Abstract
Members of the family Secoviridae are non-enveloped viruses with mono- or bipartite (RNA-1 and RNA-2) linear positive-sense ssRNA genomes with the size of the RNAs combined ranging from 9 to 13.7 kb. They are related to picornaviruses and are classified in the order Picornavirales. The majority of known members infect dicotyledonous plants and many are important plant pathogens (e.g. grapevine fanleaf virus and rice tungro spherical virus). This is a summary of the current International Committee on Taxonomy of Viruses (ICTV) report on the taxonomy of the family Secoviridae available at www.ictv.global/report/secoviridae.
Keywords: Secoviridae, ICTV report, taxonomy
Abbreviation
NTP, nucleotide triphosphate.
Full-Text
Table 1. Characteristics of the family Secoviridae.
| Typical member: | cowpea mosaic virus (RNA-1: X00206; RNA-2: X00729), species Cowpea mosaic virus, genus Comovirus |
|---|---|
| Virion | Non-enveloped, 25–30 nm in diameter with icosahedral symmetry |
| Genome | 9.0–13.7 kb of positive-sense, mono- or bipartite RNA |
| Replication | In association with intracellular membranes derived from the endoplasmic reticulum |
| Translation | Directly from genomic RNA as large polyproteins, which are cleaved by 3C-like proteinases |
| Host range | Plants (mainly dicots), transmitted mainly by insects or nematodes. Some seed transmission demonstrated |
| Taxonomy | In the order Picornavirales, family includes one subfamily with three genera, five additional genera and more than 70 species |
Virion
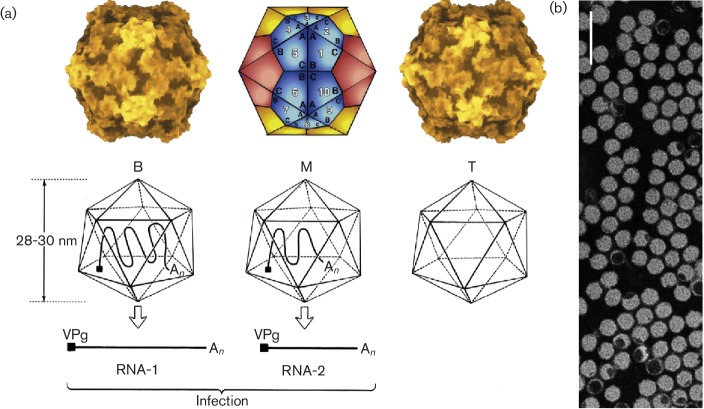
Virions are non-enveloped, 25–30 nm in diameter and exhibit icosahedral symmetry (Table 1). Many virus preparations contain empty virus particles. In the case of viruses with a bipartite genome, the two RNAs are encapsidated in separate virions (Fig. 1) [1].
Fig. 1.
Virion structure and organization. (a) Top left: molecular rendering of the cowpea mosaic virus particle. Top centre: diagrammatic representation of a T=1 lattice. A, Small capsid protein; B, C-terminal domain of the large capsid protein; C, N-terminal domain of the large capsid protein. Top right: molecular rendering of the red clover mottle virus particle. Bottom: diagram of the three types of comovirus particles with the B-particle containing one molecule of RNA-1, the M-particle containing one molecule of RNA-2 and the T-particle being empty. (b) Negative contrast electron micrograph of particles of cowpea mosaic virus. The bar represents 100 nm.
Genome
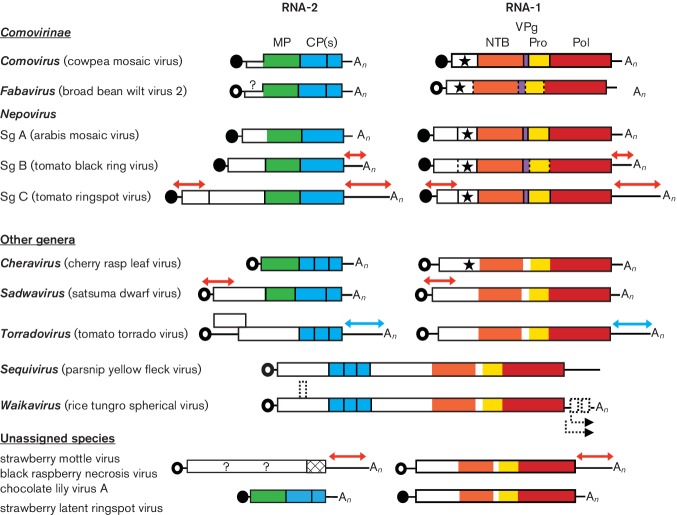
The genome consists of one or two molecules of linear positive-sense ssRNA that are covalently linked to a small protein (viral protein genome-linked, VPg; 2–4 kDa) at their 5′ end and have a 3′-terminal poly(A) tract. Each RNA encodes, in the majority of the cases, a single polyprotein (Fig. 2).
Fig. 2.
Genome organization of representative members of the family Secoviridae. Each RNA is shown with ORFs represented with boxes. Circles depict VPg molecules covalently attached at the 5′-end of the RNAs. Black circles represent VPg confirmed experimentally and open circles represent putative VPgs. Poly(A) tails are represented at the 3′-end of the RNAs when present (An). Red and blue arrows above the sequences represent regions of extensive sequence identity between RNAs 1 and 2. In the latter, for torradoviruses, this identity is also characterized by conserved indels. Protein domains with conserved motifs for the putative NTP-binding protein (NTB, shown in orange), VPg (purple), proteinase (Pro, yellow), RNA-dependent RNA polymerase (Pol, red), movement protein (MP, green) and coat protein(s) (CP, blue) are shown. The star represents a conserved motif found in the protease cofactor (Co-Pro) protein of comoviruses and in the equivalent protein of other viruses. Proteinase cleavage sites identified experimentally or deduced by sequence comparisons are indicated by solid or dotted vertical lines, respectively. Possible ORFs in the genome of waikaviruses are shown with dotted rectangles and putative subgenomic RNAs are shown by dotted arrows below the waikavirus genome. Representatives of each nepovirus subgroup (Sg A, B, C) are also depicted.
Replication
In the case of viruses with a bipartite genome, neither RNA species alone can infect plants systemically. Viral proteins are usually expressed as large polyproteins, which are cleaved by virus-encoded 3C-like proteinases. The replication block contains the domain characteristics of nucleoside triphosphate (NTP)-binding proteins (NTB or putative helicase), 3C-like proteinases (Pro) and RNA-dependent RNA polymerases (Pol) (Fig. 2). Replication occurs in association with intracellular membranes derived from the endoplasmic reticulum.
Taxonomy
Comovirus
Bipartite genome (subfamily Comovirinae). Comoviruses usually have narrow host ranges. Mosaic and mottle symptoms are characteristic. Transmission in nature is exclusively by beetles, especially members of the family Chrysomelidae. Beetles retain their ability to transmit virus for days or weeks [2].
Fabavirus
Bipartite genome (subfamily Comovirinae). Fabaviruses have wide host ranges among dicotyledonous and some families of monocotyledonous plants. Symptoms are ringspots, mottling and wilting. In nature, they are transmitted by aphids in a non-persistent manner.
Nepovirus
Bipartite genome (subfamily Comovirinae). The genus consists of >35 species that are widely distributed in temperate regions. Ringspot symptoms are characteristic. Many nepoviruses are transmitted non-persistently by longidorid nematodes. Seed and/or pollen transmission are also common. In herbaceous plants, the symptoms induced are often transient with a so-called ‘recovery’ phenomenon. The genus can be divided into subgroups (A, B, C) based on sequence and genome organization [3].
Cheravirus
Bipartite genome. Symptoms are usually mild or absent. Cherry rasp leaf virus is transmitted by nematodes in the field [4].
Sadwavirus
Bipartite genome, only one species, Satsuma dwarf virus, members of which have a wide host range. The natural mode of transmission is unknown [5].
Torradovirus
Bipartite genome. RNA-2 contains an ORF upstream and partially overlapping the large ORF. Some torradoviruses are known to be transmitted by whiteflies in a semi-persistent manner. Aphid transmission has been demonstrated for carrot torrado virus 1 [6].
Sequivirus
Monopartite genome. The natural host range of sequiviruses includes plants in several families. Transmission is by aphids in a semi-persistent manner. However, it is dependent on the presence of a helper virus in the genus Waikavirus.
Waikavirus
Monopartite genome. The natural host range of waikaviruses is usually restricted to plants within a few families. Field transmission is semi-persistent by aphids or leafhoppers. Some waikaviruses are helper viruses for the insect transmission of other viruses; for example, rice tungro spherical virus is the helper virus for rice tungro bacilliform virus (family Caulimoviridae).
Resources
Full ICTV Online (10th) Report: www.ictv.global/report/secoviridae.
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Michael J. Adams, Donald B. Smith, Richard J. Orton and Nick J. Knowles.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Sanfaçon H, Wellink J, Le Gall O, Karasev A, van der Vlugt R, et al. Secoviridae: a proposed family of plant viruses within the order Picornavirales that combines the families Sequiviridae and Comoviridae, the unassigned genera Cheravirus and Sadwavirus, and the proposed genus Torradovirus . Arch Virol. 2009;154:899–907. doi: 10.1007/s00705-009-0367-z. [DOI] [PubMed] [Google Scholar]
- 2.Lomonossoff G. Cowpea mosaic virus. In: Mahy BWJ, Van Regenmortel MH, editors. Encylopedia of Virology. 3rd ed. Oxford: Elsevier; 2008. pp. 569–574. (editors) [Google Scholar]
- 3.Fuchs M, Schmitt-Keichinger C, Sanfaçon H. A renaissance in nepovirus research provides new insights into their molecular interface with hosts and vectors. Adv Virus Res. 2017;97:61–105. doi: 10.1016/bs.aivir.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Le Gall O, Sanfaçon H, Ikegami M, Iwanami T, Jones T, et al. Cheravirus and Sadwavirus: two unassigned genera of plant positive-sense single-stranded RNA viruses formerly considered atypical members of the genus Nepovirus (family Comoviridae) Arch Virol. 2007;152:1767–1774. doi: 10.1007/s00705-007-1015-0. [DOI] [PubMed] [Google Scholar]
- 5.Iwanami T. Sadwavirus. In: Mahy BWJ, Van Regenmortel MH, editors. Encylopedia of Virology. 3rd ed. Oxford: Elsevier; 2008. pp. 523–526. (editors) [Google Scholar]
- 6.Rozado-Aguirre Z, Adams I, Collins L, Fox A, Dickinson M, et al. Detection and transmission of Carrot torrado virus, a novel putative member of the Torradovirus genus. J Virol Methods. 2016;235:119–124. doi: 10.1016/j.jviromet.2016.05.018. [DOI] [PubMed] [Google Scholar]