Abstract
Detection of chikungunya virus (CHIKV) or viral RNA is the primary laboratory test used to diagnose infection in serum collected <6 days after onset of illness. Two real-time reverse transcription–polymerase chain reaction (RT-PCR) kits are available commercially, but validity data are limited. There are 2 commercial sources of inactivated positive-control CHIKV RNA to be used with purchased primers. The Centers for Disease Control and Prevention provides viral RNA–positive controls and primer and probe nucleotide sequences for real-time RT-PCR testing. Detection of CHIKV-specific immunoglobulin M (IgM) antibody becomes a sensitive test for samples collected approximately >5 days of illness. Commercially available CHIKV IgM–detection assays include lateral flow rapid tests, IgM antibody capture enzyme-linked immunosorbent assays (MAC-ELISAs), and indirect immunofluorescence tests. Nine commercial CHIKV IgM detection assays were evaluated at 3 reference laboratories to provide guidance to public health diagnostic laboratories on their performance parameters. Sensitivity of the rapid tests and 3MAC-ELISAs was <50%, and thus these assays are not recommended. Three of the MAC-ELISA kits and 1 indirect immunofluorescence kit had comparable performance to the reference assays. In summary, commercial assays with performance comparable to reference assays are available for molecular and serological diagnosis of CHIKV infections.
Keywords: Chikungunya virus, arbovirus diagnostic testing, real-time RT-PCR, IgM antibody capture enzyme-linked immunosorbent assay (MAC-ELISA)
The introduction of Chikungunya virus (CHIKV) to the Caribbean in 2013 resulted in large outbreaks throughout the Americas [1–3]. The clinical similarity of CHIKV to other arbovirus infections makes diagnosis difficult on the basis of clinical symptoms alone, particularly as arboviruses such as dengue virus and now Zika virus may cocirculate in the same geographical regions and are transmitted by the same mosquito species [4–9]. Expansion and endemicity of CHIKV is likely, and large outbreaks of CHIKV infection may continue for the foreseeable future. CHIKV infection was classified as a reportable disease in the United States in January 2015. Public health laboratories throughout the Americas will need to build and maintain high-volume diagnostic testing capacity and will need validated and reliable commercial CHIKV diagnostic assays to respond to these increased diagnostic testing responsibilities.
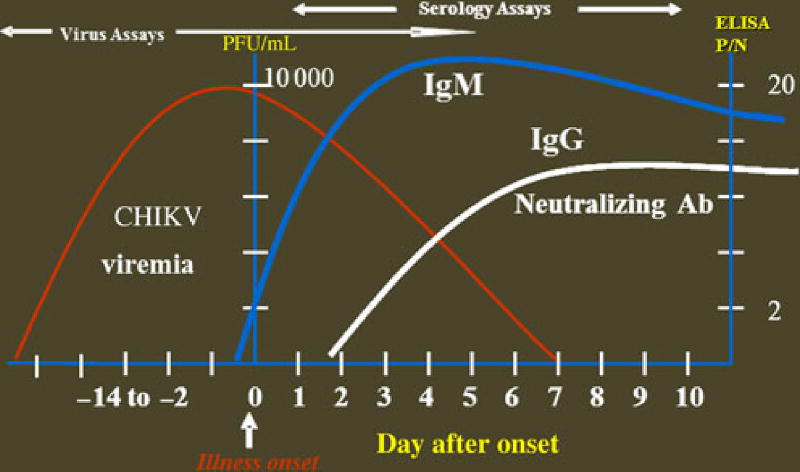
Laboratory diagnosis of CHIKV infection is accomplished by serologic methods, virus isolation, and viral RNA detection by reverse transcription–polymerase chain reaction (RT-PCR). The testing algorithm developed at the Center for Disease Control and Prevention (CDC) to diagnose CHIKV infections is based on characteristics of CHIKV infection and the timing of specimen collection (Figure 1). CHIKV replicates rapidly to high titers in the host, and viral RNA generally can be detected by real-time RT-PCR in the first week after onset of clinical illness. Previously, in travelers to India who had been infected with CHIKV, viral RNA was detectable by real-time RT-PCR in samples collected out to 8 days after onset of illness [10]. Immunoglobulin M (IgM) antibodies elicited in the immune response are normally detectable in serum by days 5–7 after onset of illness. In the study of India travelers, CHIKV-specific IgM was detected in 1 patient on the day of illness onset but was not detectable in the rest until 9 days after onset of illness [10].
Figure 1.
Time course of chikungunya virus (CHIKV) viremia and immune response. Limit of detection (LOD) of real-time reverse transcription–polymerase chain reaction (RT-PCR) assay is approximately 100 plaque-forming units (PFU)/mL (approximately 1 RNA transcript/reaction); the LOD of the immunoglobulin M (IgM) antibody capture enzyme-linked immunosorbent assay (ELISA) positive-to-negative ratio (P/N) is >2. Abbreviations: Ab, antibody; IgG, immunoglobulin G.
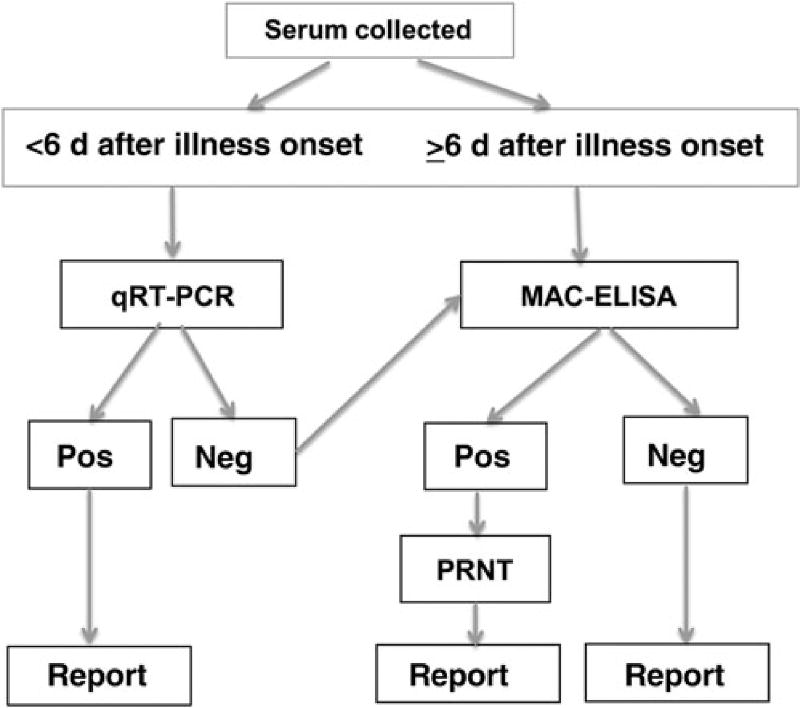
In the CHIKV testing algorithm developed by the CDC arbovirus diagnostic laboratory, samples collected <6 days after onset of illness are first tested by CHIKV real-time RT-PCR (Figure 2). Nucleotide sequences of 2 sets of primers and probes are listed in Table 1 [10–12]. The 3855 primer/probe set was designed to detect the East Central South African (ECSA) genotypes in travelers returning from India, and is broadly reactive, detecting both Asian and ECSA genotypes [10]. The 856 primer/ probe set is designed specifically for the Asian genotype CHIKV strain currently in the Caribbean and is slightly more sensitive for the Asian genotype than the 3855 set. Because of the higher sensitivity, the 856 set is useful for confirming positive results for specimens with low CHIKV RNA levels. Each laboratory should keep these specificity and sensitivity data in mind when designing an appropriate testing algorithm (ie, which sets to use and in what order). Specimens with positive results of tests that use both sets of primers and probes are considered to be confirmed CHIKV-positive specimens. If any of the negative controls test positive, the entire run is invalidated. Failure of the positive control to generate a positive result also invalidates the entire run.
Figure 2.
The Centers for Disease Control and Prevention diagnostic testing algorithm for detection of chikungunya virus (CHIKV) infection. Serum is collected from patients meeting the clinical case definition of fever and arthralgia who have returned from a region where CHIKV is endemic or CHIKV infection is epidemic. All samples with positive and equivocal real-time quantitative reverse transcription–polymerase chain reaction (qRT-PCR) results are repeated with a second set of primer/ probes for confirmation. Specimens with positive results of tests using both sets of primers and probes are considered to be confirmed CHIKV-positive specimens. Samples with positive or equivocal immunoglobulin M capture enzyme-linked immunosorbent assay (MAC-ELISA) results are confirmed to be positive if plaque reduction neutralizing testing (PRNT) yields positive results. Abbreviations: Neg, negative result; Pos, positive result.
Table 1.
Chikungunya Virus (CHIKV) Oligonucleotide Primers and Probes Used in Centers for Disease Control and Prevention Real-time Reverse Transcription–Polymerase Chain Reaction Assays Designed to Detect CHIKV Asian Genotype Strains
| Primer/Genome 5′ Position | Nucleotide Sequence |
|---|---|
| CHIKV3855F | GAGCATACGGTTACGCAGATAG |
| CHIKV3957Ca | TACTGGTGATACATGGTGGTTTC + TGCTGGTGACACATGGTGGTTTC |
| CHIKV3886FAM (probe)a | ACGAGTAATCTGCGTACTGGGACGTA + ACGAGTCATCTGCGTATTGGGACGCA |
| CHIK856F | ACCATCGGTGTTCCATCTAAAG |
| CHIK962C | GCCTGGGCTCATCGTTATT |
| CHIK908FAM (probe) | ACAGTGGTTTCGTGTGAGGGCTAC |
The 3855 primer/probe set is broadly reactive, detecting both Asian and the East/Central/South African genotypes. The 856 set is designed specifically for the Asian genotype CHIKV strain currently in the Caribbean and is slightly more sensitive for the Asian genotype than the 3855 set. The threshold of detection of both sets of primer/probes is approximately 1 RNA transcript.
Primer CHIKV3957C is produced by mixing equal volumes of the 100 µM stock solutions of the 2 individual primers; the CHIKV3886FAM probe is an equal mixture of the 25 µM individual probe stocks.
At the CDC, samples collected on or after day 6 of illness and samples with negative real-time RT-PCR results are tested by the CHIKV IgM antibody capture enzyme-linked immunosorbent assay (MAC-ELISA) [13, 14]. Positive MAC-ELISA results are confirmed by the 90% plaque reduction neutralization test [10, 15]. A sample with positive or equivocal MAC-ELISA results with a neutralizing titer is classified as a confirmed CHIKV-positive sample; those with negative results of plaque reduction neutralization testing are considered to have nonspecific reactivity (ie, to be negative for CHIKV).
There are numerous commercial molecular and serological CHIKV diagnostic assays available; those available as of June 2015 are listed in Table 2. Performance of the RealStar Chikungunya RT-PCR Kit (Altona Diagnostics, Hamburg, Germany) was assessed by Panning et al in the premarket format and was found to have a sensitivity (approximately 5.3 RNA copies/ reaction) and specificity comparable to that of the in-house assay [16]. The product handbook of the genesig Chikungunya Nonstructural protein 2 standard (nsp2) RT-PCR kit (Primer-design, Southampton, United Kingdom) states that the test is sensitive to <100 RNA copies of target; however, no validity studies have been published in the literature. Inactivated CHIKV RNA–positive controls, to be used with published, validated primers and probes, are available through Vircell Microbiologists (Granada, Spain) and Zeptometrix (Buffalo, New York). Both positive-control RNAs, produced from CHIKV ECSA genotype strains, are detectable by the CDC primer/probe sets in Table 1 and the article by Lanciotti et al [10] (data not shown). RNA lysate positive controls prepared from an Indian (ECSA genotype) or Caribbean (Asian genotype) isolate are available from the CDC upon request (available at: http://www.cdc.gov/ncezid/dvbd/specimensub/index.html).
Table 2.
Sources of Commercially Manufactured Chikungunya Virus (CHIKV) Diagnostic Assays as of June 2015
| Manufacturer | Location | Assay Name and Format | Reference | Samples/Tests per Kit, No. |
|---|---|---|---|---|
| CHIKV real-time RT-PCR kit | ||||
| Altona Diagnostics | Germany | RealStar Chikungunya RT-PCR Kit | 011013 | 96 reactions |
| Primerdesign | United Kingdom | genesig Chikungunya Non structural protein 2 standard (nsp2) RT-PCR kit | CHIKV-EASY | 150 reactions |
| CHIK RNA positive control (inactivated) | ||||
| Vircell | Spain | Amplirun CHIKV RNA control (strain S27) | MBC099 | Approximately 1000 runs |
| Zeptometrix | USA | NATtrol inactivated virus (strain India 2006) | NATCHIKV-ST | Approximately 200 runs |
| Plate MAC-ELISA | ||||
| IBL International | Germany | CHIK IgM micro-capture ELISA | RE58841 | 91 |
| CTK Biotech | USA/China | RecombiLISA CHIK IgM Test | E0315 | 91 |
| Genway | Germany | CHIKV IgM µ-capture ELISA | 40-521-475066 | 91 |
| Abcam | Germany | Anti-CHIKV IgM human ELISA kit | ab177848 | 91 |
| SD Diagnostics | Korea | CHIKa IgM ELISA | 16EK10 | 91 |
| Euroimmun | Germany | Anti-CHIKV ELISA (IgM) | EI293a-9601M | 93 |
| Inbios | USA | CHIKjj Detect MAC-ELISA | CHKM-C | 92 |
| CHIKV IgM detection rapid test | ||||
| CTK Biotech | USA | On-site CHIK IgM Combo Rapid test | R0066C | 30 |
| SD Diagnostics | Korea | SD BIOLINE Chikungunya IgM | 46FK10 | 25 |
| CHIKV IgM immunofluorescence assay | ||||
| Euroimmun | Germany | Anti-CHIKV IIFT (IgM) | Fl293a-1010 G/M | 50 |
Abbreviations: IgM, immunoglobulin M; IIFT, indirect immunofluorescence test; MAC-ELISA, IgM antibody capture enzyme-linked immunosorbent assay; RT-PCR, reverse transcription–polymerase chain reaction.
External evaluations of the CHIKV IgM detection assays listed in Table 2 have primarily focused on the rapid tests, as they are attractive options for testing at the point of care and/or elsewhere in resource-limited settings, where CHIKV outbreaks have primarily occurred [17–22] However, the performance of the rapid tests has been disappointing owing to low sensitivity. Therefore, to provide guidelines and recommendations to diagnostic laboratories on the use of these commercial assays, a comprehensive side-by-side comparison of the available (as of July 2015) commercial serological assays was performed at the CDC, the Public Health Agency of Canada National Microbiology Laboratory (NML), and the Caribbean Public Health Agency (CARPHA) in Trinidad-Tobago. With the exception of the CHIK IgM micro-capture ELISA kit manufactured by IBL International, all the CHIKV IgM detection assays listed in Table 2 were evaluated at the CDC, with a reference panel of 70–90 archived, well-characterized sera from CHIK outbreaks in Yap and the Caribbean and from travelers to India and the Philippines that had been submitted to the CDC arbovirus diagnostic laboratory. Positive control sera containing IgM to other alpha-viruses (Venezuelan equine encephalitis, eastern equine encephalitis, O′nyong-nyong, and Mayaro viruses) and other arboviruses (dengue virus) were included in the sample panel to calculate specificity. An in-house positive control was included in every run. At the NML, the performance of the Euroimmun ELISA was compared to in-house assays, using samples submitted to the NML for diagnostic testing from travelers to the Caribbean and other regions where CHIKV is circulating. CARPHA evaluated 4 kits (Abcam, Inbios, and Euroimmun MAC-ELISAs and the Euroimmun indirect immunofluorescence test [IIFT]), using samples submitted from the CARPHA member countries in the Caribbean that had been sent to the CDC for testing. The results of these evaluations have been published elsewhere and are not reported here [23]. However, conclusions of the CDC evaluations are as follows. The Euroimmun and Inbios CHIKV MAC-ELISA kits had the highest accuracy (99% and 100%, respectively) and reproducibility. The Euroimmun IIFT also had high performance (96% accuracy), but a technician skilled in immunofluorescence assay techniques is required, and a higher proportion of samples had equivocal results, necessitating retesting. There was considerable lot-to-lot variation with the Abcam ELISA kits. The initial Abcam kits evaluated had results highly concordant with CDC results; other lots had low sensitivity when used to test the same samples. All runs of all lots were valid according to the criteria in the kit instructions (ie, the kit positive and cutoff control ODs were within the acceptable range). The CDC notified Abcam of the lot-to-lot variability in performance and the problem with the assay controls, after which the manufacturer replaced the controls with human serum containing CHIKV-specific IgM and improved quality control and quality assurance procedures. Because the kit components were modified, the reformatted Abcam ELISA kit needed to be re-evaluated at the CDC. The reformatted Abcam kits were evaluated with as many of the same samples as had been included in the previous Abcam kit evaluations; results were 99% concordant with CDC results. The 2 rapid tests and the remaining plate-based CHIKV MACELISA kits all had unacceptably low performance (sensitivity, <50%), including false-negative results with the CDC in-house positive control serum. Thus, the Inbios and Euroimmun MAC-ELISAs and the Euroimmun IIFT have performance comparable to the reference standard CHIKV MAC-ELISA [13, 14]. The rereleased Abcam MAC-ELISA containing CHIKV-specific IgM human serum controls also has high performance. However, any further changes to the composition of the Abcam kit will invalidate these results and necessitate reevaluation.
In summary, there are numerous validated, commercially manufactured molecular and serological CHIKV diagnostic assays available from which laboratories can choose. The CDC also provides CHIKV RNA–positive controls and primer/probe nucleotide sequences for real-time RT-PCR testing. Nine of 10 commercial CHIKV IgM detection assays were evaluated at the CDC, but only the Inbios and Euroimmun MAC-ELISAs, the Euroimmun IIFT, and the reformatted Abcam MAC-ELISA had acceptable performance. The 2 rapid tests and the 3 remaining plate-based CHIKV MAC-ELISA kits lacked sensitivity and are not recommended for use in their current format. Information on kit prices, the availability of the kits in the United States, and whether the kits have received Food and Drug Administration approval is not provided here, as this information is continually changing and would quickly become outdated and inaccurate.
The CDC CHIKV diagnostic testing algorithm shown in Figure 2 can be used to guide CHIKV testing at public health laboratories. Laboratories with only real-time RT-PCR or MAC-ELISA capacity should test all received samples at all days after onset of illness with the test that they have. Negative results should be interpreted as inconclusive, and the laboratory should consider sending the sample to a reference laboratory where comprehensive testing can be done.
Acknowledgments
We thank Robert Lanciotti, chief of the Center for Disease Control and Prevention (CDC) Arboviral Diseases Branch diagnostic laboratory, for providing the primer and probe nucleotide sequences; Amanda Panella, Janeen Laven, and Olga Kosoy, from the CDC Arboviral Diseases Branch laboratory, for diagnostic testing of the samples used in the evaluation; and the commercial kit manufacturers, for making the assays available for evaluation.
Footnotes
Presented in part: Gaps and Opportunities in Chikungunya Research: Expert Consultation on Chikungunya Disease in the Americas, Rockville, Maryland, 30 June–2 July 2015.
Disclaimer. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the CDC.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Cassadou S, Boucau S, Petit-Sinturel M, Huc P, Leparc-Goffart I, Ledrans M. Emergence of chikungunya fever on the French side of Saint Martin island, October to December 2013. Euro Surveill. 2014;19:13–6. doi: 10.2807/1560-7917.es2014.19.13.20752. [DOI] [PubMed] [Google Scholar]
- 2.Lepiniec L, Dalgarno L, Huong VT, Monath TP, Digoutte JP, Deubel V. Geographic distribution and evolution of yellow fever viruses based on direct sequencing of genomic cDNA fragments. J Gen Virol. 1994;75:417–23. doi: 10.1099/0022-1317-75-2-417. [DOI] [PubMed] [Google Scholar]
- 3.Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49:942–8. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 4.Kuno G. A re-Examination of the history of etiologic confusion between dengue and chikungunya. PLoS Negl Trop Dis. 2015;9:e0004101. doi: 10.1371/journal.pntd.0004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers AM. Chikungunya. Clin in Lab Med. 2010;30:209–19. doi: 10.1016/j.cll.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Powers AM, Roehrig JT. Alphaviruses. Methods Mol Biol. 2011;665:17–38. doi: 10.1007/978-1-60761-817-1_2. [DOI] [PubMed] [Google Scholar]
- 7.Sahadeo N, Mohammed H, Allicock OM, et al. Molecular characterisation of chikungunya virus infections in Trinidad and comparison of clinical and laboratory features with dengue and other acute febrile cases. PLoS Negl Trop Dis. 2015;9:e0004199. doi: 10.1371/journal.pntd.0004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso CW, Paploski IA, Kikuti M, et al. Outbreak of exanthematous illness associated with Zika, chikungunya, and dengue viruses, Salvador, Brazil. Emerg Infect Dis. 2015;21:2274–6. doi: 10.3201/eid2112.151167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musso D, Cao-Lormeau VM, Gubler DJ. Zika virus: following the path of dengue and chikungunya? Lancet. 2015;386:243–4. doi: 10.1016/S0140-6736(15)61273-9. [DOI] [PubMed] [Google Scholar]
- 10.Lanciotti RS, Kosoy OL, Laven JJ, et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13:764–7. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanciotti RS, Lambert AJ. Phylogenetic analysis of chikungunya virus strains circulating in the Western Hemisphere. Am J Trop Med Hyg. 2016;94:800–3. doi: 10.4269/ajtmh.15-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanciotti RS, Valadere AM. Transcontinental movement of Asian genotype chikungunya virus. Emerg Infect Dis. 2014;20:1400–2. doi: 10.3201/eid2008.140268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J Clin Microbiol. 2000;38:1823–6. doi: 10.1128/jcm.38.5.1823-1826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin DA, Noga A, Kosoy O, Johnson AJ, Petersen LR, Lanciotti RS. Evaluation of a diagnostic algorithm using immunoglobulin M enzyme-linked immunosorbent assay to differentiate human West Nile virus and St. Louis Encephalitis virus infections during the 2002 West Nile cirus epidemic in the United States. Clin Diagn Lab Immunol. 2004;11:1130–3. doi: 10.1128/CDLI.11.6.1130-1133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaty BJ, Calisher CH, Shope RE. Arboviruses. In: Lennette ELD, Lennette E, editors. Diagnostic procedures for viral, rickettsial, and chlamydial infections. Washington, DC: American Public Health Association; 1995. pp. 189–212. [Google Scholar]
- 16.Panning M, Hess M, Fischer W, Grywna K, Pfeffer M, Drosten C. Performance of the RealStar chikungunya virus real-time reverse transcription-PCR kit. J Clin Microbiol. 2009;47:3014–6. doi: 10.1128/JCM.01024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blacksell SD, Tanganuchitcharnchai A, Jarman RG, et al. Poor diagnostic accuracy of commercial antibody-based assays for the diagnosis of acute chikungunya infection. Clin Vaccine Immunol. 2011;18:1773–5. doi: 10.1128/CVI.05288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosasih H, Widjaja S, Surya E, et al. Evaluation of two IgM rapid immunochromatographic tests during circulation of Asian lineage chikungunya virus. Southeast Asian J Trop Med Public Health. 2012;43:55–61. [PubMed] [Google Scholar]
- 19.Litzba N, Schuffenecker I, Zeller H, et al. Evaluation of the first commercial chikungunya virus indirect immunofluorescence test. J Virol Methods. 2008;149:175–9. doi: 10.1016/j.jviromet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Prat CM, Flusin O, Panella A, Tenebray B, Lanciotti R, Leparc-Goffart I. Evaluation of commercially available serologic diagnostic tests for chikungunya virus. Emerg Infect Dis. 2014;20:2129–32. doi: 10.3201/eid2012.141269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rianthavorn P, Wuttirattanakowit N, Prianantathavorn K, Limpaphayom N, Theamboonlers A, Poovorawan Y. Evaluation of a rapid assay for detection of IgM antibodies to chikungunya. Southeast Asian J Trop Med Public Health. 2010;41:92–6. [PubMed] [Google Scholar]
- 22.Yap G, Pok KY, Lai YL, et al. Evaluation of chikungunya diagnostic assays: differences in sensitivity of serology assays in two independent outbreaks. PLoS Negl Trop Dis. 2010;4:e753. doi: 10.1371/journal.pntd.0000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson BW, Goodman CH, Holloway K, de Salazar PM, Valadere AM, Drebot M. Evaluation of commercially available chikungunya virus immunoglobulin M detection assays. Am J Trop Med Hyg. 2016;95:182–92. doi: 10.4269/ajtmh.16-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]