Abstract
Iflaviridae is a family of small non-enveloped viruses with monopartite, positive-stranded RNA genomes of approximately 9–11 kilobases. Viruses of all classified species infect arthropod hosts, with the majority infecting insects. Both beneficial and pest insects serve as hosts, and infections can be symptomless (Nilaparvatalugens honeydew virus 1) or cause developmental abnormalities (deformed wing virus), behavioural changes (sacbrood virus) and premature mortality (infectious flacherie virus). The host range has not been examined for most members. The most common route of infection for iflaviruses is the ingestion of virus-contaminated food sources. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the Iflaviridae, which is available at www.ictv.global/report/iflaviridae.
Keywords: Iflaviridae, ICTV Report, Taxonomy
Virion
Virions are roughly spherical and exhibit icosahedral symmetry with a diameter of 22–30 nm. Virions have no envelope and no distinctive surface structures (Table 1, Fig. 1).
Table 1. Characteristics of the family Iflaviridae.
| Typical member: | infectious flacherie virus (AB000906), species Infectious flacherie virus, genus Iflavirus |
|---|---|
| Virion | Non-enveloped, 22–30 nm-diameter virions |
| Genome | 9–11 kb of positive-sense, non-segmented RNA |
| Replication | Cytoplasmic within viral replication complexes formed from a variety of host cellular membranes |
| Translation | Directly from genomic RNA containing an internal ribosomal entry site (IRES) |
| Host range | Arthropoda |
| Taxonomy | Member of the order Picornavirales; >10 species in the single genus Iflavirus |
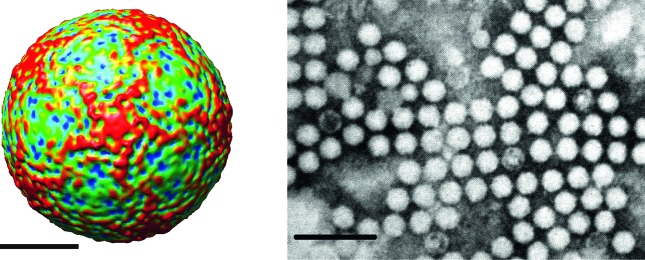
Fig. 1.
(Left) Surface view of the virion of infectious flacherie virus along a five-fold axis reconstructed by cryo-electron microscopy. The bar represents 10 nm (courtesy of J. Hong). (Right) Negative contrast electron micrograph of the isometric particles of an isolate of infectious flacherie virus. The bar represents 100 nm (courtesy of H. Bando).
Genome
Iflaviruses possess single-stranded, positive-sense, non-segmented RNA genomes with a single open reading frame (ORF). The ORF is translated directly into a polyprotein that is subsequently processed to yield structural, i.e. capsid (N-terminal region) and non-structural (C-terminal region) proteins [1]. The 5′ end of the genome bears a covalently linked protein, VPg, which plays a role in RNA replication.
Replication
Replication occurs in the host cell cytoplasm. The coding regions for capsid proteins, arranged in the order VP2–VP4–VP3–VP1, are often preceded by a region encoding a short leader protein (L) of unknown function that is removed from VP2 before capsid assembly. VP4 is analogous to the VP4 present in some dicistroviruses and, in the case of infectious flacherie virus, is present as a minor structural component of the capsid. The non-structural proteins include an RNA helicase, a 3C-like cysteine protease and an RNA-dependent RNA polymerase (Fig. 2). Evidence suggests that translation in some iflaviruses is mediated by an internal ribosomal entry site (IRES) located in the 5′ UTR [2]. The viral RNA is infectious and serves as both genomic and viral mRNA. The mechanisms of polyprotein processing and the effects on host cell macromolecular synthesis during infection have not been well studied for the members of this family.
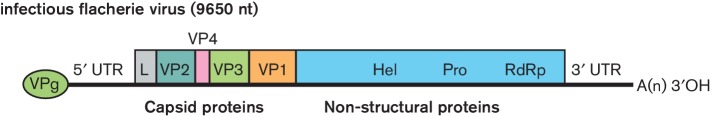
Fig. 2.
Genome structure of infectious flacherie virus. The genome encodes a single polyprotein that is auto-catalytically cleaved into three major structural proteins (VP1, VP2 and VP3) and non-structural proteins used in replication. The structural proteins are encoded in the 5′-proximal region of the genome and the non-structural proteins are encoded in the 3′-proximal region. The 5′ end of the genome bears a covalently linked protein, VPg, which plays a role in RNA replication.
Taxonomy
Currently, members of the family are placed together within a single genus, Iflavirus. However, phylogenetic analysis of the complete translated genomes of iflaviruses shows that a number of distinct clades are present. These may be separated taxonomically into different genera in the near future as more virus sequences become available.
All member viruses have been isolated from arthropods, primarily insects. Beyond the original host descriptions, the host range of most members has not been examined. However, honeybee iflaviruses, deformed wing virus, Varroa destructor virus 1, slow bee paralysis virus and sacbrood virus have been shown to infect other Apis species, as well as several Bombus species [3]. Deformed wing virus, Varroa destructor virus 1 and slow bee paralysis virus can also be vectored to honeybees by parasitic mites (Varroa and Tropilaelaps genera). Varroa and Tropilaelaps mites are also capable of serving as hosts for deformed wing virus and Varroa destructor virus 1. Deformed wing virus and Varroa destructor virus 1 are also vertically and sexually transmitted in honey bees. The most common route of infection for iflaviruses is the ingestion of virus-contaminated food sources. Trophallaxis in social insects facilitates intra-colonial virus dispersal [4]. In addition to the gut, gonads, fat body, muscle, brain and glandular tissues also have been shown to be a target for several iflaviruses. Once the virus gains entry to the host cell, the infection process is rapid, with progeny virus being produced in hours [5].
Resources
Full ICTV Online (10th) Report: www.ictv.global/report/iflaviridae.
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Michael J. Adams, Donald B. Smith, Richard J. Orton and Nick J. Knowles.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Ye S, Xia H, Dong C, Cheng Z, Xia X, et al. Identification and characterization of Iflavirus 3C-like protease processing activities. Virology. 2012;428:136–145. doi: 10.1016/j.virol.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ongus JR, Roode EC, Pleij CW, Vlak JM, van Oers MM. The 5′ non-translated region of Varroa destructor virus 1 (genus Iflavirus): structure prediction and IRES activity in Lymantria dispar cells. J Gen Virol. 2006;87:3397–3407. doi: 10.1099/vir.0.82122-0. [DOI] [PubMed] [Google Scholar]
- 3.McMahon DP, Fürst MA, Caspar J, Theodorou P, Brown MJ, et al. A sting in the spit: widespread cross-infection of multiple RNA viruses across wild and managed bees. J Anim Ecol. 2015;84:615–624. doi: 10.1111/1365-2656.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Miranda JR, Genersch E. Deformed wing virus. J Invertebr Pathol. 2010;103:S48–S61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 5.van Oers MM. Genomics and biology of iflaviruses. In: Asgari S, Johnson K, editors. Insect Virology. Norfolk: Academic Press; 2010. pp. 231–250. [Google Scholar]