Abstract
Regulatory T cells promote cancer by suppressing anti-tumor immune responses. We found that anti-LAP antibody which targets the latency-associated peptide (LAP)/TGF-β complex on Tregs and other cells enhances anti-tumor immune responses and reduces tumor growth in models of melanoma, colorectal carcinoma and glioblastoma. Anti-LAP decreases LAP+ Tregs, tolerogenic dendritic cells and TGF-β secretion, and is associated with CD8+ T cell activation. Anti-LAP increases infiltration of tumors by cytotoxic CD8+ T cells and reduces CD103+ CD8 T cells in dLNs and spleen. We identified a role for CD103+ CD8 T cells in cancer. Tumor-associated CD103+ CD8 T cells have a tolerogenic phenotype with increased expression of CTLA-4 and IL-10 and decreased expression of IFN-γ, TNF-α, and granzymes. Adoptive transfer of CD103+ CD8 T cells promotes tumor growth whereas CD103 blockade limits tumorigenesis. Thus, anti-LAP targets multiple immunoregulatory pathways and represents a potential approach for cancer immunotherapy.
INTRODUCTION
Classic CD4+ Tregs are identified by the intracellular marker Foxp3 (1, 2). However, targeting classic Tregs for treatment in humans is hampered by the expression of Foxp3 and surface Treg markers on activated cells. Other types of Tregs have also been described including Tr1 and Th3 cells (3, 4) although they are not as well understood or characterized as classic Foxp3+ Tregs. We have been interested in Tregs that express TGF-β on their surface complexed to latency-associated peptide (LAP), which identifies regulatory CD4+ T cells that have been described in the models of oral tolerance and autoimmunity (3, 5, 6) and are increased in cancer. In colorectal cancer (CRC), LAP+ CD4 tumor-infiltrating lymphocytes (TILs) are 50-fold more suppressive than FOXP3+ CD4 T cells. In head and neck cancer, LAP is up-regulated on FOXP3+ CD4 T lymphocytes (7). TGF-β is secreted in the tumor microenvironment by different cells and has an important role in dampening the anti-tumor immune response (8, 9). In cancer, TGF-β controls cell growth, induces angiogenesis, tumor cell invasion and promotes immune suppression (10). LAP and TGF-β are translated as one precursor polypeptide from the Tgfb1 gene and undergoes cleavage by furin, which separates the N-terminal LAP protein portion from TGF-β. TGF-β is then reassembled with LAP to form a small latent complex (SLC) that retains TGF-β in its inactive form on the cell surface. The SLC is then deposited on the cell surface bound to the LAP membrane receptor GARP or embedded in the extracellular matrix (11–13). We utilized anti-LAP antibodies that we developed (14) to investigate LAP targeting as cancer immunotherapy.
RESULTS
Anti-LAP monoclonal antibody decreases tumor growth in models of melanoma, glioblastoma and colorectal carcinoma
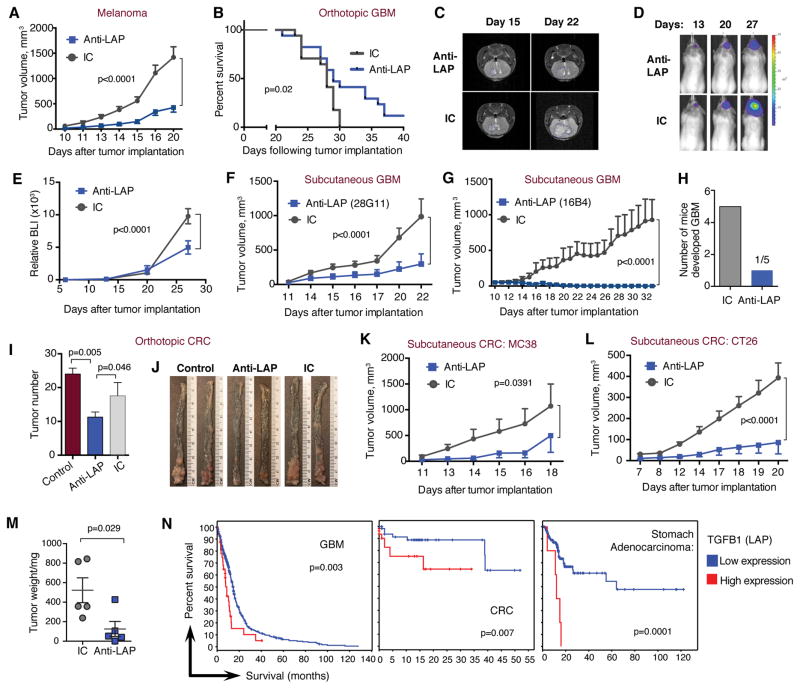
We utilized a mouse monoclonal anti-LAP antibody (14) in orthotopic and flank syngeneic tumor models. Anti-LAP reduced tumor growth in B16 melanoma (Fig. 1A) and in both intracranial (orthotopic) (Fig. 1B–E and fig. S1A) and sub-cutaneous (Fig. 1F and G) glioblastoma (GL261) models. Anti-LAP also affected established B16 tumors (fig. S1B). In glioblastoma, an early therapeutic effect was observed as only rare tumor cells were observed at two weeks whereas all control mice developed solid tumors by this time (Fig. 1H and fig. S1C). In CRC, anti-LAP reduced tumor number in the azoxymethane (AOM)/Dextran Sulfate Sodium Salt (DSS) orthotopic model of spontaneously induced CRC, (Fig. 1I, J and fig. S1D and E) and in two sub-cutaneous CRC models, MC38 and CT26 (Fig. 1K–M). We employed The Cancer Genome Atlas (TCGA) dataset to study the relationship between the expression of the LAP/TGF-β encoding gene, TGFB1, and its associated genes (THBS1/TSP-1, LRRC32/GARP, HSPA5/GRP78, and LTBP1/2) with cancer patient survival. We found that the relatively high expression of these genes based on z score was associated with poorer patient survival (Fig. 1N and fig. S2).
Figure 1. Therapeutic effect of anti-LAP in cancer models.
(A) B16 melanoma tumor volume over time in WT mice treated with either anti-LAP (TW7-28G11 clone) or IC antibodies (n=6 (anti-LAP) and n=7 (IC), at the last time point). Data are representative of at least three independent experiments. (B) Survival curves of WT mice with intracranial GL261 GBM treated with anti-LAP clone TW7-16B4 (n=17; data are combined from two independent experiments; log-rank test). Intracranial tumor growth measured by MRI (representative images) on days 15 and 22 (C) and by bioluminescence (BLI; representative images) (D) and measured as relative BLI on days 13, 20 and 27 (E; n=6 (anti-LAP) and n=7 (IC), at the last time point). (F, G) Tumor volume of subcutaneous GL261 GBM implanted in WT mice and treated with anti-LAP clone TW7-28G11 (F, n=5) or anti-LAP clone TW7-16B4 (G, n=8). (H) Early therapeutic effect of anti-LAP on intracranial GL261 GBM. Number of mice developed solid tumors following anti-LAP treatment. (I, J) Orthotopic AOM/DSS induced CRC in WT mice treated with anti-LAP; tumor number (I) and representative images of CRC colons (J) are shown (n=9 (anti-LAP and IC) and n=5 (control); one-way ANOVA). Data are representative of two independent experiments. (K–L) Tumor volume of subcutaneous MC38 (K, n=5) and CT26 (L, n=5) CRC models in WT mice treated with anti-LAP. Data are representative of two independent experiments. (M) CT26 tumor weight measured at day 20 (n=5; two-tailed t-test). (N) Percent survival in patients with relatively high or low mRNA expression of TGFB1 (LAP) based on z score. The “high” expression group was determined based on pre-computed z score value from gene expression (greater than 0.005-0.5) while the “low” group consisted of the remaining patients in the dataset. Graphs and p values were downloaded from The Tumor Cancer Genome Atlas (TCGA) dataset via cBioPortal (52, 53). Error bars, mean±s.e.m. Two-way ANOVA (A, E–G, K, and L) was used for p value calculations. P values for the last time points are shown.
Anti-LAP decreases LAP+ CD4 T cells and blocks the release of TGF-β
Potential mechanisms of anti-LAP effects include reduction of LAP+ T cells and/or blocking TGF-β release from the small latent complex (fig. S3A). Increased numbers of splenic LAP+ T cells in animals with B16 melanoma were reduced following anti-LAP treatment (Fig. 2A, fig. S3B, and S3C) as were the frequency of LAP+ T cells in the tumor and draining lymph nodes (dLNs) (Fig. 2A). Different non-competing antibodies were used for anti-LAP treatment (clone TW7-28G11) and for measuring LAP+ cells (clone TW7-16B4) (fig. S3D). To determine whether anti-LAP blocked release of membrane bound TGF-β we utilized P3U1 cells that over-express the Tgfb1 gene and secrete TGF-β when LAP is activated. Both 16B4 and 28G11 anti-LAP clones reduced the release of TGF-β (Fig. 2B). Thus, anti-LAP decreases LAP+ cells and blocks TGF-β release from the cell.
Figure 2. Modulation of LAP+ CD4 T cells following anti-LAP treatment.
(A) Frequency of LAP+ T cells in naïve and anti-LAP or IC treated B16 melanoma-bearing mice. Mice were treated with anti-LAP clone TW7-28G11 and LAP+ T cells measured with a non-competing anti-LAP clone (TW7-16B4) by flow cytometry in spleen (n=8), dLN and tumor (n=5). Data are from at least three independent experiments. (B) Active TGF-β release from P3U1 cells expressing mouse LAP/TGF-β, treated with anti-LAP clones TW7-16B4 and TW7-28G11 or IC, measured by ELISA (n=3); for more details see Materials and Methods. Representative of three independent experiments. (C) Expression of indicated immune markers in LAP+ vs. LAP− T cells in spleen, dLN and tumor of B16 melanoma-bearing mice by flow cytometry (n=5); representative of two independent experiments. (D) qRT-PCR analysis of Lag3 and Tigit in LAP+ and LAP− T cells isolated from naïve or B16 tumor-bearing mice (n=3). (E) Heat map of differentially expressed genes in LAP+ and LAP− T cells isolated from naïve or B16 tumor-bearing mice ordered by Euclidian distance based hierarchical clustering (n=3). (F, G) In vitro suppression of naïve CD4+ T cell proliferation by LAP+ T cells sorted from spleens and dLNs of melanoma-bearing mice. Representative histograms of proliferation of responder CD4+ T cells (F) and percent suppression (G) are shown. Foxp3+ cells served as controls. Indicated samples were treated with TGF-β receptor inhibitor (TGFBRI), DMSO control, anti-TGF-β or IC antibody (n=4–11; combined data from four experiments, normalized to the level of suppression of LAP+ cells in spleen). Error bars, mean±s.e.m. One-way ANOVA (A, left panel; B and G) and two-tailed t-test (A, middle and right panels; C and D) were used for p value calculations.
LAP+ CD4 T cells from tumor-bearing mice have suppressive properties
We measured markers associated with Tregs (Foxp3), exhausted T cells (Lag3, PD1, PD-L1, Tim3) and CD103 in TILs from B16 melanoma mice on both LAP+ and LAP− T cells. Expression of these markers was increased on LAP+ vs. LAP− T cells (Fig. 2C and fig. S3E). A similar tolerogenic phenotype was observed for LAP+ Tregs from dLNs and spleens of tumor-bearing mice (Fig. 2C, fig. S3F and S3G). We also measured gene expression and found that cancer-associated genes, including Lag3, Tigit, and Vcam were expressed at higher levels in LAP+ vs. LAP− T cells (Fig. 2D and fig. S4A). Interestingly, Irf4 that has been shown to promote effector function of Tregs (15) was also overexpressed in LAP+ T cells (fig. S4A). Using the Nanostring Pan Cancer Immunology code set, we found 480 genes differentially expressed between B16 melanoma and control mice (Fig. 2E). Among them, genes associated with effector Treg function, such as Aire, Gata3, Irf4, Foxp3, Stat3, Tgfb1, Tgfb2, Entpd1 (CD39), Itgae (CD103), Il10, Gzma, Gzmb and cancer-associated T cell markers, such as Havcr2 (Tim3), Ctla4, Tigit, and Lag3 were expressed at a higher level in LAP+ T cells in naïve mice. These genes were further upregulated on LAP+ T cells in tumor-bearing mice (Fig. 2E). On the other hand, genes associated with T cell activation, including Il6ra, Pin1 and Mapk14 were downregulated in LAP+ T cells in tumor-bearing mice (Fig. 2E).
Since Foxp3 is a marker of Tregs in mice, we analyzed the relationship between LAP and Foxp3 in B16 tumor-bearing mice. We performed principle component analysis (PCA) on the Nanostring-based gene expression data in LAP+/−Foxp3+/− CD4 T cell subsets. We found that PC1 was associated with the variance between Foxp3+/− generated datasets, whereas PC2 was associated with the variance between LAP+/− generated datasets. Thus, in addition to the differences between Foxp3+/−, we found LAP+ T cell subsets clustered differently from LAP− T cells in both Treg and non-Treg CD4 populations (fig. S4B). We analyzed the distribution of LAP+/−Foxp3+/− T cell subsets in vivo and found that most LAP+ T cells were Foxp3+ both in the periphery and in the tumor (fig. S4C). LAP+ T cells from both dLNs and spleens of tumor-bearing mice reduced the proliferation of responder CD4+ T cells in vitro (Fig. 2F and 2G). Blocking TGF-β signaling with either TGF-β receptor inhibitor or anti-TGF-β mAb (Fig. 2G) reduced the suppression, indicating that LAP+ T cells suppressed in vitro through a TGF-β-dependent mechanism.
Anti-LAP treatment modulates dendritic cell subsets in the spleen
Antigen-presenting cells play a key role in anti-tumor immunity. Since anti-LAP blocks the secretion of TGF-β which is known to interfere with the maturation of splenic antigen-presenting cells (16), we investigated anti-LAP on dendritic cells (DCs) in the spleen in the B16 melanoma model. We measured CD11c-Hi/CD11b-Int (subsequently referred to as CD11c-Hi) and CD11c-Int/CD11b-Hi (CD11c-Int) cell subsets. CD11c-Hi cells were increased following anti-LAP whereas CD11c-Int cells were reduced (Fig. 3A and 3B). We also measured splenic DCs in the GBM model and observed a similar effect (fig. S5A and S5B). In GBM, tumors grow slowly and the effect of anti-LAP on splenic CD11c/CD11b DCs in GBM was observed with long-term anti-LAP treatment (>3 weeks). CD11c-Int cells express higher levels of LAP vs. CD11c-Hi cells (Fig. 3C and fig. S5C), which suggests that the CD11c-Int cells are more tolerogenic. Because the function of these two DC subsets is not well defined, we characterized their inflammatory properties. We found that both MHCII and CD86 were expressed at higher levels on CD11c-Hi vs. CD11c-Int cells (Fig. 3D and fig. S5D). We then sorted these two DC subsets, stimulated them with LPS or anti-CD40 and measured cytokine expression. We found that Il10 was expressed at higher levels and Il12 at lower levels in the CD11c-Int subset (Fig. 3E), indicating a more tolerogenic phenotype as compared to CD11c-Hi cells. To determine whether these DC subsets affected CD8 T cells, we co-cultured them with labeled CD8+ naïve T cells and measured cytokine secretion and growth. We found increased expression of IFN-γ and TNF-α in cells co-cultured with the CD11c-Hi subset (Fig. 3F) demonstrating that CD11c-Hi cells promote an effector phenotype in CD8+ cells. Furthermore, CD11c-Hi cells supported CD8+ T cell survival to a greater extent than the CD11c-Int subset (Fig. 3G and fig. S5E). Finally, we found that anti-LAP treatment decreased LAP+ CD11c-Int cells (Fig. 3H and fig. S5F) and reduced the expression of the tolerance-associated proteins PD-L1 and CD103 on CD11c-Int cells (Fig. 3I and fig. S5G). This is presumably secondary to a reduction of TGF-β by anti-LAP (Fig. 2B), since both genes could be up-regulated by TGF-β (17, 18). Thus, anti-LAP increased dendritic cells with a pro-inflammatory phenotype and decreased DCs with an anti-inflammatory phenotype in the spleen. We found that membrane LAP expression was reduced on CD11c+ cells in spleen, dLN and tumor after anti-LAP treatment (Fig. 3J and fig. S5H) indicating that anti-LAP may also affect DCs in the tumor microenvironment. Of note, we did not identify CD11c/CD11b subsets in dLN or tumor (fig. S5I).
Figure 3. Modulation of dendritic cell subsets following anti-LAP treatment.
(A, B) Effect of anti-LAP treatment on CD11c-Hi/CD11b-Int and CD11c-Int/CD11b-Hi DCs in spleen (shown as contour dot plots (A) and quantified as cell frequencies (B) (n=3; representative of two independent experiments). (C) Expression of LAP on CD11c-Hi and CD11c-Int DCs. Fluorescence minus one (FMO) control was used as a negative control for staining (n=6; representative of two experiments). (D) Expression of MHCII and CD86 on CD11c-Hi and CD11c-Int DCs (n=3; representative of two experiments). (E) Il12 and Il10 expression measured by qRT-PCR following stimulation of CD11c-Int and CD11c-Hi cells sorted from the spleen and treated with LPS or anti-CD40. (F, G) CD8+ T cells were co-cultured with CD11c-Int or CD11c-Hi for three days and IFN-γ and TNF-α measured in the supernatants (F) and live CD8+ T cells (G) quantified by flow cytometry (n=4; representative of three experiments). (H, I) Expression of LAP (H; n=4), PD-L1 and CD103 (I; n=6) on splenic CD11c-Int cells from tumor-bearing mice following anti-LAP treatment; representative of three experiments. (J) Expression of LAP on CD11c+ cells from spleen, dLN and tumor of B16 tumor-bearing mice following anti-LAP treatment (n=4–5; data are from two independent experiments). Error bars, mean±s.e.m. Two-tailed t-test was used for p value calculations.
Anti-LAP treatment enhances anti-tumor adaptive immune responses
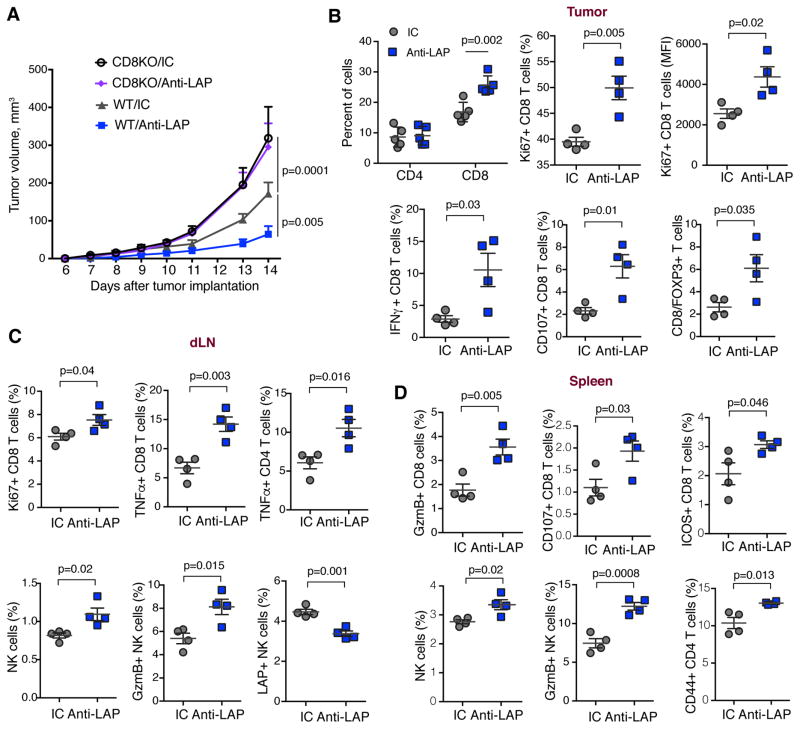
To test whether CD8+ T cells were required for the therapeutic effect of anti-LAP, we implanted the B16 melanoma in CD8-deficient mice. and found that the therapeutic effect of anti-LAP was abolished (Fig. 4A). Consistent with this, the therapeutic effect of anti-LAP was also reversed in animals treated with anti-CD8 (fig. S6A). No difference was observed in CD4-deficient mice (fig. S6B). When we analyzed TILs from mice implanted with the B16 melanoma, we found an increase in infiltrating CD8+ T cells following anti-LAP whereas CD4+ T cells did not change (Fig. 4B and fig. S6C). Similar results were observed in the intracranial GBM model (fig. S6D). Intratumoral CD8+ T cells expressed higher levels of the proliferation marker Ki67, the pro-inflammatory cytokine IFN-γ and the degranulation marker CD107 (Fig. 4B and fig. S6E). Anti-LAP treatment also increased the ratio of CD8+ T cells to Foxp3+ Tregs in the tumor in both B16 melanoma and intracranial GL261 GBM models (Fig. 4B, fig. S6E and S6F). We then examined the dLNs and spleen of B16 melanoma-bearing mice. In dLN, anti-LAP enhanced the proliferation of CD8+ T cells, increased the levels of TNF-α in CD8+ and CD4+ T lymphocytes, and increased NK cells and the levels of granzyme B they express (Fig. 4C and fig. S6G). In the spleen, we observed higher levels of granzyme B, CD107 and ICOS on CD8+ T cells after anti-LAP treatment, demonstrating a stronger effector phenotype of cytotoxic T cells following treatment. In addition, the frequency of NK cells and the expression of granzyme B by NK cells were increased. Furthermore, the CD44 activation marker was up-regulated on CD4+ T cells (Fig. 4D and fig. S6H). Of note, the percentage of LAP+ CD8 T cells was very low in the spleen, dLN and tumor and did not change with anti-LAP treatment (fig. S6I and S6J) suggesting that these cells do not play a significant role in the anti-tumor effect of anti-LAP. Taken together, these results demonstrate that anti-LAP affects adaptive immune responses both systemically and within the tumor driving them to a more inflammatory phenotype.
Figure 4. Effect of anti-LAP treatment on the adaptive immune response.
(A) Tumor volume measured over time in WT and CD8KO mice implanted with B16 melanoma cells and treated with either anti-LAP or IC antibodies (n=5 (CD8KO/anti-LAP and WT/anti-LAP) and n=4 (CD8KO/IC and WT/IC)). Data are representative of two independent experiments. (B–D) Analysis of the immune response in tumor (B), dLN (C) and spleen (D) of B16 tumor-bearing mice by flow cytometry (n=4). Data are representative of two independent experiments. Error bars, mean±s.e.m. Two-way ANOVA (A) and two-tailed t-test (B–D) were used for p value calculations. P values for the last time point are shown.
Anti-LAP treatment affects tolerogenic CD103+ CD8 T cells
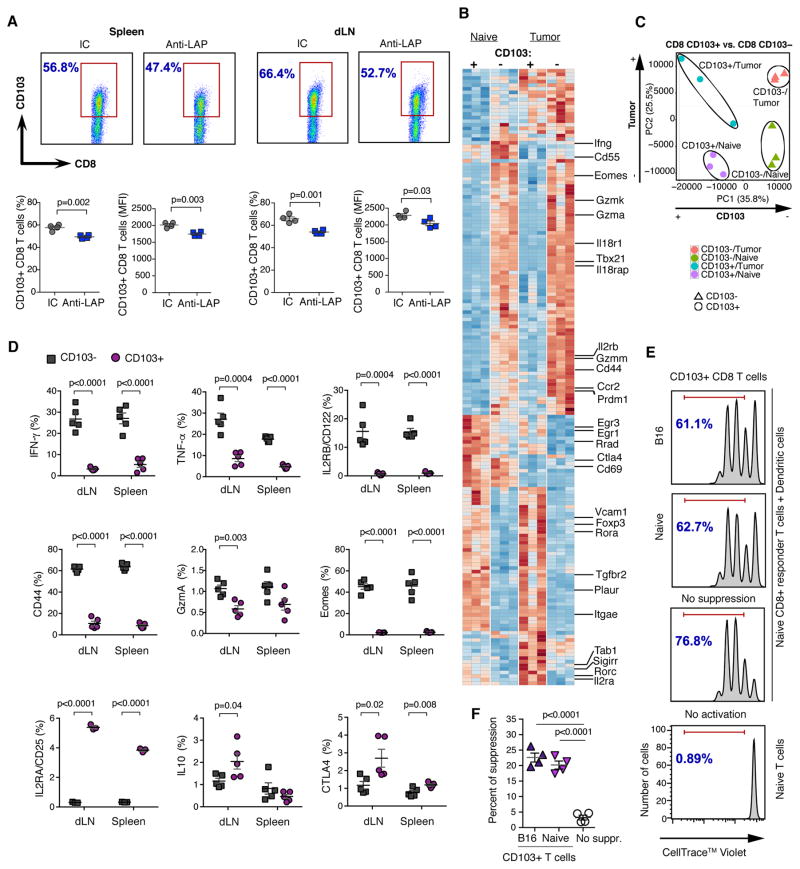
We consistently found a reduction of CD103+ CD8 T cells in both the spleen and dLNs following anti-LAP treatment (Fig. 5A). TGF-β plays an important role in the induction of CD103+ CD8 T cells (19), which may explain why anti-LAP reduces their number. Because the frequency of infiltrating CD103+ CD8 T cells in B16 tumors was very low (fig. S7A and S7B), we focused on CD103+ CD8 T cells in the periphery. We measured gene expression in CD103+ and CD103− CD8 T cells from the dLNs and spleen of naïve and B16 melanoma-bearing mice using the Nanostring Pan Cancer Immunology code set. We found 171 differentially expressed genes between groups (Fig. 5B), among them, activation and effector markers, including Cd44, Gzma, Gzmm, Gzmk, Il2rb, Prdm1, Il18r1, Tbx21, Eomes, and Ccr2; these genes were specifically overexpressed in CD103− CD8 T cells in naïve mice and were further upregulated in tumor-bearing mice. On the other hand, negative regulators of T cell activation including Egr3, Ctla4, and Tgfbr2 were higher in CD103+ CD8 T cells. Importantly, the Treg associated genes Il2ra, Foxp3, and Rorc (20–22) were up-regulated in CD103+ CD8 T cells in tumor-bearing mice. Interestingly, tumor suppressor genes, such as Erg1 and Rrad, were down-regulated in CD103+ cells from tumor-bearing mice vs. naïve mice, whereas oncogenes, such as Plaur and Vcam were up-regulated, suggesting that the tumor itself may further modulate the CD103+ T cell subset. Of note, in the intracranial GBM model, CD103+ CD8 T cells infiltrate the tumor and anti-LAP reduced these cells both in the tumor and systemically (fig. S7C).
Figure 5. CD103+ CD8 T cells are decreased by anti-LAP and possess regulatory properties.
(A) CD103+ CD8 T cells after anti-LAP treatment in spleen and dLNs by flow cytometry. Dot plot graphs (upper panels) and statistical calculations (lower panels) for cell frequencies and MFI are shown (n=4). Data are representative of at least three independent experiments. (B) Heat map showing differentially expressed genes in CD103+ and CD103− CD8 T cells from either naïve or B16 tumor-bearing mice (n=3). (C) PCA analysis of the global gene expression profiles shown in (B). (D) Phenotypic characterization of CD103+ and CD103− CD8 T cells in dLNs and spleen of B16 melanoma-bearing mice by flow cytometry (n=3–5; data are combined from two independent experiments). (E, F) In vitro suppression of naïve CD8+ T cell proliferation by CD103+ CD8 T cells isolated from naïve or melanoma-bearing mice. (E) Representative histograms of proliferation of responder CD8+ T cells and (F) percent of suppression (n=4; data are representative of three independent experiments). Error bars, mean±s.e.m. Two-tailed t-test (A and D) and one-way ANOVA (F) were used for p value calculations.
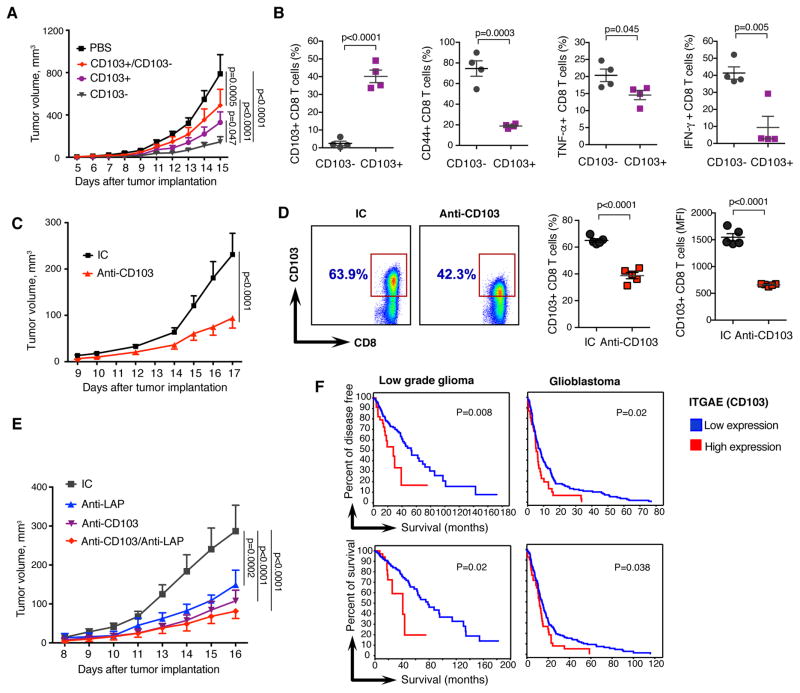
To further investigate CD103+ and CD103− CD8 T cell subsets in naïve vs. tumor conditions, we performed a PCA analysis on the global gene signature and found differential clustering of CD103+ vs. CD103− CD8 T cell subsets in both naïve and tumor conditions (Fig. 5C). PC1 mainly accounts for the variance between CD103+/− generated datasets, whereas PC2 accounts for the variance between tumor/naive generated datasets. Thus, CD103+ marks a CD8 T cell population that is different both in naïve mice as well as under tumor conditions. We then measured protein expression of activation markers IFN-γ, TNF-α, GzmA, CD44, Eomes, IL18R, IL2RB, IL2, CD107, and Ly6C on CD103+ vs. CD103− CD8 T cells in spleen and dLNs of melanoma-bearing mice. We found that CD103+ CD8 cells expressed lower levels of these markers (Fig. 5D, fig. S7D and S7E). KLRG1 was also decreased on CD103+ CD8 T cells. On the other hand, IL-10, CTLA4, and CD25/IL2RA were up-regulated on CD103+ CD8 T cells from dLNs consistent with the regulatory phenotype of CD103+ CD8 T cells. We then found that CD103+ CD8 T cells isolated from either B16-bearing mice or naïve mice suppressed CD8+ T cell proliferation (Fig. 5E and 5F). Suppression was mediated by the PD1/PD-L1 axis as it was blocked by anti-PD1 or anti-PD-L1 antibodies (fig. S7F). Consistent with this, CD103+ CD8 T cells expressed higher level of PD-L1 then CD103− CD8 T cells (fig. S7G and S7H). Of note, anti-PD1 and anti-LAP antibodies had comparable effects on B16 tumor growth (fig. S7I). We then examined the in vivo tumor-promoting role of CD103+ CD8 T cells by adoptively transferring CD103+ or CD103− CD8 T cells from B16 melanoma-bearing mice to CD8-deficient animals. We found greater tumor growth in animals that received CD103+ CD8 T cells and smaller tumor growth in animals that received CD103− CD8 T cells (Fig. 6A and fig. S8A). Co-transfer of CD103+/CD103− CD8 T cells increased tumor growth compared to the transfer of CD103− CD8 T cells alone. Without T cell transfer (PBS group) there was greater tumor growth compared to all the CD8+ T cell transfer groups demonstrating that CD8+ T cells control tumor growth. The increased tumor growth that we observed in the PBS group as compared to the CD103+ CD8 T cell group could be explained by the stability of CD103+ CD8 T cells after adoptive transfer: only 40% of cells remain CD103+ CD8 T cells (Fig. 6B).
Figure 6. CD103+ CD8 T cells have pro-tumorigenic properties.
(A) B16 melanoma tumor volumes in mice adoptively transferred with PBS (n=4), CD103+ (n=5), CD103− (n=4) or CD103+/CD103− (n=4) CD8 T cells from B16 melanoma mice to CD8KO mice. Data are representative of two independent experiments. (B) Expression of CD103, CD44, TNF-α and IFN-γ in mice after adoptive transfer of CD103+ CD8 T cells as compared to CD103− CD8 T cells by flow cytometry (n=4). (C) Tumor volumes measured over time in B16 melanoma model treated with anti-CD103 (clone M290; n=10). (D) Expression of CD103 on CD8 cells in dLNs of mice from C by flow cytometry with a non-competing 2E7 clone of anti-CD103. Both frequency of CD103+ CD8 T cells and MFI are presented (n=5). (E) Tumor volumes measured over time in B16 melanoma model treated with IC, anti-CD103, anti-LAP or combined anti-CD103+anti-LAP (n=5). (F) Percent survival in patients with relatively high or low mRNA expression of ITGAE (CD103) based on z score (details are as in Fig. 1N). Results for low grade glioma and glioblastoma are shown. Error bars, mean±s.e.m. Two-way ANOVA (A, C, and E) and two-tailed t-test (B and D) were used for p value calculations. P values for the last time points are shown.
We isolated these cells at the end of the experiment and found they maintained expression of CD103 and lower levels of pro-inflammatory genes (Fig. 6B and fig. S8B). We observed a similar effect when CD103+ or CD103− CD8 T cells were adoptively transferred from untreated mice (fig. S8C). To further investigate the role of CD103 we treated B16 melanoma and MC38 CRC-bearing mice with anti-CD103 antibody, which primarily targets CD103+ CD8 T cells (fig. S8D). We found that anti-CD103 treatment reduced tumor growth (Fig. 6C and fig. S8E) and was associated with reduction of CD103+ CD8 T cells (Fig. 6D). We then asked whether combined targeting of CD103 and LAP would have a synergistic effect; we treated tumor-bearing mice simultaneously with anti-CD103 plus anti-LAP. We did not observe a decrease of B16 tumor growth as compared to single antibody treatment with either anti-CD103 or anti-LAP (Fig. 6E). Consistent with our findings, in patients with both high and low grade gliomas, high CD103 expression was associated with shorter survival (Fig. 6F). Of note, CD103 is also expressed on CD4+ Tregs, which can contribute to a poorer prognosis. To address the potential role of LAP on CD103+ CD8 T cell function, we examined surface LAP expression and found that the frequency of LAP+CD103+ CD8 T cells was very low and there was no increased LAP expression on CD103+ vs. CD103− CD8 T cells (fig. S8F).
Anti-LAP treatment combined with antigen specific vaccination enhances tumor immunotherapy and improves immune memory
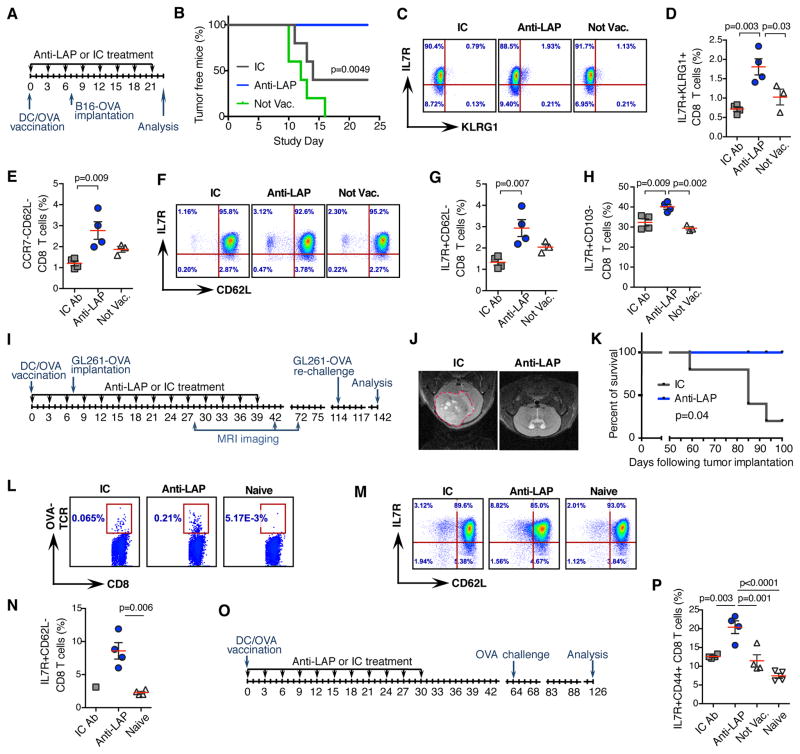
Because anti-LAP enhances the maturation of antigen presenting cells, we investigated combining anti-LAP with antigen-specific vaccination. We employed B16 melanoma cells that express ovalbumin (B16-OVA) and treatment with DCs loaded with ovalbumin (Fig. 7A). In this model, ovalbumin serves as a tumor-associated antigen. One week following vaccination with OVA-loaded DCs, mice were implanted with B16-OVA and treated with anti-LAP every third day. No mice vaccinated and treated with anti-LAP developed tumors whereas 60% of mice treated with IC and all mice in control group developed tumors (Fig. 7B). We then asked whether anti-LAP affected immune memory following DC vaccination. We used a series of markers for activation and memory of CD8+ T cells including IL7R, KLRG1, CCR7, and CD62L. Based on previously reported CD8+ T cell memory markers (23–25), we found an increase of effector memory-like CD8+ T cells in dLNs of anti-LAP treated mice (Fig. 7C–G and fig. S9A). Consistent with our results above, IL7R+CD103− CD8 T cells were increased in mice treated with anti-LAP (Fig. 7H and fig. S9B).
Figure 7. Anti-LAP treatment combined with antigen specific vaccination enhances tumor immunotherapy and improves immune memory.
(A) Study design. (B) Percentage of tumor free mice (n=5; log-rank test). Data are representative of two independent experiments. (C, D) Accumulation of memory-like CD8+ T cells, based on IL7R and KLRG1 expression in anti-LAP treated mice. Representative flow cytometry dot plots (C) and quantification (D) are shown (n=4 (anti-LAP and IC) and n=3 (not vaccinated)). (E) Percentage of memory-like CD8+ T cells, based on CCR7 and CD62L expression in anti-LAP treated mice (n=4 (anti-LAP and IC) and n=3 (not vaccinated)). (F, G) Frequency of memory-like CD8+ T cells, based on IL7R and CD62L expression in anti-LAP treated mice. Representative flow cytometry dot plots (F) and quantification (G) are shown (n=4 (anti-LAP and IC) and n=3 (not vaccinated)). (H) Percentage of IL7R+CD103− CD8 T cells (n=4 (anti-LAP and IC) and n=3 (not vaccinated)). (I) Study design. (J) Representative MRI images of mice treated with either IC or anti-LAP. (K) Survival curves of mice pre-vaccinated with DC-OVA, implanted with intracranial GBM and treated with anti-LAP (n=5; log-rank test). (L) Frequencies of OVA-TCR-specific CD8+ T cells (stained with H-2Kb OVA Pentamer) in mice treated with anti-LAP. (M, N) Accumulation of memory-like CD8+ T cells in anti-LAP treated mice. Representative flow cytometry dot plots (M) and quantification (N) are shown (n=4; two-tailed t-test). (O, P) Mice vaccinated with DC-OVA and treated by anti-LAP develop immune memory. (O) Study design. (P) Accumulation of memory-like IL7R+CD44+ CD8 T cells in anti-LAP treated mice by flow cytometry (n=4). Error bars, mean±s.e.m. One-way ANOVA (D, E, G, H, and P) was used for p value calculations.
We also investigated the intracranial GBM model in which glioma cells expressing ovalbumin (GL261-OVA) were implanted (Fig. 7I). One week following vaccination with OVA-loaded DCs, mice were implanted with GL261-OVA and treated with anti-LAP. Disease onset was delayed and, based on MRI imaging, none of anti-LAP treated mice developed tumors (Fig. 7J and K). On day 114, we re-challenged mice that did not develop tumors by implanting GL261-OVA subcutaneously and followed them for an additional month. None of these mice developed tumors, indicating that they had developed antigen specific immunity against the tumor. We investigated the immune response against OVA in surviving mice and found that anti-LAP treated mice developed increased numbers of both OVA-specific CD8 cells (Fig. 7L) and memory cells as measured by IL-7R and CD62L markers (Fig. 7M and 7N). To investigate the contribution of anti-LAP to immune memory we vaccinated mice with DCs loaded with OVA and treated them with anti-LAP for 4 weeks (Fig. 7O). A month later, we re-challenged the mice with a small number of subcutaneously injected GL261-OVA cells. Two months later, we analyzed CD8+ T cells and found specific up-regulation of IL7R+CD44+ CD8 T cells in anti-LAP treated mice (Fig. 7P and fig. S9C) indicating that anti-LAP supports anti-tumor memory. Thus, combination therapy with anti-LAP improved the immune response to antigen-specific DC vaccination and enhanced immune memory.
Discussion
Although targeting Tregs is an important avenue to boost tumor immunity, this approach has been limited due to a lack of drugable Treg targets and lack of specificity for Tregs (26, 27). We found that targeting LAP may be an effective way to affect Tregs and boost tumor immunity since the LAP/TGF-β complex identifies a subset of highly suppressive Tregs that are up-regulated in human malignancies (7, 28, 29). Consistent with multiple roles of TGF-β, we found increased CTL responses, reduction of tolerogenic CD103+ CD8 T cells, activation of NK cells, maturation of DCs and improved immune memory following anti-LAP treatment. In humans, LAP+Foxp3+ T cells are more suppressive than LAP-Foxp3+ T cells (28). Consistent with this, anti-LAP did not affect Foxp3+ T cell numbers in our studies. Foxp3 can also be transiently expressed on activated effector T cells in humans (30) and the accumulation of a Foxp3-lo population, represented by non-Treg cells, correlates with better survival of CRC patients than Foxp3-hi cells (31). These studies may explain different roles of Tregs in CRC reported by investigators.
We found that CD103+ CD8 T cells have a tolerogenic immune profile, exhibit suppressive properties and have a tumor-promoting role in vivo as compared to CD103− CD8 T cells. Anti-LAP treatment reduced CD103+ CD8 T cells, presumably because it decreases bioavailable TGF-β, which regulates the generation of CD103+ CD8 T cells (18, 32, 33). Indeed, TGF-β has been demonstrated to regulate the generation of CD103+ CD8 T cells (18, 32, 33). Furthermore, we found that direct targeting of CD103 by an anti-CD103 antibody that reduces CD103+ CD8 T cells in mice similar to what we observed with anti-LAP also had a therapeutic effect in the B16 melanoma and MC38 CRC models. Anti-CD103 antibody appears to act systemically in the B16 melanoma model as only a few CD103+ CD8 T cells infiltrate the tumor in this model. Interestingly, combinatorial treatment with anti-LAP and anti-CD103 did not result in a synergistic therapeutic effect indicating that LAP and CD103 pathways overlap. A previous study of anti-CD103 did not show a therapeutic effect in the CT26 model of CRC (34). Different roles for CD103+ CD8 T cells have been reported. Some studies report increased effector function against cancer cells (19, 32, 35, 36) whereas others demonstrate that CD103+ CD8 T cells could be regulatory in transplantation models and autoimmunity (37–41). Our study supports these later observations and extends them to cancer. It is possible that CD103+ CD8 T cells kill cancer cells in the tumor environment while suppressing T cell growth systemically. Of note, CD103 has been described as a marker of CD4+ regulatory cells and is present on tolerogenic DCs (2, 34, 42–44).
LAP is not only expressed on CD4+ T cells but also on CD8 cells, γδ T cells, NK cells, B cells and DCs (45–48). Thus, the anti-tumor effect of anti-LAP could be related to multiple targets. Dendritic cells play a key role in tumor antigen-specific vaccination and we found that anti-LAP plus DC vaccination enhanced the anti-tumor effects of DC vaccination. Interestingly, immature DCs express higher levels of LAP and we found that LAP+ DCs in humans have suppressive properties (45).
Although we demonstrate the therapeutic efficacy of anti-LAP antibody in a range of models, it is known that models do not always predict responses in humans. Furthermore, the subcutaneous models we studied do not mimic the natural tumor environment in humans. Nonetheless, LAP+ cells are increased in human cancer, possess tolerogenic function, and predict a poor prognosis in human cancer (7, 28, 29). Thus, despite the limitations of animal models, targeting LAP+ cells is consistent with the importance of TGF-β, and Tregs in the physiology of cancer in humans.
In summary, in addition to targeting Tregs, our results demonstrate a more complex process as anti-LAP also modulates DCs that express surface LAP, blocks TGF-β and decreases tolerogenic CD103+ CD8 T cells (fig. S10). Anti-LAP acts on multiple populations to promote anti-tumor immunity by increasing the activity of CD8+ T effector cells and enhancing immune memory. Consistent with our findings, LAP and CD103 expression in human cancer is associated with a poorer prognosis, providing an important translational link between our results and human disease and making anti-LAP an attractive candidate for cancer immunotherapy.
Materials and Methods
Study Design
Our objective was to investigate the therapeutic effect of anti-LAP antibody in models of cancer and characterize its effect on immune function. We used mice and primary cells and cell lines to address immunologic mechanism. Cages were randomly assigned to different treatment groups. Tumor size and weight loss were the major factors for ending data collection. All data were included in analysis, though in rare situations, clear outliers were excluded. Experimental replication is indicated in the figure legends. Although the study was not blinded, some in vivo experiments were performed independently by investigators in other laboratories. Data were collected using methods that provide numerical values (calipers, scales, bioluminescence imaging for tumor size measurement; flow cytometer for assessing protein expression or cell proliferation, real-time PCR instrument and nSolver digital analyzer for mRNA expression measurement). Animal pre-clinical studies were reported in accordance with the ARRIVE guidelines.
Animals
C57BL/6, BALB/c, CD4 (B6.129S2-Cd4tm1Mak/J) and CD8 (B6.129S2-Cd8atm1Mak/J)-deficient 6–8 week male mice were purchased from the Jackson Laboratories. Foxp3-GFP reporter mice were housed in a conventional specific pathogen-free facility at the Harvard Institutes of Medicine. All experiments were carried out in accordance with guidelines prescribed by the Institutional Animal Care and Use Committee at Harvard Medical School.
Antibody Treatments
Mice were treated with either anti-LAP or IC mAbs prepared in PBS. Mouse anti-LAP monoclonal antibodies were isolated from hybridoma generated in-house. Two clones were employed for in vivo treatments: TW7-28G11 (IgG2b) and TW7-16B4 (IgG1). Respective ICs, MPC-11 (IgG2b) and MOPC-21 (IgG1), anti-CD103 (clone M290) and anti-PD1 (RMP-1) were purchased from BioXCell. As a standard treatment, antibodies were administered intraperitoneallly, 10 mg/kg every third day following tumor implantation. In some experiments, mice were treated i.p. with 100 μg per mouse of anti-CD8β (clone Lyt-3.2; BioXCell) Ab or IC (rat IgG1, HRPN; BioXCell) on days –1, 7, and 14 after B16F10 implantation.
Statistical analysis
Two sample t-test was used to compare two groups, one-way ANOVA with Tukey’s adjustment for multiple comparisons was used to compare more than two groups, and two-way ANOVA was used to compare two and more groups over time. Survival curves were compared using a log-rank test. Two-sided alpha level of 0.05 was used for all tests. Analyses were completed using GraphPad Prism version 7.0a. Details of each analysis are included in the source data in supplementary materials.
Supplementary Material
Acknowledgments
We thank B. Healy for providing consultation on statistical analysis. A. Anderson, S. Kurtulus, I. Mascanfroni, D. Hu and C. Baecher-Allan for assistance and helpful discussion. D. Kozoriz for cell sorting, P. C. Gokhale for support with GBM model experiments, F. von Glen, R. Griffin, V. Kannan and A. Gurunathan for their technical assistance.
Funding: Supported by the National Institute of Health (R21NS090163 to H. L. Weiner) and Innovation Discovery Grant (from BWH to H. L. Weiner).
Footnotes
Author contributions: G.G., A.P.C., G.M., H.L., R.M.R., V.K., and H.L.W. designed all experiments. G.G., A.P.C., R.M.R., B.K., T.V., N.S., S.R., L.G., P.K., A.J.L., T.M., and M.A.M. performed all experiments. A.M. analyzed gene expression data. G.G., A.M., and B.K. performed statistical analyses. A.M.C.F., H.L., and V.K. provided input for the manuscript. G.G. and H.L.W. supervised the experiments and wrote the manuscript.
Competing interests: A patent related to this study has been filed (PCT/US16/13408, titled “Treatment of cancer with anti-LAP monoclonal antibodies”, by H.L.W., G.G., and A.P.C.). H.L.W. and G.G. are consulting Tilos Therapeutics company, which develops anti-LAP for clinical application. The other authors declare no competing interests.
References and Notes
- 1.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 2.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 3.Ochi H, Abraham M, Ishikawa H, Frenkel D, Yang K, Basso AS, Wu H, Chen ML, Gandhi R, Miller A, Maron R, Weiner HL. Oral CD3-specific antibody suppresses autoimmune encephalomyelitis by inducing CD4+ CD25− LAP+ T cells. Nat Med. 2006;12:627–635. doi: 10.1038/nm1408. [DOI] [PubMed] [Google Scholar]
- 4.Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev. 2006;212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen ML, Yan BS, Bando Y, Kuchroo VK, Weiner HL. Latency-associated peptide identifies a novel CD4+CD25+ regulatory T cell subset with TGFbeta-mediated function and enhanced suppression of experimental autoimmune encephalomyelitis. J Immunol. 2008;180:7327–7337. doi: 10.4049/jimmunol.180.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Cunha AP, Wu HY, Rezende RM, Vandeventer T, Weiner HL. In vivo anti-LAP mAb enhances IL-17/IFN-gamma responses and abrogates anti-CD3-induced oral tolerance. Int Immunol. 2015;27:73–82. doi: 10.1093/intimm/dxu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jie HB, Gildener-Leapman N, Li J, Srivastava RM, Gibson SP, Whiteside TL, Ferris RL. Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. Br J Cancer. 2013;109:2629–2635. doi: 10.1038/bjc.2013.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Travis MA, Sheppard D. TGF-beta activation and function in immunity. Annu Rev Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Zhu J, Dong X, Shi M, Lu C, Springer TA. GARP regulates the bioavailability and activation of TGFbeta. Mol Biol Cell. 2012;23:1129–1139. doi: 10.1091/mbc.E11-12-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280:7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- 13.Stockis J, Colau D, Coulie PG, Lucas S. Membrane protein GARP is a receptor for latent TGF-beta on the surface of activated human Treg. Eur J Immunol. 2009;39:3315–3322. doi: 10.1002/eji.200939684. [DOI] [PubMed] [Google Scholar]
- 14.Oida T, Weiner HL. TGF-beta induces surface LAP expression on murine CD4 T cells independent of Foxp3 induction. PLoS One. 2010;5:e15523. doi: 10.1371/journal.pone.0015523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program coopts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Tang H, Guo Z, An H, Zhu X, Song W, Guo J, Huang X, Chen T, Wang J, Cao X. Splenic stroma drives mature dendritic cells to differentiate into regulatory dendritic cells. Nat Immunol. 2004;5:1124–1133. doi: 10.1038/ni1130. [DOI] [PubMed] [Google Scholar]
- 17.Song S, Yuan P, Wu H, Chen J, Fu J, Li P, Lu J, Wei W. Dendritic cells with an increased PD-L1 by TGF-beta induce T cell anergy for the cytotoxicity of hepatocellular carcinoma cells. Int Immunopharmacol. 2014;20:117–123. doi: 10.1016/j.intimp.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 18.Mokrani M, Klibi J, Bluteau D, Bismuth G, Mami-Chouaib F. Smad and NFAT pathways cooperate to induce CD103 expression in human CD8 T lymphocytes. J Immunol. 2014;192:2471–2479. doi: 10.4049/jimmunol.1302192. [DOI] [PubMed] [Google Scholar]
- 19.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA. TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med. 2005;201:1647–1657. doi: 10.1084/jem.20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voo KS, Wang YH, Santori FR, Boggiano C, Wang YH, Arima K, Bover L, Hanabuchi S, Khalili J, Marinova E, Zheng B, Littman DR, Liu YJ. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A. 2009;106:4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos PD, Rudensky AY. Treg cells in cancer: a case of multiple personality disorder. Sci Transl Med. 2012;4:164fs144. doi: 10.1126/scitranslmed.3005283. [DOI] [PubMed] [Google Scholar]
- 23.Obar JJ, Jellison ER, Sheridan BS, Blair DA, Pham QM, Zickovich JM, Lefrancois L. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J Immunol. 2011;187:4967–4978. doi: 10.4049/jimmunol.1102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obar JJ, Lefrancois L. Early signals during CD8 T cell priming regulate the generation of central memory cells. J Immunol. 2010;185:263–272. doi: 10.4049/jimmunol.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiser J, Banerjee A. Effector, Memory, and Dysfunctional CD8(+) T Cell Fates in the Antitumor Immune Response. J Immunol Res. 2016;2016:8941260. doi: 10.1155/2016/8941260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Workman CJ, Vignali DA. Targeting regulatory T cells in tumors. FEBS J. 2016;283:2731–2748. doi: 10.1111/febs.13656. [DOI] [PubMed] [Google Scholar]
- 27.Pere H, Tanchot C, Bayry J, Terme M, Taieb J, Badoual C, Adotevi O, Merillon N, Marcheteau E, Quillien VR, Banissi C, Carpentier A, Sandoval F, Nizard M, Quintin-Colonna F, Kroemer G, Fridman WH, Zitvogel L, Oudard SP, Tartour E. Comprehensive analysis of current approaches to inhibit regulatory T cells in cancer. Oncoimmunology. 2012;1:326–333. doi: 10.4161/onci.18852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scurr M, Ladell K, Besneux M, Christian A, Hockey T, Smart K, Bridgeman H, Hargest R, Phillips S, Davies M, Price D, Gallimore A, Godkin A. Highly prevalent colorectal cancer-infiltrating LAP(+) Foxp3(−) T cells exhibit more potent immunosuppressive activity than Foxp3(+) regulatory T cells. Mucosal Immunol. 2014;7:428–439. doi: 10.1038/mi.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahalingam J, Lin YC, Chiang JM, Su PJ, Fang JH, Chu YY, Huang CT, Chiu CT, Lin CY. LAP+CD4+ T cells are suppressors accumulated in the tumor sites and associated with the progression of colorectal cancer. Clin Cancer Res. 2012;18:5224–5233. doi: 10.1158/1078-0432.CCR-12-0211. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 31.Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 32.Hadley GA, Bartlett ST, Via CS, Rostapshova EA, Moainie S. The epithelial cell-specific integrin, CD103 (alpha E integrin), defines a novel subset of alloreactive CD8+ CTL. J Immunol. 1997;159:3748–3756. [PubMed] [Google Scholar]
- 33.Ling KL, Dulphy N, Bahl P, Salio M, Maskell K, Piris J, Warren BF, George BD, Mortensen NJ, Cerundolo V. Modulation of CD103 expression on human colon carcinoma-specific CTL. J Immunol. 2007;178:2908–2915. doi: 10.4049/jimmunol.178.5.2908. [DOI] [PubMed] [Google Scholar]
- 34.Anz D, Mueller W, Golic M, Kunz WG, Rapp M, Koelzer VH, Ellermeier J, Ellwart JW, Schnurr M, Bourquin C, Endres S. CD103 is a hallmark of tumor-infiltrating regulatory T cells. Int J Cancer. 2011;129:2417–2426. doi: 10.1002/ijc.25902. [DOI] [PubMed] [Google Scholar]
- 35.Webb JR, Wick DA, Nielsen JS, Tran E, Milne K, McMurtrie E, Nelson BH. Profound elevation of CD8+ T cells expressing the intraepithelial lymphocyte marker CD103 (alphaE/beta7 Integrin) in high-grade serous ovarian cancer. Gynecol Oncol. 2010;118:228–236. doi: 10.1016/j.ygyno.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Djenidi F, Adam J, Goubar A, Durgeau A, Meurice G, de Montpreville V, Validire P, Besse B, Mami-Chouaib F. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol. 2015;194:3475–3486. doi: 10.4049/jimmunol.1402711. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Lan Q, Lu L, Chen M, Xia Z, Ma J, Wang J, Fan H, Shen Y, Ryffel B, Brand D, Quismorio F, Liu Z, Horwitz DA, Xu A, Zheng SG. Phenotypic and functional characteristic of a newly identified CD8+ Foxp3− CD103+ regulatory T cells. J Mol Cell Biol. 2014;6:81–92. doi: 10.1093/jmcb/mjt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uss E, Rowshani AT, Hooibrink B, Lardy NM, van Lier RA, ten Berge IJ. CD103 is a marker for alloantigen-induced regulatory CD8+ T cells. J Immunol. 2006;177:2775–2783. doi: 10.4049/jimmunol.177.5.2775. [DOI] [PubMed] [Google Scholar]
- 39.Keino H, Masli S, Sasaki S, Streilein JW, Stein-Streilein J. CD8+ T regulatory cells use a novel genetic program that includes CD103 to suppress Th1 immunity in eye-derived tolerance. Invest Ophthalmol Vis Sci. 2006;47:1533–1542. doi: 10.1167/iovs.04-1454. [DOI] [PubMed] [Google Scholar]
- 40.Lu L, Yu Y, Li G, Pu L, Zhang F, Zheng S, Wang X. CD8(+)CD103(+) regulatory T cells in spontaneous tolerance of liver allografts. Int Immunopharmacol. 2009;9:546–548. doi: 10.1016/j.intimp.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Mattoscio M, Nicholas R, Sormani MP, Malik O, Lee JS, Waldman AD, Dazzi F, Muraro PA. Hematopoietic mobilization: Potential biomarker of response to natalizumab in multiple sclerosis. Neurology. 2015;84:1473–1482. doi: 10.1212/WNL.0000000000001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Annacker O, Coombes JL, Malmstrom V, Uhlig HH, Bourne T, Johansson-Lindbom B, Agace WW, Parker CM, Powrie F. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J Exp Med. 2005;202:1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott CL, Aumeunier AM, Mowat AM. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol. 2011;32:412–419. doi: 10.1016/j.it.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Gandhi R, Anderson DE, Weiner HL. Cutting Edge: Immature human dendritic cells express latency-associated peptide and inhibit T cell activation in a TGF-beta-dependent manner. J Immunol. 2007;178:4017–4021. doi: 10.4049/jimmunol.178.7.4017. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Morgan R, Chen C, Cai Y, Clark E, Khan WN, Shin SU, Cho HM, Al Bayati A, Pimentel A, Rosenblatt JD. Mammary-tumor-educated B cells acquire LAP/TGF-beta and PD-L1 expression and suppress anti-tumor immune responses. Int Immunol. 2016;28:423–433. doi: 10.1093/intimm/dxw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezende RM, da Cunha AP, Kuhn C, Rubino S, M’Hamdi H, Gabriely G, Vandeventer T, Liu S, Cialic R, Pinheiro-Rosa N, Oliveira RP, Gaublomme JT, Obholzer N, Kozubek J, Pochet N, Faria AM, Weiner HL. Identification and characterization of latency-associated peptide-expressing gammadelta T cells. Nat Commun. 2015;6:8726. doi: 10.1038/ncomms9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen ML, Yan BS, Kozoriz D, Weiner HL. Novel CD8+ Treg suppress EAE by TGF-beta- and IFN-gamma-dependent mechanisms. Eur J Immunol. 2009;39:3423–3435. doi: 10.1002/eji.200939441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohlfest JR, Andersen BM, Litterman AJ, Xia J, Pennell CA, Swier LE, Salazar AM, Olin MR. Vaccine injection site matters: qualitative and quantitative defects in CD8 T cells primed as a function of proximity to the tumor in a murine glioma model. J Immunol. 2013;190:613–620. doi: 10.4049/jimmunol.1201557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahuja SS. In vitro generation of functional human and murine dendritic cells. Methods Mol Biol. 2001;156:67–77. doi: 10.1385/1-59259-062-4:67. [DOI] [PubMed] [Google Scholar]
- 51.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 52.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.