Abstract
Peroxisomes participate in lipid metabolism, and are a major source of ROS in the cell. Their importance in cellular energy balance and redox homeostasis is well-established, as is the need to maintain peroxisome homeostasis to prevent pathologies associated with too few, or too many, of these organelles. How cells regulate peroxisome number has remained somewhat elusive. Recently, the tumor suppressors ATM and TSC, which regulate mTORC1 signaling, have been localized to peroxisomes. When activated by peroxisomal ROS, ATM signals to TSC to repress mTORC1 signaling and increase autophagic flux in cells, and also phosphorylates the peroxisomal protein PEX 5 to target peroxisomes for selective autophagy (pexophagy), providing a mechanism for regulation of peroxisomal homeostasis using ROS as a rheostat.
Peroxisomes: The “Late Bloomers” of the Cell Biology World
Peroxisomes were the last of the major organelles to be discovered following identification by De Duve in the late 60s [1,2], and only very recently found their place in the world of cell signaling. Similar to mitochondria, peroxisomes are capable of autonomous replication, although unlike mitochondria, peroxisomes do not contain their own DNA. Peroxisomes are highly metabolic organelles involved in several key cellular functions, including bile acid synthesis, glyoxylate and dicarboxylate metabolism, D-amino acid metabolism and β-oxidation of branched and very long chain fatty acids (VLCFAs). Importantly, D-amino acid metabolism and peroxisomal β-oxidation result in the production of reactive oxygen and nitrogen species (ROS and RNS) [3,4], which in excess, can cause cellular damage, and trigger catabolic functions such as autophagy [5–7]. It has been estimated that peroxisomes contribute approximately 35% of the ROS generated in the cell [8]. As autonomously replicating organelles, maintaining the balance between peroxisome biogenesis (to support key metabolic processes), and degradation (to limit excess ROS production) is critical for normal cellular homeostasis.
Maintaining Peroxisome Homeostasis
Excess ROS has been linked to over 150 diseases, including atherosclerosis, diabetes, cancer, and neurodegenerative diseases [9,10]. To prevent excessive production of ROS, cells must maintain peroxisome homeostasis by balancing peroxisome biogenesis with degradation. Peroxisome biogenesis occurs by importation of proteins to the peroxisome by import receptors that recognize their cargo via peroxisome targeting signal (PTS) sequences in a process very analogous to nuclear localization via nuclear localization signals. Peroxisome proteins (PEX proteins) such as PEX5 that function as import receptors, recognize PTS sequences in proteins destined for the peroxisome, and deliver them to this organelle [11,12]. These peroxisome import receptors are essential for the assembly of functional peroxisomes [11]. Mutations in these import receptors cause defects in peroxisome biogenesis and/or an absence of peroxisomes. Such peroxisome biogenesis disorders (PBDs) [11,13] differ in severity, the worst being Zellweger syndrome, which results in a complete absence of functional peroxisomes, and is lethal in the first months of life [13]. While the importance of maintaining peroxisome homeostasis is clear, mechanisms that specifically, and appropriately, allow the cell to recognize and remove excessive or aberrantly functioning peroxisomes to prevent pathologies associated with too few or too many of these organelles have remained elusive.
Autophagy is Important for Maintaining Peroxisome Homeostasis
Autophagy is a catabolic process in which cells deliver, in-bulk, cytoplasmic components for degradation to the lysosome. Autophagy plays a pivotal role in cell survival during starvation, and also participates in normal cellular functions, including selective autophagy of organelles. Selective autophagy of peroxisomes (pexophagy) is thought to be the major pathway by which excess peroxisomes are eliminated [14–18]. Selective autophagy is accomplished via adapter proteins involved in target recognition and recruitment of the phagophore membrane. These adapters include p62 and NBR1, which contain both an LC3-interacting region [17] that binds to LC3-associated with the nascent phagophore, and a ubiquitin-associated [17] domain that binds to monoubiquitinated lysine residues in the target [19]. The autophagy adapters p62 and NBR1 have both been implicated in autophagy of peroxisomes [20,21], although the peroxisomal proteins recognized by these adapters, and the signaling pathways responsible for regulation of pexophagy, have remained elusive.
Cell Signaling Pathways that Localize and Function at the Peroxisome
Until recently, the best-known connection between cell signaling and peroxisome biology was the regulated transcription of genes required for peroxisome biogenesis [22,23]. For example, peroxisome proliferator activated receptors (PPARs) bind ligands, such as the hypolipidemic thioldiozinediones (TZDs), to transcriptionally upregulate genes that promote peroxisome biogenesis. Studies investigating the “peroxisome proteome” have characterized over 85 different proteins found in peroxisomes using combined biochemical fractionation and mass specrometry approaches [24–26]. However, these studies have primarily identified enzymes involved in metabolic processes carried out by peroxisomes, rather than proteins specifically linked to canonical cell signaling pathways, although several kinases involved in metabolism, such as mevalonate and phosphomevalonate kinase, have been determined to be peroxisomal proteins [27]. The first report about peroxisome as signaling organelles appear by Dixit et al., in which they demonstrated that mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, Cardif, or VISA) localized at peroxisome apart from mitochondria and plays role in antiviral innate immunity [28]. The same group further expanded the role of peroxisome in innate immune response and reported that peroxisomes increases expression of type III interferon in response to diverse pathogenic stimuli [29].
Our group has now shown that the peroxisome is indeed an important a site for cross-talk between signaling pathways in which the TSC-2 (tuberous sclerosis complex 2) and ATM (ataxia telangiectasia mutated) tumor suppressors participate. An important function for ATM outside the nucleus was established by our group in 2010, when we reported that cytoplasmic ATM was activated in response to oxidative stress [30]. In this report, we showed that ATM phosphorylated LKB in response to exogenous or endogenous ROS, and later reactive nitrogen species (RNS) [31] to activate AMPK and TSC2. As a result, mTORC1 signaling was shut off, decreasing protein synthesis (due to decreased phosphorylation of ribosomal S6 kinase) and activating autophagy (mTORC1 is a known repressor of autophagy). These data established a new function for ATM in the cytoplasm, and identified a new signaling pathway in which both ATM and TSC2 participated to regulate autophagy in response to oxidative stress.
These early studies were soon followed by data that the TSC signaling node (TSC1, TSC2 and Rheb) was localized to the peroxisome [32]. Peroxisome targeting sequences were identified on both TSC1 and TSC2, which if mutated, abrogated their localization to the peroxisome. Data were also obtained that when localized to the peroxisome, the TSC tumor suppressor was activated by peroxisomal ROS, using drugs such as fibrate Wy-14643, which increase ROS production by peroxisomes [32]. In the liver, PPAR activation by fibrates results in an imbalance between PPAR-mediated transcription of peroxisomal ROS generating enzymes and ROS scavenging enzymes, leading to elevated production of peroxisomal ROS. For example, in response to fibrates, ROS generating enzymes, such as fatty acyl CoA oxidase, are increased in the rodent liver 10–30 fold, whereas ROS scavenging enzymes, such as catalase, are upregulated only 1–2 fold [4]. This elevated ROS is thought to contribute to hepatocarcinogenesis observed in response to fibrates [33,34], a hallmark of the adverse affect of these drugs in rodent models. As expected, when TSC2 was activated by peroxisomal ROS, repression of mTORC1 occurred, which increased autophagic flux in the cell.
Most recently, we have shown that ATM localization to the peroxisome mediates activation of the TSC tumor suppressor and specifically targets peroxisomes for pexophagy in response to ROS [35]. An initial report from the Watters group had localized ATM to the peroxisome, and identified a putative peroxisome import signal at the carboxyterminus of this kinase [36]. We confirmed ATM localization to the peroxisome, and went on to show this was the site for a functional interaction between the ATM and TSC tumor suppressors. When activated by peroxisomal ROS, ATM signals via LKB1 and AMPK to phosphorylate and activate TSC2, repressing mTORC1 and increasing autophagic flux. Importantly, ATM also phosphorylates the peroxisomal protein PEX5 on peroxisomes at S141, triggering PEX5 ubiquitination by the peroxisomal E3-ligase, PEX2/10/12. This ubiquitination of PEX5 at L209 creates a binding site for the autophagy adapter protein p62, which recognizes ubiquitinated peroxisomes via its ubiquitin-binding domain and tethers them to the autophagophore via its LC3-binding domain to target them for pexophagy.
A New Era for Cell Signaling in Peroxisome Biology
The discovery that the TSC and ATM signaling nodes localize and function at the peroxisome opens new opportunities for understanding how the biology of this organelle is regulated by these, and possibly other, cell signaling pathways. In particular, ATM phosphorylation of PEX5 opens the possibility that phosphorylation of this import receptor by ATM, or possibly other kinases, may regulate peroxisomal biogenesis as well as destruction. Furthermore, since ATM activation of TSC2 is mediated by the LKB1 and AMPK kinases, it is possible that these kinases may also function at the peroxisome to phosphorylate peroxisomal proteins resident at this organelle.
Figure 1.
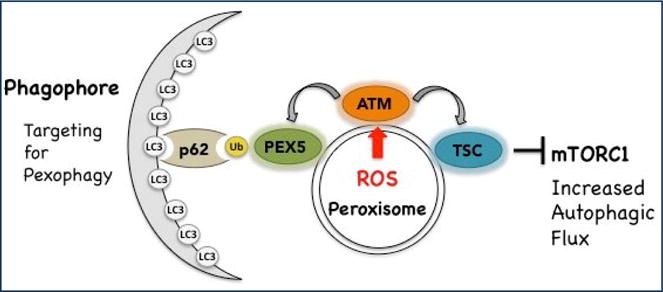
Schematic diagram depicting the localization of ATM and TSC at peroxisome. In response to peroxisomal ROS, ATM is activated and signals through TSC to suppress mTORC1. It also phosphorylates PEX5, leading to PEX5 ubiquitination and selective autophagy (pexophagy). Targeting of peroxisomes for pexophagy is mediated by binding of the adapter protein p62 to ubquitinated PEX5 and LC3 on the phagophore forming the nascent autophagosome.
The TSC and ATM Tumor Suppressors.
TSC2 functions as a negative regulator of mTORC1 signaling. TSC2 and its activation partner, TSC1 form a GTPase activating protein (GAP) that resides at endomembranes and regulates Rheb, which is required for mTORC1 signaling. Thus, TSC1/2 functions as a “brake” on mTORC1 signaling. The TSC tumor suppressor itself is regulated by upstream signals that can activate or repress its GAP activity. TSC is activated by AMP kinase (AMPK) phosphorylation in response to energy stress, and inactivated when TSC2 is phosphorylated by other kinases in response to growth factor signaling, such as AKT. AKT inactivates TSC by creating a 14-3-3 binding site, which removes TSC2 from the membrane, sequestering it in the cytosol and relieving inhibition of Rheb.
ATM is a DNA repair kinase, and “first responder” to double-strand breaks in DNA. When activated by DNA damage, ATM triggers several downstream signaling cascades that initiate DNA repair, cell cycle arrest and apoptosis. Although ATM’s role in the nucleus as a DNA repair kinase is well-known, data have begun to emerge that this kinase also plays an important, albeit poorly defined, role in cellular metabolism. ATM regulates cellular ROS level and plays role in carbon metabolism, adipocyte differentiation and glucose homeostasis, insulin resistance, cardiac remodeling and induction of autophagy. Due to its role in various metabolic activities, ATM is emerging as a therapeutic target for various diseases such as diabetes, cancer and neuronal degeneration.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant R01 CA143811 to C.L.W., and the Robert A. Welch Endowed Chair in Chemistry (BE-0023).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1**.de Duve C. The peroxisome in retrospect. Annals of the New York Academy of Sciences. 1996;804:1–10. doi: 10.1111/j.1749-6632.1996.tb18603.x. Informative retrospective on peroxisome biology from the eminent cell biologist who discovered these organelles. [DOI] [PubMed] [Google Scholar]
- 2.Leighton F, Poole B, Beaufay H, Baudhuin P, Coffey JW, Fowler S, De Duve C. The large-scale separation of peroxisomes, mitochondria, and lysosomes from the livers of rats injected with triton WR-1339. Improved isolation procedures, automated analysis, biochemical and morphological properties of fractions. J Cell Biol. 1968;37:482–513. doi: 10.1083/jcb.37.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. An comprehensive review of how pharmacologic agents that target the PPAR receptor increase peroxisome activity and production of reactive oxygen species that can damage cells. [DOI] [PubMed] [Google Scholar]
- 4.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochimica et biophysica acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Alexander A, Walker CL. Differential localization of ATM is correlated with activation of distinct downstream signaling pathways. Cell cycle. 2010;9:3685–3686. doi: 10.4161/cc.9.18.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell death and differentiation. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 7.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivashchenko O, Van Veldhoven PP, Brees C, Ho YS, Terlecky SR, Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Molecular biology of the cell. 2011;22:1440–1451. doi: 10.1091/mbc.E10-11-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fransen M, Nordgren M, Wang B, Apanasets O. Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim Biophys Acta. 2012;1822:1363–1373. doi: 10.1016/j.bbadis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weller S, Gould SJ, Valle D. Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet. 2003;4:165–211. doi: 10.1146/annurev.genom.4.070802.110424. [DOI] [PubMed] [Google Scholar]
- 14.Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 15.Farre JC, Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Till A, Lakhani R, Burnett SF, Subramani S. Pexophagy: the selective degradation of peroxisomes. Int J Cell Biol. 2012;2012:512721. doi: 10.1155/2012/512721. Overview of the process of pexophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 21.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Rout MP. The peroxisome: a production in four acts. The Journal of cell biology. 2008;181:185–187. doi: 10.1083/jcb.200803126. A brief overview of signaling pathways and kinases related to peroxisome biogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleem RA, Knoblach B, Mast FD, Smith JJ, Boyle J, Dobson CM, Long-O’Donnell R, Rachubinski RA, Aitchison JD. Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. The Journal of cell biology. 2008;181:281–292. doi: 10.1083/jcb.200710009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronemeyer T, Wiese S, Ofman R, Bunse C, Pawlas M, Hayen H, Eisenacher M, Stephan C, Meyer HE, Waterham HR, et al. The proteome of human liver peroxisomes: identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PLoS One. 2013;8:e57395. doi: 10.1371/journal.pone.0057395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiese S, Gronemeyer T, Ofman R, Kunze M, Grou CP, Almeida JA, Eisenacher M, Stephan C, Hayen H, Schollenberger L, et al. Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol Cell Proteomics. 2007;6:2045–2057. doi: 10.1074/mcp.M700169-MCP200. [DOI] [PubMed] [Google Scholar]
- 26*.Schluter A, Fourcade S, Domenech-Estevez E, Gabaldon T, Huerta-Cepas J, Berthommier G, Ripp R, Wanders RJ, Poch O, Pujol A. PeroxisomeDB: a database for the peroxisomal proteome, functional genomics and disease. Nucleic Acids Res. 2007;35:D815–822. doi: 10.1093/nar/gkl935. An important resource for predicting peroxisome targeting sequences in proteins, and catalog of proteins known to localize to this organelle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.http://www.peroxisomedb.org.
- 28**.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668681. doi: 10.1016/j.cell.2010.04.018. Seminal findings that first reported a function for peroxisomes as a signaling platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. Describes a new cytoplasmic function for ATM activation in response to ROS that suppresses mTORC1 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2950–E2957. doi: 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nature cell biology. 2013;15:1186–1196. doi: 10.1038/ncb2822. First demonstration that a cell signaling pathway contained a node that resided at the peroxisome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misra P, Viswakarma N, Reddy JK. Peroxisome Proliferator-Activated Receptor-alpha Signaling in Hepatocarcinogenesis. Subcell Biochem. 2013;69:77–99. doi: 10.1007/978-94-007-6889-5_5. [DOI] [PubMed] [Google Scholar]
- 34.Yeldandi AV, Rao MS, Reddy JK. Hydrogen peroxide generation in peroxisome proliferatorinduced oncogenesis. Mutation research. 2000;448:159–177. doi: 10.1016/s0027-5107(99)00234-1. [DOI] [PubMed] [Google Scholar]
- 35**.Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. The first report that the ATM kinase functions at the peroxisome, and regulates pexophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Watters D, Kedar P, Spring K, Bjorkman J, Chen P, Gatei M, Birrell G, Garrone B, Srinivasa P, Crane DI, et al. Localization of a portion of extranuclear ATM to peroxisomes. The Journal of biological chemistry. 1999;274:34277–34282. doi: 10.1074/jbc.274.48.34277. The first report that ATM localizes to peroxisomes. [DOI] [PubMed] [Google Scholar]


