics. 2015;10:247–257. doi: 10.1080/15592294.2015.1017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Brocardo PS, Budni J, Kaster MP, Santos AR, Rodrigues AL. Folic acid administration produces an antidepressant-like effect in mice: evidence for the involvement of the serotonergic and noradrenergic systems. Neuropharmacology. 2008;54:464–473. doi: 10.1016/j.neuropharm.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, Yang XF, Wang H. Regulation of homocysteine metabolism and methylation in human and mouse tissues. Faseb J. 2010;24:2804–2817. doi: 10.1096/fj.09-143651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chocyk A, Bobula B, Dudys D, Przyborowska A, Majcher-Maslanka I, Hess G, Wedzony K. Early-life stress affects the structural and functional plasticity of the medial prefrontal cortex in adolescent rats. Eur J Neurosci. 2013;38:2089–2107. doi: 10.1111/ejn.12208. [DOI] [PubMed] [Google Scholar]
- Clinton SM, Watson SJ, Akil H. High novelty-seeking rats are resilient to negative physiological effects of the early life stress. Stress. 2014;17:97–107. doi: 10.3109/10253890.2013.850670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Valentino RJ, Lucki I. Assessing substrates underlying the behavioral effects of antidepressants using the modified rat forced swimming test. Neuroscience and Biobehavioral Reviews. 2005;29:547–569. doi: 10.1016/j.neubiorev.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Czyrak A, Rogoz Z, Skuza G, Zajaczkowski W, Maj J. Antidepressant activity of S-adenosyl-L-methionine in mice and rats. J Basic Clin Physiol Pharmacol. 1992;3:1–17. doi: 10.1515/jbcpp.1992.3.1.1. [DOI] [PubMed] [Google Scholar]
- Daniels WM, Pietersen CY, Carstens ME, Stein DJ. Maternal separation in rats leads to anxiety-like behavior and a blunted ACTH response and altered neurotransmitter levels in response to a subsequent stressor. Metab Brain Dis. 2004;19:3–14. doi: 10.1023/b:mebr.0000027412.19664.b3. [DOI] [PubMed] [Google Scholar]
- Delle Chiaie R, Pancheri P, Scapicchio P. Efficacy and tolerability of oral and intramuscular S-adenosyl-L-methionine 1,4-butanedisulfonate (SAMe) in the treatment of major depression: comparison with imipramine in 2 multicenter studies. The American journal of clinical nutrition. 2002;76:1172S–1176S. doi: 10.1093/ajcn/76/5.1172S. [DOI] [PubMed] [Google Scholar]
- Doherty TS, Forster A, Roth TL. Global and gene-specific DNA methylation alterations in the adolescent amygdala and hippocampus in an animal model of caregiver maltreatment. Behavioural brain research. 2016;298:55–61. doi: 10.1016/j.bbr.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Giles WH, Felitti VJ, Dube SR, Williams JE, Chapman DP, Anda RF. Insights into causal pathways for ischemic heart disease: adverse childhood experiences study. Circulation. 2004;110:1761–1766. doi: 10.1161/01.CIR.0000143074.54995.7F. [DOI] [PubMed] [Google Scholar]
- Franklin TB, Saab BJ, Mansuy IM. Neural mechanisms of stress resilience and vulnerability. Neuron. 2012;75:747–761. doi: 10.1016/j.neuron.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, Huang Y, Young D, Jacob ST. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006;136:1522–1527. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Galaxy: a platform for interactive large-scale genome analysis. Genome research. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Beedle AS. Early life events and their consequences for later disease: a life history and evolutionary perspective. Am J Hum Biol. 2007;19:1–19. doi: 10.1002/ajhb.20590. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haj-ali V, Mohaddes G, Babri SH. Intracerebroventricular insulin improves spatial learning and memory in male Wistar rats. Behavioral neuroscience. 2009;123:1309–1314. doi: 10.1037/a0017722. [DOI] [PubMed] [Google Scholar]
- Hajszan T, Dow A, Warner-Schmidt JL, Szigeti-Buck K, Sallam NL, Parducz A, Leranth C, Duman RS. Remodeling of hippocampal spine synapses in the rat learned helplessness model of depression. Biological psychiatry. 2009;65:392–400. doi: 10.1016/j.biopsych.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallschmid M, Benedict C, Schultes B, Born J, Kern W. Obese men respond to cognitive but not to catabolic brain insulin signaling. Int J Obes (Lond) 2008;32:275–282. doi: 10.1038/sj.ijo.0803722. [DOI] [PubMed] [Google Scholar]
- Hardy ML, Coulter I, Morton SC, Favreau J, Venuturupalli S, Chiappelli F, Rossi F, Orshansky G, Jungvig LK, Roth EA, Suttorp MJ, Shekelle P. S-adenosyl-L-methionine for treatment of depression, osteoarthritis, and liver disease. Evidence report/technology assessment. 2003:1–3. [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O’Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nature biotechnology. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Newport DJ, Wagner D, Wilcox MM, Miller AH, Nemeroff CB. The role of early adverse experience and adulthood stress in the prediction of neuroendocrine stress reactivity in women: a multiple regression analysis. Depression and anxiety. 2002;15:117–125. doi: 10.1002/da.10015. [DOI] [PubMed] [Google Scholar]
- Herman JP, Mueller NK. Role of the ventral subiculum in stress integration. Behavioural brain research. 2006;174:215–224. doi: 10.1016/j.bbr.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Herman JP, Prewitt CM, Cullinan WE. Neuronal circuit regulation of the hypothalamo-pituitary-adrenocortical stress axis. Critical reviews in neurobiology. 1996;10:371–394. doi: 10.1615/critrevneurobiol.v10.i3-4.50. [DOI] [PubMed] [Google Scholar]
- Herman JP, Schafer MK, Young EA, Thompson R, Douglass J, Akil H, Watson SJ. Evidence for hippocampal regulation of neuroendocrine neurons of the hypothalamo-pituitary-adrenocortical axis. J Neurosci. 1989;9:3072–3082. doi: 10.1523/JNEUROSCI.09-09-03072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer MA, Brunelli SA, Masmela J, Shair HN. Maternal interactions prior to separation potentiate isolation-induced calling in rat pups. Behavioral neuroscience. 1996;110:1158–1167. doi: 10.1037//0735-7044.110.5.1158. [DOI] [PubMed] [Google Scholar]
- Hulshof HJ, Novati A, Sgoifo A, Luiten PG, den Boer JA, Meerlo P. Maternal separation decreases adult hippocampal cell proliferation and impairs cognitive performance but has little effect on stress sensitivity and anxiety in adult Wistar rats. Behavioural brain research. 2011;216:552–560. doi: 10.1016/j.bbr.2010.08.038. [DOI] [PubMed] [Google Scholar]
- Huo Y, Li J, Qin X, Huang Y, Wang X, Gottesman RF, Tang G, Wang B, Chen D, He M, Fu J, Cai Y, Shi X, Zhang Y, Cui Y, Sun N, Li X, Cheng X, Wang J, Yang X, Yang T, Xiao C, Zhao G, Dong Q, Zhu D, Wang X, Ge J, Zhao L, Hu D, Liu L, Hou FF Investigators C. Efficacy of folic acid therapy in primary prevention of stroke among adults with hypertension in China: the CSPPT randomized clinical trial. JAMA. 2015;313:1325–1335. doi: 10.1001/jama.2015.2274. [DOI] [PubMed] [Google Scholar]
- Huot RL, Plotsky PM, Lenox RH, McNamara RK. Neonatal maternal separation reduces hippocampal mossy fiber density in adult Long Evans rats. Brain research. 2002;950:52–63. doi: 10.1016/s0006-8993(02)02985-2. [DOI] [PubMed] [Google Scholar]
- Ishii D, Matsuzawa D, Matsuda S, Tomizawa H, Sutoh C, Shimizu E. Methyl donor-deficient diet during development can affect fear and anxiety in adulthood in C57BL/6J mice. PloS one. 2014;9:e105750. doi: 10.1371/journal.pone.0105750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joca SR, Padovan CM, Guimaraes FS. Activation of post-synaptic 5-HT(1A) receptors in the dorsal hippocampus prevents learned helplessness development. Brain research. 2003;978:177–184. doi: 10.1016/s0006-8993(03)02943-3. [DOI] [PubMed] [Google Scholar]
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kember RL, Dempster EL, Lee TH, Schalkwyk LC, Mill J, Fernandes C. Maternal separation is associated with strain-specific responses to stress and epigenetic alterations to Nr3c1, Avp, and Nr4a1 in mouse. Brain Behav. 2012;2:455–467. doi: 10.1002/brb3.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman IA. Organization of brain somatomotor-sympathetic circuits. Exp Brain Res. 2008;187:1–16. doi: 10.1007/s00221-008-1337-5. [DOI] [PubMed] [Google Scholar]
- Kimble DP. Hippocampus and internal inhibition. Psychol Bull. 1968;70:285–295. doi: 10.1037/h0026470. [DOI] [PubMed] [Google Scholar]
- Kleinridders A, Cai W, Cappellucci L, Ghazarian A, Collins WR, Vienberg SG, Pothos EN, Kahn CR. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3463–3468. doi: 10.1073/pnas.1500877112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 2014;80:115–132. doi: 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Konycheva G, Dziadek MA, Ferguson LR, Krageloh CU, Coolen MW, Davison M, Breier BH. Dietary methyl donor deficiency during pregnancy in rats shapes learning and anxiety in offspring. Nutr Res. 2011;31:790–804. doi: 10.1016/j.nutres.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Frontiers in psychiatry. 2013;4:78. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd CO, Huot RL, Thrivikraman KV, Nemeroff CB, Plotsky PM. Long-term adaptations in glucocorticoid receptor and mineralocorticoid receptor mRNA and negative feedback on the hypothalamo-pituitary-adrenal axis following neonatal maternal separation. Biol Psychiatry. 2004;55:367–375. doi: 10.1016/j.biopsych.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Ladd CO, Thrivikraman KV, Huot RL, Plotsky PM. Differential neuroendocrine responses to chronic variable stress in adult Long Evans rats exposed to handling-maternal separation as neonates. Psychoneuroendocrinology. 2005;30:520–533. doi: 10.1016/j.psyneuen.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Lajud N, Roque A, Cajero M, Gutierrez-Ospina G, Torner L. Periodic maternal separation decreases hippocampal neurogenesis without affecting basal corticosterone during the stress hyporesponsive period, but alters HPA axis and coping behavior in adulthood. Psychoneuroendocrinology. 2012;37:410–420. doi: 10.1016/j.psyneuen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- LaSalle JM. A genomic point-of-view on environmental factors influencing the human brain methylome. Epigenetics. 2011;6:862–869. doi: 10.4161/epi.6.7.16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas M, Bredewold R, Neumann ID, Veenema AH. Maternal separation interferes with developmental changes in brain vasopressin and oxytocin receptor binding in male rats. Neuropharmacology. 2010;58:78–87. doi: 10.1016/j.neuropharm.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Lutz PE, Turecki G. DNA methylation and childhood maltreatment: from animal models to human studies. Neuroscience. 2014;264:142–156. doi: 10.1016/j.neuroscience.2013.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DM, Parker KJ. Stress inoculation-induced indications of resilience in monkeys. J Trauma Stress. 2007;20:423–433. doi: 10.1002/jts.20265. [DOI] [PubMed] [Google Scholar]
- Maccari S, Krugers HJ, Morley-Fletcher S, Szyf M, Brunton PJ. The consequences of early-life adversity: neurobiological, behavioural and epigenetic adaptations. Journal of neuroendocrinology. 2014;26:707–723. doi: 10.1111/jne.12175. [DOI] [PubMed] [Google Scholar]
- Malkesman O, Braw Y, Maayan R, Weizman A, Overstreet DH, Shabat-Simon M, Kesner Y, Touati-Werner D, Yadid G, Weller A. Two different putative genetic animal models of childhood depression. Biol Psychiatry. 2006;59:17–23. doi: 10.1016/j.biopsych.2005.05.039. [DOI] [PubMed] [Google Scholar]
- Mariotti F, Hammiche A, Blouet C, Dare S, Tome D, Huneau JF. Medium-term methionine supplementation increases plasma homocysteine but not ADMA and improves blood pressure control in rats fed a diet rich in protein and adequate in folate and choline. European journal of nutrition. 2006;45:383–390. doi: 10.1007/s00394-006-0611-1. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, Nugent BM. At the frontier of epigenetics of brain sex differences. Frontiers in behavioral neuroscience. 2015;9:221. doi: 10.3389/fnbeh.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Meaney MJ, Szyf M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain research. 2008;1237:12–24. doi: 10.1016/j.brainres.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNay EC, Ong CT, McCrimmon RJ, Cresswell J, Bogan JS, Sherwin RS. Hippocampal memory processes are modulated by insulin and high-fat-induced insulin resistance. Neurobiol Learn Mem. 2010;93:546–553. doi: 10.1016/j.nlm.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Diorio J, Francis D, Widdowson J, LaPlante P, Caldji C, Sharma S, Seckl JR, Plotsky PM. Early environmental regulation of forebrain glucocorticoid receptor gene expression: implications for adrenocortical responses to stress. Developmental neuroscience. 1996;18:49–72. doi: 10.1159/000111395. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues in Clinical Neuroscience. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger Y, Bettscheider M, Murgatroyd C, Spengler D. Sex differences in brain epigenetics. Epigenomics. 2010;2:807–821. doi: 10.2217/epi.10.60. [DOI] [PubMed] [Google Scholar]
- Michelini LC, Stern JE. Exercise-induced neuronal plasticity in central autonomic networks: role in cardiovascular control. Experimental physiology. 2009;94:947–960. doi: 10.1113/expphysiol.2009.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir SA, Nagy-Szakal D, Dowd SE, Szigeti RG, Smith CW, Kellermayer R. Prenatal methyl-donor supplementation augments colitis in young adult mice. PloS one. 2013;8:e73162. doi: 10.1371/journal.pone.0073162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischoulon D, Fava M. Role of S-adenosyl-L-methionine in the treatment of depression: a review of the evidence. The American journal of clinical nutrition. 2002;76:1158S–1161S. doi: 10.1093/ajcn/76/5.1158S. [DOI] [PubMed] [Google Scholar]
- Molina-Hernandez M, Tellez-Alcantara NP, Olivera-Lopez JI, Jaramillo MT. The folic acid combined with 17-beta estradiol produces antidepressant-like actions in ovariectomized rats forced to swim. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35:60–66. doi: 10.1016/j.pnpbp.2010.08.022. [DOI] [PubMed] [Google Scholar]
- Monroy E, Hernandez-Torres E, Flores G. Maternal separation disrupts dendritic morphology of neurons in prefrontal cortex, hippocampus, and nucleus accumbens in male rat offspring. Journal of chemical neuroanatomy. 2010;40:93–101. doi: 10.1016/j.jchemneu.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Moosavi M, Naghdi N, Choopani S. Intra CA1 insulin microinjection improves memory consolidation and retrieval. Peptides. 2007;28:1029–1034. doi: 10.1016/j.peptides.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Moosavi M, Naghdi N, Maghsoudi N, Zahedi Asl S. The effect of intrahippocampal insulin microinjection on spatial learning and memory. Horm Behav. 2006;50:748–752. doi: 10.1016/j.yhbeh.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C. Epigenetic programming of neuroendocrine systems during early life. Experimental physiology. 2014;99:62–65. doi: 10.1113/expphysiol.2013.076141. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nam H, Clinton SM, Jackson NL, Kerman IA. Learned helplessness and social avoidance in the Wistar-Kyoto rat. Frontiers in behavioral neuroscience. 2014;8:109. doi: 10.3389/fnbeh.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, Grigorenko EL. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Development and psychopathology. 2012;24:143–155. doi: 10.1017/S0954579411000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet DH. Modeling depression in animal models. Methods Mol Biol. 2012;829:125–144. doi: 10.1007/978-1-61779-458-2_7. [DOI] [PubMed] [Google Scholar]
- Palma-Gudiel H, Cordova-Palomera A, Leza JC, Fananas L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neurosci Biobehav Rev. 2015;55:520–535. doi: 10.1016/j.neubiorev.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Papakostas GI, Cassiello CF, Iovieno N. Folates and S-adenosylmethionine for major depressive disorder. Canadian journal of psychiatry. Revue canadienne de psychiatrie. 2012;57:406–413. doi: 10.1177/070674371205700703. [DOI] [PubMed] [Google Scholar]
- Pare WP, Redei E. Depressive behavior and stress ulcer in Wistar Kyoto rats. Journal of physiology, Paris. 1993;87:229–238. doi: 10.1016/0928-4257(93)90010-q. [DOI] [PubMed] [Google Scholar]
- Park CR, Seeley RJ, Craft S, Woods SC. Intracerebroventricular insulin enhances memory in a passive-avoidance task. Physiology & behavior. 2000;68:509–514. doi: 10.1016/s0031-9384(99)00220-6. [DOI] [PubMed] [Google Scholar]
- Paternain L, Martisova E, Campion J, Martinez JA, Ramirez MJ, Milagro FI. Methyl donor supplementation in rats reverses the deleterious effect of maternal separation on depression-like behaviour. Behavioural brain research. 2016;299:51–58. doi: 10.1016/j.bbr.2015.11.031. [DOI] [PubMed] [Google Scholar]
- Pickering C, Gustafsson L, Cebere A, Nylander I, Liljequist S. Repeated maternal separation of male Wistar rats alters glutamate receptor expression in the hippocampus but not the prefrontal cortex. Brain research. 2006;1099:101–108. doi: 10.1016/j.brainres.2006.04.136. [DOI] [PubMed] [Google Scholar]
- Plotsky PM, Meaney MJ. Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain research. Molecular brain research. 1993;18:195–200. doi: 10.1016/0169-328x(93)90189-v. [DOI] [PubMed] [Google Scholar]
- Plotsky PM, Thrivikraman KV, Nemeroff CB, Caldji C, Sharma S, Meaney MJ. Long-term consequences of neonatal rearing on central corticotropin-releasing factor systems in adult male rat offspring. Neuropsychopharmacology. 2005;30:2192–2204. doi: 10.1038/sj.npp.1300769. [DOI] [PubMed] [Google Scholar]
- Plum L, Schubert M, Bruning JC. The role of insulin receptor signaling in the brain. Trends Endocrinol Metab. 2005;16:59–65. doi: 10.1016/j.tem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Karpf AR, James SR, Melnyk S, Han T, Tryndyak VP. Epigenetic alterations in the brains of Fisher 344 rats induced by long-term administration of folate/methyl-deficient diet. Brain research. 2008;1237:25–34. doi: 10.1016/j.brainres.2008.07.077. [DOI] [PubMed] [Google Scholar]
- Porsolt RD, Le Pichon M, Jalfre M. Depression: a new animal model sensitive to antidepressant treatments. Nature. 1977;266:730–732. doi: 10.1038/266730a0. [DOI] [PubMed] [Google Scholar]
- Porter JP, Brody MJ. Neural projections from paraventricular nucleus that subserve vasomotor functions. The American journal of physiology. 1985;248:R271–281. doi: 10.1152/ajpregu.1985.248.3.R271. [DOI] [PubMed] [Google Scholar]
- Rana S, Pugh PC, Jackson N, Clinton SM, Kerman IA. Inborn stress reactivity shapes adult behavioral consequences of early-life maternal separation stress. Neurosci Lett. 2015;584:146–150. doi: 10.1016/j.neulet.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S, Pugh PC, Katz E, Stringfellow SA, Lin CP, Wyss JM, Stauss HM, White CR, Clinton SM, Kerman IA. Independent effects of early-life experience and trait aggression on cardiovascular function. American journal of physiology. Regulatory, integrative and comparative physiology. 2016;311:R272–286. doi: 10.1152/ajpregu.00505.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reger MA, Watson GS, Frey WH, 2nd, Baker LD, Cholerton B, Keeling ML, Belongia DA, Fishel MA, Plymate SR, Schellenberg GD, Cherrier MM, Craft S. Effects of intranasal insulin on cognition in memory-impaired older adults: modulation by APOE genotype. Neurobiology of aging. 2006;27:451–458. doi: 10.1016/j.neurobiolaging.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Reger MA, Watson GS, Green PS, Baker LD, Cholerton B, Fishel MA, Plymate SR, Cherrier MM, Schellenberg GD, Frey WH, 2nd, Craft S. Intranasal insulin administration dose-dependently modulates verbal memory and plasma amyloid-beta in memory-impaired older adults. J Alzheimers Dis. 2008;13:323–331. doi: 10.3233/jad-2008-13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensel MA, Wilcoxen TE, Schoech SJ. The influence of nest attendance and provisioning on nestling stress physiology in the Florida scrub-jay. Horm Behav. 2010;57:162–168. doi: 10.1016/j.yhbeh.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Roberts SH, Tranter R. Higher dose L-methylfolate may be an effective adjunctive therapy for adults with major depression who have inadequate response to SSRIs. Evidence-based mental health. 2013;16:75. doi: 10.1136/eb-2013-101269. [DOI] [PubMed] [Google Scholar]
- Robin S, Maupoil V, Groubatch F, Laurant P, Jacqueson A, Berthelot A. Effect of a methionine-supplemented diet on the blood pressure of Wistar-Kyoto and spontaneously hypertensive rats. The British journal of nutrition. 2003;89:539–548. doi: 10.1079/BJN2002810. [DOI] [PubMed] [Google Scholar]
- Robin S, Maupoil V, Laurant P, Jacqueson A, Berthelot A. Effect of a methionine-supplemented diet on the blood pressure of Sprague-Dawley and deoxycorticosterone acetate-salt hypertensive rats. The British journal of nutrition. 2004;91:857–865. doi: 10.1079/BJN20041116. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams-Dodd F, Lipska BK, Weinberger DR. Neonatal lesions of the rat ventral hippocampus result in hyperlocomotion and deficits in social behaviour in adulthood. Psychopharmacology (Berl) 1997;132:303–310. doi: 10.1007/s002130050349. [DOI] [PubMed] [Google Scholar]
- Sanchez MM, Ladd CO, Plotsky PM. Early adverse experience as a developmental risk factor for later psychopathology: evidence from rodent and primate models. Developmental Psychopathology. 2001;13:419–449. doi: 10.1017/s0954579401003029. [DOI] [PubMed] [Google Scholar]
- Santarelli S, Lesuis SL, Wang XD, Wagner KV, Hartmann J, Labermaier C, Scharf SH, Muller MB, Holsboer F, Schmidt MV. Evidence supporting the match/mismatch hypothesis of psychiatric disorders. Eur Neuropsychopharmacol. 2014;24:907–918. doi: 10.1016/j.euroneuro.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Santilli F, Davi G, Patrono C. Homocysteine, methylenetetrahydrofolate reductase, folate status and atherothrombosis: A mechanistic and clinical perspective. Vascul Pharmacol. 2016;78:1–9. doi: 10.1016/j.vph.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Schaible TD, Harris RA, Dowd SE, Smith CW, Kellermayer R. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Human molecular genetics. 2011;20:1687–1696. doi: 10.1093/hmg/ddr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton RC, Sloan Manning J, Barrentine LW, Tipa EV. Assessing Effects of l-Methylfolate in Depression Management: Results of a Real-World Patient Experience Trial. The primary care companion to CNS disorders. 2013:15. doi: 10.4088/PCC.13m01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman AD, Petty F. Neurochemical basis of the action of antidepressants on learned helplessness. Behavioral and neural biology. 1980;30:119–134. doi: 10.1016/s0163-1047(80)91005-5. [DOI] [PubMed] [Google Scholar]
- Shishkina GT, Kalinina TS, Berezova IV, Bulygina VV, Dygalo NN. Resistance to the development of stress-induced behavioral despair in the forced swim test associated with elevated hippocampal Bcl-xl expression. Behavioural brain research. 2010;213:218–224. doi: 10.1016/j.bbr.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Simmons RK, Stringfellow SA, Glover ME, Wagle AA, Clinton SM. DNA methylation markers in the postnatal developing rat brain. Brain research. 2013;1533:26–36. doi: 10.1016/j.brainres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomi SJ. Early stress and adult emotional reactivity in rhesus monkeys. Ciba Found Symp. 1991;156:171–183. doi: 10.1002/9780470514047.ch11. discussion 183–178. [DOI] [PubMed] [Google Scholar]
- Suri D, Bhattacharya A, Vaidya VA. Early stress evokes temporally distinct consequences on the hippocampal transcriptome, anxiety and cognitive behaviour. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2014;17:289–301. doi: 10.1017/S1461145713001004. [DOI] [PubMed] [Google Scholar]
- Suri D, Veenit V, Sarkar A, Thiagarajan D, Kumar A, Nestler EJ, Galande S, Vaidya VA. Early stress evokes age-dependent biphasic changes in hippocampal neurogenesis, BDNF expression, and cognition. Biol Psychiatry. 2013;73:658–666. doi: 10.1016/j.biopsych.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M, Weaver IC, Champagne FA, Diorio J, Meaney MJ. Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Frontiers in Neuroendocrinology. 2005;26:139–162. doi: 10.1016/j.yfrne.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Telensky P, Svoboda J, Blahna K, Bures J, Kubik S, Stuchlik A. Functional inactivation of the rat hippocampus disrupts avoidance of a moving object. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5414–5418. doi: 10.1073/pnas.1102525108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JM, Sonuga-Barke EJ, Morgan AR, Cornforth CM, Turic D, Ferguson LR, Mitchell EA, Waldie KE. The catechol-O-methyltransferase (COMT) Val158Met polymorphism moderates the effect of antenatal stress on childhood behavioural problems: longitudinal evidence across multiple ages. Developmental medicine and child neurology. 2012;54:148–154. doi: 10.1111/j.1469-8749.2011.04129.x. [DOI] [PubMed] [Google Scholar]
- Tomizawa H, Matsuzawa D, Ishii D, Matsuda S, Kawai K, Mashimo Y, Sutoh C, Shimizu E. Methyl-donor deficiency in adolescence affects memory and epigenetic status in the mouse hippocampus. Genes Brain Behav. 2015;14:301–309. doi: 10.1111/gbb.12207. [DOI] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review. Biol Psychiatry. 2016;79:87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD. bioRxiv beta - The Preprint Server for Biology. Cold Spring Harbor Laboratory; 2014. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. [Google Scholar]
- Uddin M, Sipahi L, Li J, Koenen KC. Sex differences in DNA methylation may contribute to risk of PTSD and depression: a review of existing evidence. Depression and anxiety. 2013;30:1151–1160. doi: 10.1002/da.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Doelen RH, Arnoldussen IA, Ghareh H, van Och L, Homberg JR, Kozicz T. Early life adversity and serotonin transporter gene variation interact to affect DNA methylation of the corticotropin-releasing factor gene promoter region in the adult rat brain. Development and psychopathology. 2015;27:123–135. doi: 10.1017/S0954579414001345. [DOI] [PubMed] [Google Scholar]
- van der Doelen RH, Kozicz T, Homberg JR. Adaptive fitness; early life adversity improves adult stress coping in heterozygous serotonin transporter knockout rats. Mol Psychiatry. 2013;18:1244–1245. doi: 10.1038/mp.2012.186. [DOI] [PubMed] [Google Scholar]
- Wallace RB, Kaplan R, Werboff J. Hippocampus and behavioral maturation. Int J Neurosci. 1977;7:185–200. doi: 10.3109/00207457709147209. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Grant RJ, Meaney MJ. Maternal behavior regulates long-term hippocampal expression of BAX and apoptosis in the offspring. Journal of Neurochemistry. 2002a;82:998–1002. doi: 10.1046/j.1471-4159.2002.01054.x. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proceedings of the National Academy of Sciences of the USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Szyf M, Meaney MJ. From maternal care to gene expression: DNA methylation and the maternal programming of stress responses. Endocr Res. 2002b;28:699. doi: 10.1081/erc-120016989. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SS, Zhou YM, Li D, Lun YZ. Dietary methyl-consuming compounds and metabolic syndrome. Hypertens Res. 2011;34:1239–1245. doi: 10.1038/hr.2011.133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.