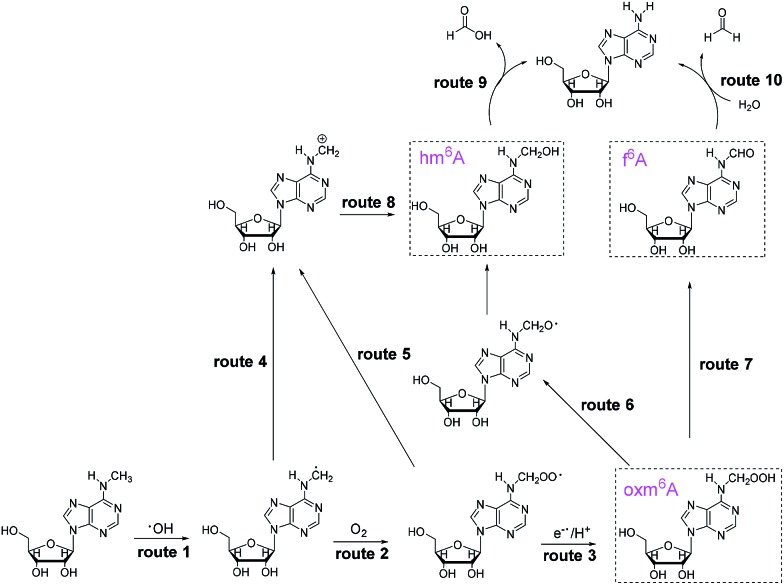
 A new route is found in the chemical oxidation of N6-methyladenosine using a H2O2/bicarbonate system through the N6-hydroperoxymethyladenosine (oxm6A) intermediate.
A new route is found in the chemical oxidation of N6-methyladenosine using a H2O2/bicarbonate system through the N6-hydroperoxymethyladenosine (oxm6A) intermediate.
Abstract
N 6-Methyladenosine (m6A) represents a relatively abundant modification in eukaryotic RNA. Because m6A has similar properties to adenosine and a low reactivity, limited research has been focused on this nucleoside. In this study, we revealed an important intermediate in the oxidation of m6A through the bicarbonate-activated peroxide system. Over the course of oxidation, we found a new mechanism in which N6-hydroxymethyladenosine (hm6A), N6-formyladenosine (f6A) and N6-hydroperoxymethyladenosine (oxm6A) were intermediate products, and adenosine was the final product. In this study, oxm6A was isolated using HPLC and characterized by mass spectrometry, NMR and diphenyl-1-pyrenylphosphine (DPPP) fluorescence detection. This study provides a new modified nucleoside and demonstrates oxidative demethylation of m6A by reactive oxygen species at the nucleobase level and in RNA strands.
N 6-Methyladenosine represents the most abundant modification in the mRNA of higher eukaryotes, present at a frequency of approximately three sites on each mRNA.1 m6A is also present on tRNA, rRNA and lnRNA.2 This modification plays an important role in the regulation of gene expression.3 Since its discovery last century,4 m6A has been the object of relatively few studies. Recently, fat mass- and obesity-associated proteins (FTO)5 and AlkBH5 6 were found to be m6A demethylases, indicating a novel regulatory mechanism in mammalian cells. Two new modifications, N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A), have been found to participate in the FTO-mediated demethylation process, which may influence RNA–protein interactions and regulate gene expression.7 In addition, transcriptome-wide profiling of m6A in mRNA and lnRNA has revealed new insights into the role of RNA modification.8 These developments have renewed interest in the investigation of this particular, distinctive modification. Therefore, we aspire to use a chemical method to differentiate m6A from A.
Hydrogen peroxide is a widely used oxidant with a high content of active oxygen,9 but its relatively slow oxidizing rate limits its usage. Bicarbonate is present in cells and serum at high concentrations, ranging from 14.7–25 mM,10 and plays an important role in biological oxidation.11 H2O2 and NH4HCO3 are environmentally friendly reagents; H2O2 produces only water as a by-product, and NH4HCO3 easily decomposes to NH3, CO2, and H2O. The reaction conditions are mild at natural pH values.
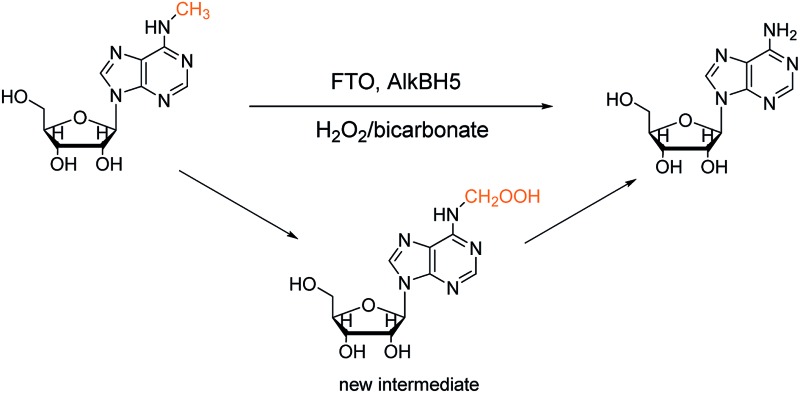
Owing to its high reactivity towards secondary amines, we considered whether the oxidant could react with m6A. Surprisingly, instead of producing N-oxides, demethylated adenosine was produced, and the presence of several intermediates in the reaction system suggested a potential mechanism in the chemical reaction (Scheme 1). These results suggest that H2O2/bicarbonate can act as a reactive oxygen species (ROS) for demethylation. In this study, we determine a key intermediate in the demethylation process, and we investigate the underlying mechanism.
Scheme 1. Proposed mechanism of the demethylation process, and structures of oxidation products.
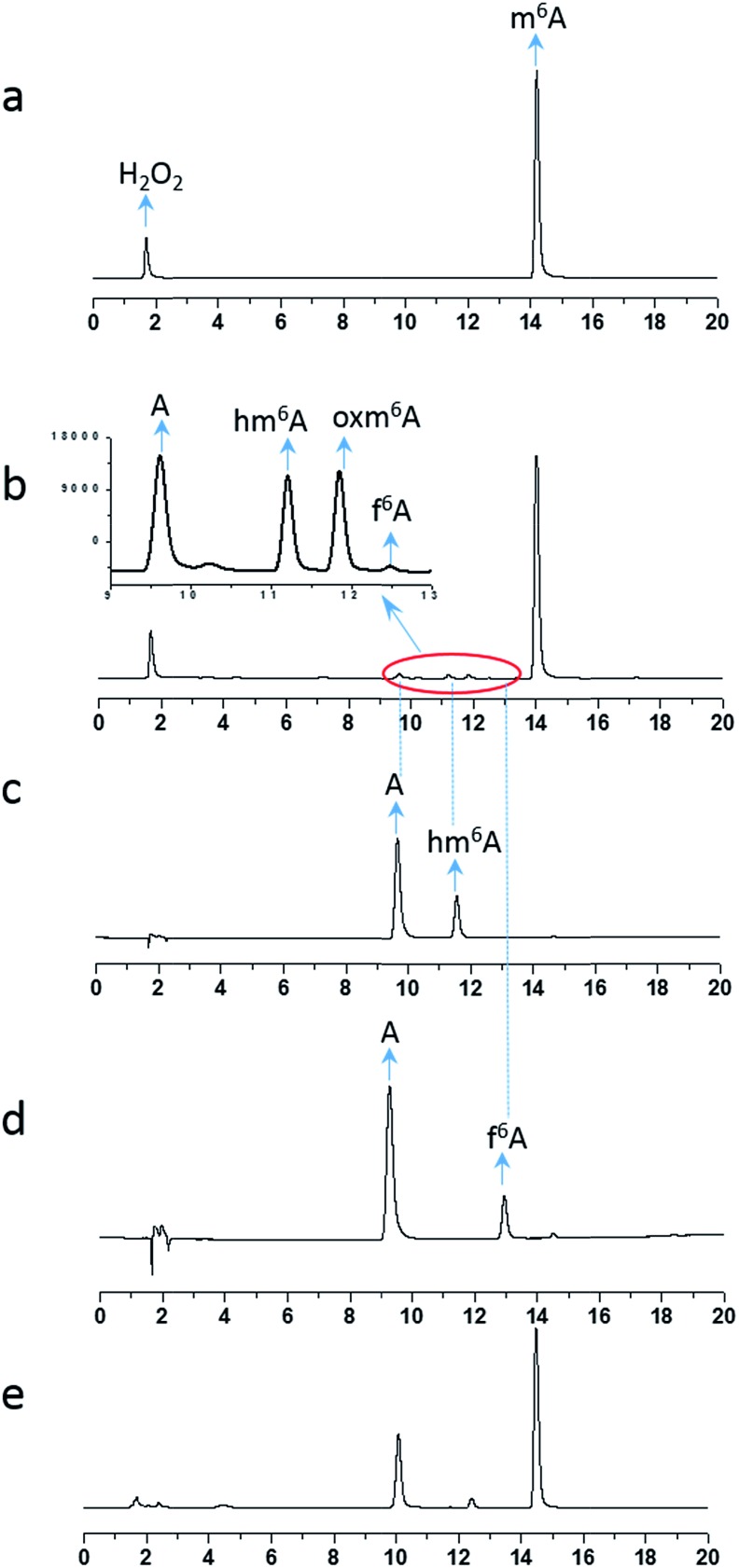
To investigate the demethylation process, we used high-performance liquid chromatography (HPLC) to monitor the reaction (UV detector at 260 nm). When a 2 mM aliquot of m6A was treated with 200 mM H2O2 and 1 M NH4HCO3 at 37 °C for one hour, four products were formed: A, hm6A, oxm6A and f6A (Fig. 1). The LC-MS data showed masses corresponding to A (267.9), hm6A (297.8), oxm6A (313.8) and f6A (295.9), successively in the positive-ion mode (Fig. S1†). Product A was further characterized by 1H and 13C NMR (see ESI†). To confirm the occurrence of hm6A and f6A, these compounds were synthesized according to reported procedures.7 An equilibrium reaction between adenosine and formaldehyde produced hm6A (Scheme 1, Route 9). Further HPLC analysis indicated that the synthesized hm6A and f6A have the same retention times as the reported hm6A and f6A, respectively (Fig. 1b–d). We found that hm6A and f6A were unstable and could decompose to A (adenosine) during HPLC analysis (Fig. 1c and d). N6-Hydroperoxymethyladenosine (oxm6A) was found to be a new intermediate, in addition to hm6A and f6A, during the demethylation of m6A (Fig. 1b). When we incubated the m6A with bicarbonate or H2O2 alone, no reaction was observed (Fig. S2 and S3†).
Fig. 1. HPLC chromatograph of 2 mM m6A incubated with 200 mM H2O2 and 1 M NH4HCO3 at 37 °C for 0 h (a), 1 h (b) and 24 h (e). As shown in the HPLC profiles, when the reaction proceeded for 1 h, one major product (A) was produced, accompanied by three intermediates (hm6A, f6A, oxm6A). The synthesized hm6A (c) and f6A (d) standards have the same retention time as two of the new peaks in the reaction mixture. Because hm6A and f6A are unstable, they can coexist with A during HPLC analysis.
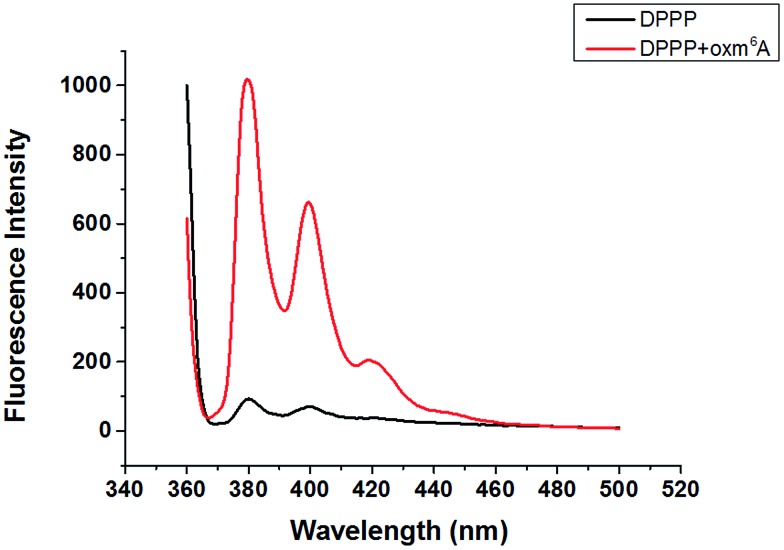
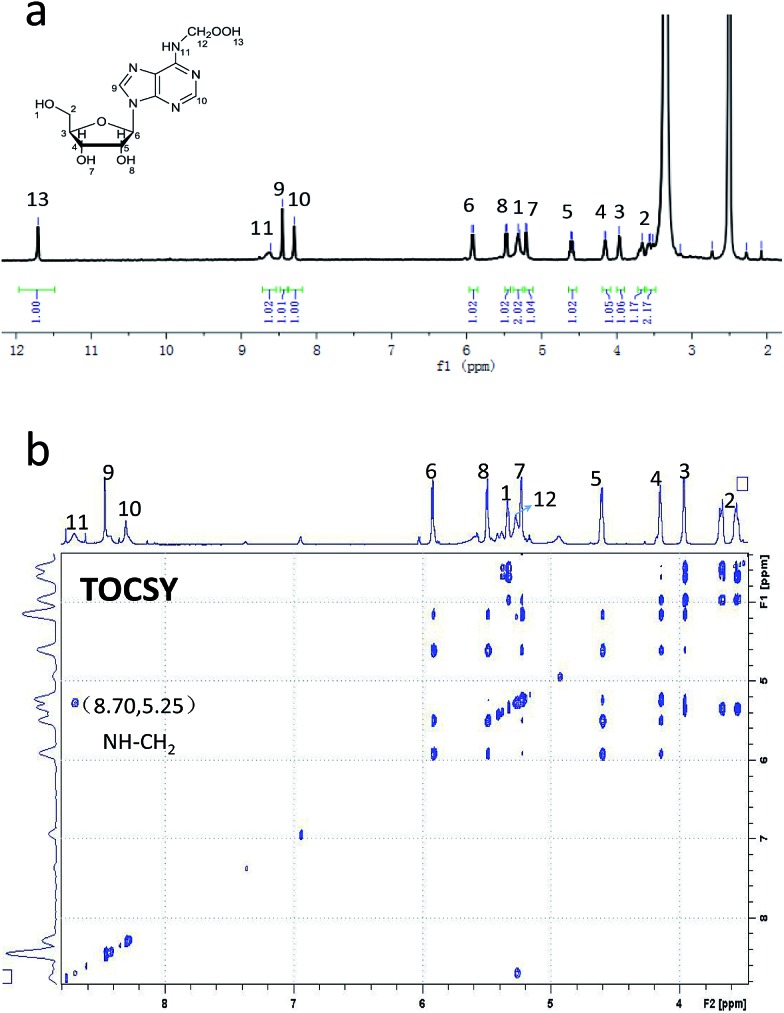
Diphenyl-1-pyrenylphosphine (DPPP), as a fluorescent reagent, can be used for hydroperoxide determinations.12 When we incubated the intermediate with DPPP in the presence of butylated hydroxytoluene (BHT) at 37 °C for 1 h, the fluorescence increased, indicating the formation of a hydroperoxide intermediate (Fig. 2). Further characterization of oxm6A was achieved using high-resolution mass spectrometry, 1H NMR, 13C NMR and TOCSY (ESI, Fig. 3, S4 and S5†), with the corresponding chemical structures shown in Scheme 1. To confirm the chemical shifts of the protons in N–H and OO–H, 1H NMR was performed in DMSO-d6 and in D2O. In the DMSO-d6 solution, the chemical shifts of the protons were 8.61 ppm (–N–H) and 11.71 ppm (–OOH) (Table 1). To confirm our hypothesis, we changed the solution to D2O, where deuterons can be incorporated at the N–H and O–H positions because of hydrogen–deuterium (H/D) exchange behavior. As we expected, these two protons disappeared in the D2O solution (Fig. 3). We then used total correlation spectroscopy (TOCSY) to show the H–H correlation; the TOCSY spectrum was acquired using a 600 MHz Bruker Avance II spectrometer equipped with a 5 mm triple resonance cryoprobe. The pulse sequence was DIPSI2ETGP. The relaxation delay was 1 s, with 8 acquisitions per increment, and a spectral width of 8 × 8 ppm and time domain of 2k × 176 were used. In the spectrum, the NH proton had a cross peak with CH2 at δ (8.66, 5.26 ppm), further confirming the oxm6A structure. When we analysed the reaction mixture using LC-MS, we detected a relatively small mass signal of 311.8; this finding may indicate the generation of another intermediate, N6-carboxyladenosine, in a relatively low yield (Fig. S1c†). Meanwhile, our control experiments indicated that adenosine, uridine, cytidine and guanosine were stable in the H2O2/bicarbonate solution at concentrations of 200 mM H2O2 and 1 M NH4HCO3 (Fig. S6†) after one hour.
Fig. 2. Fluorescence emission spectra (λex = 352 nm) of DPPP in the presence of (a) and in the absence of oxm6A (b) after incubation with BHT at 37 °C for 60 min.
Fig. 3. 1H NMR spectrum (a) and TOCSY spectrum (b) of oxm6A in DMSO-d6.
Table 1. 1H chemical shifts (δ, ppm) of oxm6A in DMSO-d6 at room temperature.
| Number | Chemical shift (δ, ppm) | Number | Chemical shift (δ, ppm) | Number | Chemical shift (δ, ppm) |
| 1 | 5.33 | 6 | 5.91 | 11 | 8.66 |
| 2 | 3.68 | 7 | 5.23 | 12 | 5.26 |
| 3 | 3.96 | 8 | 5.50 | 13 | 11.72 |
| 4 | 4.14 | 9 | 8.46 | ||
| 5 | 4.61 | 10 | 8.31 |
Because our goal was to fully investigate the mechanism of m6A demethylation, we extended the reaction time to 24 hours. After 24 hours, we found that only A (primary product) and a small amount of oxm6A were present (Fig. 1e), whereas hm6A and f6A disappeared. This result suggested that hm6A and f6A were converted into A (Scheme 1, Routes 9 and 10).
To investigate the behaviour of oxm6A, it was separated from the reaction mixture, incubated in HEPES buffer (50 mM, pH 7.4) at 37 °C and then analysed by HPLC every 2 h. We found that the amount of A increased at the expense of oxm6A (Fig. S7 in the ESI†), and it had a half-time of approximately 8.5 h (Fig. S8†), which was more stable than hm6A and f6A (approximately 3 h).
In the H2O2/NH4HCO3 system, the hydroxyl radical was trapped by 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) to give a signal using Electron Paramagnetic Resonance (EPR) (Fig. S9†). In the reaction system, the addition of DMSO, a hydroxyl radical scavenger, dramatically decreased the chemical demethylation level of m6A (Fig. S10†). We speculate that the reaction underwent a hydroxyl radical mechanism. A hydroxyl radical abstracted a hydrogen atom from a methyl group to yield a carbon radical, which could then bind with O2 to form oxm6A (Scheme 1, Routes 2 and 3) or bind with ˙OH to form hm6A (Scheme 1, Routes 4 and 8), parallel to the decomposition mechanism for 5′-hydroperoxymethyluracil and 5′-hydroperoxymethylcytosine, as proposed by Richard Wagner's group.13 To confirm the possibility of the ˙OH radical mechanism, we used Fenton-type reagents to react with m6A. The formation of hm6A, oxm6A, f6A and A was also observed using LC-MS analysis, confirming the reaction mechanism (Fig. S18†). Under identical experimental conditions but with the addition of a small amount of (NH4)2Fe(SO4)2 in the H2O2/NH4HCO3 reaction mixture, the reaction rate markedly increased (Fig. S11†). As the reaction is based on the hydroxyl radical mechanism, and Fe2+ as well as Cu2+ have great influences on the reaction, we therefore used Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES) to investigate the presence of iron(ii) and copper(ii) in the H2O2/bicarbonate reaction system. No signals were observed, and both the concentration of Fe2+ and Cu2+ were lower than 10 ng mL–1, indicating the reaction could proceed with just a bicarbonate-activated peroxide system (the optimized operating conditions are shown in Table S1†). In the demethylation process, two pathways are shown. A hydroxyl radical attacks the methyl radical to form hm6A (Scheme 1, Routes 4 and 8) and O2 attacks the methyl radical to form oxm6A (Scheme 1, Routes 2 and 3). The oxm6A and its peroxide radical can decompose to hm6A (Scheme 1, Routes 5, 8 and 6) and f6A (Scheme 1, Route 7), and we propose that the new route in the demethylation process would improve the efficiency of the demethylation reaction compared to just attacking the methyl radical by a hydroxyl radical.
Next, because m6A is preferentially present in the consensus sequence RRm6ACH (R is A/G and H is A/C/U),14 to examine whether the reaction occurs in RNA oligos, we prepared a 9-mer oligoribonucleotide (5′-CUGGm6ACUGG-3′) containing one m6A site and treated it with 10 mM H2O2 and 100 mM bicarbonate at 37 °C for 48 h. Because RNA may decompose in a high concentration of H2O2, we decreased the concentration of H2O2 and NH4HCO3. After the reaction, the oligo RNA was analysed using MALDI-TOF mass spectrometry as shown in Fig. S12.† We found a m6A –14 Da peak, representing a demethylation product, as well as a +14 Da peak and a +17 Da peak, which may correspond to N6-formyladenosine and N6-hydroxymethyladenosine intermediates in the demethylation pathway, respectively. At natural pH levels (pH 7.4), hm6A, oxm6A and f6A were relatively stable, but an alkaline phosphate digestion may accelerate their decomposition. Therefore, to verify the presence of hm6A, oxm6A and f6A in the oligo RNA after the reaction, we used RNase T1 followed by nuclease P1 to digest the oligo RNA,7 then analysed the reaction using LC-MS. In this analysis, RNase T1 can selectively digest the phosphodiester bond after G. We successfully detected the formation of A, hm6A, oxm6A, and f6A in the digested nucleoside, similar to our proposed mechanism for a single nucleoside (Fig. S13†).
To explore the reaction kinetics of the oxidation, two micrograms of oligo RNA were incubated with 100 μM H2O2 and 300 μM NH4HCO3 at 37 °C for 30 h in six parallel experiments, followed by digestion with nuclease P1 and alkaline phosphate. The amount of A generated from m6A was quantified using LC-MS every 3 hours (the calibration curve is shown in the ESI, Fig. S14†). As depicted in Fig. S15,† the A content exhibited a strong linear relationship with reaction time over a period of 30 hours. After adding Fe2+ to the H2O2/NH4HCO3 mixture and incubating it with oligo RNA, HPLC analysis of the enzymatically digested nucleosides in RNA showed the presence of demethylated adenosine with a decreased level of m6A after oxidation for 1 h (Fig. S16†).
Although FTO-mediated oxidation of m6A may decrease the level of m6A in vitro, no in vitro experiments have been reported in which a chemical reagent was used to demethylate m6A. We explored whether m6A in genomic RNA is a substrate of H2O2/NH4HCO3in vitro. Total RNA was extracted from Hela cells using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Four micrograms of genomic RNA was incubated with 100 μM H2O2 and 1 mM NH4HCO3 at 37 °C for 12 h. After digestion with nuclease P1 and alkaline phosphatase, the solution was analysed by LC-MS. The results showed a decrease in the m6A/A ratio by 10% in the genomic RNA (Fig. S17†), indicating that the reagents demethylated m6A in vitro.
Conclusions
In conclusion, we reported a new chemical method for the oxidative demethylation of m6A and determined an important intermediate in the reaction system. Three intermediates, N6-hydroxymethyladenosine (hm6A), N6-formyladenosine (f6A), and N6-hydroperoxymethyladenosine (oxm6A), were characterized, and the mechanism underlying the decomposition was illustrated. We also determined that the reaction could occur in oligo RNA and genomic RNA in vitro. H2O2 is a reactive oxygen species that is endogenously produced during normal metabolism15 and immune responses,16 and a high concentration of bicarbonate is found in cells and serum. Thus, this route may occur in vivo and play a role in cells. ROS have been proven to directly react with genomic DNA in a chemical reaction.17 Recently, reports have shown that ROS can induce the oxidative conversion of 5mC to 5hmC in a TET dioxygenase-dependent manner,18 indicating ROS regulate the enzymatic catalytic reaction. We propose that the oxm6A was formed through direct oxidation by ROS in vivo, just like the nucleoside analogues formed in RNA induced by Fenton-type reagents.17b Further study is in progress to study the presence and biological function of oxm6A in vivo. The discovery of the new intermediate oxm6A and the chemical route for the demethylation of m6A to A may offer new insight into the study of m6A.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program) (2012CB720600, 2012CB720603), the National Science Foundation of China (nos 91213302, 21432008, 91413109) and the National Grand Program on Key Infectious Disease (2012ZX10003002-014). The oligo RNA was kindly offered by He's Group at the University of Chicago. The TOCSY spectrum was collected by Prof. Maili Liu and Dr Xiongjie Xiao at Wuhan Institute of Physics and Mathematics. We thank Prof. Bin Hu and Dr Man He for the ICP-OES service.
Footnotes
†Electronic supplementary information (ESI) available: Methods, figures and spectra are included. See DOI: 10.1039/c5sc00484e
References
- Bokar J. A., The Biosynthesis and Functional Roles of Methylated Nucleosides in Eukaryotic mRNA, In Fine-tuning of RNA Functions by Modification and Editing, H. Grosjean, Topics in Current Genetics 12, Springer-Verlag, Berlin Heidelberg, 2005, pp. 141–177. [Google Scholar]
- (a) Saneyoshi M., Harada F., Nishimura S. Biochim. Biophys. Acta. 1969;190:264–273. doi: 10.1016/0005-2787(69)90078-1. [DOI] [PubMed] [Google Scholar]; (b) lwanami Y., Brown G. Arch. Biochem. Biophys. 1968;126:8–15. doi: 10.1016/0003-9861(68)90553-5. [DOI] [PubMed] [Google Scholar]
- (a) Pan T. Trends Biochem. Sci. 2013;11:8–17. [Google Scholar]; (b) Jia G., Fu Y., He C. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Meyer K. D., Jaffrey S. R. Mol. Cell. Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Fu Y., Dominissini D., Rechavi G., He C. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- (a) Littlefield J. W., Dunn D. B. Biochem. J. 1958;70(4):642. doi: 10.1042/bj0700642. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hall R. H. Biochemistry. 1965;4:661–670. doi: 10.1021/bi00880a008. [DOI] [PubMed] [Google Scholar]; (c) Brookes P., Lawley P. D. J. Chem. Soc. 1962;254:1348–1351. [Google Scholar]; (d) Desrosiers R., Friderici K., Rottman F. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y. G., He C. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., Vagbo C. B., Shi Y., Wang W. L., Song S. H., Lu Z., Bosmans R. P. G., Dai Q., Hao Y. J., Yang X., Zhao W. M., Tong W. M., Wang X. J., Bogdan F., Furu K., Fu Y., Jia G., Zhao X., Liu J., Krokan H. E., Klungland A., Yang Y. G., He C. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Jia G., Pang X., Wang R. N., Wang X., Li C. J., Smemo S., Dai Q., Bailey K. A., Nobrega M. A., Han K. L., Cui Q., He C. Nat. Commun. 2013;4:1798–1806. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Divon M. S., Ungar L., Osenberg S., Cesarkas K., Hirsch J. J., Amariglio N., Kupiec M., Sorek R., Rechavi G. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]; (b) Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon R. A. Top. Curr. Chem. 1993;164:21–34. [Google Scholar]
- Arai H., Berlett B. S., Chock P. B., Stadtman E. R. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10472–10477. doi: 10.1073/pnas.0504685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Song H., Li J., Jiang M., Li Y., Zhou J., Guo Z. Biochemistry. 2012;51:4580–4589. doi: 10.1021/bi300486j. [DOI] [PubMed] [Google Scholar]
- Meguro H., Akasaka K., Ohrui H., Becke A. D. Methods Enzymol. 1990;186:157–161. doi: 10.1016/0076-6879(90)86105-5. [DOI] [PubMed] [Google Scholar]
- Madugundu G. S., Cadet J., Wagner J. R. Nucleic Acids Res. 2014;42:7450–7460. doi: 10.1093/nar/gku334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Schibler U., Kelley D. E., Perry R. P. J. Mol. Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]; (b) Wei C. M., Moss B. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- Chance B., Sies H., Boveris A. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- Babior B. M., Woodman R. C. Semin. Hematol. 1990;27:247–259. [PubMed] [Google Scholar]
- (a) Bienvenu C., Wagner J. R., Cadet J. J. Am. Chem. Soc. 1996;118:11406–11411. [Google Scholar]; (b) Cao H., Wang Y. Nucleic Acids Res. 2007;35:4833–4844. doi: 10.1093/nar/gkm497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babior B. M., Woodman R. C. Semin. Hematol. 1990;27:247–259. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.