Abstract
RNA polymerase (RNAP) in cells must transcribe supercoiled DNA whose torsional state is constantly changing, but how RNAP deals with DNA supercoiling remains elusive. We report direct measurements of individual E. coli RNAPs as they transcribed supercoiled DNA. We found that a resisting torque slowed RNAP and increased its pause frequency and duration. RNAP was able to generate 11 ± 4 pN•nm (mean ± SD) of torque before stalling, sufficient to melt DNA of arbitrary sequence and establishing RNAP as a more potent torsional motor than previously known. A stalled RNAP was able to resume transcription upon torque relaxation and transcribing RNAP was resilient to transient torque fluctuations. These results provide a quantitative framework for understanding how dynamic modification of DNA supercoiling regulates transcription.
DNA supercoiling is a regulator of gene expression (1–5). RNAP must transcribe supercoiled DNA and transcription elongation in turn generates DNA supercoiling. As RNAP moves along the helical groove of DNA, it generates (+) DNA supercoiling ahead and (−) DNA supercoiling behind (the “twin supercoiled domain model”) (1, 3–6). DNA supercoiling is broadly present during transcription (3–5). Active transcription can accumulate dynamic DNA supercoiling on DNA templates that are not bound by topological constraints (3), and in the presence of a normal complement of topoisomerases in vivo (4). However, little is known about some basic properties of the interplay between transcription and DNA supercoiling. We have developed an assay to directly monitor RNAP translocation in real time as it worked under a defined torque. An RNAP was torsionally anchored to the surface of a coverslip, and either the downstream or upstream end of the DNA template was torsionally anchored to the bottom of a nanofabricated quartz cylinder held in an angular optical trap (AOT) (Fig. 1A and fig. S1) (7–11). An AOT allows simultaneous control and measurement of rotation, torque, displacement, and force of the trapped cylinder (8–11). Analysis of these measurements allowed for the determination of the RNAP position on the DNA template as it transcribed under torque (11).
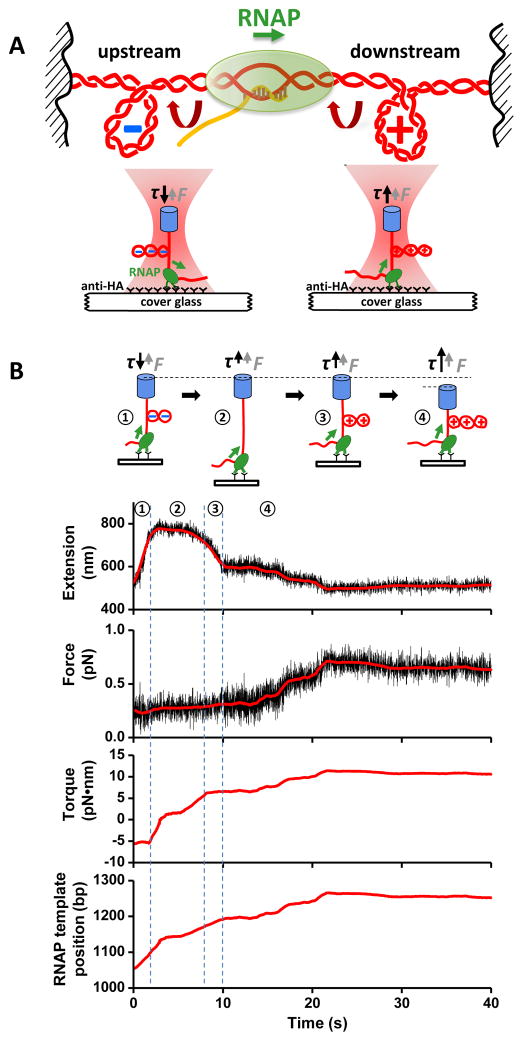
Figure 1. Stall torque experiments.
(A) (Top) A cartoon depicting the “twin-supercoiled domain” model (1). (Bottom) Experimental configuration that mimics the “twin-supercoiled domain” model for transcription against (−) supercoiling upstream or (+) supercoiling downstream.
(B) A representative set of data for downstream stall torque measurements. After the introduction of NTPs, the force on the DNA was clamped at a low value while DNA was mechanically unwound to form a (−) plectoneme. Subsequent translocation of RNAP neutralized the (−) plectoneme (➀ and ➁) and resulted in (+) plectoneme formation (➂). The force clamp was then turned off (➃). RNAP translocation increased the force (directly measured) and the corresponding torque (derived) (11) until reaching a stall (< 1 bp/s for 20–50 s). Data were filtered: extension to 200 Hz (black) and 1 Hz (red), and force to 40 Hz (black) and 1 Hz (red). The RNAP template position is defined as the distance of RNAP from the transcription start site (in bp).
We investigated how RNAP stalled as it worked against (+) supercoiling downstream or (−) supercoiling upstream. Before the cylinder was trapped, RNAP translocation could be directly visualized by rotation of a tethered cylinder (movie S1). Once trapped, the cylinder’s orientation was controlled by the AOT. RNAP translocation rotated the DNA, forming a (+) plectoneme in downstream stalling experiments (Fig. 1B, fig. S5A) or a (−) plectoneme in upstream stalling experiments (fig. S4, fig. S5B). Resisting torque build-up eventually led to transcription stalling. Our method was inspired by previous magnetic tweezers-based studies to monitor transcription and amplify its detection (12–14), but is distinct from those studies in its real-time transcription elongation detection and/or flexible torque control and readout.
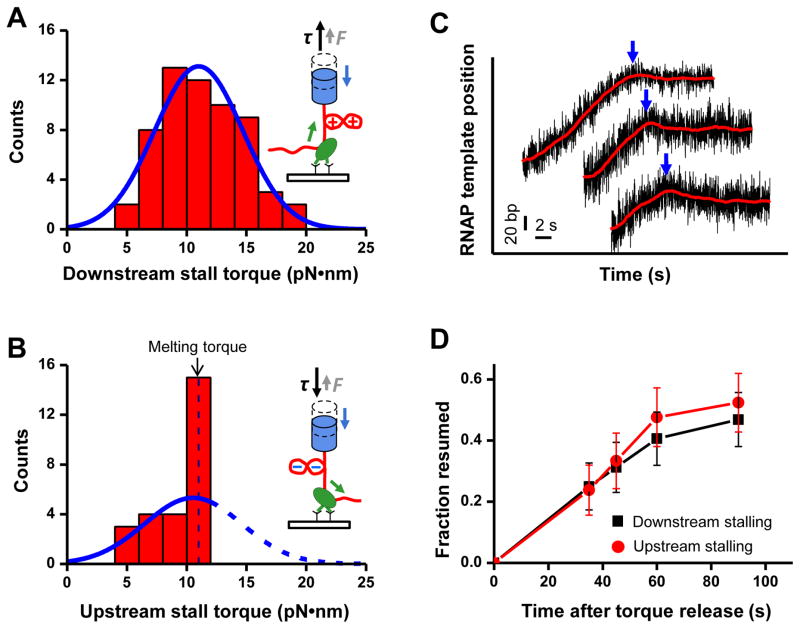
The measured downstream stall torque distribution is well fit by a Gaussian function, yielding a mean torque of 11.0 ± 3.7 pN•nm (mean ± SD) with the largest measured value being ~18 pN•nm (Fig. 2A, fig. S6A). This mean torque is sufficient to create (+) plectonemic DNA under the low forces used in our experiments. In contrast, the upstream stall torque distribution shows an asymmetry (Fig. 2B, fig. S6B). Unlike (+) supercoiled DNA, which can sustain a much higher torque before structural changes, (−) supercoiled DNA undergoes a transition at 10.5 pN•nm consistent with melting (fig. S3) (11). The upstream stall torque distribution shows a singular peak immediately before a sharp cutoff near the DNA melting torque, and approximately 60% of RNAPs were stalled between 10–12 pN•nm. These data indicate that RNAP is able to generate an upstream torque sufficient to alter DNA structure. The upstream data were fit with a Gaussian function, yielding a Gaussian centered at 10.6 ± 4.1 pN•nm, comparable to the downstream stall torque (Fig. 2B). The spreads in the measured stall torque distributions are attributed to DNA sequence variations and single molecule stochasticity, according to a thermal-ratchet kinetic model for transcription elongation that we previously developed (15–17).
Figure 2. Transcription stalling and resumption.
(A) The distribution of the measured downstream stall torques. The smooth blue curve is a fit with a Gaussian function, yielding a mean of 11.0 ± 3.7 pN•nm (mean ± SD).
(B) The distribution of measured upstream stall torques. The smooth curve is a fit with a Gaussian function assuming that the peaked fraction generated torques of at least 10 pN•nm, yielding a mean of 10.6 ± 4.1 pN•nm (mean ± SD).
(C) Example traces showing RNAP reverse translocation upon stalling. Both axes are shifted for clarity. For each trace, the arrow indicates the entry into a stall.
(D) Fraction of RNAPs that resumed transcription after torque release versus time. After stalling, torque on RNAP was relaxed and transcription was detected by an experiment similar to that shown in ➀ of Figure 1B.
Thus RNAP is fully capable of generating torque sufficient to melt DNA of arbitrary sequence (11), not just AT-rich sequences that are prone to melting (3, 4, 11). The strong (−) supercoiling generated by RNAP may facilitate initiation of transcription from adjacent promoters (18), binding of regulatory proteins (3, 4), and initiation of replication (19).
We found that in some traces RNAP reverse translocated upon stalling (Fig. 2C). This reverse motion suggests that torque may induce stalling via backtracking, during which RNAP translocates back along the template DNA and displaces the 3′ transcript from the active site preventing RNA synthesis (20–22).
In vivo, torsional stress accumulated by RNAP may be relaxed by either the arrival of a topoisomerase at the DNA template or by DNA rotation. We found that stalled RNAPs gradually resumed transcription following torque release (Fig. 2D). At 90 s after torque release, ~ 50% of stalled RNAPs had resumed transcription. Thus in vivo torque relaxation should allow a large fraction of stalled RNAPs to resume transcription, preventing them from becoming obstacles or inducing DNA damage that disrupts genome stability (23).
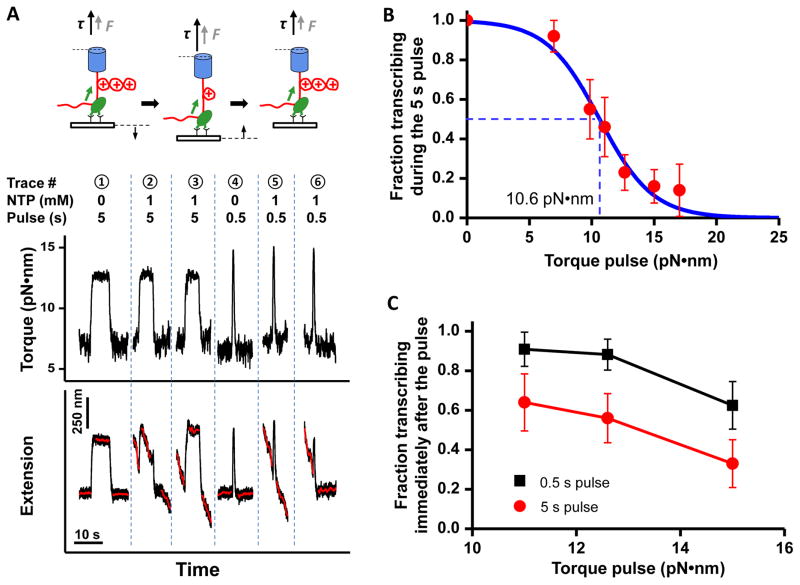
In vivo, torsional stress in local DNA segments may be present transiently due to actions of motor proteins and dynamic reconfiguration of topological domains. However, it is not known how these sudden changes in torsional stress might influence a transcribing RNAP. We thus carried out “transient torque pulse” experiments to determine how RNAP responded to a brief exposure of a resisting torque on a time scale comparable to those of topoisomerases (24–26) (0.5 s or 5 s) (Fig. 3A). We found that the fraction of active RNAPs during the 5 s pulse decreased as the torque was jumped to an increasingly higher value (Fig. 3B). The characteristic cutoff torque was 10.6 ± 4.0 pN•nm, a value similar to the mean stall torque. A significantly larger fraction of RNAPs was able to transcribe immediately (within 5 s) after the 0.5 s pulse than after the 5 s pulse (Fig. 3C), indicating that a 0.5 s torque pulse does not give sufficient time for RNAP to backtrack substantially. Thus RNAP can effectively resist transient torque fluctuations (< 0.5 s), but is unable to withstand prolonged exposure to a large torque without stalling or arresting.
Figure 3. Transcription response to a transient torque pulse.
(A) (Top) A cartoon illustrating steps of the “torque pulse” experiments and (Bottom) representative traces of data. RNAP initially transcribed under a low downstream torque of approximately +7 pN•nm, and then was subjected to a higher torque pulse for either 5 s or 0.5 s before restoration of the initial low torque. Traces 1 and 4 are controls. The extension and time axes are shifted for clarity.
(B) The probability of maintaining active transcription during the 5 s torque pulse. The blue solid line is a fit to a Boltzmann function): f = 1/[1+e(τ−τc)/τ0], where τc is the characteristic cutoff torque.
(C) The probability of resuming transcription immediately (within 5 s) after the torque pulse.
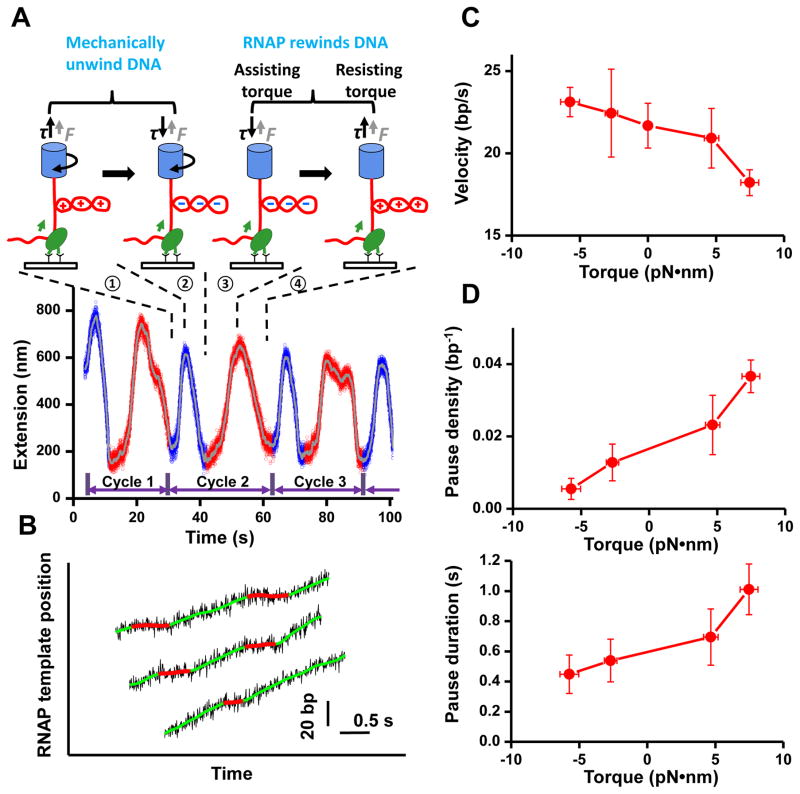
We investigated the torque-velocity relation which characterizes how the transcription speed is regulated by torque (Fig. 4A). To maintain a constant torque, we monitored transcription in the presence of a DNA plectoneme under a small and constant force. The measured transcription traces showed that continuous elongation was interrupted by frequent pausing (Fig. 4B, fig. S7). Because of the sensitivity of the assay, it was possible to resolve pauses as short as 0.2 s. By analyzing the velocity between pauses, we obtained the torque-velocity relationship of RNAP. Figure 4C shows how the transcription rate increased with an assisting torque and decreased with a resisting torque. In addition, both pause density and duration decreased with an assisting torque and increased with a resisting torque (Fig. 4D).
Figure 4. Determination of transcription torque-velocity relationship.
(A) A representative set of data for transcription measurement under a constant torque. Transcribing RNAP, under a small and constant tension of 0.15 pN, was subjected to multiple cycles of resisting and assisting torque. For each cycle, the downstream DNA was mechanically unwound to remove any (+) plectoneme (➀) and create a (−) plectoneme (➁). Subsequent RNAP transcription was assisted by the (−) DNA supercoiling (➂), until the generation of (+) supercoiling, which hindered transcription (➃). In the presence of a plectoneme, the torque on the DNA was constant for a given force (9) (fig. S3) and RNAP velocity was derived from the slope of the extension versus time curve (11). Also, we define a resisting torque to be (+) and an assisting torque to be (−). Data were filtered to 200 Hz (blue and red) and 1 Hz (grey).
(B) Representative transcription traces under a torque of +7.5 pN•nm. Continuous transcription (green smoothed data) was interrupted by pauses (red smoothed data), each of which is indicated by a red line.
(C) Transcription torque-velocity relationship. Transcription velocity was obtained by weighting each transcript position equally and the resulting velocity reflected primarily transcription rates between pauses (11, 29).
(D) Pause density (top) and duration (bottom) as a function of torque. A pause is defined as having a duration of ≥0.2 s at a given nucleotide position (11). Zero-torque data (fig. S8) had lower sensitivity to transcription due to lack of plectoneme in DNA, precluding detection of pauses of 0.2–2 s in duration, and were thus excluded from pause analysis.
We show that RNAP can generate torque, which in turn regulates transcription rate and pausing, and that excessive torque accumulation leads to transcription stalling and DNA structural alteration. A transcription-generated supercoiling wave can propagate through DNA to provide action at a distance, not only to alter DNA structure (3, 4) but also to potentially alter or dissociate bound proteins (3, 4, 27). Torsion generated by eukaryotic RNAP may alter chromatin fiber and evict histones (4, 27, 28), and torsional relaxation by chromatin may in turn facilitate transcription (28).
Supplementary Material
Acknowledgments
We thank members of the Wang lab for critical reading of the manuscript. We especially thank R.M. Fulbright for purification of the RNAP, and S. Forth, Y. Yang, M.Y. Sheinin, J.T. Inman, and R.A. Forties for assistance with single molecule assays, data acquisition, data analysis, and figure preparation. We wish to acknowledge support from an NIH grant (GM059849 to M.D.W) and an NSF grant (MCB-0820293 to M.D.W).
Footnotes
References and Notes
- 1.Liu LF, Wang JC. Proc Natl Acad Sci USA. 1987;84:7024. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travers A, Muskhelishvili G. Nat Rev Micro. 2005;3:157. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 3.Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. Nat Struct Mol Biol. 2004;11:1092. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 4.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. Nat Struct Mol Biol. 2008;15:146. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto K, Hirose S. J Cell Sci. 2004;117:3797. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 6.Wu HY, Shyy S, Wang JC, Liu LF. Cell. 1988;53:433. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 7.La Porta A, Wang MD. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 8.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nat Meth. 2007;4:223. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 9.Forth S, et al. Phys Rev Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheinin MY, Forth S, Marko JF, Wang MD. Phys Rev Lett. 2011;107:108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supplementary material on Science Online.
- 12.Harada Y, et al. Nature. 2001;409:113. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 13.Revyakin A, Ebright RH, Strick TR. Proc Natl Acad Sci USA. 2004;101:4776. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revyakin A, Liu C, Ebright RH, Strick TR. Science. 2006;314:1139. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai L, Shundrovsky A, Wang MD. J Mol Biol. 2004;344:335. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 16.Bai L, Fulbright RM, Wang MD. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 17.Bai L, Wang MD. J Stat Mech. 2010;2010:P12007. doi: 10.1088/1742-5468/2010/12/P12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilley DMJ, Higgins CF. Mol Microbiol. 1991;5:779. doi: 10.1111/j.1365-2958.1991.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 19.Kowalski D, Eddy MJ. EMBO J. 1989;8:4335. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komissarova N, Kashlev M. Proc Natl Acad Sci USA. 1997;94:1755. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Nature. 2003;426:684. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galburt EA, et al. Nature. 2007;446:820. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 23.Dutta D, Shatalin K, Epshtein V, Gottesman Max E, Nudler E. Cell. 2011;146:533. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strick TR, Croquette V, Bensimon D. Nature. 2000;404:901. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 25.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Nature. 2005;434:671. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 26.Gore J, et al. Nature. 2006;439:100. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levchenko V, Jackson B, Jackson V. Biochemistry. 2005;44:5357. doi: 10.1021/bi047786o. [DOI] [PubMed] [Google Scholar]
- 28.Lavelle C. Biochimie. 2007;89:516. doi: 10.1016/j.biochi.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Adelman K, et al. Proc Natl Acad Sci USA. 2002;99:13538. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.