Abstract
Treatment of chronic myeloid leukemia (CML) with imatinib mesylate and other second/third generation c-Abl specific tyrosine kinase inhibitors (TKIs) has significantly extended patient survival1. However, TKIs primarily target differentiated cells and do not eliminate leukemic stem cells (LSCs)2–4. Therefore, targeting minimal residual disease, to prevent acquired resistance and/or disease relapse requires identification of novel LSC-selective target(s) that can be exploited therapeutically5,6. Given that malignant transformation involves cellular metabolic changes, which may in turn render the transformed cells susceptible to specific assaults in a selective manner7, we searched for such vulnerabilities in CML LSCs. We performed metabolic analyses on both stem cell-enriched (CD34+ and CD34+CD38-) and differentiated (CD34-) patient derived CML cells, and compared their signature with that of normal counterparts. Combining stable isotope-assisted metabolomics with functional assays, we demonstrate that primitive CML cells rely on upregulated oxidative metabolism for their survival. We also show that combination-treatment of imatinib with tigecycline, an antibiotic that inhibits mitochondrial protein translation, selectively eradicates CML LSCs, both in vitro and in a xenotransplantation model of human CML. Our findings provide a strong indication for investigating the employment of TKIs in combination with tigecycline to treat CML patients with minimal residual disease.
CML is a myeloproliferative disorder brought about by the chromosomal translocation t(9;22)(q34;q11) in a hematopoietic stem cell (HSC)8,9 that drives the expansion of a leukemic clone via BCR-ABL expression, a chimeric onco-protein with a constitutive tyrosine kinase activity10. Prolonged treatment with TKIs to sustain remission is often associated with drug toxicity and/or acquired resistance, and entails high economic costs. On the other hand, rapid relapse in half of the patients is seen after treatment discontinuation11–13. Therefore, to obtain potential curative treatments that effectively eradicate CML LSCs, we specifically studied patient-derived stem cell-enriched CD34+ CML cells. In culture, proliferating untreated CD34+ primary CML cells rapidly lose surface CD34 expression (Supplementary Fig. 1a). Imatinib treatment primarily targets differentiated CD34- CML cells for apoptosis, leading to enrichment of more primitive CD34+ cells (Supplementary Fig. 1a,b). Consequently, imatinib decreases the efficiency of primary CML progenitor cells to form colonies in a short term colony forming cell (CFC) assay but, in line with the resistance of CML stem cells to TKI treatment, it does not affect the colony forming capacity of CD34+ cell in a long-term culture-initiating cell (LTC-IC) assay (Supplementary Fig. 1c,d).
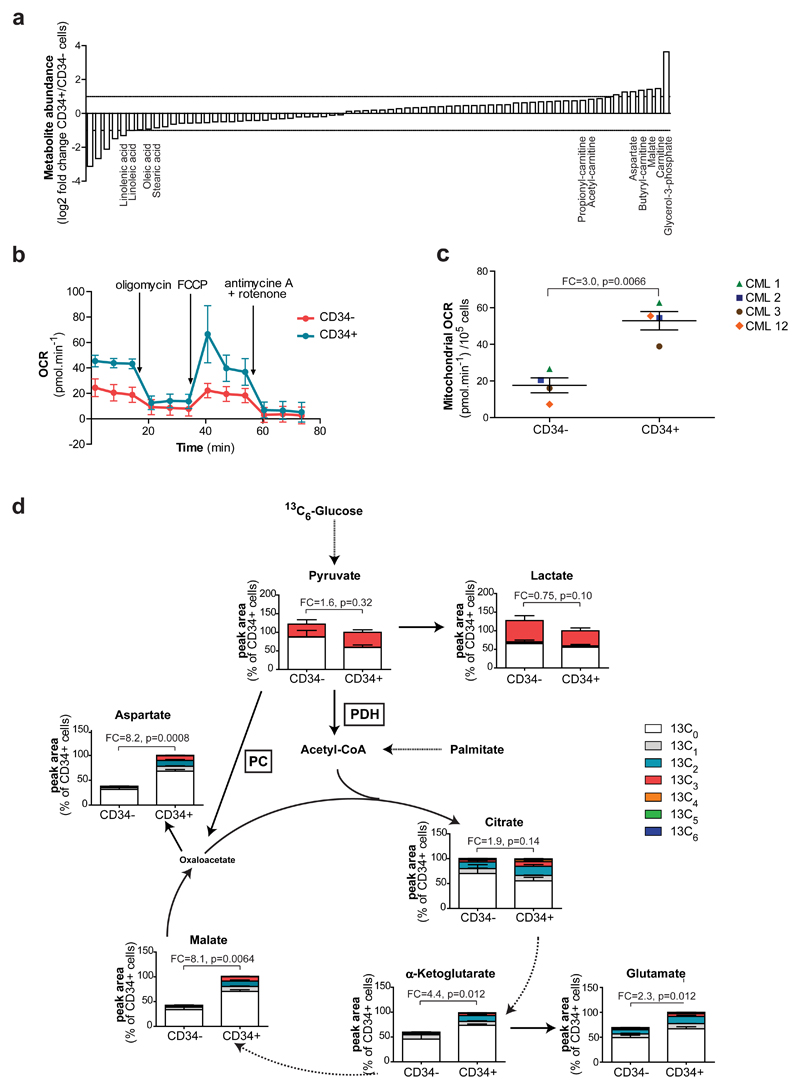
Since stem cells can exhibit different metabolic traits compared to their corresponding differentiated cells14–16, we metabolically profiled CD34+ and CD34- CML cells derived from four patients by recording the steady-state levels of 70 metabolites central to glucose, nucleotide, amino acid, fatty acid and energy metabolism, through liquid chromatography-mass spectrometry (LC-MS). The pattern of metabolites in stem cell-enriched population compared to differentiated CML cells revealed a potential increase in lipolysis and fatty acid oxidation; we found an increase in glycerol-3-phosphate, carnitine and acylcarnitine derivatives, as well as a decrease in free fatty acids, such as oleic and stearic acids (Fig. 1a and Supplementary Table 1). Of note, fatty acid oxidation has been associated with the maintenance of HSCs and potentially with leukemogenesis17,18. In order to validate and further substantiate these findings, leukemic cells were cultured for 24 hours in the presence of uniformly 13C16-labeled palmitate, and 13C isotopic enrichment in different palmitate-derived metabolites was measured by LC-MS. Substantial enhancement in palmitate-derived carbon in tricarboxylic acid (TCA) cycle metabolites and TCA cycle-derived amino acids was recognised in CD34+ leukemic cells in comparison to the differentiated CD34- cells of the same patient (Supplementary Fig. 2a). Furthermore, the steady state levels of these metabolites were increased in stem cell-enriched CML populations, while lactate levels were decreased (Fig. 1a, Supplementary Fig. 2a and Supplementary Table 1). The steady state levels of aspartate was recently recognised as bona-fide indicators of mitochondrial oxidative capacity19–21. Accordingly, CML cells derived from four patients presented on average a 3.0-fold increase in mitochondrial oxygen consumption rates in CD34+ cells compared to patient-matched CD34- cells. Moreover, the complete decrease in oxygen consumption upon ATP synthase inhibition with oligomycin demonstrated that the increased oxygen consumption is tightly linked to ATP production in these cells (Fig. 1b,c). However, the increase in the steady state levels of TCA cycle metabolites and the derived amino acids could not be solely explained by an increase in fatty acid oxidation, as the production of acetyl coenzyme A (CoA) from palmitate does not support a net production of TCA cycle metabolites (anaplerosis).
Figure 1. Primitive CML cells show an increase in oxidative metabolism compared to differentiated counterparts.
(a) Comparative steady-state metabolomics analysis of patient-matched CD34+ and CD34- CML cells measured by LC-MS. Mean, n=4 patients. (b) Representative respirometry output in CD34+ and CD34- CML cells. Mean ± S.D. (c) Basal mitochondrial oxygen consumption rate (OCR) of CD34+ and CD34- CML cells. Mean ± S.E.M. n=4 patient samples. (d) Relative isotopologue distribution of indicated metabolites in CD34+ and CD34- CML cells measured by LC-MS following 24 hours incubation with 13C6-labeled glucose. Acetyl-CoA could not be detected by LC-MS in our experimental conditions. Mean ± S.E.M. n=3 patient samples. FC, fold change of glucose-derived (13C ≥ 2) metabolite abundance relative to CD34- CML cells. PDH, Pyruvate dehydrogenase; PC, Pyruvate carboxylase. P-values were calculated with paired Student’s t-test.
To study oxidative metabolism and anaplerosis in more detail, glucose, an anaplerosis enabling metabolite, was traced in leukemic cells cultured for 24 hours with uniformly 13C6-labeled glucose. This revealed that in CD34+ CML cells, TCA cycle metabolites contained a significantly larger fraction of isotopologues with 2, 3 or more 13C atoms compared to those in CD34- cells (Fig. 1d). The larger fractions of labeled TCA cycle-derived glutamate and aspartate observed in CD34+ CML cells confirmed their higher oxidative and anaplerotic activity. Concurrently, CD34+ cells demonstrated a noticeable (although not significant) decrease in glucose-derived lactate (13C3-lactate) indicating that pyruvate is indeed more effectively shunted towards the TCA cycle (Fig. 1d). Increased acetyl CoA levels produced from fatty acid oxidation are predicted to increase the anaplerotic activity of pyruvate carboxylase (PC), but potentially to inhibit pyruvate dehydrogenase (PDH) activity. We therefore estimated the relative metabolic contribution of both enzymes in 13C6-glucose-labeled CD34+ compared to CD34- CML cells by assessing the isotopic distribution of the nearest detected relevant product (13C3-aspartate for PC and 13C2-citrate for PDH). While no change in PDH activity was detected between the two CML cell subsets, the relative activity of PC was significantly higher in CD34+ CML cells, further confirming their increased oxidative anaplerotic metabolism (Supplementary Fig. 2b,c).
To investigate whether this oxidative phenotype is unique to primitive CML cells, we assessed the metabolic profile of CD34+ cells from four CML patients and compared with normal hematopoietic CD34+ cells from four donors. Higher steady state levels of carnitine and acylcarnitine derivatives and lower levels of free fatty acids, such as oleic and linolenic acids, were detected in CD34+ CML cells (Supplementary Fig. 3a and Supplementary Table 2). In line with this, the 13C enrichment of citrate (FC=2.0; p=0.033), glutamate (FC=1.8; p=0.09) and aspartate (FC=2.3; p=0.043) from 13C16-palmitate was higher in CD34+ CML cells in comparison to CD34+ normal cells, demonstrating that the increase in fatty acid oxidation observed previously is selective to CD34+ CML cells (Supplementary Fig. 3b-d). Following incubation with 13C6-glucose, the enrichment of glucose-derived 13C isotopes in citrate, glutamate and aspartate was significantly higher in CD34+ CML cells compared to their normal counterparts which, combined with a significant increase in both PC and PDH relative activity, demonstrated a selective increase in glucose oxidation and anaplerosis in the leukemic cells (Fig. 2a-c, Supplementary Fig. 3e,f). Moreover, the mitochondrial respiration of leukemic CD34+ samples was on average 3.3-fold higher than that of CD34+ normal cells (Fig. 2d,e). As CD34+ cell pools contain both stem and progenitor cells, we next verified our findings in a CD34+CD38- CML cell population; a rare quiescent sub-population (~5% of total CD34+) that is further enriched for LSCs. Flow cytometry analysis of mitochondrial content and mitochondrial membrane potential suggested that CML LSCs possess increased mitochondrial oxidative functions compared to normal HSCs (Fig. 2f-i). Moreover, 13C6-glucose incubation of CD34+CD38- cells isolated from two CML patients revealed that CD34+CD38- CML cells contained increased levels of 13C isotopologues for citrate, aspartate and glutamate compared to CD34+CD38- normal cells, confirming that CML LSCs have an increased oxidative metabolism (Fig. 2j-l and Supplementary Fig. 3g-i).
Figure 2. Enhanced mitochondrial metabolic activity in primitive CML cells compared to normal undifferentiated hematopoietic cells.
(a-c) Relative isotopologue distribution of (a) citrate, (b) glutamate and (c) aspartate in CD34+ CML and CD34+ normal cells measured by LC-MS following 24 hours incubation with 13C6-labeled glucose. Mean ± S.E.M. n=5 patient and normal samples. FC, Fold change of glucose-derived (13C ≥ 2) metabolite abundance relative to CD34+ normal cells. P-values were calculated by unpaired Student’s t-test. (d) Representative respirometry output in CD34+ CML and CD34+ normal cells. Mean ± S.D. (e) Basal mitochondrial OCR. Mean ± S.E.M. n=9 patient samples and n=4 normal samples. P-values were calculated by unpaired Student’s t-test. (f) Representative histograms of Mitotracker Green-labeled CD34+CD38- CML cells (blue) and CD34+CD38- normal cells (red). (g) Mitochondrial content of CD34+CD38- CML cells and CD34+CD38- normal cells was determined from the geometric mean of Mitotracker Green-labeled cells. Mean ± S.E.M. n=3 patient and 3 normal samples. P-values were determined by one sample t-test. (h) Representative histograms of TMRM-labeled CD34+CD38- CML (blue) and CD34+CD38- normal (red) cells. (i) Mitochondrial membrane potential of CD34+CD38- CML cells and CD34+CD38- normal cells was determined from the geometric mean of TMRM-labeled cells. Mean ± S.E.M. n=3 patient and 3 normal samples. FC, fold change relative to normal cells. P-values were determined by one sample t-test. (j-l) Relative isotopologue distribution of (j) citrate, (k) glutamate and (l) aspartate in CD34+CD38- CML and CD34+CD38- normal cells measured by LC-MS following 24 hours incubation with 13C6-labeled glucose. n=1 patient sample.
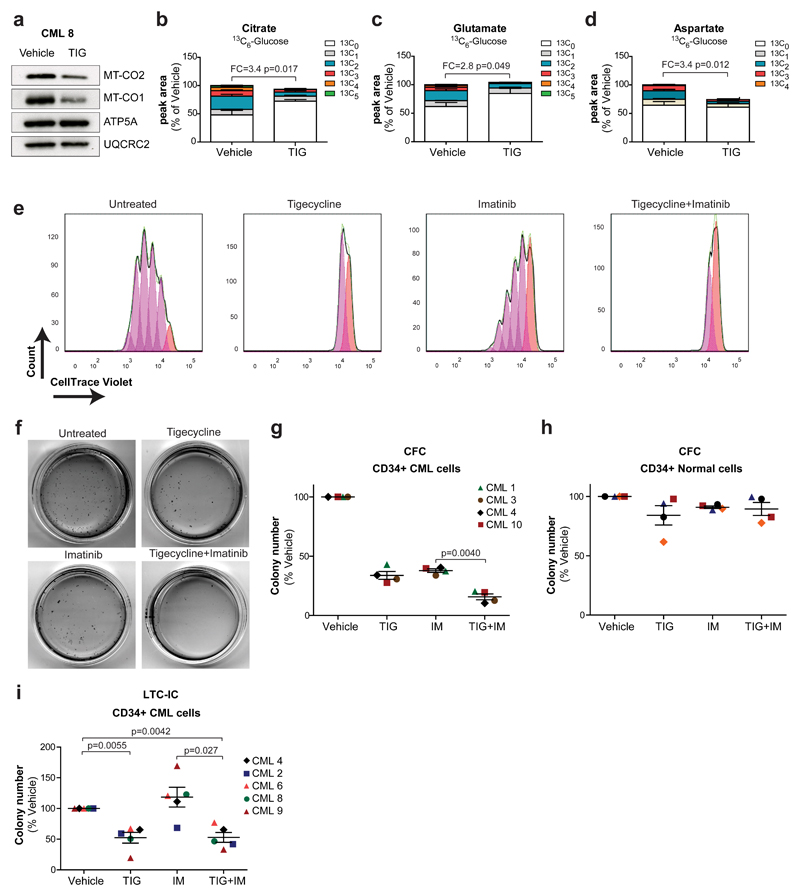
These findings suggest that in primitive CML cells, mitochondrial oxidative metabolism is crucial for production of energy and anabolic precursors and that restraining their mitochondrial functions may have a therapeutic benefit. Tigecycline is an FDA-approved antibiotic, which inhibits bacterial protein synthesis. Due to the similarity between mitochondrial and bacterial ribosomes, it also inhibits the synthesis of mitochondria-encoded proteins, all of which are required for the oxidative phosphorylation machinery22. Previous reports demonstrated therapeutic efficacy of tigecycline against cancer cells, including primary acute myeloid leukemic cells23 and the oxidative subtype of diffuse large B-cell lymphoma22. First, we demonstrated that tigecycline is capable of inhibiting the translation of the mitochondrial-encoded proteins MT-CO1 and MT-CO2, but not the nuclear-encoded mitochondrial proteins ATP5A or UQCRC2 (Fig. 3a and Supplementary Fig. 4a). This was associated with a compensatory increase in MT-CO1 and MT-CO2 mRNA levels (Supplementary Fig. 4b). In line with this, tigecycline treatment significantly impaired mitochondrial respiration of CD34+ CML cells (Supplementary Fig. 4c). Primary CD34+ CML cells were then cultured with 13C6-glucose for 24 hours in the absence or presence of tigecycline. A robust and significant decrease in glucose oxidation was noted in tigecycline-treated cells, with a 2.8-3.4-fold decrease in the incorporation of 13C isotopes into citrate, glutamate and aspartate (Fig. 3b-d). Tigecycline had a broad effect on cellular metabolism as illustrated by a combined decrease in respiration and extracellular acidification rate (Supplementary Fig. 4c,d) indicative of a concurrent impairment of oxidative phosphorylation and glycolysis. This was associated with a decrease in both PDH and PC relative activity (Supplementary Fig. 4e,f). Similarly, CD34+ CML cells treated with tigecycline displayed a significant decrease in the fraction of isotopologues with 2 or more 13C atoms following incubation with 13C5-glutamine and 13C16-palmitate (Supplementary Fig. 4g-l). In tandem to blocking oxidative metabolism, tigecycline treatment decreased the overall steady-state levels of aspartate (Fig. 3d), in support of the anaplerotic role of oxidative metabolism in the LSCs-enriched population.
Figure 3. Inhibition of aberrant oxidative metabolism targets CML progenitors and LSCs.
(a) Protein expression in CD34+ CML cells following a 72 hours in vitro treatment with tigecycline (2.5 µM). n=1 patient sample. (b-d) Relative isotopologue distribution of (b) citrate, (c) glutamate and (d) aspartate in CD34+ CML cells measured by LC-MS following 24 hours incubation with 13C6-labeled glucose in the presence or absence of tigecycline (2.5 µM). Mean ± S.E.M. n=3 patient samples. (e) Representative flow cytometry histograms obtained from cellular division tracking of CellTrace Violet-stained CD34+ CML cells following 72 hours treatment with vehicle only, tigecycline (2.5 µM), imatinib (2 µM) and combination (2.5 µM + 2 µM). (f) Representative images of colonies and (g) colony numbers following 3 days drug treatment of CD34+ CML cells. Mean ± S.E.M. n=4 patient samples. (h) Colony number of CD34+ normal cells following 72 hours drug treatment with the indicated drugs. Mean ± S.E.M. n=4 normal samples. (i) Number of colonies measured by LTC-IC assay in CD34+ CML cells. Mean ± S.E.M. n=5 patient samples. FC, Fold change of glucose-derived (13C ≥ 2) metabolite abundance relative to tigecycline-treated CD34+ CML cells. TIG, tigecycline; IM, imatinib; Combo, combination. P-values were calculated by paired Student’s t-test.
Oxidative phosphorylation and anaplerosis are essential for growth and proliferation. Labeling of CD34+ CML cells with a fluorescent cell division tracker revealed that tigecycline alone, or in combination with imatinib, strongly impaired proliferation of primary CD34+ CML cells, whereas imatinib alone had only a moderate effect, in line with its preferential effect on differentiated CD34- cells (Fig. 3e). Furthermore, treatment with imatinib or tigecycline alone decreased the number of short-term CML CFC, with their combined application effectively eliminating colony formation (Fig. 3f,g). This effect on colony growth correlated with an increase in cell death, measured by Annexin V staining (Supplementary Fig. 5a,b). Importantly, neither drug, alone or in combination, had a significant effect on normal, non-leukemic CFCs, affirming a potential therapeutic window (Fig. 3h). Thus far, our studies suggest that the combined inhibitory effect of tigecycline and imatinib on CFCs may result from the effect of the drugs on two distinct populations, where imatinib targets more mature progenitors, while tigecycline targets the more oxidative, long-term LSCs. To test this hypothesis, LTC-IC assay was performed: CD34+ CML cells were treated once with imatinib or tigecycline, alone or in combination, followed by liquid culture for 5 weeks prior to placing them in semi-solid medium. This ensures selective measurements of the functional capacity of long-term LSCs, since short-term progenitor cells lose their colony forming potential during the 5 weeks culture. This stringent in vitro stem cell assay revealed that while imatinib was ineffective, tigecycline-mediated inhibition of oxidative metabolism significantly decreased CML LSC potential (Fig. 3i). Similar findings were observed when the mitochondrial complex I inhibitor phenformin was used (Supplementary Fig. 5c,d).
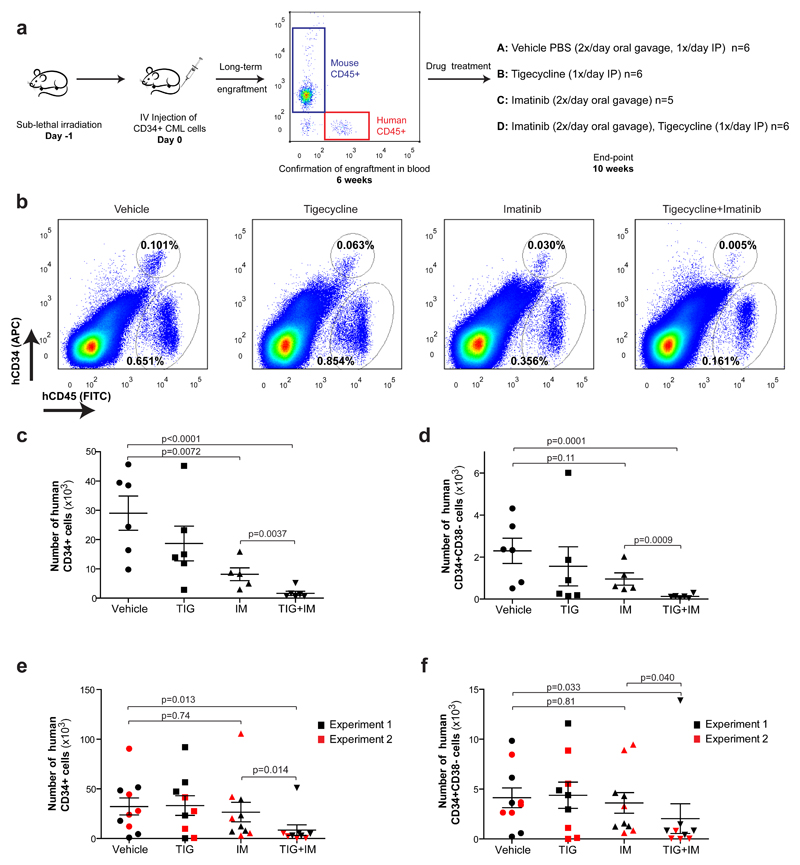
To further assess the clinical relevance of these findings, we moved to a robust xenotransplantation model of human CML. Sub-lethally irradiated immuno-compromised mice were transplanted with CD34+ human CML cells. Six weeks later, engraftment was assessed by recording the percentage of cells expressing human leukocyte common antigen (CD45+) from the total peripheral leukocytes in the blood (Fig. 4a). After ensuring equivalent and sufficient engraftment of human cells in all mice (Supplementary Fig. 6a), mice were treated daily from 6 weeks post-transplantation for a period of 4 weeks with vehicle only, tigecycline (escalating doses of 25-100 mg.kg-1: see Methods), imatinib (100 mg.kg-1) or both drugs combined (Fig. 4a). In all experimental arms, no changes in body or spleen weight or signs of toxicity were observed during treatment and total bone marrow cellularity was unaffected, confirming excellent tolerability (Supplementary Fig. 6b-d). Importantly, the decreased level of mitochondria-encoded proteins was used as a pharmacodynamic biomarker to demonstrate on-target action of tigecycline in vivo (Supplementary Fig. 6e). Following treatment, bone marrow cells were extracted and analysed by flow cytometry for the expression of the human antigens CD45 (leukocytes), CD34 (progenitors and stem cells) and CD38 (to distinguish between CD34+CD38+ progenitors and CD34+CD38- LSCs). The majority of bone marrow cells were non-leukemic host cells (human CD45-) and, as indicated above, were unaffected by the treatment (Fig. 4b and Supplementary Fig. 7a). In contrast, the total number of CML-derived CD45+ cells in the bone marrow was decreased, though marginally, in tigecycline treated mice and significantly in mice treated with imatinib (Supplementary Fig. 7b). Importantly, in the combination arm, the CML burden was further decreased, and when only undifferentiated bone marrow CD45+CD34+ CML cells were analysed, these results were even more pronounced (Fig. 4b,c and Supplementary Fig. 7a,b). The most striking effect though was seen within the more primitive human LSCs population. Whereas imatinib alone only marginally (and insignificantly) decreased the number of CD45+CD34+CD38- CML cells, the combination treatment eliminated 95% of these cells (Fig. 4d).
Figure 4. Inhibition of oxidative metabolism eliminates xenotransplanted human CML LSCs.
(a) Diagram of experimental design. The pre-treatment engraftment levels of CML cells in mice were assessed by monitoring the percentage of human CD45+ circulating leukocytes using flow cytometry. (b) Representative analyses of human CD45 and CD34 expression in murine bone marrow was used to assess engrafted CML cells following the indicated treatment. (c) Number of human CD34+ and (d) human CD34+CD38- CML cells remaining in the bone marrow following in vivo drug treatment. (e) Number of human CD34+ and (f) human CD34+CD38- CML cells remaining in the bone marrow following 2 (experiment 1) or 3 (experiment 2) weeks of drug discontinuation. n≥5 mice per treatment arm. TIG, tigecycline; IM, imatinib. P-values were calculated by unpaired Student’s t-test on logarithmically transformed variables to meet the assumption of normality.
Experiments using cord blood CD34+ cells showed that both imatinib and tigecycline, either alone or in combination, had a marginal effect on engrafted normal blood cells (Supplementary Fig. 8a-c). Finally, an additional two cohorts of mice were transplanted with CD34+ CML cells and treated as before. To investigate whether the combination of tigecycline and imatinib slowed the rate of relapse, both cohorts were then left untreated for an additional two (experiment 1) or three (experiment 2) weeks. Bone marrow analysis following drug withdrawal demonstrated that while mice treated with imatinib as a single agent showed signs of relapse (with the number of leukemic cells similar to untreated mice), the vast majority of mice treated with the combination of tigecycline and imatinib sustained low numbers of LSCs in the bone marrow (Fig. 4e,f).
Taken together, our findings indicate that primitive CML stem/progenitor cells are highly susceptible to the inhibition of oxidative phosphorylation, while CD34+ normal cells are not. This previously unknown metabolic vulnerability shown here represents a therapeutic target for treatment with the FDA-approved mitochondrial translation inhibitor tigecycline. Finally, in vivo, combining tigecycline treatment with the standard-of-care drug, imatinib, produces a selective cytotoxic effect on CD34+ CML and on more primitive LSCs at clinically administrable doses.
Methods
Reagents
Imatinib mesylate was purchased from LC Laboratories. Stock solution of 1 mM imatinib was prepared in sterile distilled water and stored at 4°C. Tigecycline powder (T3324-1, LKT Laboratories) was stored at 4°C and solutions of 5 mM were freshly prepared in dimethylsulfoxide (DMSO; Sigma-Aldrich) the day of the experiment. 13C6-labeled glucose, 13C5-labeled glutamine, 13C16-labeled palmitate were obtained from Cambridge Isotope Laboratories.
Primary samples
CML samples were leukapheresis products from patients in chronic phase CML at the time of diagnosis, with informed consent in accordance with the Declaration of Helsinki and approval of the National Health Service (NHS) Greater Glasgow Institutional Review Board. Normal samples were; i) bone marrow products from healthy donors, ii) surplus cells collected from femoral head bone marrow, surgically removed from patients undergoing hip replacement (with written patient consent and approval from the NHS Greater Glasgow and Clyde Biorepository), or iii) leukapheresis products from patients with non-myeloid Ph-negative haematological disorders. CD34+ cells were isolated using CD34 MicroBead Kit or CliniMACS (both Miltenyi Biotec); the flow through consisting of CD34- cells. For isolation of the CD34+CD38- population, CD34+ samples were stained with anti-human CD34 (APC; BD Biosciences) and anti-human CD38 (PerCP; Biolegend) and sorted using a FACSAria™ Fusion Cell sorter (BD Biosciences).
Cell culture
Primary samples were cultured at 37°C and 5% CO2 in serum-free media (SFM) consisting of Iscove's modified Dulbecco medium (Sigma-Aldrich) supplemented with bovine serum albumin (BSA)/insulin/transferrin (BIT9500; StemCell Technologies), 1 mM streptomycin/penicillin, 0.1 mM 2-mercaptoethanol, and a physiological growth factor (PGF) cocktail comprising of 0.2 ng/mL SCF/GM-CSF/MIP-α, 1.0 ng/mL G-CSF/IL6 (PeproTech EC Ltd) and 0.05 ng/mL LIF (StemCell Technologies).
Drug treatment
Imatinib, tigecycline and phenformin in vitro treatments were performed at a concentration of 2 µM, 2.5 µM and 20 µM respectively unless stated otherwise.
Oxygen consumption rate (OCR) measurements
OCR was measured using the Seahorse XF96 analyzer (Seahorse Bioscience, Billerica, MD). Primary cells were suspended in XF Assay Medium supplemented with 25 mM glucose, 1 mM pyruvate and PGF cocktail. 60,000-100,000 cells were seeded per well of a Seahorse XF96 cell culture plate (35 μl volume) pre-coated with Cell-tak (Fisher Scientific). Cells were left to adhere for a minimum of 30 minutes (min) in a CO2-free incubator at 37°C, after which 140 µl of XF Assay Medium were added into each well. The plate was left equilibrating for 10 min in the CO2-free incubator before being transferred to the Seahorse XF96 analyzer. Measurement of OCR was done at baseline and following sequential injections of a) oligomycin (1 µM), an ATP synthase inhibitor b) FCCP (1.6 µM), a mitochondrial uncoupler c) antimycin A (1 µM) and rotenone (1 µM; all Sigma-Aldrich) a complex III and complex I inhibitor respectively. This enabled us to measure the OCR coupled to ATP production, as well as the maximal and the mitochondrial OCR respectively. OCRs were normalized by cell number.
Cellular division tracking
CD34+ CML cells were stained with 1 μM CellTrace Violet (CellTrace™ Violet Cell Proliferation Kit; Life Technologies) for 30 min at 37°C. The reaction was quenched by adding cell culture media containing 10% fetal bovine serum. Cells were then suspended in SFM supplemented with PGF cocktail and treated with tigecycline (2.5 µM) and imatinib (2 µM) as indicated. After 3 days, CellTrace™ Violet staining was assessed by flow cytometry (BD FACSVerse™).
Colony Forming Cell (CFC) assay
Primary cells were plated in SFM supplemented with PGF cocktail in the presence of indicated drugs. After 3 days, cells from each condition were transferred in methylcellulose-based medium (Methocult H4034 Optimum, StemCell Technologies) in duplicate and colonies were manually counted after 12-14 days.
Human long-term culture initiating-cell (LTC-IC) assay
CD34+ CML cells were plated onto irradiated stromal layers (M2-10B4 and S1/S1) in the presence of indicated drugs in medium for long-term culture of human cells (MyeloCult™ H5100, StemCell Technologies). Cells were kept for a minimum of 5 weeks with weekly half-media change. At this point, the remaining viable progenitors were assessed by CFC assay.
Mitochondrial content and membrane potential
Mitochondrial content was assessed by co-staining CML and normal CD34+ cells with 100 nM Mitotracker Green (MTG; Thermo Fisher Scientific) with anti-human CD34 (APC) and anti-human CD38 (PerCP) antibodies for 30 min at room temperature. For mitochondrial membrane potential assessments, cells were stained with 100 nM Tetramethylrhodamine, methyl ester (TMRM; Thermo Fisher Scientific) and anti-human CD34 (APC), anti-human CD38 (PerCP) antibodies. Fluorescence levels of TMRM and MTG were analysed by flow cytometry.
Western blot analysis
CD34+ CML cells were lysed in RIPA buffer and total protein concentration was measured with a Pierce BCA kit (Thermo scientific). Equal amounts of proteins (5-20 µg) were heated at 95°C for 5 min, and separated in 10% gels for SDS-PAGE. Proteins were transferred on to nitrocellulose membranes (Millipore), blocked in 5% BSA (in TBS+0.01% Tween), and incubated overnight at 4ºC with the following primary antibodies: total OXPHOS cocktail (Abcam, ab110413, 1:2000) and MT-CO2 (Thermo Fisher Scientific, A-6404, 1:2000). The membranes were then incubated with secondary HRP-linked antibodies (1:5000) for 1 hour (h) at room temperature. The Pierce enhanced chemiluminescence detection system was used (Thermo Fisher Scientific).
Cell death
CML CD34+ cells were seeded at 2 × 105 cells/ml and treated for 72 h with 2.5 µM tigecycline, 2 µM imatinib and the combination as indicated. Cell death was quantified by measuring the percentage of Annexin V (FITC; Biolegend) positive CD34+ CML cells by flow cytometry (BD FACSVerse™).
RT-QPCR
CML CD34+ cells were seeded at 2×105 cells/ml and treated for 72 h with 2.5 µM tigecycline. RNA was extracted using the RNeasy Mini Kit (Qiagen). Reverse transcription was performed with SuperScript™ VILO™ Master Mix (Thermo Fisher Scientific).
The following primers were used: MT-CO1_F CTTTTCACCGTAGGTGGCCT, MT-CO1_R AGTGGAAGTGGGCTACAACG, MT-CO2_F CCGTCTGAACTATCCTGCCC, MT-CO2_R GAGGGATCGTTGACCTCGTC, 18S_F GTAACCCGTTGAACCCCATT and 18S_R CCATCCAATCGGTAGTAGCG. cDNAs were mixed with the Fast SYBR® Green Master Mix (4385612, Applied Biosystems), 0.25 µM of each primer, and the volume adjusted to 20 µl according to the manufacturer instructions. The PCR was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems) with the following steps: 20 s at 95°C followed by 40 cycles of 3s at 95°, 30 s at 60°C. The relative quantitation of mRNA was performed by the comparative ΔΔCt method using 18S for normalization.
Intracellular metabolites extraction
Primary cells were plated in the presence of 13C6-labeled glucose, 13C5-labeled glutamine or 13C16-labeled palmitate for 24 h at a concentration of 0.5x106 cells/ml custom-formulated serum-like culture medium containing physiological concentrations of metabolites found in human plasma. After 24 h, cells were washed twice with ice-cold PBS and intracellular metabolites extracted with a cold solution of methanol, acetonitrile and water (5:3:2). The cell extracts were centrifuged at 16,000g for 10 min at 4°C and the supernatants were subjected to LC-MS analysis.
LC-MS analysis
LC-analysis was performed as described previously24,25. A Q-Exactive Orbitrap mass spectrometer (Thermo Scientific) was used together with a Thermo Scientific UltiMate 3000 HPLC system. The HPLC setup consisted of a ZIC-pHILIC column (SeQuant, 150x2.1 mm, 5 µm, Merck KGaA, with a ZIC-pHILIC guard column (SeQuant, 20x2.1 mm). The aqueous mobile phase solvent was 20 mM ammonium carbonate plus 0.1% ammonium hydroxide solution and the organic mobile phase acetonitrile. The metabolites were separated over a linear gradient from 80% organic to 80% aqueous for 15 min. The column temperature was 45°C and the flow rate 200 μL/min. The run time was 22.2 min. All metabolites were detected across a mass range of 75-1000 m/z using the Q-Exactive mass spectrometer at a resolution of 35,000 (at 200m/z), with electrospray ionization and polarity switching mode. Lock masses were used and the mass accuracy obtained for all metabolites was below 5 ppm. Data were acquired with Thermo Xcalibur software.
The peak areas of different metabolites were determined using Thermo LCquan software where metabolites were identified by the exact mass of the singly charged ion and by known retention time on the HPLC column. Commercially available standards compounds had been analysed previously to determine ion masses and retention times on the ZIC-pHILIC column. The 13C labelling patterns were determined by measuring peak areas for the accurate mass of each isotopologue of metabolites. Intracellular metabolites were normalized to cell number and volume.
Mouse engraftment
To study the in vivo engraftment of CML CD34+ cells, 1.5x106 CD34+ CML cells were transplanted via tail vein into 8 week-old sub-lethally irradiated (2.5Gy) female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ NSG mice (The Jackson Laboratory). 6 weeks post-transplant, drug treatment was started with imatinib (100 mg.kg-1; oral gavage twice daily) and tigecycline for 4 weeks (week 1: 25 mg.kg-1, week 2: 50 mg.kg-1, weeks 3-4: 100.mg kg-1; intraperitoneal once daily). At end point, bone marrow cells were stained with anti-human CD45 (FITC; BD Biosciences), CD34 (APC; BD Biosciences) and CD38 (PerCP; Biolegend) antibodies for flow cytometry analysis. Dual-fusion D-FISH was performed as previously described26 to verify engraftment of Ph+ cells. To study the in vivo engraftment of normal cells, 1x105 cord blood CD34+ cells were transplanted into 8 week-old sub-lethally irradiated (2.5Gy) female NSG mice (The Jackson Laboratory). 6 weeks post-transplant, drug treatment was initiated using the same treatment schedule and dose as for CML CD34+ cells (see above). At end point, the remaining cells were stained with anti-human CD45 (FITC; BD Biosciences), CD34 (APC; BD Biosciences) and CD38 (PerCP; Biolegend) antibodies and analysed by flow cytometry.
For the drug withdrawal experiments, 1x106 CD34+ CML cells were transplanted via tail vein into 8-10 week-old sub-lethally irradiated (2.5Gy) female NSG mice (The Jackson Laboratory). 6-8 weeks post-transplant, mice were treated with imatinib (100 mg.kg-1; oral gavage twice daily) and tigecycline (intraperitoneal once daily) for additional 3 to 4 weeks (experiment 1: week 1: 25 mg.kg-1, week 2: 50 mg.kg-1, weeks 3-4: 100.mg kg-1; experiment 2: week 1: 25 mg.kg-1, week 2: 50 mg.kg-1, week 3: 100.mg kg-1). Mice were then left untreated for an additional 2 (experiment 1) or 3 weeks (experiment 2). At end point, bone marrow cells were stained with anti-human CD45 (FITC; BD Biosciences), CD34 (APC; BD Biosciences) and CD38 (PerCP; Biolegend) antibodies for flow cytometry analysis.
Ethics
Ethical approval has been given to the research tissue bank (REC 15/WS/0077) and for using surplus human tissue in research (REC 10/S0704/60). Animal work was carried out with ethical approval from the University of Glasgow under the Animal (Scientific Procedures) Act 1986.
Statistical analyses
No statistical method was used to predetermine sample size. For in vitro experiments, a minimum of 3 patient samples were chosen as a sample size to ensure adequate power. Data obtained from each patient sample represents an independent experiment. For in vivo work, a minimum number of 2 mice per arm were used. The investigators were not blinded to allocation during experiments. Mice were allocated based on their pre-treatment engraftment levels and no method of randomization was used. All mice were cared for equally in an unbiased fashion by animal technicians and investigators. No animal was excluded from the analysis.
P values were calculated by two-tailed paired or unpaired Student’s t-test using GraphPad Prism software (GraphPad Software 5.0) as indicated in the figures legends. Where indicated, variables were transformed using the natural logarithms before t tests were performed to meet the assumption of equal variances.
Supplementary Material
Acknowledgements
We thank all patients and healthy donors who contributed samples; A. Hair for sample processing; T. Gilbey and T. Harvey for cell sorting; N. Van Den Broek and G. MacKay for technical assistance and A. King for editorial work. This study was supported by Cancer Research UK; the Cancer Research UK Glasgow Centre (C596/A18076) and the BSU facilities at the Cancer Research UK Beatson Institute (C596/A17196); MRC/AstraZeneca project grants (MR/K014854/1); the Glasgow Experimental Cancer Medicine Centre (ECMC), which is funded by Cancer Research UK and by the Chief Scientist's Office (Scotland); the Howat Foundation and Friends of Paul O’Gorman; Bloodwise Specialist Programme (14033); the Kay Kendall Leukaemia Fund (KKL501 and KKL698); Lady Tata International Award; Leuka. G.V.H. is a KKLF Intermediate Research Fellow/Leadership Fellow/John Goldman Fellow.
Footnotes
Authors Contribution
E.M.K., G.V.H. and E.G. wrote the manuscript. E.M.K., P.B., T.L.H., G.V.H. and E.G. designed experiments and interpreted data. E.M.K. and P.B. performed experiments and analysed data. A.M.M. and K.D. assisted with the in vivo work. S.T. provided the custom-formulated culture medium. T.L.H. and G.V.H. provided primary cells. G.V.H. and E.G. supervised the project. All authors reviewed the manuscript.
Competing Financial Interests
E.G. is a Founder and Shareholder of MetaboMed Ltd. T.L.H. has previously received research support from Bristol-Myers Squibb and Novartis
Additional information
Additional information regarding human samples, experimental design and material can be found on the Life Sciences Reporting Summary.
References
- 1.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 3.Corbin AS, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton A, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holyoake TL, Helgason GV. Do we need more drugs for chronic myeloid leukemia? Immunological reviews. 2015;263:106–123. doi: 10.1111/imr.12234. [DOI] [PubMed] [Google Scholar]
- 6.Abraham SA, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nature reviews. Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 8.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 9.Groffen J, et al. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 10.Konopka JB, Watanabe SM, Witte ON. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell. 1984;37:1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- 11.Rousselot P, et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood. 2007;109:58–60. doi: 10.1182/blood-2006-03-011239. [DOI] [PubMed] [Google Scholar]
- 12.Mahon FX, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 13.Ross DM, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122:515–522. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 14.Simsek T, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell stem cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takubo K, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell stem cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu WM, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell stem cell. 2013;12:62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito K, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nature reviews. Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan LB, et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardaci S, et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat Cell Biol. 2015;17:1317–1326. doi: 10.1038/ncb3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norberg E, et al. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets. Cell Death Differ. 2016 doi: 10.1038/cdd.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skrtic M, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay GM, Zheng L, van den Broek NJ, Gottlieb E. Analysis of Cell Metabolism Using LC-MS and Isotope Tracers. Methods Enzymol. 2015;561:171–196. doi: 10.1016/bs.mie.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Karvela M, et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy. 2016;12:936–948. doi: 10.1080/15548627.2016.1162359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham SA, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.