Abstract
Small endogenous vesicles called exosomes are beginning to be explored as drug delivery vehicles. The in vivo targets of exosomes are poorly understood; however, they are believed to be important in cell-to-cell communication and may play a prominent role in cancer metastasis. We aimed to elucidate whether cancer derived exosomes can be used as drug delivery vehicles that innately target tumors over normal tissue. Our in vitro results suggest that while there is some specificity towards cancer cells over “immortalized” cells, it is unclear if the difference is sufficient to achieve precise in vivo targeting. Additionally, we found that exosomes associate with their cellular targets to a significantly greater extent (> 10-fold) than liposomes of a similar size. Studies on the association of liposomes mimicking the unique lipid content of exosomes revealed that the lipid composition contributes significantly to cellular adherence/internalization. Cleavage of exosome surface proteins yielded exosomes exhibiting reduced association with their cellular targets, demonstrating the importance of proteins in binding/internalization. Furthermore, although acidic conditions are known to augment the metastatic potential of tumors, we found that cells cultured at low pH released exosomes with significantly less potential for cellular association than cells cultured at physiological pH.
Keywords: Exosomes, Liposomes, Association, Protein Cleavage, pH
Introduction
Exosomes, once thought to be nothing more than cell debris, are now believed to be an integral component of extracellular communication [1, 2]. Exosomes are characterized as 50–100 nm vehicles synthesized through the reverse budding of the late endosomal membrane, forming a multivesicular body (MVB). Fusion of MVBs with the plasma membrane releases exosomes into the extracellular environment [3], where they can interact and associate with target cells. Proteomic analysis of exosomes reveals elevated levels of adhesion proteins that promote adsorption to the cell surface [4, 5]. Exosomes have also recently been shown to attach to recipient cells via phosphatidylserine receptors [6, 7]. After exosomes associate with their target cells, they are internalized through both endocytotic [8–10] and phagocytotic [11] pathways allowing for effective delivery of functional cargo to recipient cells [12–14]. Work by Al-Nedawi et al. demonstrated the ability of exosomes derived from highly aggressive brain gliomas to transfer the mutated epidermal growth factor receptor (EGFRvIII) to cancerous cells lacking EGFRvIII, thereby endowing cells with oncogenic activity [12]. Similarly, functional mRNA found in exosomes derived from a mouse mast cell line can be transferred to human mast cells. After transfer, new mouse proteins were translated in the recipient human cells [13]. These latter findings, combined with the observation that serum from cancer patients contains elevated levels of exosomes, are consistent with a role for exosomes in tumor progression [15].
Exosomes’ innate ability to deliver their cargo to recipient cells makes them an intriguing candidate for use as a drug delivery vehicle. In fact, exosomes have recently been used to deliver siRNA to the brains of mice. Targeting to the brain was achieved through fusion of the neuron-specific RVF peptide to the lamp2b protein found on dendritic cell derived exosomes. Successful siRNA delivery resulted in a 62% knockdown in the therapeutic target [16]. In another recent study, mouse lymphoma cell-derived exosomes loaded with the anti-inflammatory drug curcumin were able to protect mice from a lipopolysaccharide septic shock challenge by delivering curcumin to activated myeloid cells. In contrast, curcumin incorporated into liposomes was only slightly more efficient than free curcumin in preventing mortality [17].
Previous studies have suggested that exosomes are more readily associated with cancer cells as compared to normal cells [18]. Considering the purported role of exosomes in intercellular communication within tumors, it follows that exosomes might be particularly effective for delivery to cancer cells, especially to the parent cell line that produced the exosomes. Tumor derived exosomes have a distinct protein and lipid composition resembling that of the cells from which they are derived, suggesting that exosomes may be uniquely suited to interact with their parent cells line in comparison to cells of various origins. In this study we quantify the association of exosomes with both cancer and immortalized cell lines, and compare this to the adherence/internalization of similarly-sized liposomes. Furthermore, we perform experiments to determine the extent to which the lipid and protein components contribute to exosome association with recipient cells.
In addition, it has been suggested by Parolini et al [18] that low extracellular pH in which cells are grown increases the propensity for exosomes to fuse with target cells. Because the tumor microenvironment is known to be acidified [19], it is possible that the physiological properties of exosomes produced under these conditions may also be affected. We rigorously test this hypothesis by comparing the association of exosomes with recipient cells harvested from cells cultured at different pHs. To further elucidate any potential role of an acidic microenvironment on exosome association, the media of recipient cells was also acidified to assess its effect on adherence/internalization.
Materials and Methods
Chemicals
Dulbecco’s Modification of Eagle’s Medium (DMEM, with 4.5 g/L glucose, L-glutamine, and sodium pyruvate), Dulbecco’s Phosphate-Buffered Saline (DPBS), Fetal Bovine Serum (FBS), Trypsin, and Penicillin-Streptomycin were all purchased from Mediatech, Inc. (Manassas, VA). Plasmocin™ was obtained from InvivoGen (San Diego, CA). FBS was ultra-centrifuged for 10 hours at 120,000 × g to remove contaminating exosomes. All media was filtered with 0.22 μm low protein binding cellulose acetate filter from Thermo Fisher Scientific, Inc. (Rockford, IL) before use. MCF-7 cells and PC3 cells were purchased from ATCC. H460 and ARPE-19 cell lines were a gift from Dr. Uday Kompella. 16HBE cells were a generous gift from Dr. Brian Day. SK-Mel-5 and MDA-MB-231 cells were a gift from Dr. David Ross. WPMY cells were a gift from Dr. Isabel Schlaepfer. Uryanl Acetate was ordered from Electron Microscopy Sciences (Hatfield, PA). Formvar/carbon-coated EM grids were made at the University of Colorado, Denver Electron Microscopy Core. Sodium phosphate monobasic monohydrate, sodium phosphate dibasic and bis-tris (Bis(2-hydroxyethyl)-amino-tris(hydroxymethyl)-methane) were purchased from Sigma-Aldrich (St. Louis, MO). Complete Mini EDTA-Free Protease inhibitor cocktail tablets were purchased from Roche (Indianapolis, IN). Anti-calnexin was purchased from Enzo Life Sciences (Farmingdale, NY). Anti-cytochrome C was purchased from Bio Legend (San Diego, CA). Anti-HSP70 was purchased from Cell Signalling Technology (Danvers, MA). Anti-HSP90 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CD63 and Anti-CD9 were purchased from Abcam (Cambridge, MA). The chemiluminescent kit, ECL™ Plus Western Blot Detection System and both secondary antibodies, ECL™ Anti-mouse IgG horseradish peroxidase linked F(ab′)2, and ECL™ Anti-rabbit IgG horseradish peroxidase linked F(ab′)2 were obtained from GE Healthcare (UK Little Chalfont Buckinghamshire). Carnation powdered milk was purchased from Nestle (Solon, OH). Millipore Immobilon Polyvinylidene Fluoride (PVDF) Transfer Membranes were purchased from Thermo Fisher Scientific, Inc. (Rockford, IL). D(+)-Sucrose, 99.7%, for biochemistry, was purchased from Acros Organics (Fairlawn, New Jersey). L-α-phosphatidylcholine (Chicken Egg) (PC), L-α-phosphatidylserine (Porcine Brain) (PS), Sphingomyelin (Porcine Brain) (SM), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). Proteinase K was obtained from Qiagen (Valencia, CA) and NEB2 buffer was purchased from New England BioLabs (Ipswich, MA). BCA Protein Assay Reagent, HPLC grade methanol and chloroform were purchased from Thermo Fisher Scientific, Inc. (Rockford, IL). DID (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) and rhodamine DHPE (Rhodamine B 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine, Triethylammonium Salt) were obtained from Invitrogen (Carlsbad, CA). Fluorescein was purchased from J.T Baker (Center Valley, PA). Hydrochloric acid was obtained from RICCA Chemical Company (Arlington, TX). Sequencing grade trypsin was purchased from Promega Corporation (Madison, WI).
Cell Culture
All cancer (PC3, MCF-7, MDA-MB-231, SK-Mel-5, H460) and non-cancer immortalized (16HBE, WPMY-1, ARPE-19) cell lines were grown in DMEM, 10% FBS, supplemented with Pen-Strep and Plasmocin™ at 5 μg/ml. Media conditions were identical across all cell lines to prevent any media related effects on the extent of exosome association with the various cell lines. All cells appeared healthy and maintained consistent doubling times under these conditions. Identical media conditions were used for experiments in which the pH of the media was shifted from pH 7.4 to pH 6.8 and 6.3, with the addition of 30 mM Bis-Tris. After the addition of Bis-Tris to the media, HCl was used to lower the pH to 6.8 and 6.3, while no HCl was added to the media of cells cultured at pH 7.4.
Exosome Isolation
Exosomes were isolated from the supernatant of PC3, MCF-7 and MDA, MB-231 cells lines. All cell lines were grown to 50% confluency in 225 cm2 BD cell culture flasks, before the media was replaced by exosome-free DMEM 10% FBS media. After 48 hours, the supernatant was collected and purified by a series of centrifugation steps: 10 min 300 × g, 20 min 15,000 × g, and 10 hours 100,000 × g. Concentrated exosomes were then washed in PBS and centrifuged at 200,000 × g for 10 hours on a sucrose density cushion. The sucrose cushion consisted of three distinct layers, 12 % sucrose, 30 % sucrose and 50% sucrose. Exosomes have been previously reported to have a density between 1.1 and 1.2 g/cm3 [13, 20, 21]. Subsequently, the 30% sucrose fraction and the top of the 50% sucrose fraction were collected, washed with PBS, and centrifuged at 100,000 × g for 10 hours. The pelleted exosomes were re-suspended in 500 μl PBS. Exosome protein content was quantified using the BCA protein assay. It is well known that proteins account for 25–75% of the weight of various biological membranes, with most membranes possessing approximately 50% protein [22]. However, the exact protein-to-lipid weight ratio of exosome has not been quantified. For our experiments we assumed exosomes to be equal parts protein and lipid by weight, thus the total weight of exosomes was estimated by doubling BCA protein assay results. While assuming a 50/50 weight ratio of proteins to lipid in exosomes may result in slightly erroneous values when comparing to the equivalent amount of liposomes, the > 10-fold differences reported in figure 3 are significantly greater than could be accounted for by more extreme protein-to-lipid ratios in exosomes. Exosomes were then labeled with specific concentrations of DID based on the weight of each sample. Unincorporated DID was removed through density gradient centrifugation as described above.
Figure 3.
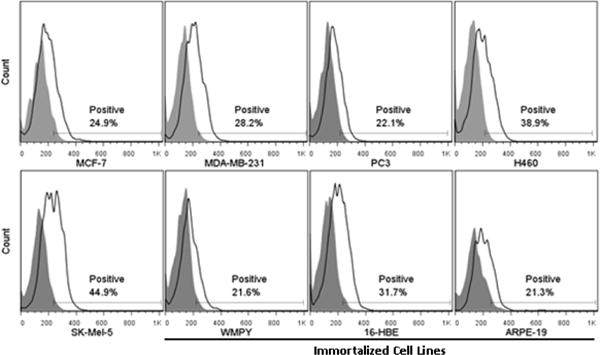
Extent of PC:Cholesterol (2:1 mole ratio) liposomes association with five cancer cell lines (MCF-7, MDA- MB-231, PC3, H460 and SK-Mel-5) and three immortalized cell lines (WMPY, 16-HBE and ARPE-19). Liposomes labeled with 0.01% DID were incubated with all cell lines for 4 hrs at 37°C. Extent of association was measured using flow cytometry. Association was quantified by monitoring the percentage of cells that had increased fluorescence when compared to control cells (“Positive”).
Electron Microscopy
Electron Microscopy was performed on a Technai G2 Bio-twin made by FEI. Isolated exosomes, re-suspended in phosphate buffer, were deposited on formvar/carbon-coated EM grids and left to dry in ambient air. Phosphate buffer containing 2.0% uranyl acetate was added to the exosome-coated grids and left for 10 minutes before washing with phosphate buffer containing 0.4% uranyl acetate.
Confocal Microscopy
Confocal microscopy was performed on a Nikon Eclipse TE2000-E microscope. Images were analyzed using Nikon EZ-C1 FreeViewer Version 390 software. Exosomes were labeled with the fluorescent molecule DID. Long-chain dialkylcarbocyanines, such as DID, are used extensively to label biological membranes [23, 24]. Five microliters of DID labeled exosomes, in PBS, were placed on a cover slip and imaged.
Fluorescent Imaging of Exosome Uptake
Fluorescent images were taken using an OperettaTM High Content Imaging System instrument (Perkin Elmer; Waltham, MA). MCF-7 cells were grown on a 96-well black walled plate made by Greiner Bio-One (Monroe, NC). Five micrograms MCF-7 exosomes in 100 μl DMEM supplemented with 10% FBS, labeled with DID, were incubated with cells for 4 hours. Cell media was subsequently removed and cells were rinsed with PBS twice. Cells were stained with Hoechst 33342 nucleic acid stain at a concentration of 10 μg/ml in PBS for 15 minutes. Cells were rinsed with PBS to remove free Hoeschst 33342 stain. Cells were then fixed for 10 minutes with 3.7% formalin. Formalin was removed by washing with PBS. Images were analyzed using Harmony 3.5.1 software manufactured by Perkin Elmer.
Western Blotting
Cells and exosomes were lysed with lysis buffer containing protease inhibitors and subsequently centrifuged for 10 min at 500 × g to remove debris. Twenty micrograms of protein from cells or exosomes were separated by SDS-PAGE. Gels were blotted onto polyvinylidene fluoride (PVDF) membranes and incubated with Tris-buffered saline (TBS) containing 5% powdered milk for one hour, followed by overnight incubation with 5% powdered milk and primary antibody. The PDVF membrane was then washed twice with TBS containing 0.05% Tween 20 before the addition of TBS with 5% powdered milk and the corresponding secondary antibody for 1 hour. Chemiluminescence was used to identify the presence of the protein of interest. Chemiluminescence was imaged using a Molecular Dynamics Storm 860.
Mass Spectrometry Analysis
One hundred μg of isolated exosomes from MCF-7 and PC3 cells were subjected to in solution digestions using performic acic. Performic acid solution was prepared in a 1:19 ratio of 30% hydrogen peroxide:formic acid. Perfromic acid solution was warmed to 55 °C for 3 minutes immediately prior to use. Three volumes of performic acid were added to the exosome sample and incubated on ice for 3 hours. The reaction was quenched using 5 volumes of ice cold double distilled water. Samples were dried to completion using a speed vacuum and resuspended in 50 μl of 50 mM ammonium bicarbonate. Sequencing grade trypsin was added to the sample in a 1:50 trypsin: protein ratio and incubated overnight at 37 °C. The next morning the samples were dried to completion using a speed vacuum and the pellet resuspended in 30 μl of 0.1% formic acid in water. Tryptic digests were separated using a 5–50% ACN gradient over 120 minutes on a C18 column (Michrocom, Agilent). MS/MS spectra were collected using the Amazon Speed ion ETD trap equipped with CaptiveSpray nanoBooster ionization source (Bruker Daltonics) at the University of Colorado School of Pharmacy Mass Spectrometry Core. Acetonitrile enriched nitrogen gas was used as a sheath gas to increase the charge state of peptide ions and enhance identifications. Data processing was performed using ProteinScape 3.1. Database searches were performed against all of the human entries in the Swiss Prot database using the Mascot Server using 0.6 Da peptide mass tolerance and 0.5 Da MS/MS tolerance allowing for 1 missed cleavage and modifications for dioxidation of methionine and trioxidation of cysteine. Protein identification was considered significant if at least 2 unique peptides were used for identification.
Sucrose Density Gradients
To identify the density of isolated exosomes, exosomes were placed on top of a discontinuous sucrose density gradient and centrifuged at 200k × g for 10 hrs. The sucrose gradient was composed of 6 distinct sucrose density layers ranging from 1.05 g/cm3 to 1.23 g/cm3.
Lipid Extraction
Lipids from PC3 cells and PC3-derived exosomes were extracted using the Bligh and Dyer method [25]. Briefly, approximately 300 μg exosomes or cells (based on BCA protein content) in 200 μl PBS were added to 1.9 ml CHCl3:MeOH 1:2 and vortexed. Subsequently, 0.625 ml CHCl3 was added and vortexed, followed by addition of 0.625 ml dH2O. Samples were then centrifuged to separate the organic and aqueous phases. The organic phase was recovered and dried in pre-weighed HPLC vials. The HPLC vials were re-weighed on a Mettler Toledo MX5 Micro Balance. The difference in weight allowed for an accurate weight of total lipids extracted from PC3 cells and PC3 derived exosomes.
Liposome Formulations
All liposome formulations were prepared by mixing lipids at specific ratios with DID in chloroform in pre-weighed 300 ul glass HPLC vials. Lipid mixtures in HPLC vials were dried under nitrogen gas and placed under vacuum to remove residual chloroform. As with exosome and cell lipid extracts, each liposome formulation in HPLC vials was re-weighed on a Mettler Toledo MX5 Micro Balance to obtain an accurate weight of total lipids. To the HPLC vials containing the dried lipids, 500 μl PBS was added, and subsequently sonicated to remove lipids from the HPLC vial walls. The lipid/PBS mixture was removed and extruded through 100 nm pore size polycarbonate membranes (Avestin, Ottawa, ON). All liposome formulations had a mean diameter of 105 ± 10 nm (data not shown).
Protein Cleavage on Exosomes
To cleave membrane bound proteins from the surface of PC3-derived exosomes labeled with DID, 200 μg exosomes (based on BCA protein content) were incubated with proteinase K at 50 μg/ml and 1× NEB2 buffer at 37°C for one hour, via a protocol modified from Escrevente et al. [9]. One microgram of control exosomes and proteinase K-treated exosomes were then immediately incubated with PC3 and MCF-7 cell lines for four hours; control experiments indicated that the low level of proteinase K had no detectable effect on cell detachment/viability. Cells were then analyzed with flow cytometry to determine the extent of exosome association.
Sizing
Exosomes and liposomes were sized using a NanoSight LM20. Exosomes were diluted 1/1000 from stock before analysis. Measurements were done in triplicate, using the same stock of exosomes. The size and standard deviation from each of the three individual measurements was then averaged.
Flow Cytometry
All flow cytometry experiments were performed on a Becton Dikinson FACScan. Briefly, between 1 and 10 ug of DID or rhodamine DHPE-labeled exosomes or liposomes were added to cells cultured in Costar® 96 well cell culture flat bottom tissue culture flasks. After four hours incubation with either exosomes or liposomes, cells were rinsed with PBS, removed by trypsinization, suspended in PBS, and put on ice. A minimum of ≥5,000 events were acquired per sample. The small amount of exosomes harvested in any individual preparation require that experiments be replicated a minimum of three times with exosomes from different preparations. While the absolute numbers for association did vary among different exosome preparations, consistent trends were observed. Results from an individual experiment are reported, and the differences were statistically analyzed with FlowJo software as detailed below. The y-axis of flow cytometry data is presented as a count of cells, where each data set is fit to have equivalent maximum peak heights in order to facilitate visual comparison between samples. Using this type of analysis, the area under the curve does not indicate the absolute number of cells analyzed. The extent of exosomes association with recipient cells was quantified by monitoring the percentage of cells that had increased fluorescence when compared to control cells (“positive”). Probability binning, built into Flowjo software, was use to statistically analyze samples of the same cell line, while t-tests were used to statistically analyze the differences among samples across multiple cell lines.
Results
Characterization of exosomes isolated from cell culture
Exosomes isolated from the supernatant of cultured MCF-7 cells were evaluated using electron microscopy. Figure 1A depicts vesicular structures, uniform in size (50 – 100 nm), and comparable to previously described exosomes [21, 26–28]. Further, the presence of HSP90, HSP70, calnexin, cytochrome c, CD9, and CD63 in MCF-7-derived exosomes was assessed by western blot analysis. Exosomes are known to be selectively enriched in the cytosolic proteins HSP90 and HSP70 and the tetraspan glycoproteins CD9 and CD63, while proteins associated with the endoplasmic reticulum and mitochondria are not actively incorporated into exosomes [26, 29–33]. MCF-7 cell lysates were positive for all proteins analyzed, while MCF-7-derived exosome samples were devoid of cytochrome c and had minimal calnexin expression (Figure 1B). The lack of ER and mitochondrial proteins in exosome preparations demonstrates that the vesicles observed with electron microscopy are isolated exosomes that are relatively free of cell debris. Post isolation, exosomes were labeled with the lipophilic fluorescent probe DID (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate). Confocal microscopy was used to visualize the incorporation of DID into exosomes (Figure 1C). To visualize the uptake of MCF-7 exosomes labeled with DID, MCF-7 exosomes were incubated with MCF-7 cells for 4 hours. Post incubation, cells were washed with PBS and the nucleus of cells were fluorescently stained. Uptake of 4T1 exosomes can be seen in Figure 1D.
Figure 1.
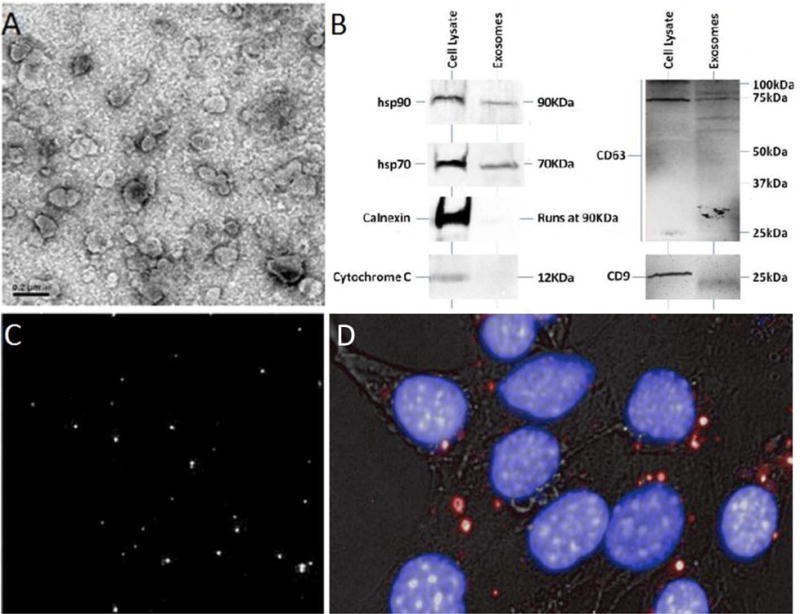
Characterization of Exosomes. A) Electron microscope image of exosomes derived from MCF-7 cells. Scale bar = 0.2 μm. B) Biochemical characterization of exosomes by western blot. MCF-7 exosomes and MCF-7 cells were lysed with lysis buffer, and debris removed. The protein content of MCF-7 exosomes and MCF-7 cell samples were quantified using the BCA protein assay. Twenty micrograms of exosome protein or cell proteins were separated using SDS-PAGE and subsequently subjected to western blotting with antibodies for HSP70, HSP90, calnexin, cytochrome c, CD9 and CD63. Isolated exosomes were positive for HSP90, HSP70, CD9 and CD63, but were void of cytochrome c and had negligible calnexin compared to cell lysate, which was positive for all proteins. CD63 proteins in both exosomes and cell lysate samples self-associated to make higher molecular weight species. The western blot demonstrates successful separation of exosomes from cells and cell debris. C) Confocal microscope image of MCF-7-derived exosomes labeled with the fluorescent dye DID. D) MCF-7 cells were incubated for 4 hours with DID-labeled exosomes (red). MCF-7 nuclei were stained with Hoeschst 33342 (Blue). Bright field images are overlaid to visualize cells.
To further characterize the presence of isolated exosomes, LC/MS was performed on both MCF-7 and PC3 exosomes. We identified 59 unique proteins in MCF-7 exosome samples (Supplemental Table 1) and 80 unique proteins from PC3 (Supplemental Table 2). To each table we have added the subcellular localization and function of each protein identified in MCF-7 and PC3 exosomes. Analysis of the data revealed the presence of the canonical markers of exosomes in both MCF-7 and PC3 exosome samples including: Heat Shock Proteins 70, 90, and 27; CD9, Ras-related protein 13 (RAB-13 and others); Annexins 1, 2, 5, and 7; Pyruvate kinase (PKM); Alpha-enolase (ENO1); Glyceraldehyde-3-phosphate dehydrogenase (GAPDH); Glucose-6-phosphate 1-dehydrogenase (G6PD); and actin [33]. Furthermore, a search of Vesiclepedia (http://microvesicles.org/) in June 2014, revealed that every protein identified, using LC/MS, in both MCF-7 and PC3 exosomes had been previously found associated with exosomes. In addition, the vast majority of proteins identified also are classified in the UniProtKB Gene Ontology Cellular Component sections as members of “extracellular vesicles exosomes”, further supporting the notion that our protein cohorts are derived from exosomes (http://www.uniprot.org/).
Comparison of exosome and liposome cellular association
The biological role of exosomes is unclear; however, one potential function is cell-to-cell communication. Numerous groups have published on the ability of exosomes to be endocytosed by both the “parent” cell line [9, 10, 34] and by cells of various origin [11, 13, 35–38]. Here we examine the extent of exosome adherence/internalization by the “parent” cell line in comparison to 105 nm liposomes (Table 1) composed of PC:Cholesterol, mole ratio 2:1. This lipid composition was chosen because it mimics that of Daunoxome, a commercial liposome product used to treat cancer. Ten micrograms of liposomes and either 1 ug or 10 ug PC3- derived exosomes labeled with 0.1% of the fluorescent dye DID were incubated with PC3 cells for 4 hours at 37°C. The degree of association was analyzed using flow cytometry (Figure 2). The higher mean fluorescence intensity of the cells incubated with 1 μg of exosomes, in comparison to cells incubated with 10 μg liposomes, suggests that considerably greater than ten times more exosomes were associated with PC3 cell than PC:Cholesterol liposomes. This would indicate the physical/chemical properties of exosomes promote association to cells, and reaffirms the notion of exosomes’ importance in cell-to-cell communication.
Table 1.
Exosome mean sizes and standard deviations are calculated from three separate measurements on the preparations used in our experiments. PC:Cholesterol (mole ratio 2:1) liposomes were extruded through 100 nm pore size polycarbonate membranes.
| Size (nm) | S.D. | |
|---|---|---|
| MCF-7 | 138 ± 5 | 55 ± 9 |
| MDA-MB-231 | 142 ± 10 | 50 ± 1 |
| PC3 | 146 ± 13 | 62 ± 7 |
| Liposome | 105 ± 10 | 35 ± 2 |
Figure 2.
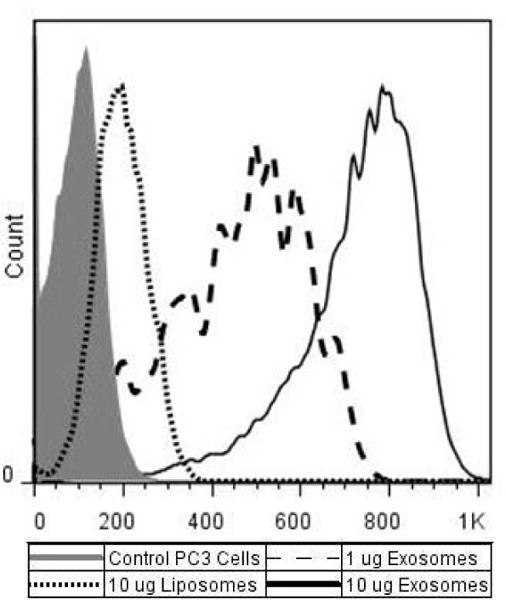
Association of Exosomes and Liposomes with PC3 cells. PC:Cholesterol (2:1 mole ratio) liposomes or PC3- derived exosomes labeled with 0.01% DID were incubated with PC3 cells for 4 hrs at 37°C. Extent of association was measured using flow cytometry. Mean autofluorescence for PC3 control cells was 102, mean fluorescent association by PC3 cells incubated with 10 μg of liposomes was 194; PC3 cells incubated with 10 μg exosomes had a mean fluorescence of 718, and cells incubated with 1 μg exosome had a mean fluorescence of 425.
Extent of exosome adherence/internalization by various cells lines
Cell-to-cell communication via exosomes may not only be important for normal cell function, but also may play a role in cancer progression and metastasis. This is consistent with the elevated exosome concentrations in the blood of cancer patients [15, 39]. To assess whether exosomes are associated with their “parent” cell line to a greater extent than by cells of various origins, and to determine if exosomes adhere/internalize with cancer cells (independent of origin) more than with “normal” immortalized cells, exosomes derived from three cancer cell lines were added back to the media of five cancer cell lines and three immortalized cell lines. Comparison of particle association with numerous cell lines is complicated by differences in cell doubling times, size, and morphology, and by the rate of endocytosis. Cells chosen for analysis were derived from a range of tissues. Because cells were derived from many different tissues, we expect their peripheral content to be distinct from each other. Additionally, cells were chosen for study based on their growth rate and physical characteristics. All eight of the cell lines used for our experiments doubled approximately every two days (data not shown). Further, each cell line’s size and morphology was scrutinized using microscopy. The cell lines selected for our experiments all grew as a monolayer, while cell lines with exceptionally large or small cell morphology were excluded from our experiments due to concerns that cells with significantly greater surface area might exhibit artificially enhanced association per cell as determined by flow cytometry.
Additionally, to ensure that any differences in the extent of exosome adherence/internalization by the various cells was not a result of varying levels of non-specific endocytosis, 10 μg of neutral (PC:Cholesterol 2:1 mole ratio) liposomes were incubated with all eight cell lines. The extent of liposome association was monitored by flow cytometry and used to assess non-specific association in each cell line (Figure 3). The eight cell lines monitored had a baseline auto-fluorescence of 129 ± 8.8. Increase in cell fluorescence, as a result of liposome association, was quantified by monitoring the percentage of cells that had increased fluorescence when compared to control cells, and was reported as percent positive. The five cancer cell lines, from a wide range of tissues, (MCF-7 – mammary gland/breast, MDA-MB-231 -mammary gland/breast, PC3 – prostate, H460 – lung, and SK-Mel-5 – skin) on average took up liposomes to a slightly greater extent than the three immortalized cell lines (WPMY – prostate/stroma, 16-HBE- bronchial epithelial cells, and ARPE-19 – retinal pigmented epithelium cells) (Table 2). We assume that these rates of liposome association reflect differences in non-specific association, and relative values are used to account for differences in binding/endocytotic activity among cell lines (see below).
Table 2.
Extent of exosome association with recipient cells as seen in Figures 3 and 4 are compiled for comparison. Adherence/internalization experiments of liposomes and exosomes were performed simultaneously to prevent day-to-day variations in the extent of association. Bolded numbers indicate association of “daughter” exosomes by the “parent” cell line. Both liposome association and exosome association values for both cancer and immortalized cell lines are averaged. Data from Figure 4 is further normalized as described in the text to account for differences in non-specific association when comparing exosome association between cancer and immortalized cell lines.
| Cell Line | Percent Positive | Normalized Percent Positive | ||||||
|---|---|---|---|---|---|---|---|---|
| Liposome | MCF-7 Exo | MDA Exo | PC3 Exo | Relative Liposome Uptake | MCF-7 Exo | MDA Exo | PC3 Exo | |
| MCF-7 | 24.9 | 34.1 | 32.4 | 16.4 | 0.9 | 38.6 | 36.7 | 18.6 |
| MDA-MB-231 | 28.2 | 40.6 | 18.0 | 17.2 | 1.0 | 40.6 | 18.0 | 17.2 |
| PC3 | 22.1 | 32.3 | 24.3 | 18.0 | 0.8 | 41.9 | 31.0 | 23.0 |
| H460 | 38.9 | 45.1 | 32.2 | 20.3 | 1.4 | 32.7 | 23.3 | 14.7 |
| SK-Mel-5 | 44.9 | 39.8 | 22.1 | 16.9 | 1.6 | 25.0 | 13.9 | 10.6 |
| WPMY | 21.6 | 29.8 | 14.7 | 9.5 | 0.8 | 38.9 | 19.2 | 12.4 |
| 16-HBE | 31.7 | 15.5 | 7.7 | 6.1 | 1.1 | 13.8 | 6.9 | 5.4 |
| ARPE-19 | 21.3 | 45.7 | 16.2 | 14.0 | 0.8 | 60.5 | 21.4 | 18.5 |
| Average | 29.2 | 35.4 | 21.0 | 14.8 | – | 36.5 | 21.3 | 15.1 |
| Average Liposome Cancer % Positive | 31.8 | Avenge Normalized Cancer % Positive | 25.7 | |||||
| Average Liposome Immortalized & Positive | 24.9 | Average Normalized Immortalized % Positive | 21.9 | |||||
| Auerage Exosome Cancer % Positive | 27.3 | Average Normalized WPMY and ARPE-19 % Positive | 28.5 | |||||
| Average Exosome Immortalized % Positive | 17.7 | |||||||
| Average Exosome WPMY and ARPE-19% Positive | 21.7 | |||||||
| Average Liposome WPMY and ARPE-19% Positive | 21.5 | |||||||
Exosomes collected from two metastatic cancer cell lines, MDA-MB-231 and PC3, and one non-metastatic cell line MCF-7, were labeled with DID. Fluorescently-labeled exosomes were added back to the media of both cancer and immortalized cells. After a four-hour incubation, cells were removed and analyzed for changes in fluorescence by flow cytometry. As described above for liposomes, association was quantified by monitoring the percentage of cells that had increased fluorescence when compared to control cells.
Analysis of the data in Figure 4 suggests that exosomes derived from MCF-7, MDA-MB-231, and PC3 cancer cell lines are not preferentially associated with their “parent” cell lines. When taken as an average, the three immortalized cell lines have decreased exosome association as compared to the five cancer cell lines, as seen in Figure 4. However, non-specific adherence/internalization by immortalized cells is also lower than that of the five cancer cell lines (Figure 3). In order to compare the extent of exosome association with the various cell lines, and account for the differences in non-specific adherence/internalization, the data were normalized by calculating the ratio of percent positive liposome adherence/internalization for all cell lines relative to MDA-MB-231 cells, and that value was used to normalize all exosome association data (Table 2). Normalization revealed that on average, the three immortalized cell lines accumulated exosomes to a lesser extent than the cancer cell lines. But, further examination shows that 16-HBE cells had significantly decreased exosome association in comparison to cancer cells, while WPMY and ARPE-19 cells had relatively similar extents of exosome association as the five cancer cell lines.
Figure 4.
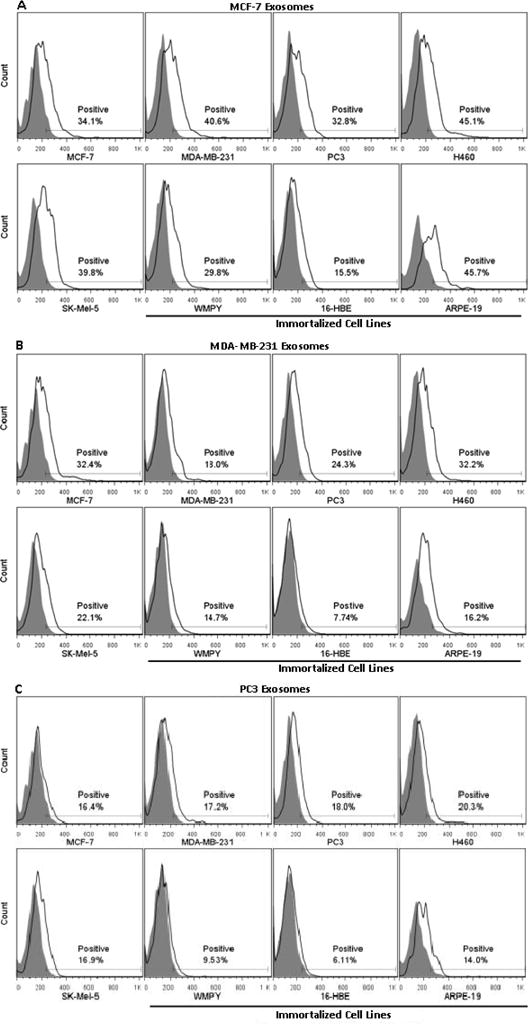
Exosome association with cancer and immortalized cell lines. Exosomes isolated from the supernatant of MCF-7, MDA-MB-231, and PC3 cell lines were added back to the media of five cancer cell lines (MCF-7, MDA- MB-231, PC3, H460 and SK-Mel-5) and three immortalized cell lines (WMPY, 16-HBE and ARPE-19). Exosomes were labeled with 0.003% DID by weight; the greater association of exosomes as compared to liposomes required lower levels of DID to avoid detector saturation. Extent of association was measured using flow cytometry and quantified by monitoring the percentage of cells that had increased fluorescence when compared to control cells (“Positive”). Results from a single experiment are presented; repeat experiments yielded virtually identical results.
Although there does not appear to be increased exosome association with their “parent” cell lines or a distinct difference in exosome association between cancer and immortalized cells, adherence/internalization of MCF-7 exosomes was significantly greater than exosomes derived from MDA-MB-231 and PC3 cells. To ensure that the increased adherence/internalization of MCF-7 exosomes was not due to increased size or density resulting in enhanced settling on monolayer cells causing more cell contact, exosomes from MCF-7, MDA-MB-231 and PC3 cells were sized using a NanoSight LM20. As seen in Table 2, exosomes derived from each cell line have similar sizes and standard deviations. Additionally, MCF-7, MDA-MB-231, and PC3 exosomes all partitioned into the sucrose gradient layer corresponding to 1.18 g/cm3 (data not shown), indicating that the isolated exosomes from the three cell lines have similar densities. These data indicated that increased association of MCF-7-derived exosomes was not likely a result of differences in their physical properties. Instead, these data suggest that the constituents of MCF-7 exosomes are responsible for their greater adherence/internalization.
An additional goal of the study was to compare the association of prostate cancer-derived exosomes with cancer and immortalized prostate cells. The data indicate that there is a significantly greater association of PC3-derived exosomes by the prostate cancer (PC3) cells as compared to the immortalized prostate (WPMY) cells (p<.05). This finding suggests that in the tumor microenvironment, PC3 exosomes may preferentially associate with cancer cells over normal cells, and thus PC3-derived exosomes may serve as selective delivery vehicles for prostate cancer therapeutics. Unfortunately, we were unable to do a similar comparison with breast cancer cell lines because immortalized breast cells (MCF-12 A and MCF-10A) did not fit the growth criteria to be included in the experiment. As a result, we cannot draw definitive conclusions regarding differential exosome adherence/internalization in breast cancer cells versus immortalized breast cells.
Influence of lipids and proteins on exosome cellular association
To elucidate why exosomes adhere/internalize to a significantly greater extent (> 10-fold) than PC:Cholesterol liposomes, we investigated how the lipid and protein components of exosomes affect adherence/internalization. A number of studies have concluded that exosomes have a unique lipid composition, distinct from the plasma membrane and organelles of the “parent” cell line [18, 24, 40–43]. Currently, no consensus on the lipid composition of exosomes derived from various cells has been reached; however, there does appear to be a number of trends conserved across the various cell types. Table 3 is a compilation of the lipid composition of exosomes reported in the literature. Evaluation of the trends reported for exosome lipid composition revealed a two-fold increase in the mole percentage of cholesterol in exosomes as compared to “parent” cells. Additionally, the mole percentage of phospholipids is altered considerably in exosomes. Exosomes also contain 2.5-fold more sphingomyelin (SM) and two-fold more anionic lipids (PS/PI) compared to their “parent” cells. Phosphatidylcholine (PC) decreases from 55% to 30% of the phospholipid content, and phosphatidylethanolamine (PE) remains relatively unchanged in exosomes with respect to the “parent” cell line.
Table 3.
Representative cholesterol and phospholipid composition of exosomes and cells. Mole percentages were compiled from the works of Wubbolts et al. 2003(37), Laulagnier et al. 2004(38) and 2005(39), Subra, Laulagnier et al. 2007(40), Trajkovic et al. 2008(41), and Parolini et al. 2009(18).
| Cholesterol | Phospholipids | |||
|---|---|---|---|---|
| Mole % Cell | 15 | 85 | ||
| Mole % Exosome | 30 | 70 | ||
| Change From Cell Membrane | 2× | .82× | ||
| Phospholipids | Sphingomyelin | PS/PI | PC | PE |
|
| ||||
| Mole % Cell | 10 | 10 | 55 | 25 |
| Mole % Exosomes | 25 | 20 | 30 | 25 |
| Change From Cell | ||||
| Membrane | 2.5× | 2× | .54× | 1× |
To assess if the lipid composition of exosomes affects their propensity for cellular association, fluorescently-labeled liposomes with specific cholesterol/phospholipid compositions mimicking either the “parent” cell lipid content or the exosome lipid composition (Table 3) were incubated with PC3 cells for 4 hrs at 37°C. Flow cytometry analysis indicated that liposomes formulated to mimic exosomes (30/70 cholesterol/phospholipid mole ratio) adhere/internalize significantly more than liposomes formulated to mimic the “parent” cell lipid composition (15/85 cholesterol/phospholipid mole ratio; Figure 5) to their cellular targets. In order to determine whether the increase in cholesterol content or the change in phospholipid ratios is responsible for the increased association, a third formulation having the phospholipid content of the “parent” cell but increased cholesterol content (30%) was evaluated. Results with the high cholesterol “parent” cell liposomes (30/70 cholesterol/phospholipid mole ratio) suggest that although the increased cholesterol content improved adherence/internalization, the phospholipids also contribute to the enhanced adherence/internalization (Figure 5A). PS receptors on phagocytic cells have been shown to bind PS on liposomes and exosomes [6, 7, 44, 45], suggesting a similar receptor-mediated interaction with PS maybe augmenting the association of liposomes and exosomes with recipient cells.
Figure 5.
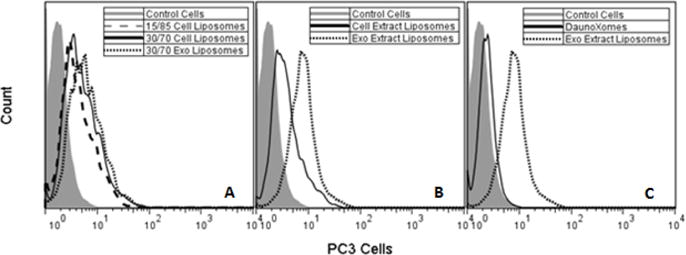
Effect of lipid content of liposome association. A) Liposomes with specific cholesterol/phospholipid content simulating the lipid content of either exosomes or cells were incubated with PC3 cells. Association of 15/85 Cell Liposomes is significantly lower than 30/70 Exosome Liposomes (p<.05). B) Liposomes prepared from the lipid extracts of PC3-derived exosomes and PC3 cells were incubated with PC3 cells. Association of the liposomes composed of exosome lipids are significantly greater (p<.05). C) Association of liposomes mimicking the commercial product DaunoXome (PC:Cholesterol 2:1 mole ratio) was compared to liposomes prepared from exosome lipid extracts. Association of liposomes prepared from exosome lipids was significantly higher (p<.05).
To further characterize the importance of the exosome lipid composition with respect to association with recipient cells, lipids from PC3 cells and PC3-derived exosomes were extracted and used to prepare fluorescently- labeled liposomes. After incubation with PC3 cells, data from flow cytometry showed that liposomes prepared from exosome lipid extracts associate to a significantly greater extent with PC3 cells compared to liposomes prepared from total cell lipid extracts (Figure 5B). Similarly, liposomes prepared from the lipid extracts of exosomes adhered/internalized significantly more than PC:Cholesterol liposomes (Figure 5C). These data further indicate that the lipid composition of exosomes contributes to their enhanced cell association.
In addition to the lipid composition, the protein content of exosomes is known to contain elevated levels of adhesion/targeting, transmembrane, and transport/fusion proteins relative to the “parent” cell [46, 47]. To investigate the importance of proteins with respect to exosome endocytosis, PC3-derived exosomes were incubated with proteinase K to cleave transmembrane proteins protruding from the surface of exosomes or proteins otherwise adhering to exosome surfaces. Incubation of PC3-derived exosomes subjected to proteinase K digestion revealed a significant decrease in their association with both PC3 and MCF-7 cells as compared to untreated PC3-derived exosomes (Figure 6). Results are similar to those seen by Escrevente et al [9]. No significant change in the size of exosomes, pre- and post-proteinase K digestion was observed (data not shown). Because we have no definitive way to measure the efficiency of proteinase K to cleave relevant proteins from the surface of exosomes, it is possible that our results are underestimating the extent to which proteins facilitate exosome association with recipient cells.
Figure 6.
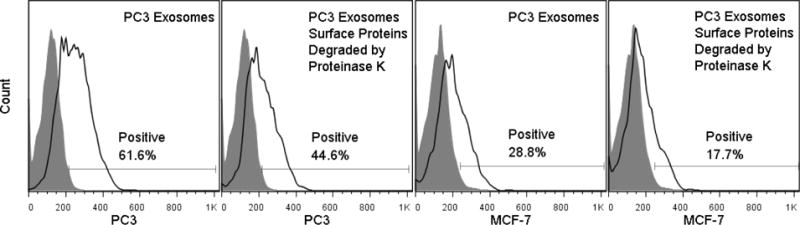
Effect of proteins on exosome association. PC3-derived exosomes were treated with proteinase K to remove surface protein moieties. PC3-derived exosomes (untreated or subjected to proteinase K) were incubated with PC3 and MCF-7 cells for 4 hrs at 37°C. Association was quantified by monitoring the percentage of cells that had increased fluorescence when compared to control cells (“Positive”). For both PC3 and MCF-7 cells, untreated PC3 exosomes were associated with their cellular targets to a significantly greater extent than PC3 exosomes subjected to proteinase K (p<.05).
pH cell culture conditions influence exosome cellular association
Tumor microenvironments are known to be acidified reaching a pH as low as 6.6 [19]; conditions known to affect numerous biological properties of cells [48–50]. This suggests that exosomes released from cells cultured below physiological pH may also have distinct properties, including their propensity for association with recipient cells. To elucidate how the pH of the microenvironment affects the properties of the “daughter” exosomes, MDA- MB-231 and PC3 cells were cultured at pH 7.4, 6.8, and 6.3 for 48 hours. Subsequently, exosomes were isolated from the media of both cell lines cultured at the three different pHs. All exosomes were characterized based on size and density to determine if differences in physical properties resulted from culture in acidic conditions. Size measurements of exosomes released from both MDA-MB-231 cells and PC3 cells cultured at all three pHs revealed no significant differences (Table 4). Additionally, exosomes derived from PC3 cells cultured at pH 7.4, 6.8, and 6.3 were placed on a sucrose gradient and centrifuged at 200k × g for 10 hrs. The majority of exosomes derived from each pH accumulated into the sucrose layer corresponding to a density of 1.18 g/cm3 (Figure 7). The density of exosomes has been previously characterized to be between 1.12 and 1.20 g/cm3 [21, 51]. The similar sizes and densities suggest that any differences in adherence/internalization do not result from altered physical properties.
Table 4.
Exosome mean sizes and standard deviations were calculated from three separate measurements on preparations harvested from cells grown at the indicated pH.
| pH 7.4 | pH 6.8 | pH 6.3 | ||||
|---|---|---|---|---|---|---|
| Size (nm) | S.D. | Size (nm) | S.D. | Size (nm) | S.D. | |
| MDA-MB-231 | 152 ± 8 | 78 ± 11 | 130 ± 8 | 69 ± 11 | 152 ± 8 | 78 ± 11 |
| PC3 | 156 ± 18 | 64 ± 10 | 151 ± 10 | 77 ± 19 | 138 ± 10 | 62 ± 20 |
Figure 7.
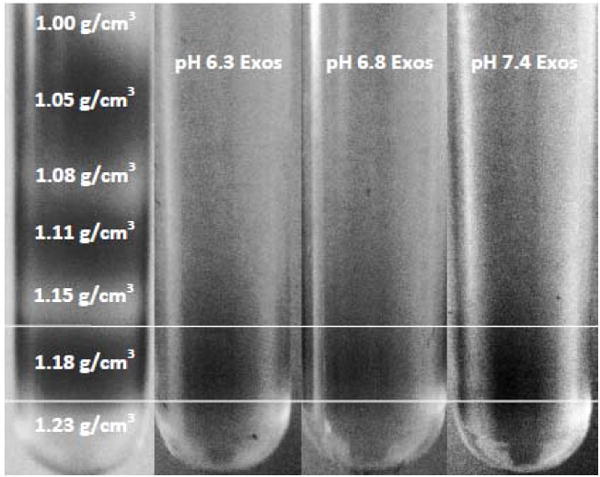
Density of isolated exosomes. Exosomes labeled with 0.01% DID in PBS loaded onto a continuous sucrose gradient and centrifuged at 200k × g for 10 hrs. The fluorescent dye, DID, allowed for visual identification of exosomes. In order to distinguish between each sucrose layer, a control sucrose gradient was run (far left) where every other sucrose layer was stained with fluorescein. Exosomes isolated from the supernatant of PC3 cells cultured at pH 6.3, 6.8 and 7.4 all partitioned into the sucrose layer corresponding to a density of 1.18 g/cm3.
Exosomes derived from MDA-MB-231 and PC3 cells cultured at pH 7.4, 6.8, and 6.3 were labeled with the fluorescent probe DID at 0.01% by weight. One microgram of labeled exosomes was added back to the media of their “parent” cell line growing at pH 7.4, 6.8, and 6.3 for 4 hrs at 37°C (Figure 8) to assess any differences in exosome adherence/internalization. Exosomes isolated from MDA-MB-231 and PC3 cells cultured at pH 7.4 were associated with their cellular targets to a slightly greater extent than exosomes isolated from cells cultured at pH 6.8. The most drastic difference was seen when comparing both pH 7.4- and pH 6.8- derived exosomes to exosomes isolated from cells cultured at pH 6.3. The pH 6.3-derived exosomes were associated significantly less than pH 7.4- and pH 6.8- derived exosomes by MDA-MB-231 cells cultured at all pHs. Taken collectively, the results identify major differences between the exosomes derived from cells cultured at pH 7.4 and pH 6.3 with respect to association with their parent cell line. In contrast, the pH in which cells are growing does not dramatically alter their association profile regardless of the conditions under which exosomes were harvested.
Figure 8.
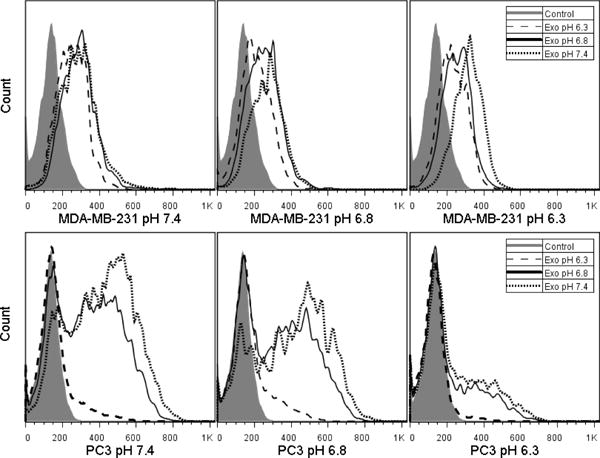
Effect of pH on exosome uptake. Top: Exosomes derived from MDA-MB-231 cells cultured at pH 7.4, 6.8 and 6.3 were incubated with MDA-MB-231 cells growing at pH 7.4, 6.8, and 6.3. Bottom: Exosomes derived from PC3 cells cultured at pH 7.4, 6.8 and 6.3 were incubated with PC3 cells growing at pH 7.4, 6.8, and 6.3. All exosomes were labeled with 0.01% DID. For each panel, association with pH 6.3 exosomes was significantly less than with pH 7.4 exosomes (p<.05).
Discussion
The scope of exosomes’ biological functions is still being unraveled, and cell-to-cell communication is frequently implicated. Previous studies have shown that exosomes are capable of delivering functional cargo in vitro [12–14] and in vivo [16, 17], leading others [47, 52, 53] to propose the use of exosomes as efficient drug delivery vehicles. Considering reports of exosome uptake by the parent cell line [9, 10, 34], and the potential to develop exosomes derived from cancer cells as tumor-homing delivery vehicles, the main goal of this study was to determine if exosomes are endowed with properties that allow for preferential association with their parent cell line or cancer cells in general. To this end, our in vitro studies have identified differences in the adherence/internalization of exosomes by cancer and immortalized cell lines. Differences in the non-specific association with the various cell lines may also affect the extent of exosome adherence/internalization, a factor that has not been considered in prior studies. Normalization of the data to account for non-specific (liposome) association suggests that exosomes are not selectively associated with a specific cell line(s). However, because exosomes associate with their cellular targets to a significantly greater extent than liposomes (at least 10-fold) in all cell lines, the adherence/internalization mechanisms may be completely different for these vehicles. While the greater association of exosomes could potentially be exploited for drug delivery, the differences in adherence/internalization between cancer and immortalized cell lines in culture may not be sufficient for cancer- derived exosomes to specifically target their “parent” cell line, suggesting that targeting moieties may need to be incorporated into exosomes to more efficiently target tumors[16].
The unique composition of exosomes [18, 24, 40–43] likely contributes to the enhanced association as compared to liposomes. Our results are consistent with this suggestion, and indicate that liposomes composed of exosomal lipids are more readily associated with recipient cells (Fig. 5). It is important to recognize that exosomes are enriched in cholesterol, SM, and anionic phospholipids, most notably PS, with a corresponding decrease in PC. Work with model membranes suggests that high cholesterol and SM concentrations endow exosomes with significant rigidity and stability [54–57]. Elevated cholesterol concentrations increase the cohesive properties of the membrane resulting in decreased permeability [54]. In addition, cholesterol and SM are known to form intermolecular hydrogen bonds [55, 56] in a 1/1 stoichiometric complex [54], forming lipid raft domains [57]. Recent studies from our laboratory have suggested that cholesterol domains increase gene delivery both in vitro [58, 59] and in vivo [60], implicating lipid domains as important modulators of intracellular trafficking. The stability and rigidity imparted by cholesterol and SM also likely enable exosomes to circulate in the blood for extended periods as cholesterol and SM together incorporated into model liposomes decrease the rate of clearance by the RES (reticuloendothelial system) [56, 61, 62]. The tight packing of the membrane also potentially inhibits the transfer of exosome lipids to high-density lipoproteins (HDL) [56]. While cholesterol and SM promote long in vivo circulation times of lipid vesicles, PS is present on the exterior of apoptotic bodies and foreign material which signals macrophages to engulf these particles [62]. Recently, PS receptors on phagocytic cells have been seen to bind PS in exosomes, which would likely compromise circulation times [6, 7]. It is interesting that work by Haque et al. reported that a neutral lipid molar ratio of 35/30/15/20 for PC/PE/SM/cholesterol, respectively, was optimal for liposome fusion and stability [63]. It is intriguing that this lipid composition closely resembles the neutral lipid molar ratios of exosomes, suggesting that nature has optimized the lipid components of exosomes for maximum in vivo stability and delivery.
We have demonstrated the importance of exosome lipids in facilitating adherence/internalization with recipient cells. However, the protein components of exosomes may also augment exosome adherence/internalization. Exosomes are enriched in transmembrane proteins, including adhesion proteins, tetraspanins, and the ICAM family of proteins. Adhesion proteins, most notably integrins, are abundant in exosomes and are important cell adhesion receptors [64, 65]. Integrins on the outer surface of B cell-derived exosomes have been shown to promote binding to infant foreskin fibroblasts [66]. Another important class of transmembrane proteins are tetraspanins, which are believed to be responsible for target cell selection and interactions [67]. Furthermore, ICAMs found on exosomes may facilitate capture at the cell surface [4, 5]. Our results demonstrating that cleavage of exosomal proteins severely diminished the extent of exosome association, suggesting that proteins also play a role in adherence/internalization of exosomes by the recipient cell (Fig. 6).
Low extracellular pH is a distinguishing phenotype of solid tumors [68]. Poor vascularization results in hypoxic regions and acidification of the tumor microenvironment [69, 70]. Hypoxic conditions lead to the release of proteins (predominately in exosomes) involved in angiogenesis, adhesion, and immune recruitment resulting in enhanced metastatic potential [69]. This is consistent with the notion that low pH selects for cancer cells with metastatic phenotypes [70, 71]. Furthermore, exosomes have been implicated to play a role in cancer metastasis [72, 73]. Thus, it is reasonable to suggest that cells cultured in acidic conditions might release exosomes with increased metastatic potential. Because we aimed to investigate the potential of exosomes as a drug delivery vehicle, our goal was to determine whether low pH-derived exosomes are associated to a greater extent than exosomes derived from cells cultured at physiological pH. Parolini et al. [18] concluded that exosomes derived from cells cultured at pH 6.0 had a greater fusion efficiency than exosomes harvested from cells cultured at physiological pH. In contrast, our results demonstrated that exosomes harvested from cells cultured at pH 7.4 associated more readily than those from cells cultured at pH 6.3. These data suggest that harvesting exosomes from cells grown under acidic conditions does not enhance their potential for drug delivery, at least in vitro. A possible explanation for the observed differences between our results and those of Parolini et al. [18] is the condition of the cells at low pH. Parolini et al. [18] reports that growth of melanoma cells is unaffected when cultured at low pH. In contrast MDA-MB-231 and PC3 cells, used in our experiments exhibit a reduced rate of doubling (data not shown) suggesting cells were not thriving in acidified conditions. These observations indicate that the behavior of our cell lines under acidified conditions differs from that used by Parolini et al., in that stressed cells may not release exosomes with a composition that enhances exosome association with recipient cells.
Conclusions
This work has highlighted the enhanced association of cancer cell-derived exosomes as compared to liposomes of a similar size by both cancer and immortalized cell lines. While we do not observe preferential association of exosomes by their parent cell line, cancer cells take up exosomes to a greater extent than immortalized cell lines. However, the enhanced adherence/internalization of exosomes by the cancer cell lines is not significant when differences in non-specific association are accounted for (presumably due to higher baseline levels of endocytosis/phagocytosis by cancer cells), but PC3 exosomes did exhibit significant preferential association to prostate cancer cells. In addition, we found that the unique lipid and protein composition of exosomes both contribute to facilitate adherence/internalization. While our results suggest that exosomes may still need to be targeted to enhance delivery specifically to cancer cells, the fact that exosomes are associated to a significantly greater extent by all cells suggests that their physical/chemical properties might be exploited to improve drug delivery. Lastly, we demonstrated that acidification of the microenvironment to which cells are exposed during exosome harvest and/or during adherence/internalization did not significantly increase association.
Supplementary Material
Acknowledgments
We would like to thank Amie Moody and Mike Miura at the University of Colorado Denver for their help in preparing and analyzing western blots. Additionally, we are grateful to John Carpenter for use of the NanoSight LM20 instrument. We would also like to thank Joe Gomez at the University of Colorado School of Pharmacy Mass Spectrometry Core for training and assistance. The authors would like to thank Brian Reid and the High Throughput and High Content Screening Core at the University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences for use of the Operetta™ High Content Imaging System instrument. Finally, we would like to thank Dot Dill and the electron microscopy core at the University of Colorado Denver. We also need to thank the University of Colorado Cancer Center Biostatistics and Bioinformatics Shared Resource Core for their help in evaluating statistical significances. This work was supported by grants 1RO1GM093287-01A1 and 1R01EB016378-01A1 from NIH, and an NIH training grant (5 T32 GM 8732).
References
- 1.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature Reviews Immunology. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 2.Camussi G, Deregibus MC, Bruno S, Grange C, Fonsato V, Tetta C. Exosome/microvesicle- mediated epigenetic reprogramming of cells. Am J Cancer Res. 2011;1:98–110. [PMC free article] [PubMed] [Google Scholar]
- 3.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends in Cell Biology. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nolte-’t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113:1977–1981. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 5.Segura E. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106:216–223. doi: 10.1182/blood-2005-01-0220. [DOI] [PubMed] [Google Scholar]
- 6.Keller S, König A-K, Marmé F, Runz S, Wolterink S, Koensgen D, Mustea A, Sehouli J, Altevogt P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer letters. 2009;278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 7.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 8.Morelli AE. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 9.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. Journal of Cellular Biochemistry. 2010;111:488–496. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 11.Feng D, Zhao W-L, Ye Y-Y, Bai X-C, Liu R-Q, Chang L-F, Zhou Q, Sui S-F. Cellular Internalization of Exosomes Occurs Through Phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 12.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 13.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Aliotta JM, Pereira M, Johnson KW, de Paz N, Dooner MS, Puente N, Ayala C, Brilliant K, Berz D, Lee D. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Experimental Hematology. 2010;38:233–245. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic Oncology. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 17.Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D, Zhang H-G. A Novel Nanoparticle Drug Delivery System: The Anti-inflammatory Activity of Curcumin Is Enhanced When Encapsulated in Exosomes. Molecular Therapy. 2010;18:1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. Microenvironmental pH Is a Key Factor for Exosome Traffic in Tumor Cells. Journal of Biological Chemistry. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Seminars in Radiation Oncology. 2004;14:198–206. doi: 10.1016/j.semradonc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Fevrier B. Cells release prions in association with exosomes. Proceedings of the National Academy of Sciences. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stillwell W. An introduction to biological membranes : from bilayers to rafts. Elsevier/Academic Press; London; Waltham, MA: 2013. [Google Scholar]
- 23.Kamoun WS, Chae SS, Lacorre DA, Tyrrell JA, Mitre M, Gillissen MA, Fukumura D, Jain RK, Munn LL. Simultaneous measurement of RBC velocity, flux, hematocrit and shear rate in vascular networks. Nat Methods. 2010;7:655–660. doi: 10.1038/nmeth.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 25.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 26.Caby MP. Exosomal-like vesicles are present in human blood plasma. International Immunology. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 27.Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Current Opinion in Cell Biology. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 29.Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, Multhoff G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell JP, Court J, Mason MD, Tabi Z, Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. Journal of Immunological Methods. 2008;335:98–105. doi: 10.1016/j.jim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Cho J-a, Lee Y-S, Kim S-H, Ko J-K, Kim C-W. MHC independent anti-tumor immune responses induced by Hsp70-enriched exosomes generate tumor regression in murine models. Cancer letters. 2009;275:256–265. doi: 10.1016/j.canlet.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 32.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, Patel P, Selby PJ, Banks RE. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 33.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 34.Safaei R. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug- resistant human ovarian carcinoma cells. Molecular Cancer Therapeutics. 2005;4:1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 35.Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of Cell Science. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 36.Montecalvo A, Shufesky WJ, Stolz DB, Sullivan MG, Wang Z, Divito SJ, Papworth GD, Watkins SC, Robbins PD, Larregina AT, Morelli AE. Exosomes as a short-range mechanism to spread alloantigen between dendritic cells during T cell allorecognition. Journal of immunology. 2008;180:3081–3090. doi: 10.4049/jimmunol.180.5.3081. [DOI] [PubMed] [Google Scholar]
- 37.Vallhov H, Gutzeit C, Johansson SM, Nagy N, Paul M, Li Q, Friend S, George TC, Klein E, Scheynius A, Gabrielsson S. Exosomes Containing Glycoprotein 350 Released by EBV-Transformed B Cells Selectively Target B Cells through CD21 and Block EBV Infection In Vitro. The Journal of Immunology. 2010;186:73–82. doi: 10.4049/jimmunol.1001145. [DOI] [PubMed] [Google Scholar]
- 38.Lässer C, Seyed Alikhani V, Ekström K, Eldh M, Torregrosa Paredes P, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. Journal of Translational Medicine. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C, Robbins PD. The Roles of Tumor-Derived Exosomes in Cancer Pathogenesis. Clinical and Developmental Immunology. 2011;2011:1–11. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wubbolts R. Proteomic and Biochemical Analyses of Human B Cell-derived Exosomes. Potential Implications for their Function and Multiveesicular Body Formation. Journal of Biological Chemistry. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 41.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laulagnier K, Vincent-Schneider H, Hamdi S, Subra C, Lankar D, Record M. Characterization of exosome subpopulations from RBL-2H3 cells using fluorescent lipids, Blood Cells. Molecules, and Diseases. 2005;35:116–121. doi: 10.1016/j.bcmd.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 44.Fadok VA, Bratton DL, Rose DM, Pearson A, Ezekewitz RA, Henson PM. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature. 2000;405:85–90. doi: 10.1038/35011084. [DOI] [PubMed] [Google Scholar]
- 45.Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. The Journal of biological chemistry. 2001;276:1071–1077. doi: 10.1074/jbc.M003649200. [DOI] [PubMed] [Google Scholar]
- 46.Simpson RJ, Jensen SS, Lim JWE. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 47.Schiffelers R, Kooijmans, Vader, van D, Van S. Exosome mimetics: a novel class of drug delivery systems. International Journal of Nanomedicine. 2012:1525. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eagle H. The effect of environmental pH on the growth of normal and malignant cells. J Cell Physiol. 1973;82:1–8. doi: 10.1002/jcp.1040820102. [DOI] [PubMed] [Google Scholar]
- 49.Griffiths L, Dachs GU, Bicknell R, Harris AL, Stratford IJ. The influence of oxygen tension and pH on the expression of platelet-derived endothelial cell growth factor/thymidine phosphorylase in human breast tumor cells grown in vitro and in vivo. Cancer research. 1997;57:570–572. [PubMed] [Google Scholar]
- 50.Martinez-Zaguilan R, Seftor EA, Seftor RE, Chu YW, Gillies RJ, Hendrix MJ. Acidic pH enhances the invasive behavior of human melanoma cells. Clin Exp Metastasis. 1996;14:176–186. doi: 10.1007/BF00121214. [DOI] [PubMed] [Google Scholar]
- 51.Segura E, Amigorena S, Théry C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses, Blood Cells. Molecules, and Diseases. 2005;35:89–93. doi: 10.1016/j.bcmd.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 52.van Dommelen SM, Vader P, Lakhal S, Kooijmans SAA, van Solinge WW, Wood MJA, Schiffelers RM. Microvesicles and exosomes: Opportunities for cell-derived membrane vesicles in drug delivery. Journal of Controlled Release. 2011 doi: 10.1016/j.jconrel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 53.Lakhal S, Wood MJA. Exosome nanotechnology: An emerging paradigm shift in drug delivery. BioEssays. 2011;33:737–741. doi: 10.1002/bies.201100076. [DOI] [PubMed] [Google Scholar]
- 54.Needham D, Nunn RS. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys J. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McIntosh TJ, Simon SA, Needham D, Huang CH. Structure and cohesive properties of sphingomyelin/cholesterol bilayers. Biochemistry. 1992;31:2012–2020. doi: 10.1021/bi00122a017. [DOI] [PubMed] [Google Scholar]
- 56.Ishida T, Harashima H, Kiwada H. Liposome clearance. Biosci Rep. 2002;22:197–224. doi: 10.1023/a:1020134521778. [DOI] [PubMed] [Google Scholar]
- 57.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 58.Xu L, Anchordoquy TJ. Effect of cholesterol nanodomains on the targeting of lipid-based gene delivery in cultured cells. Mol Pharm. 2010;7:1311–1317. doi: 10.1021/mp100097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu L, Wempe MF, Anchordoquy TJ. The effect of cholesterol domains on PEGylated liposomal gene delivery in vitro. Ther Deliv. 2011;2:451–460. doi: 10.4155/tde.11.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, Betker J, Yin H, Anchordoquy TJ. Ligands located within a cholesterol domain enhance gene delivery to the target tissue. J Control Release. 2012;160:57–63. doi: 10.1016/j.jconrel.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen TM, Chonn A. Large unilamellar liposomes with low uptake into the reticuloendothelial system. FEBS Lett. 1987;223:42–46. doi: 10.1016/0014-5793(87)80506-9. [DOI] [PubMed] [Google Scholar]
- 62.Allen TM, Austin GA, Chonn A, Lin L, Lee KC. Uptake of liposomes by cultured mouse bone marrow macrophages: influence of liposome composition and size. Biochimica et biophysica acta. 1991;1061:56–64. doi: 10.1016/0005-2736(91)90268-d. [DOI] [PubMed] [Google Scholar]
- 63.Haque ME, McIntosh TJ, Lentz BR. Influence of Lipid Composition on Physical Properties and PEG-Mediated Fusion of Curved and Uncurved Model Membrane Vesicles: “Nature’s Own” Fusogenic Lipid Bilayer†. Biochemistry. 2001;40:4340–4348. doi: 10.1021/bi002030k. [DOI] [PubMed] [Google Scholar]
- 64.Hemler ME. Targeting of tetraspanin proteins — potential benefits and strategies. Nature Reviews Drug Discovery. 2008;7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campbell ID, Humphries MJ. Integrin Structure, Activation, and Interactions. Cold Spring Harbor Perspectives in Biology. 2011;3:a004994–a004994. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clayton A. Adhesion and signaling by B cell-derived exosomes: the role of integrins. The FASEB Journal. 2004 doi: 10.1096/fj.03-1094fje. [DOI] [PubMed] [Google Scholar]
- 67.Rana S, Zöller M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochemical Society transactions. 2011;39:559–562. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 68.Parks SK, Chiche J, Pouyssegur J. pH control mechanisms of tumor survival and growth. Journal of Cellular Physiology. 2011;226:299–308. doi: 10.1002/jcp.22400. [DOI] [PubMed] [Google Scholar]
- 69.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moellering RE, Black KC, Krishnamurty C, Baggett BK, Stafford P, Rain M, Gatenby RA, Gillies RJ. Acid treatment of melanoma cells selects for invasive phenotypes. Clinical & Experimental Metastasis. 2008;25:411–425. doi: 10.1007/s10585-008-9145-7. [DOI] [PubMed] [Google Scholar]
- 71.Rofstad EK. Acidic Extracellular pH Promotes Experimental Metastasis of Human Melanoma Cells in Athymic Nude Mice. Cancer research. 2006;66:6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- 72.Hao S, Ye Z, Li F, Meng Q, Qureshi M, Yang J, Xiang J. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp Oncol. 2006;28:126–131. [PubMed] [Google Scholar]
- 73.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, Ratajczak MZ. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. International Journal of Cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


