Abstract
Nonalcoholic fatty liver disease (NAFLD) is a growing epidemic paralleling the increase in obesity and diabetes mellitus seen in Western diet-consuming countries. As NAFLD can lead to life-threatening conditions such as cirrhosis and hepatocellular carcinoma (HCC), an understanding of factors that trigger its development and pathological progression is needed. Although by definition this disease is not associated with alcohol consumption, exposure to environmental agents that have been linked to other diseases might have a role in the development of NAFLD. Here, we focus on one class of these agents, endocrine-disrupting chemicals (EDCs), and their potential to influence the initiation and progression of a cascade of pathological conditions associated with fatty liver. Experimental studies have revealed several potential mechanisms by which EDC exposures might contribute to disease pathogenesis, including modulation of nuclear hormone receptor (NR) function and alteration of the epigenome. However, many questions remain to be addressed about the causal link between acute and chronic EDC exposure and the development of NAFLD in humans. Future studies that address these questions hold promise not only for understanding the linkage between EDC exposure and liver disease, but for elucidating the molecular mechanisms underpinning NAFLD and the development of new prevention and treatment opportunities.
Introduction
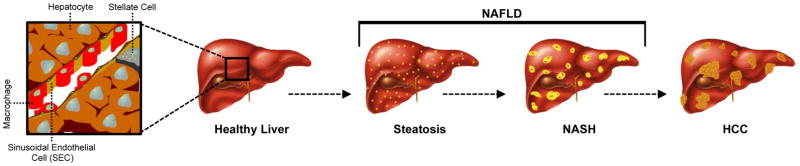
Nonalcoholic fatty liver disease (NAFLD) is characterized by simple, reversible hepatic steatosis (fatty liver) with or without additional macrophage infiltration and inflammation (steatohepatitis or nonalcoholic steatohepatitis (NASH), respectively), which can progress to irreversible fibrosis and life-threatening cirrhosis and hepatocellular carcinoma (HCC) (FIG. 1)1–6. NAFLD is the most common chronic liver disease worldwide and is especially prevalent in high-fat diet (HFD) consuming countries such as the USA7. Since its first diagnosis in the 1980s1, the incidence of NAFLD incidence has reached 30% in the USA8, paralleling increases in obesity9 (35% of adults in the USA have a body mass index (BMI) >30 kg/m2), risk of HCC6 and cardiovascular disease10, 11. Although NAFLD is most commonly spurred by overnutrition coupled with a lack of exercise, environmental factors might also contribute to the rapid rise in both obesity and NAFLD prevalence. Indeed, diets rich in fructose have now been implicated in the development of NAFLD development1.
Figure 1. Pathophysiology of NAFLD progression.
From left to right: a healthy liver is presented, that upon presentation of ‘risk factors’ such as obesity, fructose consumption and/or exposure to endocrine-disrupting chemicals, leads to lipid accumulation (depicted as small yellow dots) in the liver (steatosis), which is the first (reversible) stage of nonalcoholic fatty liver disease (NAFLD). Activation and/or recruitment of macrophages to the liver leads to nonalcoholic steatohepatitis (NASH) and eventual fibrosis. Importantly, progression to NASH and the development of fibrotic and/or cirrhotic lesions (not pictured) represent an irreversible stage of liver disease. Left untreated, a subset of cases culminates in the development of neoplastic events that give rise to hepatocellular carcinoma (HCC), with or without cirrhosis, as the final endpoint of hepatic disease progression.
One class of environmental risk factors that might promote NAFLD is chemicals that can disrupt or alter the function of endocrine and metabolic organs such as the liver, which is the central organ controlling lipid homeostasis. These chemicals are termed endocrine-disrupting chemicals (EDCs)12, 13, or more recently, metabolism-disrupting chemicals (MDCs)12. The timely interest in these compounds as potential stimulators of obesity and NAFLD, along with other risk factors such as a HFD and fructose, stems from the following: many EDCs have been mass produced over the past four decades, driven by their widespread use (for example, bisphenol A (BPA) and phthalates in plastic production)12, 13; animal ‘intervention’ studies have suggested that EDCs might cause increased adiposity or NAFLD in exposed animals12, 13; and direct measurement of EDC levels in human blood and urine has shown near ubiquitous exposures (for example, ≥95% of people in the USA have detectable levels of BPA in their urine14).
In this Review, we highlight the literature bridging the two ‘hot’ topics of NAFLD and EDCs, and posit that early-life exposure to EDCs might represent an unappreciated driver of NAFLD development and progression in adulthood. We describe basic liver physiology along with the molecular pathways that affect hepatic lipid homeostasis and how they impact NAFLD development. We then discuss various classes of EDCs that perturb hepatic lipid levels, bind to nuclear hormone receptors (NRs) and recruit transcriptional coregulators to alter the expression of lipid homeostasis genes and/or activate kinase signalling pathways, promote NAFLD in rodent models and are associated with human NAFLD, impact epigenetic modifications (that is, DNA methylation and histone modifications) and, in the setting of early-life exposures, increase susceptibility to obesity and NAFLD in adulthood.
Liver physiology
The liver is the largest glandular organ in the body. To perform its many and diverse functions, the liver relies on a compartmentalized structure. Liver architecture is composed of small hexagonal lobules, wherein each lobule is connected by a network of sinusoids formed by specialized sinusoidal endothelial cells (FIG. 1). Adjacent to the sinusoid resides hepatic stellate cells that function as a repository for lipids and vitamin A. The sinusoids traverse a collection of two other primary cell types: hepatocytes, which represent the parenchymal cell type of the liver, and Kupffer cells that represent the resident macrophage population15.
Liver response to environmental cues
Although the many different cell types in the liver demonstrate considerable phenotypic and functional heterogeneity, their cooperative molecular contributions endow the liver with the ability to interpret and respond to a number of environmental stimuli. In some situations, these environmental responses have a beneficial effect on liver function and overall health. For example, in the setting of a starvation environment in utero, the liver develops in such a way that the physiological set points for several liver functions, including gluconeogenesis, are primed for an adult environment in which nutrients are in short supply. In this context, modulation of fetal development can confer a survival advantage on offspring exposed to an environment in which resources are likely to be limited, resulting in a thrifty phenotype16. However, individuals programmed with a thrifty phenotype in utero (for example as a result of famine or placental insufficiency), who go on to develop in a nutrient-rich environment instead of the ‘anticipated’ nutrient-poor environment, are more prone to metabolic disorders. For example, prenatal exposure to famine (especially in late gestation) during the Dutch Hunger Winter (1944–1945) was associated with decreased glucose tolerance in adults17. In the same cohort, prenatal exposure to famine was associated with a more atherogenic lipid profile than those who were not exposed to famine in utero18. Early-life exposure to famine during the Great Chinese Famine (1958–1961) was associated with a sex-specific increase in the prevalence of moderate–severe NAFLD in adulthood, providing direct evidence of the link between poor fetal nutrition and perturbed liver function19.
Basis of NAFLD
As the epicenter for metabolic homeostasis, the liver performs a myriad of functions including haematopoiesis and turnover of red blood cells20, production of enzymes for blood clotting, hormone biosynthesis and turnover, protein and bile synthesis, drug metabolism, lipid metabolism, glycogen storage and release, and gluconeogenesis21. Should any of these key functions become compromised (especially lipid metabolism), several disease sequelae can result. Initial excess lipid accumulation in the liver (steatosis; a reversible step) can progress to NASH (characterized by macrophage infiltration and inflammation) and then to fibrosis and/or cirrhosis (irreversible); a subset of the latter cases advance to HCC. Together, both the ‘early’ presenting conditions of hepatic steatosis and NASH constitute NAFLD (FIG. 1)5, 10.
NAFLD is a growing problem in HFD-consuming countries such as the USA. For example, analysis of fatty liver (as assayed by ultrasound) in viral hepatitis negative patients in the U.S. National Health and Nutrition Examination Survey (NHANES) has suggested that NAFLD increased from 18% in 1998–1991 to 31% in 2011–20128. At 30%, this prevalence represents ~75–100 million people in the USA11. The large increase in NAFLD incidence over the past two decades has been accompanied by an increased risk of HCC6 and resultant deaths, as well as cardiovascular disease10, 11. Importantly, NASH, the inflammatory form of NAFLD, is currently the second leading cause of liver disease in adults scheduled for liver transplantation in the USA10. Thus, understanding the factors that trigger NAFLD is of the utmost importance to curbing the rising need for liver transplantation and later-stage lethal events such as HCC. Among the environmental factors that contribute to the development of NAFLD, early-life exposures to EDCs might represent an unappreciated risk factor to consider in addition to obesity and type 2 diabetes mellitus (T2DM)22, due to their potential to alter lipid homeostatic ‘set points’ that favour NAFLD.
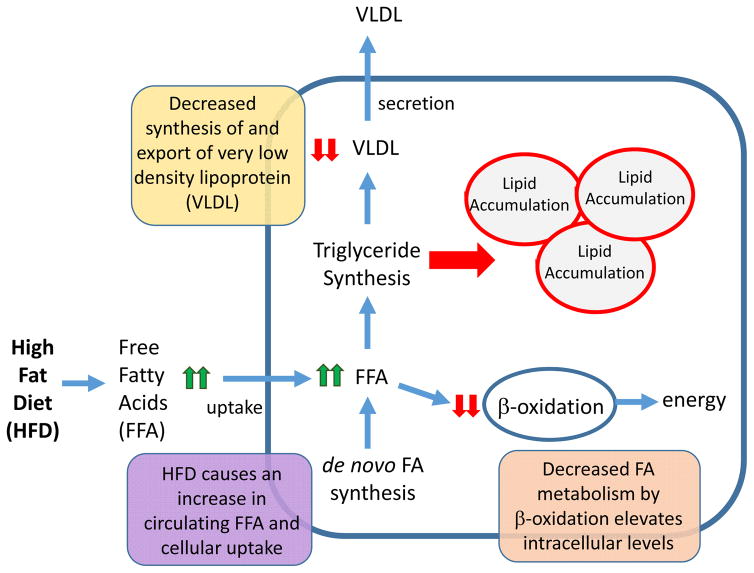
Hepatic lipid homeostasis is maintained by hepatocyte uptake and de novo synthesis of free fatty acids (FFAs), FFA disposal by oxidation or de novo triglyceride synthesis, and export of triglycerides from hepatocytes as very low density lipoprotein (VLDL) (FIG. 2). Fatty liver or steatosis develops when hepatic uptake of FFAs exceeds their oxidation and secretion as triglycerides. The consequence of aberrant lipid accumulation in the liver imposes differential cellular effects on subpopulations of hepatic cell types. In hepatocytes, uptake and/or de novo synthesis of fatty acids are disproportionally increased relative to fatty acid oxidation. This imbalance stimulates triglyceride synthesis to dispose of the excess FFAs. As triglyceride synthesis outpaces the capacity for VLDL synthesis and export, triglycerides accumulate within hepatocytes, resulting in steatosis23 (FIG. 2). Although triglycerides are not inherently hepatotoxic, aberrant hepatocyte processing of FFAs activates resident and infiltrated macrophages through Toll-like receptor 4 pathways to initiate a pro-inflammatory cascade that contributes to NAFLD3, 24. The chemokine and angiogenic signals produced from infiltrated macrophages leads to dysregulation of sinusoidal endothelial cells that form the fenestrated vasculature of the liver25. Excess hepatic lipid also serves as an activation signal for the normally quiescent stellate cells that initiates the fibrotic process that often accompanies more severe forms of liver disease such as NASH and HCC3, 4, 23 (FIG. 1).
Figure 2. Altered hepatic metabolic pathways leading to NAFLD.
The liver is central to the maintenance of whole-body lipid homeostasis. Mechanistically, uptake of dietary fats is facilitated by release of bile acids that are synthesized in the liver and secreted by the gall bladder into the intestine. Bile salts emulsify fat, creating free fatty acids (FFAs) and monoglycerides, which are rapidly absorbed by enterocytes of the intestine. In the intestine, FFAs and monoglycerides are resynthesized into triglycerides, which are packaged into chylomicrons and are taken up by the liver via receptor-mediated endocytosis. The liver is also is responsible for converting carbohydrates and protein into FFAs, which are packaged into triglycerides and exported from the liver as VLDL. The liver is also the primary source of β-oxidation that serves to metabolize FFAs to produce energy in the form of ATP, as well as to generate ketone bodies that are used as an alternative fuel source during periods of fasting. Altogether the balance between lipid uptake and release, triglyceride synthesis and β-oxidation helps to preserve energy homeostasis in the liver. Disruption of these processes by a high-fat diet (HFD) is accompanied by aberrant lipid accumulation in the liver, which leads to a cascade of pathologies ranging from steatosis to hepatocellular carcinoma. Endocrine-disrupting chemicals (EDCs) can also promote nonalcoholic fatty liver disease (NAFLD), either alone or with a HFD, by increasing FFA uptake, increasing de novo lipogenesis, decreasing triglyceride export via VLDL, and/or decreasing FFA β-oxidation.
Environmental exposures
Established risk factors for NAFLD in humans include obesity and insulin resistance or T2DM22, 26, 27, as well as specific genetic mutations that result in increased lipid synthesis and uptake, and/or decreased FFA oxidation and triglyceride export28. However, such germ-line mutations are rare and would not explain the vast majority of NAFLD cases, which points to the involvement of environmental factors in the development of this disease. For example, dietary intake of saturated fat, trans-fatty acids, carbohydrate and simple sugars (fructose and sucrose) might contribute to aberrant hepatic lipid accumulation26. Interestingly, some polyunsaturated fatty acids (PUFAs), such as n-3 PUFAs like α-linolenic acid seem to reduce NAFLD3. However, true causal relationships between these nutrients and NAFLD remain to be fully determined. Of these dietary factors, fructose has been reported to be a risk factor for human NAFLD1, 3, 29–31, but whether fructose alone (for example in the absence of obesity) can trigger or facilitate the progression of NAFLD is still debatable26. Finally, maternal diet might impact offspring susceptibility to NAFLD via changes in the neonatal or infant-gut microbiota, although again, causality remains to be firmly established2.
Of the various environmental chemical exposures that might negatively affect the liver, the growing class of EDCs has gained attention for their ability to perturb hepatic function. EDCs are defined as compounds that exert adverse health effects secondary to disruption of the endocrine system. Structurally, EDCs comprise a wide range of both natural and manmade substances that are derived from persistent organic pollutants (POPs; that is, dioxins, benzo[a]pyrene and polychlorinated biphenyls (PCBs)), organochlorines (such as dichlorodiphenyltrichloroethane (DDT) and its metabolite dichlorodiphenyldichloroethylene (DDE)), plasticizers (such as bisphenol A (BPA)), phthalates (such as di-2-ethylhexyl phthalate (DEHP)), organotins (such as tributyltin (TBT)), polyfluoroalkyls (such as perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS)) and other pesticides (such as cypermethrin (CYP), atrazine (ATZ), and carbendazim)13, 32. Mounting evidence suggests that these EDCs, many of which are also sequestered and metabolized in the liver, might contribute to the development of NAFLD.
Relevant to the NAFLD–obesity linkage, several of these EDCs including BPA, TBT, PCBs, phthalates, PFOA and PFOS are classified as ‘obesogens’, based on animal studies wherein early-life exposures promote obesity later in adulthood33. The original obesogen hypothesis proposed by Grun and Blumberg34 primarily focused on the effects of these exposures on adipocytes (that is, fat storage) and pancreatic β cells (that is, insulin secretion)34, 35. In rodent models, perinatal exposure to obesogens increases fat mass in both male and female offspring32, 34, 36. Thus, one pathway by which EDCs might impact NAFLD is through peripheral effects of obesity on adipose dysfunction, deregulation of the satiety axis in the hypothalamus, as well as liver cell autonomous effects.
Liver fat metabolism and EDCs
Nuclear receptors and coregulators
The vast majority of EDCs exert their activity as endocrine disruptors via their ability to bind NRs and thus act as NR agonists or antagonists. The liver expresses an extensive repertoire of NRs that have important roles in hepatic lipid metabolism (FIG. 3). When activated by a ligand, the classic activity of NRs involves docking at response elements in the promoter or enhancer region of target genes followed by binding of steroid receptor coactivator (SRC) complexes that recruit additional coregulators with histone-modifying enzymatic activities, such as acetylases and methylases37, 38. The concerted action of these coregulators leads to transactivation of target gene programs37. Experience with selective oestrogen receptor (ER) modulators (SERMs)39 as well as EDCs40 showed that for agonists, NRs adopt an active conformation and interact with coactivators, whereas for antagonists, receptors adopt an inactive conformation that recruits corepressors. In addition to classic genomic activity, ligand-dependent NR signalling also occurs in the cytoplasm, so-called ‘non-genomic’ signalling41, 42. This extra-nuclear NR signalling results in the activation of kinases and downstream signalling pathways, such as protein kinase B (also called AKT) and mitogen-activated protein kinases (MAPKs) that mediate biological responses independent of NR nuclear localization. Non-genomic signalling is characterized by its rapid action, and occurs independent of RNA or protein synthesis.
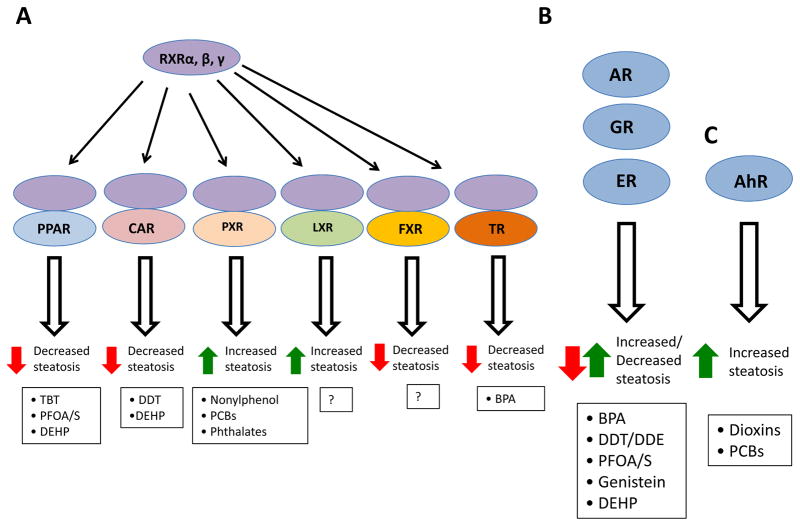
Figure 3. NR-mediated effects of EDCs on fatty liver development.
a The NR1 subfamily of nuclear hormone receptors (NRs) heterodimerize with retinoid X receptors (RXRs) to either promote (pregnane X receptor (PXR) or liver X receptor (LXR)) or inhibit (peroxisome proliferator-activated receptors (PPARs), constitutive androstane receptor (CAR), farnesoid X receptor (FXR), and thyroid receptors (TRs)) hepatic steatosis upon binding their naturally occurring agonist ligands. Select endocrine-disrupting chemicals (EDCs) known to bind these NRs and affect their activity are depicted at the bottom of the figure. For example, tributyltin (TBT) binding RXR–PPAR enhances steatosis, unlike natural free fatty acid ligands. LXR activates lipogenic genes upon binding its natural ligands (oxysterols) and promotes steatosis, but whether its activity is modulated by specific EDCs is currently unclear. b Another major class of NRs that bind EDCs is the steroid receptors such as the androgen receptor (AR), glucocorticoid receptor (GR) and oestrogen receptor (ER). Steroid hormones can either increase (glucocorticoid) or decrease (oestrogen and androgen) hepatic steatosis. Select EDCs known to bind these NRs and affect their activity are depicted at the bottom of the figure. c Aryl hydrocarbon receptor (AhR) represents the third major NR effector of EDC action in the liver. AhR binds EDCs, such as dioxins and polychlorinated biphenyls (PCBs), leading to enhanced steatosis. BPA, bisphenol A; DDE, dichlorodiphenyldichloroethylene; DDT, dichlorodiphenyltrichloroethane; DEHP, di-2-ethylhexyl phthalate; PFOA, perfluorooctanoic acid; PFOS, perfluorooctane sulfonate.
Several NRs have roles in non-genomic signalling. Perhaps the most prominent of these are the steroid hormone receptors — oestrogen receptors (ERα and ERβ), androgen receptor (AR) and progesterone receptor (PR)43, 44. In addition to these steroid receptors, accumulating evidence suggests that other NRs (for example, peroxisome proliferator-activated receptor γ (PPARγ)45, retinoid X receptor α (RXRα)46, truncated thyroid receptor α (TRα) isoforms44, 47 and retinoic acid receptors (RARα and RARγ48)) might also signal via a similar mechanism. Non-genomic signalling does have the potential to affect the genome and alter transcription, as kinases activated in non-genomic signaling pathways can phosphorylate and regulate the activity of epigenomic programmers, transcription factors and/or their associated coregulators, even in the absence of a liganded NR interacting with target genes.
Transcriptional profiling of all major isoforms of the 49 known NRs from isolated livers of 129/SvJ and C57BL/6J male mice revealed hepatic expression of 39 NRs49. A similar number of NRs (35) were identified by mass spectrometric profiling of NRs bound to their cognate DNA response elements in hepatocytes isolated from C57BL/6J male and female mice50. Of these, the NR1 subfamily that heterodimerize with retinoid X receptors (RXRα, RXRβ and RXRγ), which includes the peroxisome proliferator-activated receptors α, β and γ (PPARs), the liver X receptors α and β (LXRs), farnesoid X receptor α (FXRα), the constitutive androstane receptor (CAR), the pregnane X receptor (PXR) and thyroid receptors (TRs), have been implicated in modulation of NAFLD (FIG. 3)51–53. Activation of PPARα by its natural ligands (FFAs) increases the expression of genes encoding enzymes involved in FFA oxidation (such as CPT1 and ACOX1) and hence leads to decreased hepatic steatosis54. Similarly, xenobiotics bind and activate CAR and PXR; however, species-specific differences clearly exist55, 56. For example, 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) activates the mouse but not the human CAR57,58, while 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) activates the human, but not mouse, CAR55, 56, 59. Additionally, pregnenolone 16α-carbonitrile (PCN) preferentially activates mouse and rat PXRs over human PXR57, 60–63, but rifampicin and SR12813 are specific ligands for human but not mouse PXR57, 60–63. CAR and PXR activation decreases and increases hepatic lipid accumulation, respectively64, 65.
LXRs bind oxysterols and activate lipogenic gene programs (for example, FAS and SREBP1) that can lead to lipid accumulation in the liver66. FXR functionally responds to bile acids and induces the expression of bile acid exporter genes and NR0B2, which encodes the NR short heterodimer partner (SHP) that represses SREBP1 expression67, thereby decreasing hepatic steatosis. Due to this action, FXR agonists such as obeticholic acid and INT-767 are currently being tested in human NAFLD and NASH clinical trials2. Thyroid hormone T3, as well as synthetic TR agonists, reduce hepatic steatosis in male Fischer 344 rats fed a diet deficient in choline and methionine68 and in diabetic mice (for example, ob/ob)69, suggestive of their potential clinical usefulness. Activation of a NR that can trigger increased steatosis does not always result in increased inflammation. For example, activation of PXR induces lipogenic gene expression (for example, SCD1) and suppresses FFA-oxidation enzyme gene expression (for example, CPT1)70, yet PXR activation can also suppress the expression of inflammatory cytokines in hepatocytes treated with lipopolysaccharide71.
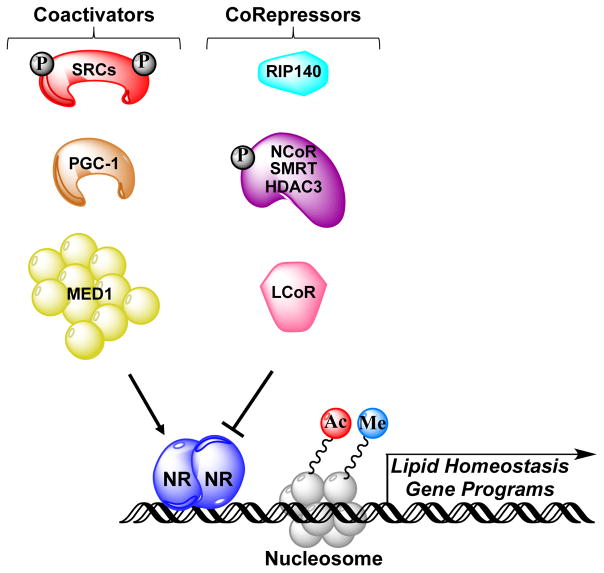
In addition to NRs that bind EDCs, coregulators that complex with liganded NRs might also have a role in NAFLD. The activity of NRs bound to endogenous ligands or EDCs is determined by the action of coregulator proteins that interact with these receptors. In the liver, a variety of coregulators have key functional roles in hepatic lipid metabolism via recruitment to ligand-activated NRs bound at genes involved in lipid homeostasis (FIG. 4). Examples of critical coactivators are the three SRC family proteins (SRC1, SRC2 and SRC3), PPARγ coactivators (PGC1α and PGC1β) and the Mediator complex subunit MED1. The key corepressors are the nuclear receptor corepressor (NCoR)–silencing mediator of retinoic acid and thyroid hormone receptor (SMRT)–histone deacetylase 3 (HDAC3) complexes and receptor-interacting protein 140 (RIP140)72.
Figure 4. Potential genomic mechanism of EDC action.
Once an endocrine-disrupting chemical (EDC) enters the liver, it is bound by specific nuclear hormone receptors (NRs). This action can either positively or negatively affect transcription of lipid homeostasis genes via specific EDC–NR complexes that recruit coactivators or corepressors to target genes. Key coactivators that modulate fatty liver progression include steroid receptor coactivators (SRCs), peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) and mediator of RNA polymerase II transcription subunit 1 (MED1), whereas nuclear receptor corepressor (NCoR)–silencing mediator of retinoic acid and thyroid hormone receptor (SMRT)–histone deacetylase 3 (HDAC3) complexes, receptor-interacting protein 140 (RIP140) and ligand-dependent corepressor (LCOR) act as corepressors. Coactivator complexes induce histone modifications associated with active gene transcription, such as acetylation (Ac) and methylation (Me), whereas corepressors generally utilize associated histone deacetylases or demethylases to remove these marks. SRCs and NCoR are subject to regulatory phosphorylation (P) events. Engagement of coregulators by EDC-bound NRs results in modulation of lipid homeostasis gene cassettes and/or reprogramming of the epigenome, which ultimately promotes NAFLD: for example, via enhanced lipogenesis gene expression and/or inhibition of free fatty acid-oxidation gene expression.
Studies in knockout mice clearly show that NR coactivators have an important role in the development of fatty liver. Genetic ablation of Src1 increased acylcarnitine levels in the fed-to-fasting transition, which is suggestive of an important role for SRC1 in regulating hepatic FFA-oxidation73. Both whole-body and liver-specific ablation of Src2 phenocopies a Von Gierke–like disease that is characterized by fasting hypoglycaemia, hepatic steatosis and increased circulating levels of triglycerides, cholesterol and FFAs74. Hepatic SRC3 mRNA and protein levels increase upon HFD feeding, and genetic ablation of Src3 protects against HFD-induced hepatic steatosis by reducing lipid accumulation and the accompanying inflammatory response75,76,77. Whole-body genetic ablation of Pgc1a/b (encoding PGC1α/β) results in increased hepatic steatosis, although liver-specific deletion is needed to confirm whether these effects are intrinsic to the liver78–80. Interestingly, PGC1α mediates the recruitment of the BAF60a subunit of the SWI/SNF chromatin-remodelling complex to PPARα-binding sites, which leads to transcriptional activation of FFA-oxidation genes; mice overexpressing BAF60a fed a HFD display reduced fatty liver81. Finally, MED1 is the key subunit of the Mediator complex that interacts with liganded NRs via its LxxLL motifs82. MED1 is required for HFD-induced hepatic steatosis as well as PPARγ-stimulated hepatic steatosis, as revealed by analysis of liver-specific Med1 knockout mice and Pparg hepatic overexpression by tail vein adenoviral injection83.
Studies in knockout mice highlight the importance of specific NR corepressors for modulating hepatic steatosis. For example, NCoR or SMRT associate with HDAC3 as part of a multi-subunit protein complex that functions generally as a NR corepressor84–86. Liver-specific ablation of Hdac3 or Ncor, but not Smrt, in mice results in hepatic steatosis87, 88. Whole-body Rip140 knockout mice are lean and resistant to HFD-induced obesity and hepatic steatosis89. Liver-specific deletion of Rip140 also reduced hepatic lipid levels in mice, which suggests that RIP140 serves as a corepressor of LXR-activated lipogenic genes such as FAS and SREBP190. Other studies have identified additional NR corepressors that regulate hepatic steatosis, such as ligand-dependent corepressor (LCOR) suppression of TRβ-induced lipogenic gene expression and hepatic steatosis in obese mice91, and small heterodimer partner interacting leucine zipper protein (SMILE) repression of LXRα-mediated SREBP1 gene expression and hepatic lipid accumulation92. Although the above mentioned animal ablation studies emphasize the importance of coregulators in modulating hepatic lipid homeostasis, information on which cell types in the liver are critical to the development of fatty liver is lacking, emphasizing the need for liver cell-type specific genetic ablation studies.
Like endogenous hormones, EDCs can activate both genomic and non-genomic actions of NRs, and induce posttranslational modifications that modulate NR and/or coregulator activity. For example, SRC3 is initially phosphorylated by oestradiol-activated kinases in the cytoplasm on a subset of conserved phosphorylation sites that subsequently leads to enhanced NR–SRC3 target gene transcription93,94. Mice harboring loss-of-function mutations in four of these conserved phosphorylation sites in SRC3 develop insulin resistance, dyslipidaemia, liver steatosis and accelerated hepatic tumorigenesis95. Furthermore, insulin (a mimic of the fed state) activates AKT resulting in phosphorylation of NCoR on Ser1460, which enhances its interaction with PPARα over LXRα, and results in repression of PPARα, decreased FFA-oxidation and enhanced hepatic lipogenesis96 (FIG. 4). Different structural classes of EDCs have been shown to modulate the activity of NRs expressed in the liver and associated with NAFLD, such as the NR1 subfamily NRs (PPARs, RXRs, PXR, CAR and TRs), steroid receptors (glucocorticoid receptor (GR), ERs and AR) and the aryl hydrocarbon receptor (AhR) (FIG. 3)32, 97, 98. POPs such as dioxins, benzo[a]pyrene and some PCBs bind and activate AhR (some also bind PPARγ and ERs); organochlorines such as DDT bind and activate CAR and ERα; the plasticizer BPA binds and activates ERs and GR, yet represses TR; organotins such as TBT bind and activate RXRs and PPARγ; polyfluoroalkyls such as PFOA and PFOS bind and activate ERs and PPARs; and phthalates such as DEHP bind and activate PPARs, CAR, PXR and GR32. Some of the EDC–NR interactions are of low affinity (for example, the equilibrium dissociation constant Kd of BPA:ERα is 0.2 μM99), whereas other interactions are much stronger (for example, the Kd of TBT:RXR or TBT:PPARγ are 12.5–20 nM100).
EDC–NAFLD link
EDC activation of NRs has been proposed to be an initiating event in the development of steatosis101, as well as promoting the transition of steatosis to steatohepatitis102. Several EDCs that function as ligands for the NRs described earlier have been shown to impact the liver and the development of NAFLD12,101,102. A 2015 review of 371 studies in federal databases suggested that 123 unique environmental chemicals are associated with NAFLD in rodents, with pesticides representing the majority (44%) and PCBs and dioxins the most potent based on lowest effect level103. To extend these data from ‘associations’ to ‘cause-and-effect’ relationships, we performed an extensive analysis of the literature with a focus on ‘interventional’ studies wherein rodents were exposed to an EDC (or mixture that might better represent environmental exposures) or vehicle control and a NAFLD phenotype scored (see Supplementary information S1 (table). These studies used several different strains of rats (for example, Sprague-Dawley, Wister, Fischer 344, Han/Wistar and Obese JCR (LA)-Leprcp (cp/cp)) and mice (for example, CD1, C57BL/6J, KM, Ldlr knockout, Apoe knockout, Std:ddY, BALB/c and ICR) exposed to different EDCs (for example, BPA, TBT, benzo[a]pyrene, dioxins (2,3,7,8-tetrachlorodibenzodioxin (TCDD) and hexachlorodibenzo-p-dioxin (HxCDD)), PCBs (77, 105, 126, 153, 126 + 118, 126 + 153, Aroclor 1260 or Aroclor 1254 (a mixture of up to 60 PCBs)), DDE, PFOA, PFOS, DEHP, pesticides (CYP, ATZ or carbendazim), or a ‘Northern contaminant mixture’ (22 compounds including 11 PCBs, DDE and PFOS)). Liver endpoints following exposure revealed increased hepatic lipid accumulation (assessed by histological analysis, Oil Red O staining or hepatic triglyceride measurements). Importantly, both perinatal (in utero) and adult animal EDC exposures showed signs of fatty liver development with different doses (see Supplementary information S1 (table)). Interestingly, combined treatment of rodents with one type of EDC followed by another class of EDC can modify the NAFLD phenotype. For example, pre-treatment of rats with TCDD led to the appearance of NAFLD, whilst DEHP reduced the TCDD-induced phenotype104. Similarly, the combination of TCDD and Aroclor 1254 seemed to enhance NAFLD in mice, compared with treatment with either EDC alone105. Although this approach has been applied in only a few experimental settings, the data are extremely relevant to humans, where exposure to more than one EDC during development and over the course of a lifetime may occur. Additional animal studies examining EDC mixtures are certainly warranted.
When combined with other risk factors for NAFLD (such as a HFD), EDCs generally exacerbate the NAFLD phenotype in exposed rodents. Two seminal studies reported that perinatal exposures to BPA (50–100 μg/kg per day) combined with a HFD after weaning (at postnatal day 21) led to male, but not female, offspring displaying more severe hepatic steatosis106, 107 and increased inflammation as well as mild fibrosis in the liver106. These data indicate that in addition to increasing hepatic lipid accumulation in the liver, EDC exposure might also trigger macrophage infiltration that can further contribute to the development of NASH (analogous to that already proposed for fructose1), although additional mechanistic studies are needed to adequately test this hypothesis. In terms of altered gene expression, the increased hepatic lipid accumulation observed with BPA treatment could be due to an imbalance of FFA uptake, synthesis or β-oxidation, and/or triglyceride export via secretion as VLDL (FIG. 2). Indeed, livers of BPA-exposed animals exhibit increased expression of a key gene involved in FFA uptake (Cd36; also known as Fat), decreased expression of genes related to triglyceride synthesis and FFA oxidation (Dgat, Agpat6, Cebpα, Cebpβ, Pck1, Acox1, Cpt1a and Cybb)107.
Relevant to the ability of EDCs to induce developmental reprogramming of the epigenome, BPA exposure altered DNA methylation and histone modifications associated with active transcription (for example, acetylation of histones H3 and H4, and trimethylation of histone H3 at lysine 36) and decreased occupancy of RNA polymerase II and critical transcription factors (C/EBPβ and SREBP1) within the Cpt1a gene107. Understanding how these epigenetic alterations are modulated by environmental exposures holds promise for understanding the increased NAFLD susceptibility caused by EDC exposures as well as the gender bias underlying the observation that female rats are refractory to BPA-induced steatosis. Some gender-bias might be EDC-specific, as in other studies both male and female mice and rats exposed to EDCs, such as TBT, polycyclic aromatic hydrocarbons (PAHs) and PCBs, displayed an observable NAFLD-like phenotype (see Supplementary information S1 (table)).
In addition to BPA, other EDCs can enhance NAFLD when promoted by a HFD (for example, treatment of mice with PFOA108 or the pesticides CYP or ATZ109). Importantly, some EDC exposures alone failed to trigger disease, with NAFLD observed only when EDC exposure was combined with a HFD. For example, treatment of male mice with PCB153 alone did not lead to NAFLD, whereas NAFLD was observed in HFD-fed animals exposed to PCB153110. Finally, some EDC exposures might not increase HFD-induced hepatic steatosis, but rather induce a NASH-like phenotype instead. An example of this phenomenon was reported for adult male mice treated with the PCB mixture Aroclor 1260111.
Taken together, the existing animal data suggest that EDC exposures might promote NAFLD, and in some cases, NASH and fibrosis as well. However, critical unsettled questions still remain with regard to exactly how an EDC exposure can promote NAFLD. Whether EDCs affect mainly hepatocytes or influence the activity and expansion and/or recruitment of hepatic macrophages, sinusoidal endothelial cells or stellate cells remains to be clarified. Although studies with the aforementioned EDCs certainly support a causal link between EDC exposure and NAFLD (see Supplementary information S1 (table)), they do not inform as to which NRs, coregulator complexes or epigenetic marks are involved in the increased disease susceptibility. Future studies should assay the appearance or loss of NAFLD in liver-specific NR knockout mice to define the NR signalling axis activated by a given EDC exposure. This knowledge could guide targeting the correct NR with a selective ligand for therapeutic purposes. In addition, as NAFLD is observed most often in experimental animal studies using a combination of EDC exposure (especially during early life) and a HFD, from a prevention standpoint, defining the interaction between EDC exposure and diets that promote this disease could be important, in addition to efforts to improve dietary habits by encouraging consumption of a low-fat diet.
In human epidemiological studies, several inherent challenges exist in comparing exposed with non-exposed populations, which makes drawing causal inferences from such studies exceedingly difficult. For example, human exposure to some EDCs, such as short-lived BPA, is nearly ubiquitous, with up to 95% of all people in the USA having detectable levels of BPA in their urine14. In the case of POPs such as dioxins and PCBs, the EDCs are very long-lived, which can result in continual exposure to animals and watersheds that humans consume. The available literature contains cross-sectional epidemiological studies which, by nature, lack the power of causal prediction. In these studies, several EDCs have been associated with either disrupted liver function (measured by levels of liver enzymes such as aspartate transaminase (AST), alanine transaminase (ALT), or γ-glutamyl transpeptidase (GGT)) or fatty liver (assayed rarely by biopsy or more frequently by ultrasound): BPA, TCDD, polychlorinated dibenzo-p-dioxins, dibenzofurans (polychlorinated dibenzodioxins and polychlorinated dibenzofurans), POPs (17 dioxins or furans and 18 PCB congeners) and PCBs (see Supplementary information S2 (table)). Importantly, one of these human association studies found that one-third of 55 men exposed to TCDD during a 10-year period had liver biopsy histologies revealing not only steatosis, but also fibrosis or macrophage infiltration112. Of note, altered enzymatic markers of liver function more correctly represent ‘liver damage’ rather than NAFLD; although elevated serum ALT and AST levels are the primary abnormality observed in patients with NAFLD, liver enzyme levels can be normal in up to 78% of patients with NAFLD113. Overall, the limited epidemiological human data to date suggest an association, but are insufficient to conclude a cause–effect relationship for EDC exposure and NAFLD in humans.
Mechanisms of EDC action
A central mechanism by which EDCs are thought to exert long-term adverse health effects is by inducing alterations in the epigenome, which due to the heritable nature of epigenetic programs, can persist across many cell generations and throughout the lifecourse. The term ‘epigenetics’ was coined to describe a process in which variations in gene expression give rise to distinct patterns of differentiation114. A more modern definition of epigenetics is the heritable alterations that regulate gene expression in the absence of changes in DNA sequence. While every cell in the human body shares essentially the same DNA sequence, epigenetic processes (sometimes refered to as “programs”) determine the phenotypic heterogeneity observed in different cell types of various tissues and organs throughout the body, and both normal and abnormal physiological function.
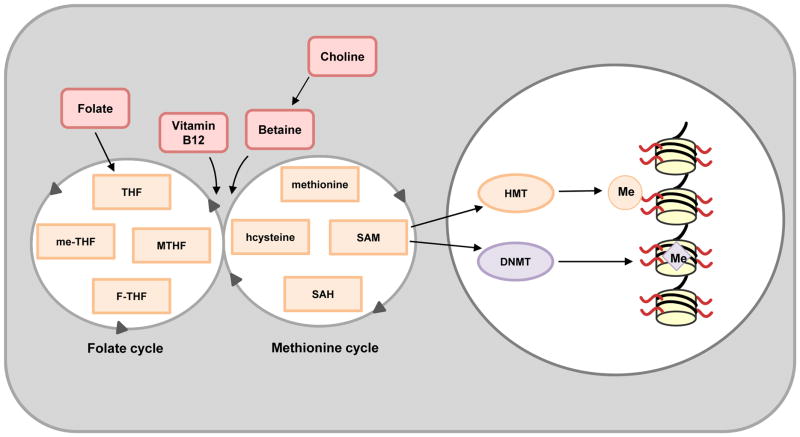
DNA methylation was the first identified molecular mechanism for epigenetic regulation of gene expression115, 116 and occurs by enzymatic transfer of a methyl group to cytosine bases of DNA, giving rise to 5-methylcytosine. The addition of methyl groups is the function of DNA methyltransferases (DNMTs), whereas removal of these methyl groups, and formation of oxidized derivatives of 5-methylcytosine, is the function of ten-eleven translocation (TET) enzymes (FIG. 5 and 6). DNA methylation alters the conformation of DNA via the action of methyl-binding proteins that inhibit transactivation of gene expression by preventing binding of transcription factors to promoters117–119 and via recruitment of chromatin-remodelling complexes that lock DNA in a closed chromatin structure120.
Figure 5. Epigenomic action of ‘writers’ of DNA or histone methylation.
Specific arginine and lysine residues on histone tails are methylated by distinct histone methyltransferases (HMTs), whereas DNA methylation occurs via the action of DNA methyltransferases (DNMTs). Both HMTs and DNMTs utilize S-Adenosyl methionine (SAM) as their methyl (Me) donor. SAM is created from methionine and its levels are influenced by methionine and interconnected folate cycles. Importantly, high folate maternal diets have been shown to affect DNA methylation patterns in rodent offspring163–165. F-THF, 10-formyltetrahydrofolate; me-THF, 5,10-methylene-THF; MTHF, 5-methyltetrahydrofolate; SAH, S-Adenosyl-L-homocysteine; THF, tetrahydrofolate.
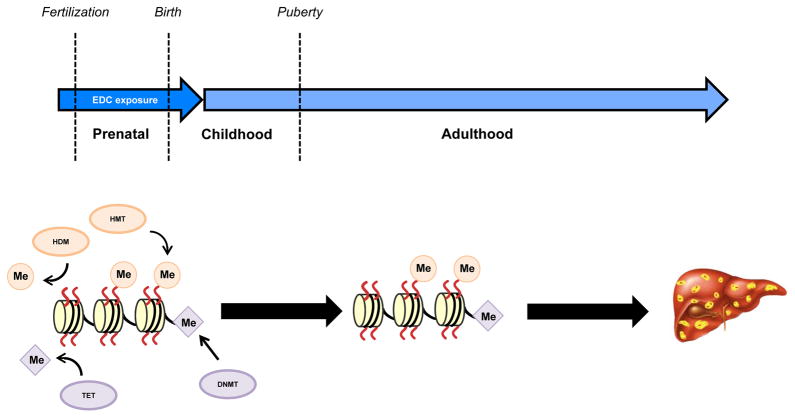
Figure 6. Early-life exposure to EDCs trigger the development of NAFLD.
Exposure to endocrine-disrupting chemicals (EDCs) during the prenatal period (a critical ‘window of susceptibility’) can result in changes to the liver epigenome that influence susceptibility to liver disease in adulthood. The activity of epigenetic ‘writers’ of DNA (DNA methyltransferases; DNMTs) or histone (histone methyltransferases; HMTs) methyl marks (Me) or ‘erasers’ of these heritable marks (ten-eleven translocation (TET) or histone demethylases (HDMs), respectively) can be influenced by a prenatal EDC exposure, which changes their activity and alters the epigenome. Such epigenetic reprogramming could confer a propensity to develop nonalcoholic fatty liver disease (NAFLD) in adulthood via reprogrammed expression of genes involved in lipid homeostasis.
Histone proteins are stably associated with DNA and form the basic scaffolding structure for DNA — the nucleosome121. The four core histone proteins (H2A, H2B, H3 and H4) are subject to post-translational modification of their N-terminal region (known as the histone ‘tail’), which protrudes out from the nucleosome and provides a platform for the assembly of protein complexes and proteins that ‘read’ epigenetics marks on these tails. Post-translational modification of histones modulates chromatin conformation and gene expression by altering the binding sites for proteins that regulate gene expression, or by facilitating the formation of secondary chromatin structure that controls chromatin accessibility. Combinations of post-translational modifications are both variable and dynamic, generating a ‘histone code’ or complex language for transcriptional regulation122, 123. Like DNA methylation, methylation of histones is a stable epigenetic modification and patterns of histone methylation can be epigenetically inherited across cell divisions and the lifecourse.
Histone methylation is regulated by the action of histone methyltransferases (HMTs) and histone demethylases (HDMs) that add or remove methyl groups, respectively (FIG. 6). Histone methylation is associated with both transcriptional activation and repression depending on the specific residue modified. Histones can also be acetylated (a transient modification compared to methylation) by histone acetyltransferase (HAT) enzymes; acetylation is associated with open chromatin conformation and activation of gene expression124. Conversely, deacetylation of histones by histone deacetylases (HDACs) promotes condensation of chromatin and repression of transcription125, 126.
Methyl groups for both DNA and histone methylation are derived from one-carbon metabolism and utilize the same methyl donor, S-Adenosyl methionine (SAM). Both DNMTs and HMTs transfer methyl groups from SAM to cytosine in DNA and lysine or arginine residues on histone tails, respectively, forming the byproduct S-Adenosyl-L-homocysteine (SAH; FIG. 5). The liver has a major role in SAM metabolism, with SAM biosynthesis and degradation regulated by the enzymes methionine adenosyltransferase (MAT) and glycine-N-methyltransferase (GNMT), respectively127. Maintenance of SAM homeostasis is necessary for liver health and to prevent injury and HCC4, 128. For example, Mat1a knockout (chronic SAM deficiency) in mice results in increased susceptibility to steatosis in response to a choline-deficient diet and spontaneous development of NASH129. Gnmt knockout mice (chronic SAM excess) also develop liver steatosis, fibrosis and HCC concordant with increased DNA and histone methylation130. Interestingly, the observed liver phenotype and DNA hypermethylation in Gnmt knockout mice can be reversed upon treatment with nicotinamide, which markedly reduces SAM levels131. Furthermore, children with mutations in GNMT exhibit mild to moderate liver disease132, 133. Collectively, these data support a role for disruption of SAM homeostasis in the development of liver disease.
In adult rodents, exposure to a methyl-deficient diet (MDD) results in increased hepatic steatosis and alteration of DNA methylation134–137. For example, MDD causes hypermethylation of Ahcy, the gene that encodes the enzyme responsible for hydrolysis of SAH, and thus increased SAH levels137. Interestingly, individual mouse strains exhibit differential susceptibility to MDD-induced liver disease, and this difference might be due to inter-strain epigenetic differences. For example, in the WSB/EiJ strain that exhibits severe NASH-like liver injury compared to the A/J strain that exhibits mild NAFLD-like liver injury in response to MDD135, 136, increased DNA methylation at gene promoters and increased Dnmt1 and Dnmt3a expression136 have been reported in the more susceptible strain, consistent with the hypothesis that epigenetic alterations might have a role in modulating NAFLD susceptibility.
Few studies to date have examined the role of histone modifications in NAFLD, but evidence exists that an imbalance between HATs and HDACs might have a role in the progression of NAFLD138. For example, liver-specific knockout of the HDAC gene Sirt1, increases susceptibility to HFD-induced hepatic steatosis139. In addition, HDAC3 has been shown to control hepatic lipogenesis in a circadian fashion and deletion of Hdac3 causes hepatic steatosis140. Furthermore, adult mice fed a HFD exhibit altered histone acetylation at genes involved in the inflammatory response141. Studies in primates have examined the epigenetic effects of maternal diet on liver disease in offspring. Maternal HFD alters histone acetylation in the livers of offspring, with a concomitant increase in lipogenic gene expression and a decrease in HDAC1 and SIRT1 expression and activity142, 143. Interestingly, these effects can be abrogated with diet reversal143.
A similar lack of data exists for alterations in DNA methylation that are associated with NAFLD, although nutrient modulation of DNA methylation in the context of obesity has been demonstrated in the agouti mouse model144, 145. Constitutive, ectopic agouti transcription (due to altered DNA methylation) results in a yellow coat phenotype, as well as increased susceptibility to diabetes mellitus, obesity and tumorigenesis146, 147. Maternal nutrient supplementation with the phyto-oestrogen (and EDC) genistein alters coat color and protects offspring from obesity by modifying the fetal epigenome148. Supplementation with genistein (or folic acid) also counteracts BPA-mediated DNA hypomethylation in early development in this mouse model149. These studies support the use of the agouti mouse as a biosensor for the study of epigenomic modulation by the environment144, including future studies aimed at examining the link between DNA methylation and the development of NAFLD. In humans, only a handful of studies have reported gene-specific alterations in DNA methylation in patients with advanced NAFLD compared with mild NAFLD150, 151, highlighting the need for additional research in this area.
Liver disease and environmental exposures across the lifecourse
Although adverse environmental exposures can occur at any time along the lifecourse to increase the risk of disease, the perinatal period might represent a window of particular vulnerability152. For example, in the context of rodent models of nutritional modulation, a maternal energy-rich diet is associated with the development of NAFLD in offspring153–161. In addition, as mentioned earlier, studies in humans have shown that fetal exposure to famine ‘mis-matched’ with a nutrient-rich adult environment19, 162 is associated with the development of hepatic steatosis. Similarly, perinatal exposure to EDCs can result in adult susceptibility to development of NAFLD in rodent models (see Supplementary information S1 (table)), although only a limited number of these studies examined epigenetic alterations that could be responsible for NAFLD susceptibility. To date, most studies have focused on epigenetic alterations associated with the disease itself, rather than a change in susceptibility of the liver that precedes disease onset. Therefore it remains an attractive, but untested, hypothesis that early-life exposure to EDCs might increase the risk of liver disease by altering patterns of DNA and/or histone methylation, and thereby changing physiological ‘set-points’ in the liver to reprogram hepatic gene expression programs to promote NAFLD (FIG. 6).
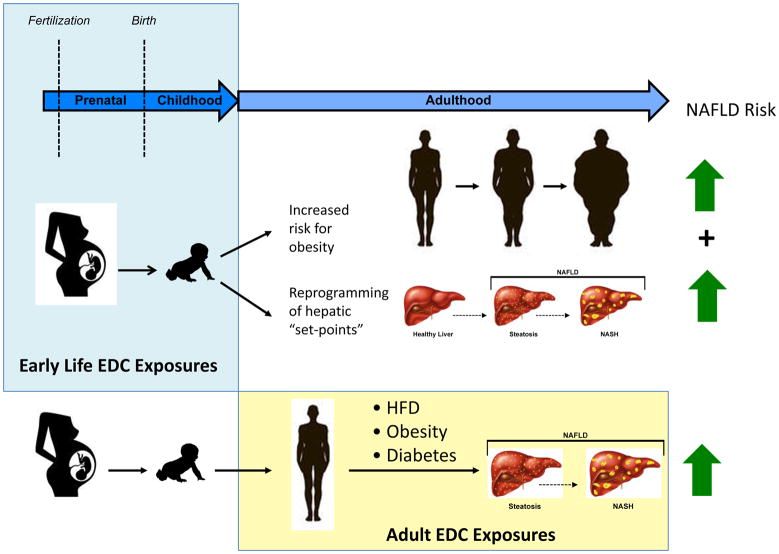
An intriguing, but underexplored aspect of the EDC–NAFLD linkage is the interplay between EDC exposure, obesity and NAFLD. As obesity is a known risk factor for NAFLD, early-life exposure to EDCs that act as ‘obesogens’ could increase susceptibility to NAFLD via increasing susceptibility to obesity. Early-life EDC exposures could also deliver a ‘double hit’, by altering both liver physiological set-points in concert with other physiological changes that increase the propensity for obesity. Alternatively, susceptibility to the NAFLD-promoting effects of later-life EDC exposure could be enhanced in obese individuals. Just as obesity combined with alcohol increases the risk of fatty liver disease, so could the physiological effects of other risk factors such as obesity, a HFD and T2DM combined with EDC exposure. Investigating the impact of early-life EDC exposures, as well as later-life exposures in the setting of other risk factors such as obesity, are therefore important future areas of study for understanding the potential contribution of EDCs to the development and progression of NAFLD (FIG. 7).
Figure 7. EDCs and NAFLD risk across the life-course.
Endocrine-disrupting chemicals (EDCs) can alter susceptibility to develop nonalcoholic fatty liver disease (NAFLD) via early-life effects that increase susceptibility to obesity and alter hepatic ‘set-points that favour the development of fatty liver, and later-life effects that contribute to the development of liver disease alone or in combination with other NAFLD risk factors such as diet, diabetes mellitus and/or obesity. HFD, high-fat diet; NASH, nonalcoholic steatohepatitis.
Conclusions
NAFLD, the fastest growing and most prevalent liver disease worldwide, represents a spectrum of diseases from simple steatosis to steatohepatitis that can progress to fatal cirrhosis and HCC. In addition to obesity and fructose as risk factors for NAFLD development and progression, certain environmental exposures to chemicals such as EDCs might increase susceptibility to NAFLD and/or co-operate with a high-fat Western diet to promote development of this disease. One mechanism of EDC action involves physical binding to NRs, which then can recruit coregulator proteins (either coactivators or corepressors) to modulate transcription of hepatic lipid homeostasis gene expression programs to favour NAFLD. In addition, early-life EDC exposures can impact the epigenome, altering DNA methylation and/or histone modifications, to affect metabolic reprogramming via altered expression of hepatic lipid pathway genes. Such reprogramming of the epigenome during development in response to nutrient availability is well established; EDC exposure in early-life might similarly reprogram hepatic lipid homeostasis gene programs toward a metabolic ‘set point’ that promotes NAFLD. Additionally, EDC exposure in adulthood might also contribute to NAFLD in combination with other prevalent predisposing factors, such as diets rich in fat, a BMI >30 kg/m2 and T2DM. We hope this Review will encourage more mechanistic studies aimed at better understanding how EDC exposures impact the epigenome to alter the expression of genes associated with hepatic lipid metabolism, which in turn promote the development of NAFLD.
Supplementary Material
Supplementary information S1 (table): EDCs reported to induce a NAFLD phenotype in rodents.
Supplementary information S2 (table): EDCs associated with liver dysfunction and potential NAFLD in humans.
Review criteria.
Full-text papers written in English cited in this review were selected based on Pubmed searches using the following search terms: “endocrine disruptor or EDC”, “fatty liver or NAFLD”, “epigenetic”, “mice”, “rat”, “human”, “NASH”, “lipogenesis”, “reprogramming” without a publication year cutoff. Additional references found in these publications provided additional papers to search and cite. Many of cited studies on nuclear receptors, coregulators and epigenetics came from our collective background knowledge.
Key points.
Nonalcoholic fatty liver disease (NAFLD) is a growing epidemic in countries consuming a Western diet, and can lead to irreversible cirrhosis and hepatocellular carcinoma
Exposure to endocrine-disrupting chemicals (EDCs) in early life could represent a ‘new’ risk factor for the development of NAFLD later in life
EDCs mechanism of action involves both modulation of nuclear hormone receptor (NR) function via coregulator proteins and alteration of the epigenome (DNA methylation and histone modification)
Animal model studies suggest causality between certain early-life EDC exposures and NAFLD presentation later in life
Studies are needed to define causality of an EDC exposure in humans with development of NAFLD, as well as to develop new prevention and treatment regimes
Acknowledgments
The authors acknowledge Kevin Phillips, Milton Feingold and David Moore for helpful discussions on NAFLD, and the National Institute of Environmental Health Sciences (NIEHS) for support (grants 1R01ES023206, P30ES023512 and U01ES026719 to C.L.W.).
Glossary terms
- NAFLD
nonalcoholic fatty liver disease
- HCC
hepatocellular carcinoma
- EDCs
endocrine-disrupting chemicals
- NR
nuclear hormone receptor
- NASH
nonalcoholic steatohepatitis
- HFD
high-fat diet
- BMI
body mass index
- MDCs
metabolism-disrupting chemicals
- BPA
bisphenol A
- NRs
nuclear hormone receptors
- NHANES
National Health and Nutrition Examination Survey
- T2DM
type 2 diabetes mellitus
- FFAs
free fatty acids
- VLDL
very low density lipoprotein
- PUFAs
polyunsaturated fatty acids
- POPs
persistent organic pollutants
- PCBs
polychlorinated biphenyls
- DDT
dichlorodiphenyltrichloroethane
- DDE
dichlorodiphenyldichloroethylene
- DEHP
di-2-ethylhexyl phthalate
- TBT
tributyltin
- PFOA
perfluorooctanoic acid
- PFOS
perfluorooctane sulfonate
- CYP
cypermethrin
- ATZ
atrazine
- SRC
steroid receptor coactivator
- SERMs
selective oestrogen receptor modulators
- MAPKs
mitogen-activated protein kinases
- ER
oestrogen receptor
- AR
androgen receptor
- PR
progesterone receptor
- PPAR
peroxisome proliferator-activated receptor
- RXR
retinoid X receptor
- TR
thyroid receptor
- RAR
retinoic acid receptor
- LXR
liver X receptor
- FXR
farnesoid X receptor
- CAR
constitutive androstane receptor
- PXR
pregnane X receptor
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- PCN
pregnenolone 16α-carbonitrile
- SHP
short heterodimer partner
- PGC1
PPARγ coactivator 1
- MED1
Mediator complex subunit MED1
- NCoR
nuclear receptor corepressor
- SMRT
silencing mediator of retinoic acid and thyroid hormone receptor
- HDAC
histone deacetylase
- RIP140
receptor-interacting protein 140
- LCOR
ligand-dependent corepressor
- SMILE
SHP interacting leucine zipper protein
- GR
glucocorticoid receptor
- AhR
aryl hydrocarbon receptor
- Kd
equilibrium dissociation constant
- TCDD
2,3,7,8-tetrachlorodibenzodioxin
- HxCDD
hexachlorodibenzo-p-dioxin
- PAHs
polycyclic aromatic hydrocarbons
- AST
aspartate transaminase
- ALT
alanine transaminase
- GGT
γ-glutamyl transpeptidase
- DNMTs
DNA methyltransferases
- TET
ten-eleven translocation
- HMTs
histone methyltransferases
- HDMs
histone demethylases
- HAT
histone acetyltransferase
- HDACs
histone deacetylases
- SAM
S-Adenosyl methionine
- SAH
S-Adenosyl-L-homocysteine
- MAT
methionine adenosyltransferase
- GNMT
glycine-N-methyltransferase
- MDD
methyl-deficient diet
Biographies
Charles E. Foulds is an Assistant Professor in the Department of Molecular and Cellular Biology and the Center for Precision Environmental Health at Baylor College of Medicine (BCM), Houston, Texas, USA. He received a PhD in Chemistry from the University of Oregon, Eugene, Oregon, USA, for work on TATA-binding proteins and did postdoctoral training at the University of Utah, Salt Lake City, Utah, USA, on ETS transcription factors and at BCM with Bert W. O’Malley on nuclear receptor coregulators. As a faculty member, his current research interest involves proteomics of DNA-bound nuclear receptor–coregulator complexes, with a special emphasis on understanding how endocrine-disrupting chemicals can perturb these complexes.
Lindsey S. Treviño is an Instructor at Baylor College of Medicine in the Center for Precision Environmental Health, Houston, Texas, USA. She received a PhD in Reproductive Physiology from Cornell University, Ithaca, New York, USA. Her research is focused on understanding the molecular basis of how early-life exposure to endocrine-disrupting chemcials (EDCs) reprograms the epigenome to promote the development of metabolic diseases such as cancer, obesity and diabetes mellitus in adulthood. She is a Future Leaders Advancing Research in Endocrinology (FLARE) Fellow, a Keystone Symposia Fellow and a National Institutes of Health Future Research Leader (NIH FRL).
Brian York is an Associate Professor in the Department of Molecular and Cellular Biology and the Dan L. Duncan Cancer Center at Baylor College of Medicine, Houston, Texas, USA. He received his PhD in biochemistry from the University of Kentucky, Lexington, Kentucky, USA, and completed postdoctoral training with Bert W. O’Malley. The research focus of the York laboratory is to elucidate the metabolic events involved in transcriptional reprogramming of the liver during the spectrum of hepatic pathology that begins with benign hepatic steatosis but progresses to nonalcoholic fatty liver disease, nonalcoholic steatohepatitis and ultimately to hepatocellular carcinoma.
Cheryl L. Walker is Director of the Center for Precision Environmental Health and Professor in the Departments of Molecular and Cellular Biology and Medicine at Baylor College of Medicine, Houston, Texas, USA. She received her PhD from the University of Texas Southwestern Medical School, Dallas, Texas, USA and completed her postdoctoral training at the National Institute of Environmental Health Sciences, North Carolina, USA. A major research focus of the Walker laboratory is on gene–environment interactions that contribute to disease pathogenesis, and on elucidating how epigenomic plasticity creates a vulnerability to environmental exposures that cause metabolic disease, obesity and cancer.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
C.E.F., L.S.T., B.Y. and C.L.W. researched data for the article, made substantial contributions to discussions about the content, wrote the article and reviewed and/or edited the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
References
- 1.Lim JS, Mietus-Snyder M, Valente A, Schwarz JM, Lustig RH. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nat Rev Gastroenterol Hepatol. 2010;7:251–264. doi: 10.1038/nrgastro.2010.41. [DOI] [PubMed] [Google Scholar]
- *2.Wesolowski SR, Kasmi KC, Jonscher KR, Friedman JE. Developmental origins of NAFLD: a womb with a clue. Nat Rev Gastroenterol Hepatol. 2016;14:81–96. doi: 10.1038/nrgastro.2016.160. This review presents the hypothesis that early-life environmental signals, such as distinct nutritional signals, may predispose an individual to NAFLD in later life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wree A, Broderick L, Canbay A, Hoffman HM, Feldstein AE. From NAFLD to NASH to cirrhosis — new insights into disease mechanisms. Nat Rev Gastroenterol Hepatol. 2013;10:627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 4.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- *5.Hardy T, Oakley F, Anstee QM, Day CP. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu Rev Pathol. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. This review describes the liver pathophysiology compromsing NAFLD. [DOI] [PubMed] [Google Scholar]
- 6.White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol. 2012;10:1342–1359.e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–90. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 8.Ruhl CE, Everhart JE. Fatty liver indices in the multiethnic United States National Health and Nutrition Examination Survey. Aliment Pharmacol Ther. 2015;41:65–76. doi: 10.1111/apt.13012. [DOI] [PubMed] [Google Scholar]
- 9.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satapathy SK, Sanyal AJ. Epidemiology and natural history of nonalcoholic fatty liver disease. Semin Liver Dis. 2015;35:221–235. doi: 10.1055/s-0035-1562943. [DOI] [PubMed] [Google Scholar]
- 11.Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- *12.Heindel JJ, et al. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol. 2016 doi: 10.1016/j.reprotox.2016.10.001. http://dx.doi.org/10.1016/j.reprotox.2016.10.001. This timely review describes EDCs that affect metabolic “set-points” and suggest they should now be called “MDCs”. [DOI] [PMC free article] [PubMed]
- *13.Gore AC, et al. EDC-2: The Endocrine Society’s second scientific statement on endocrine-disrupting chemicals. Endocr Rev. 2015;36:E1–E150. doi: 10.1210/er.2015-1010. The authoritative review of the collective literature on EDCs and their effects on various target organs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calafat AM, et al. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ Health Perspect. 2005;113:391–395. doi: 10.1289/ehp.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desmet VJ. In: The liver: biology and pathobiology. 3. Arias IM, et al., editors. Raven Press; 1994. [Google Scholar]
- 16.Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 17.Ravelli AC, et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 18.Roseboom TJ, et al. Plasma lipid profiles in adults after prenatal exposure to the Dutch famine. Am J Clin Nutr. 2000;72:1101–1106. doi: 10.1093/ajcn/72.5.1101. [DOI] [PubMed] [Google Scholar]
- *19.Wang N, et al. Exposure to famine in early life and nonalcoholic fatty liver disease in adulthood. J Clin Endocrinol Metab. 2016;101:2218–2225. doi: 10.1210/jc.2016-1076. This paper highlights important human data during the Great Chinese Famine that lack of nutrition in early life appeared to predispose for NAFLD in later life. [DOI] [PubMed] [Google Scholar]
- 20.Erslev AJ. In: The Liver: Biology and Pathobiology. Arias IM, et al., editors. Raven Press; 1994. pp. 1227–1234. [Google Scholar]
- 21.Kuntz E, Kuntz HD. In: Hepatology — textbook and atlas. Kuntz E, Kuntz HD, editors. Springer; 2008. pp. 35–76. [Google Scholar]
- 22.Saponaro C, Gaggini M, Gastaldelli A. Nonalcoholic fatty liver disease and type 2 diabetes: common pathophysiologic mechanisms. Curr Diab Rep. 2015;15:607. doi: 10.1007/s11892-015-0607-4. [DOI] [PubMed] [Google Scholar]
- 23.Jou J, Choi SS, Diehl AM. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:370–379. doi: 10.1055/s-0028-1091981. [DOI] [PubMed] [Google Scholar]
- 24.De Taeye BM, et al. Macrophage TNF-α contributes to insulin resistance and hepatic steatosis in diet-induced obesity. Am J Physiol Endocrinol Metab. 2007;293:E713–E725. doi: 10.1152/ajpendo.00194.2007. [DOI] [PubMed] [Google Scholar]
- 25.Malaguarnera L, Madeddu R, Palio E, Arena N, Malaguarnera M. Heme oxygenase-1 levels and oxidative stress-related parameters in non-alcoholic fatty liver disease patients. J Hepatol. 2005;42:585–591. doi: 10.1016/j.jhep.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 26.Papandreou D, Andreou E. Role of diet on non-alcoholic fatty liver disease: An updated narrative review. World J Hepatol. 2015;7:575–582. doi: 10.4254/wjh.v7.i3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves HL, Zaki MY, Day CP. Hepatocellular carcinoma in obesity, type 2 diabetes, and NAFLD. Dig Dis Sci. 2016;61:1234–1245. doi: 10.1007/s10620-016-4085-6. [DOI] [PubMed] [Google Scholar]
- 28.Hooper AJ, Adams LA, Burnett JR. Genetic determinants of hepatic steatosis in man. J Lipid Res. 2011;52:593–617. doi: 10.1194/jlr.R008896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang X, et al. Fructose consumption as a risk factor for non-alcoholic fatty liver disease. J Hepatol. 2008;48:993–999. doi: 10.1016/j.jhep.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vos MB, Lavine JE. Dietary fructose in nonalcoholic fatty liver disease. Hepatology. 2013;57:2525–2531. doi: 10.1002/hep.26299. [DOI] [PubMed] [Google Scholar]
- 31.Basaranoglu M, Basaranoglu G, Sabuncu T, Senturk H. Fructose as a key player in the development of fatty liver disease. World J Gastroenterol. 2013;19:1166–1172. doi: 10.3748/wjg.v19.i8.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casals-Casas C, Desvergne B. Endocrine disruptors: from endocrine to metabolic disruption. Annu Rev Physiol. 2011;73:135–162. doi: 10.1146/annurev-physiol-012110-142200. [DOI] [PubMed] [Google Scholar]
- 33.Heindel JJ, Newbold R, Schug TT. Endocrine disruptors and obesity. Nat Rev Endocrinol. 2015;11:653–661. doi: 10.1038/nrendo.2015.163. [DOI] [PubMed] [Google Scholar]
- 34.Grun F, Blumberg B. Perturbed nuclear receptor signaling by environmental obesogens as emerging factors in the obesity crisis. Rev Endocr Metab Disord. 2007;8:161–171. doi: 10.1007/s11154-007-9049-x. [DOI] [PubMed] [Google Scholar]
- 35.Alonso-Magdalena P, Quesada I, Nadal A. Endocrine disruptors in the etiology of type 2 diabetes mellitus. Nat Rev Endocrinol. 2011;7:346–353. doi: 10.1038/nrendo.2011.56. [DOI] [PubMed] [Google Scholar]
- 36.Vom Saal FS, Nagel SC, Coe BL, Angle BM, Taylor JA. The estrogenic endocrine disrupting chemical bisphenol A (BPA) and obesity. Mol Cell Endocrinol. 2012;354:74–84. doi: 10.1016/j.mce.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Foulds CE, et al. Proteomic analysis of coregulators bound to ERα on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 40.Routledge EJ, White R, Parker MG, Sumpter JP. Differential effects of xenoestrogens on coactivator recruitment by estrogen receptor (ER) α and ERβ. J Biol Chem. 2000;275:35986–35993. doi: 10.1074/jbc.M006777200. [DOI] [PubMed] [Google Scholar]
- 41.Levin ER. Cell localization, physiology, and nongenomic actions of estrogen receptors. J Appl Physiol. 2001;91:1860–1867. doi: 10.1152/jappl.2001.91.4.1860. [DOI] [PubMed] [Google Scholar]
- 42.Trevino LS, Weigel NL. Phosphorylation: a fundamental regulator of steroid receptor action. Trends Endocrinol Metab. 2013;24:515–524. doi: 10.1016/j.tem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammes SR, Levin ER. Minireview: Recent advances in extranuclear steroid receptor actions. Endocrinology. 2011;152:4489–4495. doi: 10.1210/en.2011-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammes SR, Davis PJ. Overlapping nongenomic and genomic actions of thyroid hormone and steroids. Best Pract Res Clin Endocrinol Metab. 2015;29:581–593. doi: 10.1016/j.beem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endo Y, et al. Thiazolidinediones enhance sodium-coupled bicarbonate absorption from renal proximal tubules via PPARγ-dependent nongenomic signaling. Cell Metab. 2011;13:550–561. doi: 10.1016/j.cmet.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 46.Zhang XK, et al. Regulation of the nongenomic actions of retinoid X receptor-α by targeting the coregulator-binding sites. Acta Pharmacol Sin. 2015;36:102–112. doi: 10.1038/aps.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis PJ, Goglia F, Leonard JL. Nongenomic actions of thyroid hormone. Nat Rev Endocrinol. 2016;12:111–121. doi: 10.1038/nrendo.2015.205. [DOI] [PubMed] [Google Scholar]
- 48.Al Tanoury Z, Piskunov A, Rochette-Egly C. Vitamin A and retinoid signaling: genomic and nongenomic effects. J Lipid Res. 2013;54:1761–1775. doi: 10.1194/jlr.R030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bookout AL, et al. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q, et al. In-depth proteomic characterization of endogenous nuclear receptors in mouse liver. Mol Cell Proteomics. 2013;12:473–484. doi: 10.1074/mcp.M112.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Cave MC, et al. Nuclear receptors and nonalcoholic fatty liver disease. Biochim Biophys Acta. 2016;1859:1083–1099. doi: 10.1016/j.bbagrm.2016.03.002. This review highlights the key NRs implicated in NAFLD progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballestri S, Nascimbeni F, Romagnoli D, Baldelli E, Lonardo A. The role of nuclear receptors in the pathophysiology, natural course, and drug treatment of NAFLD in humans. Adv Ther. 2016;33:291–319. doi: 10.1007/s12325-016-0306-9. [DOI] [PubMed] [Google Scholar]
- 53.Gross B, Pawlak M, Lefebvre P, Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat Rev Endocrinol. 2017;13:36–49. doi: 10.1038/nrendo.2016.135. [DOI] [PubMed] [Google Scholar]
- 54.Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab. 2012;23:351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chai SC, Cherian MT, Wang YM, Chen T. Small-molecule modulators of PXR and CAR. Biochim Biophys Acta. 2016;1859:1141–54. doi: 10.1016/j.bbagrm.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore LB, et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–7. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 58.Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–8. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maglich JM, et al. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–83. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 60.Jones SA, et al. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 61.Lehmann JM, et al. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blumberg B, et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertilsson G, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A. 1998;95:12208–13. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong B, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106:18831–6. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, et al. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. 2006;281:15013–20. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schultz JR, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–8. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe M, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perra A, et al. Thyroid hormone (T3) and TRβ agonist GC-1 inhibit/reverse nonalcoholic fatty liver in rats. FASEB J. 2008;22:2981–2989. doi: 10.1096/fj.08-108464. [DOI] [PubMed] [Google Scholar]
- 69.Martagon AJ, Lin JZ, Cimini SL, Webb P, Phillips KJ. The amelioration of hepatic steatosis by thyroid hormone receptor agonists is insufficient to restore insulin sensitivity in ob/ob mice. PLoS ONE. 2015;10:e0122987. doi: 10.1371/journal.pone.0122987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez-Velazquez JA, Carrillo-Cordova LD, Chavez-Tapia NC, Uribe M, Mendez-Sanchez N. Nuclear receptors in nonalcoholic fatty liver disease. J Lipids. 2012;2012:139875. doi: 10.1155/2012/139875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun M, Cui W, Woody SK, Staudinger JL. Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug Metab Dispos. 2015;43:335–343. doi: 10.1124/dmd.114.062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 2014;20:26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.York B, et al. Research resource: tissue- and pathway-specific metabolomic profiles of the steroid receptor coactivator (SRC) family. Mol Endocrinol. 2013;27:366–380. doi: 10.1210/me.2012-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chopra AR, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma X, et al. Deletion of steroid receptor coactivator-3 gene ameliorates hepatic steatosis. J Hepatol. 2011;55:445–452. doi: 10.1016/j.jhep.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 76.Stashi E, York B, O’Malley BW. Steroid receptor coactivators: servants and masters for control of systems metabolism. Trends Endocrinol Metab. 2014;25:337–347. doi: 10.1016/j.tem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rollins DA, Coppo M, Rogatsky I. Minireview: nuclear receptor coregulators of the p160 family: insights into inflammation and metabolism. Mol Endocrinol. 2015;29:502–517. doi: 10.1210/me.2015-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leone TC, et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci USA. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lelliott CJ, et al. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li S, et al. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bai L, et al. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology. 2011;53:1164–1174. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guenther MG, et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 85.Yoon HG, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 87.Sun Z, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leonardsson G, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci USA. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berriel Diaz M, et al. Nuclear receptor cofactor receptor interacting protein 140 controls hepatic triglyceride metabolism during wasting in mice. Hepatology. 2008;48:782–791. doi: 10.1002/hep.22383. [DOI] [PubMed] [Google Scholar]
- 91.Song Y, et al. Ligand-dependent corepressor acts as a novel corepressor of thyroid hormone receptor and represses hepatic lipogenesis in mice. J Hepatol. 2012;56:248–254. doi: 10.1016/j.jhep.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 92.Lee JM, et al. Ursodeoxycholic acid inhibits liver X receptor alpha-mediated hepatic lipogenesis via induction of the nuclear corepressor SMILE. J Biol Chem. 2014;289:1079–1091. doi: 10.1074/jbc.M113.491522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng FF, Wu RC, Smith CL, O’Malley BW. Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Mol Cell Biol. 2005;25:8273–8284. doi: 10.1128/MCB.25.18.8273-8284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu RC, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 95.York B, et al. Reprogramming the posttranslational code of SRC-3 confers a switch in mammalian systems biology. Proc Natl Acad Sci USA. 2010;107:11122–11127. doi: 10.1073/pnas.1005262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jo YS, et al. Phosphorylation of the nuclear receptor corepressor 1 by protein kinase B switches its corepressor targets in the liver in mice. Hepatology. 2015;62:1606–1618. doi: 10.1002/hep.27907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.le Maire A, Bourguet W, Balaguer P. A structural view of nuclear hormone receptor: endocrine disruptor interactions. Cell Mol Life Sci. 2010;67:1219–1237. doi: 10.1007/s00018-009-0249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Swedenborg E, Ruegg J, Makela S, Pongratz I. Endocrine disruptive chemicals: mechanisms of action and involvement in metabolic disorders. J Mol Endocrinol. 2009;43:1–10. doi: 10.1677/JME-08-0132. [DOI] [PubMed] [Google Scholar]
- 99.Rich RL, et al. Kinetic analysis of estrogen receptor/ligand interactions. Proc Natl Acad Sci USA. 2002;99:8562–8567. doi: 10.1073/pnas.142288199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *100.Grun F, et al. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol Endocrinol. 2006;20:2141–2155. doi: 10.1210/me.2005-0367. This paper formed the basis for the “obesogen hypothesis” in that the EDC TBT promoted adipogenesis by activating PPARγ and RXRα. [DOI] [PubMed] [Google Scholar]
- 101.Mellor CL, Steinmetz FP, Cronin MT. The identification of nuclear receptors associated with hepatic steatosis to develop and extend adverse outcome pathways. Crit Rev Toxicol. 2016;46:138–152. doi: 10.3109/10408444.2015.1089471. [DOI] [PubMed] [Google Scholar]
- 102.Wahlang B, et al. Polychlorinated biphenyl-xenobiotic nuclear receptor interactions regulate energy metabolism, behavior, and inflammation in non-alcoholic-steatohepatitis. Toxicol Sci. 2016;149:396–410. doi: 10.1093/toxsci/kfv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Al-Eryani L, et al. Identification of environmental chemicals associated with the development of toxicant-associated fatty liver disease in rodents. Toxicol Pathol. 2015;43:482–497. doi: 10.1177/0192623314549960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tomaszewski KE, Montgomery CA, Melnick RL. Modulation of 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in F344 rats by di(2-ethylhexyl)phthalate. Chem Biol Interact. 1988;65:205–222. doi: 10.1016/0009-2797(88)90107-x. [DOI] [PubMed] [Google Scholar]
- *105.Shan Q, Huang F, Wang J, Du Y. Effects of co-exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin and polychlorinated biphenyls on nonalcoholic fatty liver disease in mice. Environ Toxicol. 2015;30:1364–1374. doi: 10.1002/tox.22006. This paper reported how exposure to mice with two different types of EDCs can synergize together to produce a NAFLD phenotype. [DOI] [PubMed] [Google Scholar]
- *106.Wei J, et al. Perinatal exposure to bisphenol A exacerbates nonalcoholic steatohepatitis-like phenotype in male rat offspring fed on a high-fat diet. J Endocrinol. 2014;222:313–325. doi: 10.1530/JOE-14-0356. The first of two seminal studies reporting that perinatal exposures to BPA combined with a HFD after weaning led to male, but not female, offspring displaying more severe hepatic steatosis and increased inflammation. [DOI] [PubMed] [Google Scholar]
- *107.Strakovsky RS, et al. Developmental bisphenol A (BPA) exposure leads to sex-specific modification of hepatic gene expression and epigenome at birth that may exacerbate high-fat diet-induced hepatic steatosis. Toxicol Appl Pharmacol. 2015;284:101–112. doi: 10.1016/j.taap.2015.02.021. The second seminal BPA study that also reported that perinatal BPA exposure augmented HFD-induced hepatic steaosis; BPA exposure altered DNA methylation and histone modifications of a gene encoding an important enzyme in the β-oxidation pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tan X, et al. High fat diet feeding exaggerates perfluorooctanoic acid-induced liver injury in mice via modulating multiple metabolic pathways. PLoS ONE. 2013;8:e61409. doi: 10.1371/journal.pone.0061409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jin Y, et al. Chronic exposure of mice to environmental endocrine-disrupting chemicals disturbs their energy metabolism. Toxicol Lett. 2014;225:392–400. doi: 10.1016/j.toxlet.2014.01.006. [DOI] [PubMed] [Google Scholar]
- *110.Wahlang B, et al. Polychlorinated biphenyl 153 is a diet-dependent obesogen that worsens nonalcoholic fatty liver disease in male C57BL6/J mice. J Nutr Biochem. 2013;24:1587–1595. doi: 10.1016/j.jnutbio.2013.01.009. This paper reported that some EDC exposures (such as PCB153) to a rodent do not lead to NAFLD alone, but can promote that induced by HFD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *111.Wahlang B, et al. Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicol Appl Pharmacol. 2014;279:380–390. doi: 10.1016/j.taap.2014.06.019. This paper reported some EDC exposures (such as Aroclor 1260) to a rodent might not increase HFD-induced hepatic steatosis, but rather induce a NASH-like phenotype instead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pazderova-Vejlupkova J, Lukas E, Nemcova M, Pickova J, Jirasek L. The development and prognosis of chronic intoxication by tetrachlordibenzo-p-dioxin in men. Arch Environ Health. 1981;36:5–11. doi: 10.1080/00039896.1981.10667598. [DOI] [PubMed] [Google Scholar]
- 113.Obika M, Noguchi H. Diagnosis and evaluation of nonalcoholic fatty liver disease. Exp Diabetes Res. 2012;2012:145754. doi: 10.1155/2012/145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 115.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 116.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 117.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 118.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 119.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 120.Fournier A, Sasai N, Nakao M, Defossez PA. The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief Funct Genomics. 2012;11:251–264. doi: 10.1093/bfgp/elr040. [DOI] [PubMed] [Google Scholar]
- 121.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 122.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 124.Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 125.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 126.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 127.Noureddin M, Mato JM, Lu SC. Nonalcoholic fatty liver disease: update on pathogenesis, diagnosis, treatment and the role of S-adenosylmethionine. Exp Biol Med (Maywood) 2015;240:809–820. doi: 10.1177/1535370215579161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu SC, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martinez-Chantar ML, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Varela-Rey M, et al. Fatty liver and fibrosis in glycine N-methyltransferase knockout mice is prevented by nicotinamide. Hepatology. 2010;52:105–114. doi: 10.1002/hep.23639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Augoustides-Savvopoulou P, et al. Glycine N-methyltransferase deficiency: a new patient with a novel mutation. J Inherit Metab Dis. 2003;26:745–759. doi: 10.1023/B:BOLI.0000009978.17777.33. [DOI] [PubMed] [Google Scholar]
- 133.Mudd SH, et al. Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia. J Inherit Metab Dis. 2001;24:448–464. doi: 10.1023/a:1010577512912. [DOI] [PubMed] [Google Scholar]
- *134.Pogribny IP, et al. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176–186. doi: 10.1016/j.jhep.2009.03.021. This paper provides data mechanistically linking epigenetic alterations (DNA methylation and select histone modifications) to the pathogenesis of hepatic steatosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tryndyak V, et al. Interstrain differences in the severity of liver injury induced by a choline- and folate-deficient diet in mice are associated with dysregulation of genes involved in lipid metabolism. FASEB J. 2012;26:4592–4602. doi: 10.1096/fj.12-209569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tryndyak VP, et al. Status of hepatic DNA methylome predetermines and modulates the severity of non-alcoholic fatty liver injury in mice. BMC Genomics. 2016;17:298. doi: 10.1186/s12864-016-2617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tryndyak VP, et al. Coupling global methylation and gene expression profiles reveal key pathophysiological events in liver injury induced by a methyl-deficient diet. Mol Nutr Food Res. 2011;55:411–418. doi: 10.1002/mnfr.201000300. [DOI] [PubMed] [Google Scholar]
- *138.Lee JH, Friso S, Choi SW. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients. 2014;6:3303–3325. doi: 10.3390/nu6083303. This review highlights the important concept that nutrient “signals” can influence epigenomics (DNA methylation, histone modifications) that may contribute to NAFLD progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *139.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. This paper reports that the loss of a specific HDAC activity (SIRT1) in the liver results in an NAFLD phenotype by inactivation of the PGC-1α coactivator. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mikula M, Majewska A, Ledwon JK, Dzwonek A, Ostrowski J. Obesity increases histone H3 lysine 9 and 18 acetylation at Tnfa and Ccl2 genes in mouse liver. Int J Mol Med. 2014;34:1647–1654. doi: 10.3892/ijmm.2014.1958. [DOI] [PubMed] [Google Scholar]
- *142.Aagaard-Tillery KM, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. This paper reports that a maternal HFD led to increased liver triglyceride levels and epigenome alternations (specifically histone H3K14 acetylation) in Japanese macaque fetuses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suter MA, et al. A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. FASEB J. 2012;26:5106–5114. doi: 10.1096/fj.12-212878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jirtle RL. The Agouti mouse: a biosensor for environmental epigenomics studies investigating the developmental origins of health and disease. Epigenomics. 2014;6:447–50. doi: 10.2217/epi.14.58. [DOI] [PubMed] [Google Scholar]
- 145.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 146.Miltenberger RJ, Mynatt RL, Wilkinson JE, Woychik RP. The role of the agouti gene in the yellow obese syndrome. J Nutr. 1997;127:1902S–1907S. doi: 10.1093/jn/127.9.1902S. [DOI] [PubMed] [Google Scholar]
- 147.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 148.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Murphy SK, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zeybel M, et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenetics. 2015;7:25. doi: 10.1186/s13148-015-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Barker DJ Sir Richard Doll Lecture. Developmental origins of chronic disease. Public Health. 2012;126:185–189. doi: 10.1016/j.puhe.2011.11.014. [DOI] [PubMed] [Google Scholar]
- *153.Bruce KD, et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology. 2009;50:1796–1808. doi: 10.1002/hep.23205. This paper provides a link between maternal HFD and NAFLD progression to steatohepatitis in adult rodent offspring. [DOI] [PubMed] [Google Scholar]
- 154.Elahi MM, et al. Long-term maternal high-fat feeding from weaning through pregnancy and lactation predisposes offspring to hypertension, raised plasma lipids and fatty liver in mice. Br J Nutr. 2009;102:514–519. doi: 10.1017/S000711450820749X. [DOI] [PubMed] [Google Scholar]
- 155.Gregorio BM, Souza-Mello V, Carvalho JJ, Mandarim-de-Lacerda CA, Aguila MB. Maternal high-fat intake predisposes nonalcoholic fatty liver disease in C57BL/6 offspring. Am J Obstet Gynecol. 2010;203:495.e1–8. doi: 10.1016/j.ajog.2010.06.042. [DOI] [PubMed] [Google Scholar]
- 156.Oben JA, et al. Maternal obesity during pregnancy and lactation programs the development of offspring non-alcoholic fatty liver disease in mice. J Hepatol. 2010;52:913–920. doi: 10.1016/j.jhep.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 157.Ashino NG, et al. Maternal high-fat feeding through pregnancy and lactation predisposes mouse offspring to molecular insulin resistance and fatty liver. J Nutr Biochem. 2012;23:341–348. doi: 10.1016/j.jnutbio.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 158.Li J, et al. Accumulation of endoplasmic reticulum stress and lipogenesis in the liver through generational effects of high fat diets. J Hepatol. 2012;56:900–907. doi: 10.1016/j.jhep.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 159.Mouralidarane A, et al. Maternal obesity programs offspring nonalcoholic fatty liver disease by innate immune dysfunction in mice. Hepatology. 2013;58:128–138. doi: 10.1002/hep.26248. [DOI] [PubMed] [Google Scholar]
- 160.Kjaergaard M, Nilsson C, Rosendal A, Nielsen MO, Raun K. Maternal chocolate and sucrose soft drink intake induces hepatic steatosis in rat offspring associated with altered lipid gene expression profile. Acta Physiol (Oxf) 2014;210:142–153. doi: 10.1111/apha.12138. [DOI] [PubMed] [Google Scholar]
- 161.Pruis MG, et al. Maternal western diet primes non-alcoholic fatty liver disease in adult mouse offspring. Acta Physiol (Oxf) 2014;210:215–227. doi: 10.1111/apha.12197. [DOI] [PubMed] [Google Scholar]
- 162.Chen JP, et al. Fetal and infant exposure to the Chinese famine increases the risk of fatty liver disease in Chongqing, China. J Gastroenterol Hepatol. 2016;31:200–205. doi: 10.1111/jgh.13044. [DOI] [PubMed] [Google Scholar]
- 163.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim JM, Hong K, Lee JH, Lee S, Chang N. Effect of folate deficiency on placental DNA methylation in hyperhomocysteinemic rats. J Nutr Biochem. 2009;20:172–6. doi: 10.1016/j.jnutbio.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 165.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–6. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information S1 (table): EDCs reported to induce a NAFLD phenotype in rodents.
Supplementary information S2 (table): EDCs associated with liver dysfunction and potential NAFLD in humans.