SUMMARY
Faithful gene translation depends on accurate decoding, whose structural mechanism remains a matter of debate. Ribosomes decode mRNA codons by selecting cognate aminoacyl-tRNAs delivered by EF-Tu. We present high-resolution structural ensembles of ribosomes with cognate or near-cognate aminoacyl-tRNAs delivered by EF-Tu. Both cognate and near-cognate tRNA anticodons explore the A site of an open 30S subunit, while inactive EF-Tu is separated from the 50S subunit. A transient conformation of decoding-center nucleotide G530 stabilizes the cognate codon-anticodon helix, initiating step-wise “latching” of the decoding center. The resulting 30S domain closure docks EF-Tu at the sarcin-ricin loop of the 50S subunit, activating EF-Tu for GTP hydrolysis and ensuing aminoacyl-tRNA accommodation. By contrast, near-cognate complexes fail to induce the G530 latch, thus favoring open 30S pre-accommodation intermediates with inactive EF-Tu. This work unveils long-sought structural differences between the pre-accommodation of cognate and near-cognate tRNA that elucidate the mechanism of accurate decoding.
Recognition of an mRNA codon by aminoacyl-tRNA (aa-tRNA) occurs at the decoding center (DC) in the A site of the small 30S ribosomal subunit. Aminoacyl-tRNA is delivered to the ribosome as a ternary complex (TC) with elongation factor Tu (EF-Tu) and GTP (EF-Tu•GTP•aa-tRNA). Non-cognate or near-cognate TCs dissociate quickly, whereas cognate TCs dissociate slowly and stimulate GTP hydrolysis by EF-Tu1–8. GTP hydrolysis releases EF-Tu•GDP allowing aa-tRNA accommodation into the 50S A site for peptide-bond formation. EF-Tu-dependent aa-tRNA delivery, therefore, ensures the high fidelity of aa-tRNA selection5,6,9.
The structural mechanism of aa-tRNA discrimination has been extensively studied, but key questions remain unresolved (reviewed in ref10,11). EF-Tu binds a highly conserved region of the large 50S ribosomal subunit called the sarcin-ricin loop (SRL), which prearranges the EF-Tu catalytic site for GTP hydrolysis12–19. The SRL lies >70 Å from the DC, so how tRNA recognition activates EF-Tu’s GTPase center remains unclear. Induced fit of the DC—including universally conserved nucleotides G530, A1492, and A1493 of 16S rRNA, and A1913 of 23S rRNA—was proposed to provide the structural basis for tRNA recognition1,20–22. The mechanistic model focuses on A1492 and A1493, which flip out of the central 16S helix 44 to form A-minor interactions with the cognate codon-anticodon helix, thus monitoring the Watson-Crick geometry of the first and second base pairs20. This idea, however, has been questioned by observations that the DC interacts similarly with mismatched codon-anticodon helices formed by fully-accommodated near-cognate tRNAs23. Recent computational24 and biochemical25,26 studies arrive at diverse conclusions regarding the contribution of these A-minor interactions to decoding fidelity. It therefore remains undetermined what triggers the acceptance of cognate aa-tRNA at the DC.
Biochemical and biophysical studies show that tRNA discrimination occurs prior to full accommodation. Decoding intermediates of the ribosome bound to EF-Tu•GTP•aa-tRNA result in rejection of near-cognate TC or acceptance of the cognate TC2–6,8. Thus, high-resolution structures of both cognate and near-cognate pre-accommodation intermediates would address the following questions: How does the DC discriminate between cognate and near-cognate tRNAs? How does binding of the cognate aa-tRNA anticodon in the DC coordinate with GTPase activation by the SRL?
Here we use single-particle electron cryo-microscopy (cryo-EM) to visualize the binding of cognate or near-cognate TC to 70S ribosomes. Improved classification in FREALIGN27,28 allows us to resolve multiple near-atomic-resolution structures in heterogeneous samples29. We identify six aa-tRNA decoding complexes at up to 3.2-Å resolution (Fig. 1, Extended Data Fig. 1), including five elusive pre-accommodation states not visualized previously (Fig. 1b–c, e–g). These structures report on three binding steps for TC. Together, they reveal differences between cognate and near-cognate pre-accommodation states and suggest a mechanism of aa-tRNA discrimination by the ribosome.
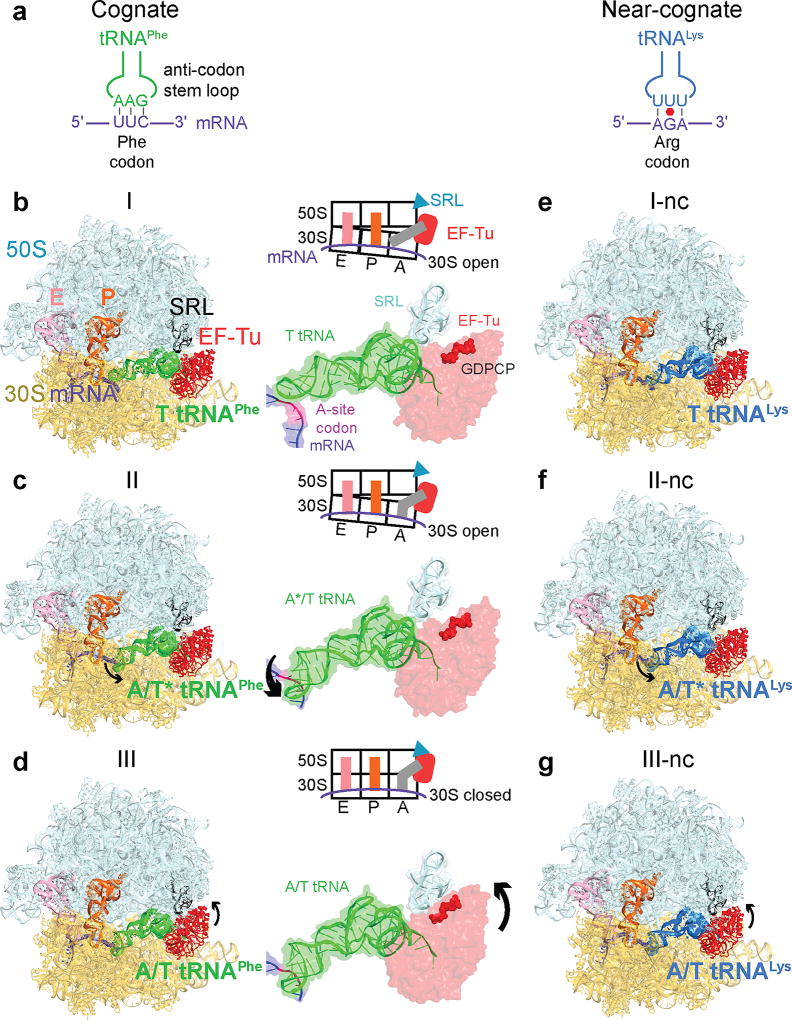
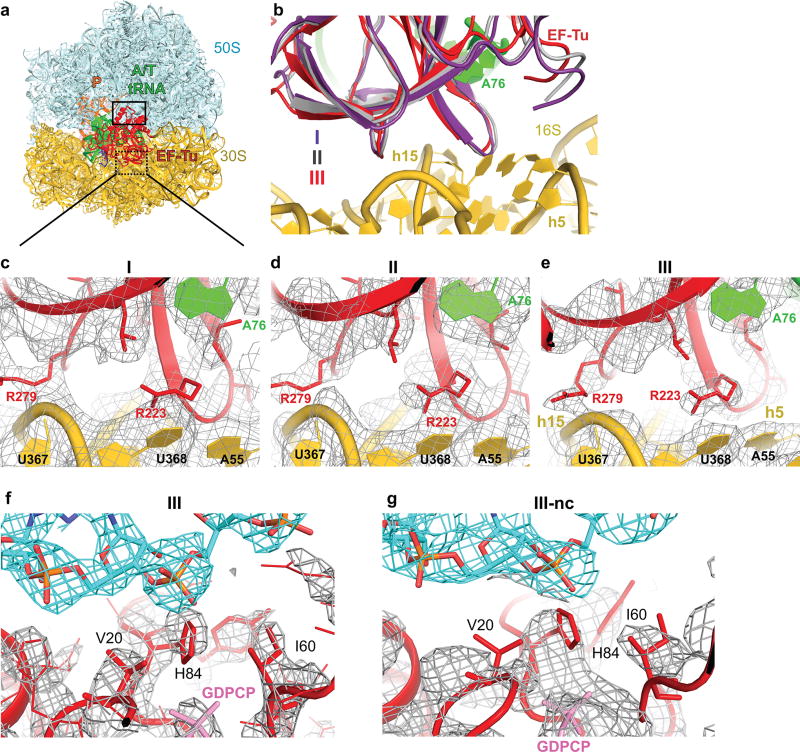
Figure 1. Cryo-EM structures of cognate or near-cognate ternary complex on the 70S ribosome.
(a) Cognate (green) and near-cognate (blue) tRNA anticodons and A-site codons used in complexes. (b) Structure I has cognate TC on the ribosome with an open 30S subunit; the anticodon and codon are not paired. (c) Structure II has cognate TC on the ribosome with an open 30S subunit; the anticodon base-pairs with the codon. (d) Structure III has cognate TC on the ribosome with a closed 30S subunit; EF-Tu docks at the sarcin-ricin loop (SRL). (e–g) Structures I-nc, II-nc, and III-nc with near-cognate TC globally resemble the cognate Structures.
Three states of cognate complex pre-accommodation
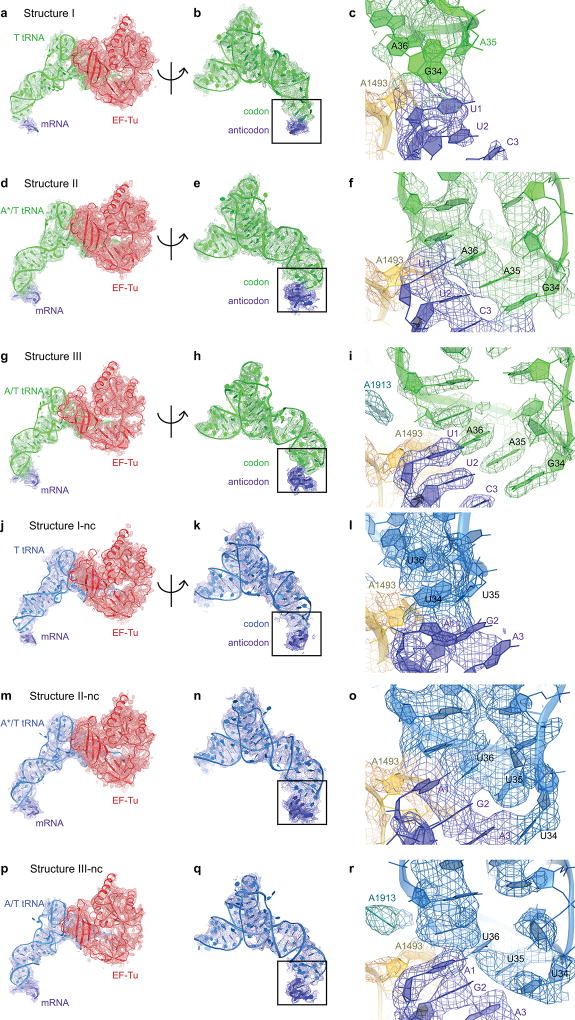
We visualized the dynamics of cognate Phe-tRNAPhe•EF-Tu•GDPCP ternary-complex on E. coli 70S ribosomes programmed with mRNA encoding a phenylalanine UUC codon in the A site. A non-hydrolyzable GTP analog, GDPCP (guanosine-5'-[(β,γ)-methyleno]triphosphate), was used to capture aa-tRNA binding states prior to GTP hydrolysis and EF-Tu dissociation15. Maximum-likelihood classification27,28 of 800,367 ribosome particles revealed three ribosome•tRNA•EF-Tu structures at 3.2- to 3.9-Å resolution: Structure I, Structure II, and Structure III (Fig. 1a–d, Extended Data Fig. 1, Extended Data Table 1). The high resolution of the maps allows detailed interpretation of interactions between the ribosome and TC (Extended Data Fig. 1–3). In Structure I, the TC binds the 30S subunit, but the anticodon does not base-pair with the codon (Fig. 2, 3), nor does EF-Tu contact the 50S subunit (Fig. 4). In Structure II, the anticodon base-pairs with the codon, while EF-Tu remains distant from the 50S subunit. In Structure III, the anticodon base-pairs with the codon, and EF-Tu contacts the SRL of the 50S subunit.
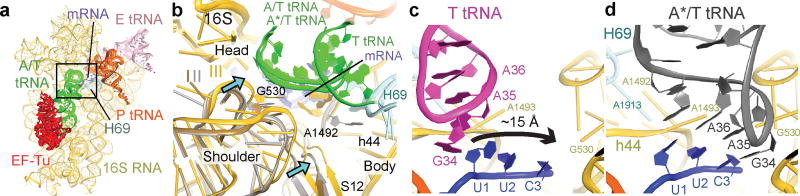
Figure 2. Interactions of cognate ternary complex with the 30S subunit.
(a) 30S subunit shown from the inter-subunit interface with the DC boxed. The 50S subunit (except for helix 69) and small-subunit proteins are omitted for clarity. (b) Conformational differences among Structures I, II, and III suggest a domain-closure pathway. (c) The anticodon in Structure I is ~15 Å from the codon-paired position near G530 (d) in Structure II.
Figure 3. Decoding-center rearrangements in Structures I to III.
(a) Disengaged DC of Structure I. (b) In Structure II, G530 adopts an intermediate position at the minor groove of the codon-anticodon helix. A1492 has partial occupancies inside and outside of helix 44 (gray). (c) In Structure III, G530 moves farther into the minor groove and hydrogen-bonds to A1492. (d–e) Cryo-EM density (gray) showing G530 (yellow) in Structures II (d) and III (e).
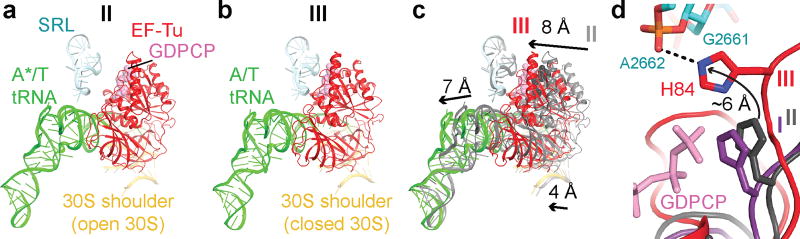
Figure 4. Activation of EF-Tu GTPase from Structure I to III.
(a) In Structures I and II (shown), EF-Tu interacts with the open 30S subunit (shoulder helices 5 and 15, shown) but is distant from the sarcin-ricin loop (SRL). (b) In Structure III, EF-Tu interacts with the closed 30S subunit and SRL. (c) Movement of the 30S shoulder shifts EF-Tu toward the SRL from Structure II (gray) to III (colored). (d) Catalytic residue His84 of EF-Tu is removed from the SRL in Structures I (purple) and II (gray) but interacts with the SRL in Structure III (colored).
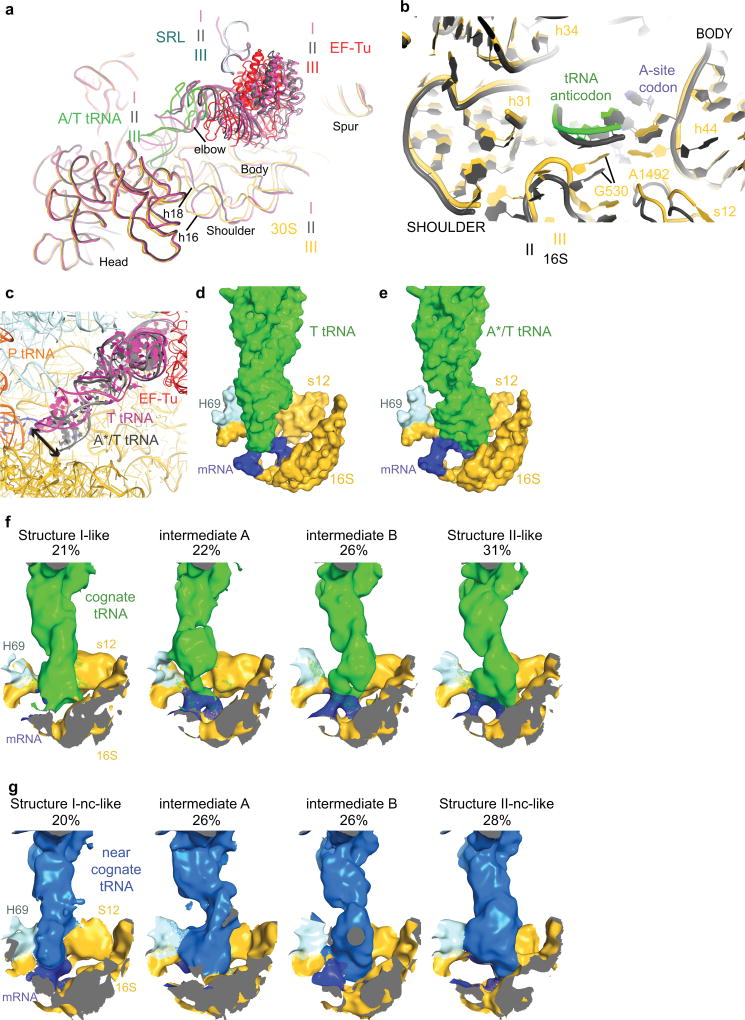
Structures I and II reveal previously unobserved states of ternary-complex binding. These structures have an open 30S-domain conformation as in ribosomes without TC20,21,30,31 or in stringent-response 70S•tRNA•RelA complexes29. In Structure III, the distance from the 30S shoulder to the body closes by 4–5 Å (Fig. 2a–b, Extended Data Figs. 4a–b, Extended Data Table 2). The conformational change in the 30S subunit from Structures I and II to Structure III coincides with 30S “domain closure”, previously inferred from comparisons of 30S structures lacking or containing A-site tRNA anticodon stem-loops (ASL)20,30.
Multiple tRNA conformations sample the A site
Structures I and II reveal different conformations of EF-Tu-bound aa-tRNA, as the ASL interrogates the A site (Fig. 1b–c, Extended Data Fig. 4c). In Structure I, the tRNA resembles the “relaxed” conformation of T tRNA (Ternary-Complex tRNA) seen in crystal structures of isolated TC32. The ASL of the tRNA reaches toward the P-site tRNA. The ASL tip is ~15 Å away from its codon-paired position (Fig. 2c, Extended Data Fig. 4d). In Structure II, the ASL is kinked toward the A-site codon, forming three Watson-Crick codon-anticodon base pairs (Fig. 2d, Extended Data Fig. 4e). The tRNA conformation resembles that of A/T tRNA (A-site / Ternary complex tRNA) characterized in 70S structures with TC14,15,33–35. Unlike in previous structures, however, the 30S subunit is open. We, therefore, distinguish this tRNA conformation as A*/T tRNA. Sub-classification of cryo-EM data revealed additional lower-resolution ASL conformations positioned between the T-tRNA and A*/T-tRNA states (Extended Data Fig. 4f; Supplementary Information).
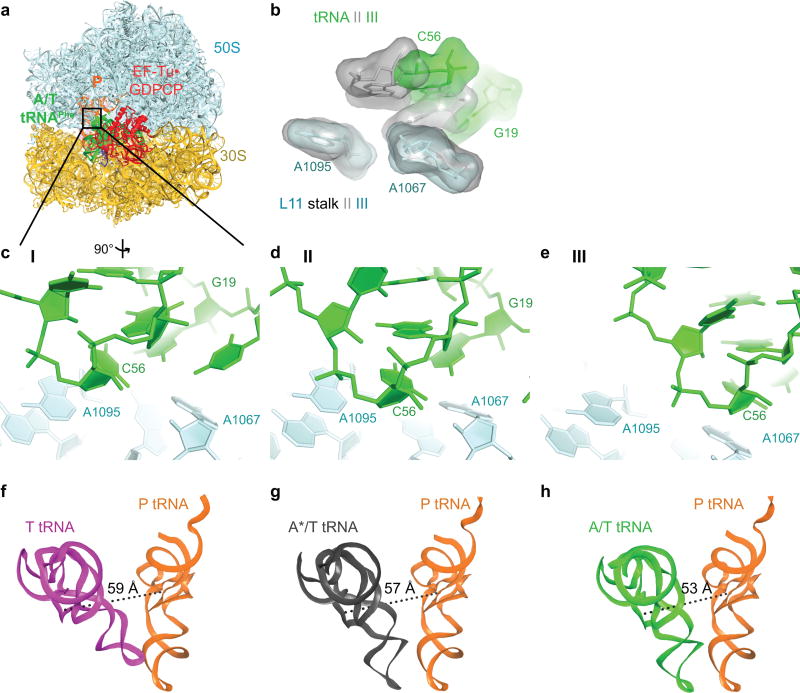
In Structure III, the tRNA adopts the A/T conformation14,15,33–35. From Structure II to III, the tRNA binds deeper into the 30S A site without a substantial conformational change. The acceptor arm shifts by 7 Å toward the P-site tRNA as the elbow slides along the L11 stalk (Extended Data Fig. 5, Supplementary Information). The shift of the tRNA is in excellent agreement with single-molecule FRET studies that monitored step-wise binding of TC8 (Supplementary Information).
G530 triggers “latching” of the DC and 30S-domain closure
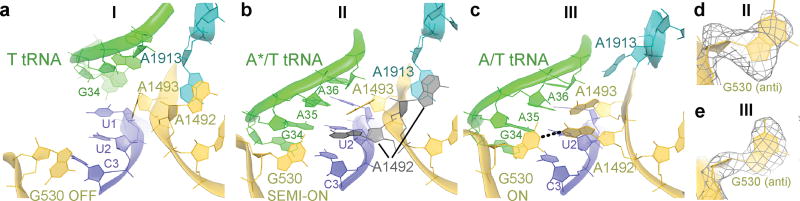
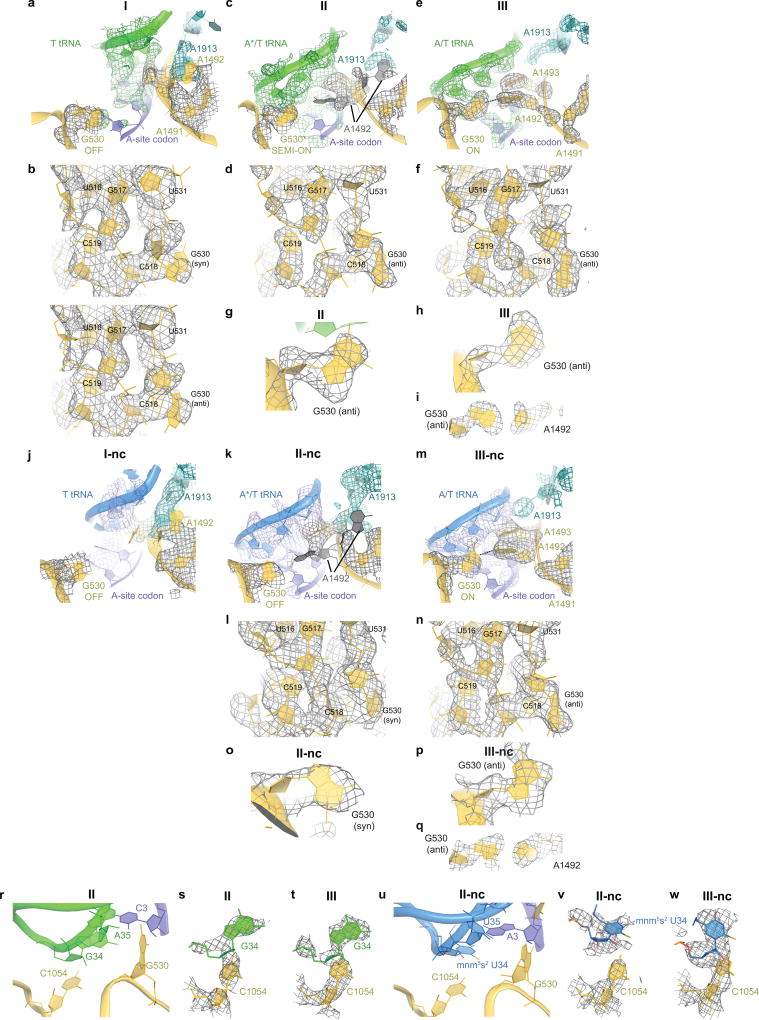
The DC provides the binding pocket for the ASL as the tRNA rearranges from the relaxed to kinked conformation. Comparing Structures I, II, and III reveals novel step-wise rearrangements of DC nucleotides in the presence of TC, suggesting that G530 at the tip of the 30S shoulder plays a central role. In Structure I, the DC nucleotides exhibit conformations found in ribosome structures with empty A sites21,30,31 (Fig. 3a and Extended Data Fig. 6a–b). In particular, G530 is separated from the anticodon and helix 44. A1492 resides inside helix 44 and stacks with A1913 from helix 69 of 23S rRNA. A1493 bulges out from helix 44. Weak density suggests that the base does not stably interact with the A-site codon but is prearranged to bind the codon-anticodon helix. Thus, the DC nucleotides in Structure I adopt the following conformations: G530 OFF, A1913 OFF, A1492 OFF and A1493 SEMI-ON.
Structure II reveals an intermediate position of G530. Strong density shows the nucleotide in the anti conformation flipped out of the 530 loop and partially within the minor groove of the codon-anticodon helix (Fig. 3b,d, Extended Data Fig. 6c–d). G530 hydrogen bonds with the ribose groups of A35 (second nucleotide) of the tRNA anticodon and C3 of the mRNA codon (Fig. 5a). Therefore, G530 in the intermediate state (SEMI-ON) stabilizes the backbone of the codon-anticodon helix. Weak densities for A1492 and A1913 indicate conformational flexibility (ON/OFF). Strong density shows A1493 contacting the minor groove of the first base pair of the codon-anticodon helix (A1493 ON).
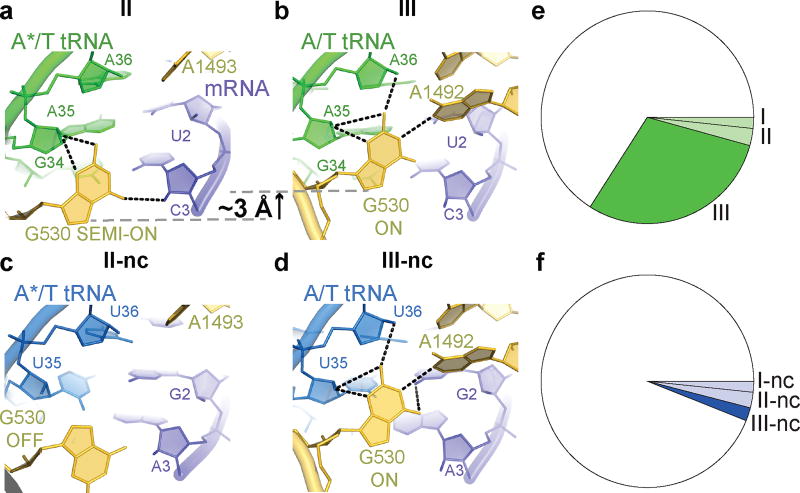
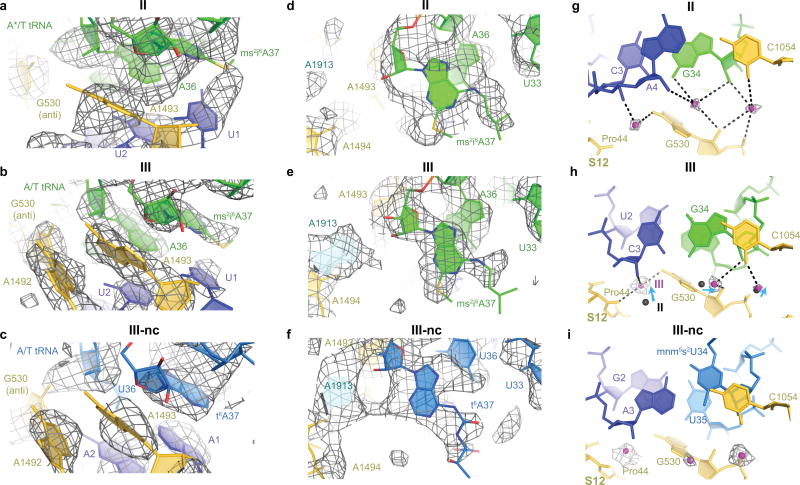
Figure 5. Differences between the cognate and near-cognate Structures.
(a) In Structure II, G530 hydrogen-bonds with the codon-anticodon backbone. (b) In Structure III, G530 moves ~3 Å, restructuring the hydrogen-bonding network. (c) In Structure II-nc, G530 does not stabilize the codon-anticodon helix. (d) In Structure III-nc, G530 stabilizes a Watson-Crick-like codon-anticodon helix. (e–f) Distributions of ribosome particles in cognate (e) and near-cognate (f) Structures.
Finally, the DC nucleotides in Structure III adopt conformations seen in A-tRNA- and A/T-tRNA-bound structures (G530/A1913/A1492/A1493 ON; Fig. 3c,e, Extended Data Fig. 6e–f)1,14,15,20–23,33. Relative to Structure II, the 530 loop and protein S12 shift closer to A1493 and A1492, as G530 moves ~3 Å further into the minor groove of the codon-anticodon helix. This shift restructures G530’s hydrogen-bond network: G530 contacts the riboses of A35 and A36 of the anticodon (at the first and second base pairs) and the base of A1492 (Fig. 5b). Thus, G530 acts as a latch that fastens the codon-anticodon helix into the DC, bringing the 530 loop of the 30S shoulder toward the body, resulting in 30S-domain closure.
30S-domain closure activates EF-Tu
TC must bind the SRL to activate EF-Tu and hydrolyze GTP, releasing EF-Tu from aa-tRNA to allow aa-tRNA accommodation12,13. The SRL prearranges the catalytic His84 of EF-Tu to coordinate the catalytic water molecule14,15. Biochemical studies show that the SRL and His84 are indispensable for GTP hydrolysis and tRNA accommodation16–18.
From Structure I to Structure III, EF-Tu progresses from the inactive-GTPase state to the activated-GTPase state. In all three structures, EF-Tu binds the shoulder of the 30S subunit at helices h5 and h15 of 16S rRNA and protein S12 (Extended Data Fig. 7). From Structures I and II (30S open) to III (30S closed), the 30S shoulder moves toward the 50S subunit by ~4 Å[0], shifting the GTPase domain of EF-Tu by ~8 Å to bind the SRL (Fig. 4). His84 binds the phosphate at A2662 of the SRL, near the terminal phosphate of GDPCP preparing for GTP hydrolysis (Fig. 4d, Extended Data Fig. 7f), as seen in the T. thermophilus 70S pre-accommodation structure15. Thus, our structures show that the decoding-center-induced movement of the 30S shoulder activates EF-Tu.
Near-cognate TC favors open 30S conformations
We next asked whether near-cognate TC has distinct pre-accommodation intermediates explaining discrimination during mRNA decoding. We repeated our experiment using a near-cognate 70S complex formed with tRNALys (UUU anticodon) and mRNA coding for arginine (AGA codon) in the A site, resulting in a G-U mismatch at the second position of the codon-anticodon helix (Fig. 1a; see Supplementary Information). A 572,417-particle dataset yielded three structures of EF-Tu-bound ribosomes at 3.8- to 4-Å resolution (Fig. 1e–g, Extended Data Figs. 1–3, Extended Data Table 1). Overall, the near-cognate structures—Structures I-nc, II-nc, and III-nc—resemble the cognate structures with aa-tRNA bound in the T (I-nc), A*/T (II-nc), and A/T (III-nc) states and the 30S in open (I-nc and II-nc) and closed (III-nc) conformations (Extended Data Fig. 6j–n, 7g; Extended Data Table 2).
In Structure II-nc, the DC differs from that in the cognate Structure II. Whereas G530 in Structure II is clearly resolved in the anti conformation and stabilizes the matched codon-anticodon helix (Fig. 5a, Extended Data Fig. 6g), G530 in Structure II-nc is less resolved, and the density is more consistent with the syn conformation (Fig. 5c, Extended Data Fig. 6k,o). The neighboring nucleotides, however, are similarly well-resolved in Structures II and II-nc, indicating an ordered 530 loop (Extended Data Fig. 6d, l). G530 (OFF) is separated from the ASL, as the latter is shifted relative to the position of the cognate anticodon (Extended Data Fig. 6r–s,u–v). While the anticodon bases interact with the bases of the codon, the G-U mismatch and third base pair deviate from Watson-Crick conformations, shifting the anticodon (Fig. 5c, Extended Data Fig. 2o).
By contrast, in the 30S-closed Structure III-nc, strong density reveals a canonical codon-anticodon helix with the G-U base pair in a tautomeric Watson-Crick-like conformation, similar to the U-A pair in Structure III (Fig. 5b, d). The DC nucleotides in Structure III-nc, including G530, are clearly resolved and adopt conformations similar to those in Structure III (Extended Data Fig. 6m–n,p–q). Thus, G530 latching is coupled with the formation of the Watson-Crick codon-anticodon helix and coincides with domain closure.
TCs formed with tRNAPhe and tRNALys exhibit similar binding affinities to their respective codons36,37 and accuracies of initial selection38, allowing us to compare near-cognate and cognate samples. The distribution of ribosomes with cognate and near-cognate TC differed (Fig. 5e–f). Whereas cognate TC bound to 34% of ribosomes, near-cognate TC bound to 7% of ribosomes—despite assembling samples with 2.5-fold more near-cognate than cognate TC—consistent with the lower affinity of near-cognate TC1–8. Moreover, the cognate complex predominantly samples the closed 30S state (87% of EF-Tu-bound ribosomes), but the near-cognate complex prefers the open 30S states (Structures I-nc and II-nc, 67% of EF-Tu-bound ribosomes) (Fig. 5f). The identities and distributions of these structures are consistent with biochemical, structural, and biophysical data (Supplementary Information). Thus, cognate TCs favor an ordered DC, closed 30S subunit, and activated EF-Tu, whereas the near-cognate TCs favor a disengaged DC, open 30S subunit and inactive EF-Tu.
A mechanism for accurate mRNA decoding
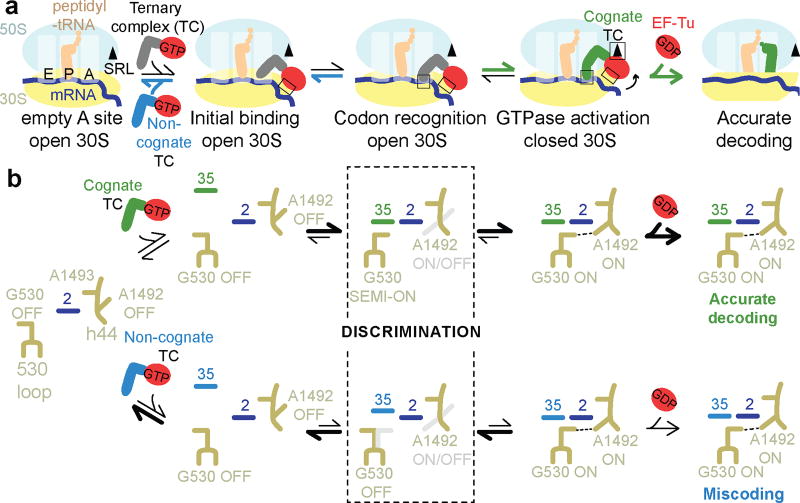
Our work reveals the elusive structures of pre-accommodation intermediates that coincide with the biochemically identified steps4–6,8,12–15: initial codon-independent binding of TC (Structures I and I-nc), codon recognition (Structures II and II-nc), and GTPase activation (Structures III and III-nc). The ensembles of cognate and near-cognate structures provide the structural basis for the initial selection of aa-tRNA (Fig. 6; Video S1).
Figure 6. Structural mechanism of initial aminoacyl-tRNA selection during mRNA decoding.
In the early step(s) of mRNA decoding, TC binds the ribosome via EF-Tu at the 30S shoulder and the tRNA elbow at the L11 stalk (Structure I or I-nc). The open 30S forces the EF-Tu GTPase domain away from the SRL. In the A site, the ASL samples the codon and the DC (Structures I/I-nc and II/II-nc). Formation of the matched codon-anticodon helix (Structure II) is stabilized by interactions between the RNA backbone of the helix and G530 in an intermediate (SEMI-ON) conformation. Engagement of G530 is, thus, independent of the nucleotide identities of the Watson-Crick base pairs (Fig. 5), consistent with uniform affinities of distinct aa-tRNAs to their cognate codons36,37. Stabilization of G530 SEMI-ON, however, may be modulated by tRNA modifications at position 34 and magnesium (Supplementary Information, Extended Data Fig. 8). Bolstered by G530, ASL binding next to helices 44 and 69 destabilizes A1913 stacking on A1492 within helix 44. As A1913 interacts with the ASL, A1492 flips into the minor groove of the codon-anticodon helix, and G530 shifts to “latch” the DC, resulting in domain closure (Structure III). The 30S shoulder moves toward the 50S subunit, docking EF-Tu at the SRL, arranging the EF-Tu active site for GTP hydrolysis. GTPase activation is a rate-limiting step of pre-accommodation4,5, and is physically distinct from GTP hydrolysis13. Structures II and III suggest that GTPase activation results from 30S-domain closure pushing EF-Tu toward the SRL. Indeed, the antibiotic paromomycin, which stabilizes DC nucleotides G530, A1492, and A1493 in the “ON” conformation favoring 30S-domain closure1,20,22, accelerates GTPase activation for both cognate and near-cognate complexes3. The critical structural role of G530 is emphasized by mutational studies, which showed that G530 is indispensable for EF-Tu-dependent aa-tRNA binding39, EF-Tu GTPase activation40 and translation efficiency41–43 (Supplementary Information).
Our structures show that domain closure requires Watson-Crick base pairing at the first two codon-anticodon positions (Structures III and III-nc), as previously proposed based on 70S crystal structures with mismatched tRNA bound in the absence of EF-Tu23. The mismatched codon-anticodon helix in Structure II-nc demonstrates that near-cognate tRNA is inefficient in stabilizing the G530 SEMI-ON state required to initiate 30S-domain closure. The high energetic cost of the Watson-Crick-like G-U base pair—which requires a keto-enol tautomerization44—shifts the conformational equilibrium toward the open 30S states, favoring departure of near-cognate TC. Nevertheless, some near-cognate TCs sample the less favorable Watson-Crick-like conformation and progress to domain closure and EF-Tu activation (Structure III-nc). A small fraction of pre-accommodated near-cognate aa-tRNAs then escapes the EF-Tu-independent proofreading5,6,9,38 leading to rare miscoding.
Our work reveals a key role for G530 in decoding and demonstrates that accurate aa-tRNA selection is achieved by physical separation of EF-Tu from the SRL at early steps of pre-accommodation. We recently reported similar step-wise 30S-domain closure in the presence of deacyl-tRNA and RelA, demonstrating the universality of the tRNA acceptance mechanism29. While we were finalizing this manuscript, a cryo-EM study reported pre-activation states of SelB, a specialized elongation factor that binds an mRNA stem-loop structure and delivers a selenocysteine-tRNA to a UGA stop codon45. Although the resolution of the open-30S structures was limited to ~5 Å, and a WC-paired codon-anticodon intermediate (similar to Structure II) was not observed, the overall mechanism of SelB GTPase activation via the 30S-domain closure appears similar to that of EF-Tu. A lower-resolution (~8 Å) study of the eukaryotic elongation factor EF1A bound to the 80S ribosome suggested that codon recognition and GTPase-activation states differ in positions of aa-tRNA and GTPase domain46. Together, these findings indicate that translation fidelity throughout all domains of life results from direct control of the GTPase by the DC.
Methods
Preparation of E. coli 70S ribosome bound with the cognate or near-cognate ternary complex
E. coli EF-Tu (tufB gene) with a C-terminal hexahistidine tag was overexpressed and purified essentially as described47. The crude E. coli EF-Tu-containing lysate was passed through a His-Trap-HP column. The column was washed with Wash Buffer (50 mM HEPES•KOH, pH 7.5, 60 mM ammonium chloride, 7 mM magnesium chloride, 15 mM imidazole, 500 mM KCl, 5% glycerol) and EF-Tu was eluted with a linear gradient of Wash Buffer mixed with Elution Buffer (50 mM HEPES•KOH, pH 7.5, 60 mM ammonium chloride, 7 mM magnesium chloride, 250 mM imidazole, 5% glycerol). The purity of EF-Tu in eluted fractions was assessed by SDS-PAGE and agarose gel electrophoresis. The purest fractions (>95%) were concentrated and exchanged into EF-Tu storage buffer (50 mM HEPES•KOH, pH 7.5, 60 mM ammonium chloride, 7 mM MgCl2, 10% glycerol, 6 mM β-mercaptoethanol, 20 µM GDP), flash-frozen in liquid nitrogen, and stored at −80°C.
30S and 50S ribosomal subunits were prepared from MRE600 E. coli as described48,49 and stored in Buffer A (20 mM Tris, pH 7, 10.5 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA, 6 mM β-mercaptoethanol) at −80°C. S100 extract was prepared as described50. tRNAfMet, tRNAPhe, and tRNALys (ChemBlock) were charged with their cognate amino acids using the S100 extract and the aminoacylation of tRNAs was confirmed by polyacrylamide gel electrophoresis as described51. mRNA containing the Shine-Dalgarno sequence and a linker to place the AUG codon in the P site and the desired codon in the A site were synthesized by IDT DNA and had the following sequences: Phe cognate complex: GGC AAG GAG GUA AAA AUG UUC AAA AAA; Lys near-cognate complex: GGC AAG GAG GUA AAA AUG AGA AAA AAA (see Supplementary Information).
The 70S•mRNA•fMet-tRNAfMet•EF-Tu•GDPCP•aminoacyl-tRNA complexes were prepared as follows. Heat-activated (42°C) 30S ribosomal subunits (4 µM) were mixed with 50S ribosomal subunits (4 µM) and with the cognate or near-cognate mRNA (20 µM) (all final concentrations) in Reaction Buffer (20 mM HEPES•KOH, pH 7.5, 20 mM magnesium chloride, 150 mM ammonium chloride, 2 mM spermidine, 0.1 mM spermine, 6 mM β-mercaptoethanol) for 45 minutes at 37°C. A two-fold molar excess of fMet-tRNAfMet was added to the ribosomal subunits and incubated for 5 minutes at 37°C, resulting in the 70S•mRNA•fMet-tRNAfMet complexes. To prepare the isolated ternary complex with Phe-tRNAPhe, 2 µM EF-Tu was pre-incubated with 1 mM GDPCP (Jena Bioscience) for 5 minutes at 37°C and then was supplemented with 2 µM Phe-tRNAPhe and incubated for 1 minute at 37°C. For the tRNALys ternary complex, 2.5 µM EF-Tu was pre-incubated with 1 mM GDPCP (Jena Bioscience) for 5 minutes at 37°C and then was supplemented with 2.5 µM Lys-tRNALys and incubated for 1 minute at 37°C. Subsequently, the ternary complexes were chilled on ice and mixed with the 70S•mRNA•fMet-tRNAfMet complexes resulting in the following concentrations for the cognate complex: 250 nM 50S; 250 nM 30S; 1.25 µM mRNA; 500 nM fMet-tRNAfMet; 1 µM EF-Tu; 500 µM GDPCP, and 1 µM Phe-tRNAPhe; for the near-cognate complex: 125 nM 50S; 125 nM 30S; 625 nM mRNA; 250 nM fMet-tRNAfMet; 1.25 µM EF-Tu; 500 µM GDPCP, and 1.25 µM Lys-tRNALys. The complexes were equilibrated on ice for at least 5 minutes prior to application to cryo-EM grids.
Grid Preparation
Holey-carbon grids (C-flat 1.2–1.3, Protochips) were coated with a thin layer of carbon and glow discharged with 20 mA with negative polarity for 45 s in an EMITECH K100× glow discharge unit. 2 µL of 70S•ternary complex sample was applied to each grid. After a 10-second incubation, the grids were blotted for 2 to 4 s at 4°C and ~95% humidity, and plunged into liquid ethane using a CP3 cryo plunger (Gatan Inc.).
Electron Microscopy
Data for the cognate complex and near-cognate complex were collected on a Titan Krios electron microscope (FEI) operating at 300 kV and equipped with K2 Summit direct electron detector (Gatan Inc.) using 0.5- to 2.2-µm underfocus. For the cognate complex, a dataset of 800,367 particles from 3028 videos was collected automatically using SerialEM52. 50 frames per video were collected at 1 e−/Å2 per frame for a total dose of 50 e−/Å2 on the sample. For the near-cognate complex, a dataset of 572,417 particles from 1773 videos of 30 frames each was collected. The videos for the near-cognate complex were taken with 1 e−/Å2 per frame for a total dose of 30 e−/Å2 on the sample. For both datasets, the super-resolution pixel size was 0.82 Å on the sample.
Image processing
Particles were extracted from aligned video sums as follows. Videos were processed using IMOD53 to decompress frames and apply the gain reference. Videos were drift-corrected using unblur54. Magnification anisotropy of the video sums was corrected with mag_distortion_estimate and mag_distortion_correct55. CTFFIND356 was used to determine defocus values. Particles were automatically picked from 10×-binned images using Signature57 with a ribosome reference (18 representative reprojections of the EM databank map 100358, which was low-pass filtered to 50 Å). 480 × 480-pixel boxes with particles were extracted from super-resolution-aligned and magnification-anisotropy-corrected images, and the stack and FREALIGN parameter file were assembled in EMAN259. To speed up processing, binned image stacks were prepared using resample.exe, which is part of the FREALIGN distribution27.
High-resolution map refinement and reconstruction
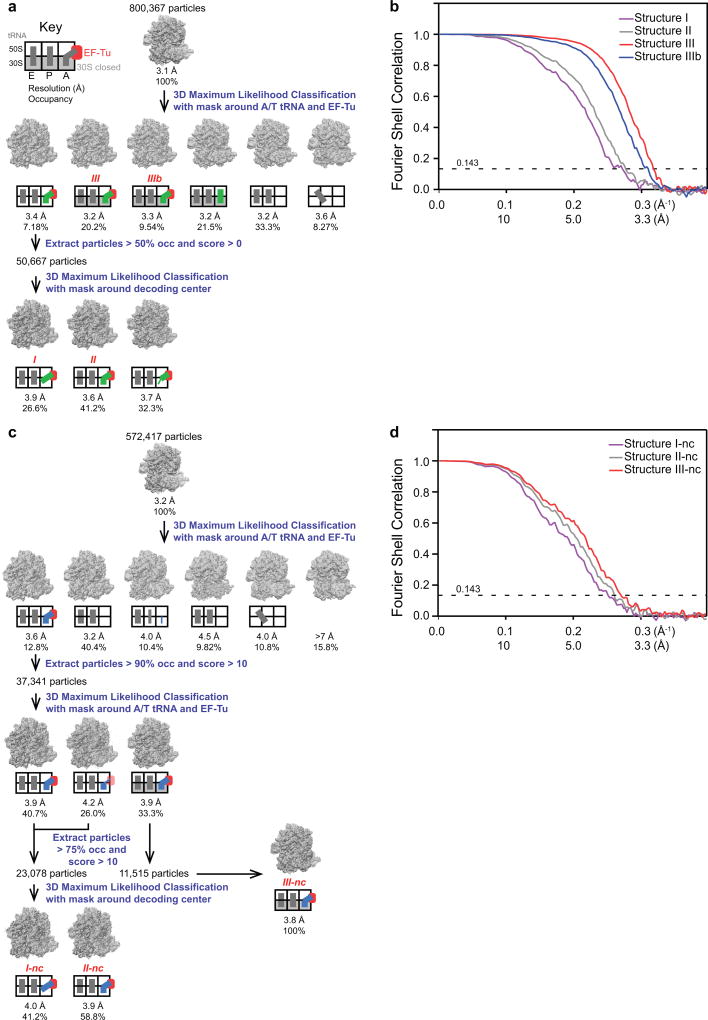
FREALIGN v9 (versions 9.07–9.11) was used for all steps of refinement and reconstruction27 (Extended Data Fig. 1). A 6×-binned image stack was initially aligned to a ribosome reference (EM databank map 1003,58) using three rounds of mode 3 (global search) alignment, including data in the resolution range from 300 Å to 30 Å. Next, the 2×-binned, and later the unbinned image stacks were successively aligned against the common reference using mode 1 (local refinement), including data up to a high-resolution limit (6 Å for the cognate ternary complex or 8 Å for near-cognate ternary complex), whereupon the resolution of the common reference stopped improving. Subsequently, the refined parameters were used for classification of 4×-binned stacks into 6 classes in 50 rounds using a spherical (60-Å radius) focus mask around EF-Tu and A/T tRNA, including resolutions from 300 to 8 Å during classification. This procedure yielded three EF-Tu-containing classes for the cognate complex and one for the near-cognate complex (Extended Data Fig. 1).
Further processing of the cognate complex was as follows. The Structure III map at 3.2-Å resolution was obtained from the 6-model classification described above at 1× binning. A related map, Structure IIIb, also had a closed 30S conformation and activated EF-Tu at the SRL, but a disordered L11 stalk. This class consisted of 72,533 particles and was not used for structure modeling and refinements. Finally, 50,667 particles belonging to the open-30S class were extracted using merge_classes.exe, including particles with >50% occupancy and scores >0. The resulting substack was subjected to further classification with a focused mask (30-Å radius) around the DC. Using three classes separated Structure I and Structure II from a third class in which the anticodon was disordered. The final maps for the Structures I and II were prepared from these classes, using 50% of particles with highest scores.
Further processing of the near-cognate complex proceeded as follows. 37,341 particles belonging to the single class bound with EF-Tu from the 6-model classification described above, were extracted using merge_classes.exe using thresholds of >90% occupancy and scores >10. The particles were classified again for 50 rounds using the same 60-Å-wide focus mask around EF-Tu and A/T tRNA, including resolutions from 300 to 8 Å during classification. This classification separated the near-cognate GTPase-activated state (Structure III-nc) from two maps with an open 30S subunit. Particles belonging to Structure III-nc were extracted using merge_classes.exe using thresholds of >75% occupancy and scores >10 and 50% of them with the highest score were used to prepare the final Structure III-nc map. Particles belonging to the classes with the open 30S subunit were extracted using merge_classes.exe using thresholds of >75% occupancy and scores >10. The resulting substack of 23,078 particles was subjected to a 2-model classification with a focused mask (30-Å radius) around the DC. The final maps for Structures I-nc and II-nc were prepared from these classes, using 50% of particles with highest scores.
We report the percentages of the particles that belong to Structures I, II or III or Structures I-nc, II-nc, or IIInc in Fig. 5e–f. The percentages were calculated using all particles assigned to the corresponding classes shown in Extended Data Fig. 1 (Structure III comprises particles assigned to both 30S-domain-closed classes III and IIIb, which only differ in the L11 stalk occupancy, as described above).
The maps used for structure refinements were sharpened by applying negative B-factors of up to −100 Å2 using bfactor.exe (included with the FREALIGN distribution27). FSC curves were calculated by FREALIGN for even and odd particle half-sets. frealign_calc_stats was used to derive the number of particles assigned to each class. Blocres was used to assess local resolution of unfiltered and unmasked volumes using a box size of 60 pixels, step size of 10 pixels, and resolution criterion of FSC value at 0.14360.
Model building and refinement
The high-resolution cryo-EM structure of the 70S•tRNA•EF-Tu•GDP•kirromycin complex (PDB: 5AFI)33, excluding EF-Tu, A/T-,P-, and E-site tRNAs, was used as a starting model for structure refinements. The starting structural models for fMet-tRNAfMet in the P and E sites were adopted from the 70S•RF2•tRNA crystal structure61. We could not distinguish the identity of E-site tRNA (tRNAfMet or tRNAPhe for cognate complex or tRNAfMet or tRNALys for near-cognate complex) due to lower than average resolution of this part of the cryo-EM maps, likely due to conformational flexibility suggested by further classification. Since tRNAfmet was used in the absence of EF-Tu and is likely to bind the E site upon deacylation, we modeled the E-site tRNA as tRNAfMet. The starting model for A/T Phe-tRNAPhe in Structures II and III was taken from PDB: 5AFI33. The starting model for Phe-tRNAPhe in Structure I was from the crystal structure of the isolated T. aquaticus ternary complex (PDB: 1TTT)32. For the near-cognate structures, the starting model for Lys-tRNALys was from the crystal structure of the ribosome with a near-cognate tRNALys in the A site (PDB: 5IB8)62. The T. aquaticus ternary complex (PDB: 1TTT)32 was used for homology modeling of E. coli EF-Tu employing SWISS-PROT63 and deriving the initial structure of GDPCP. A homology model was similarly created for E. coli L1 utilizing the crystal structure of the isolated T. thermophilus L1 stalk (PDB: 3U4M)64.
All Structures were domain-fitted using Chimera65 and refined using real-space simulated-annealing refinement using RSRef66,67 against corresponding maps. Atomic electron scattering factors68 were used during refinement. Local structural elements that differed between structures, such as the DC, were manually fitted into cryo-EM maps prior to refinement. Refinement parameters, such as the relative weighting of stereochemical restraints and experimental energy term, were optimized to produce the optimal structure stereochemistry, real-space correlation coefficient and R-factor, which report on the fit of the model to the map69. Secondary-structure restraints, comprising hydrogen-bonding restraints for ribosomal proteins and base-pairing restraints for RNA molecules were employed as described70. The structures were next refined using phenix.real_space_refine71 followed by a round of refinement in RSRef applying harmonic restraints to preserve protein backbone geometry66,67. Ions were modeled as Mg2+ in Structure III, filling the difference-map peaks (using CNS)72 residing next to oxygen atoms. Phenix was used to refine B-factors of the models against their respective maps71. The resulting structural models have good stereochemical parameters, characterized by low deviation from ideal bond lengths and angles and agree closely with the corresponding maps as indicated by high correlation coefficients and low real-space R factors (Extended Data Table 1). Structure quality was validated using MolProbity73.
The cryo-EM maps for Structure I and Structure II-nc did not allow unambiguous visual assignment of the G530 conformation. To interpret a predominant conformation, we prepared two ribosome models with G530 in the alternative conformations, syn and anti, and refined the complete ribosome structures independently against corresponding maps. Following the refinements, a preferred fit was assessed based on the local real-space correlation coefficient (CC, calculated only for G530 non-hydrogen atoms). The local CC in Structure I suggests G530-syn and G530-anti fit nearly equally well (CC=0.67 vs. CC=0.66, respectively, Extended Data Fig. 6b). The local CC in Structure II-nc suggests a better fit for G530-syn (CC=0.63), whereas a refined G530-anti yields the moderately lower CC of 0.57. In Structure II, whose density unambiguously shows G530-anti (Extended Data Fig. 8d,g), this preferred conformation yields the CC of 0.71, whereas a refined G530-syn fits poorly and exhibits the CC of 0.44.
Calculation of distance differences using smFRET data
We used published results to estimate the real-time change in distance between accommodating aminoacyl-tRNA and P-site bound tRNA during EF-Tu-mediated tRNA accommodation. An absolute distance between two labeled residues in smFRET experiment is difficult to measure, but distance differences can be derived from relative changes between two FRET efficiency values75. A low-FRET state of 0.35 in ref8 or 0.33 in ref76 and mid-FRET state of 0.58 or 0.4376 were reported during aminoacyl-tRNA accommodation when residue 47 of the accommodating tRNA was labeled with acceptor dye and residue 8 of P-site tRNA was labeled with donor dye. Using the equation in ref75 to deduce distance differences from FRET efficiencies and assuming R0 of 55 Å for the Cy3/Cy5 FRET pair, we calculate that the accommodating tRNA is ~4–6 Å farther from P-site tRNA in the low-FRET state than in the mid-FRET state.
Data Availability
The models generated and analyzed during the current study are available from the RCSB Protein Data Bank: 5UYK (Structure I), 5UYL (Structure II), 5UYM (Structure IIII), 5UYN (Structure I-nc), 5UYP (Structure II-nc) and 5UYQ (Structure III-nc). The cryo-EM maps used to generate models are available from the Electron Microscopy Database: EMBD-8615 (Structure I), EMBD-8616 (Structure II), EMBD-8617 (Structure IIII), EMBD-8618 (Structure I-nc), EMBD-8619 (Structure II-nc) and EMBD-8620 (Structure III-nc).
Extended Data
Extended Data Figure 1. Overview of classification procedures and resolution curves for all Structures.
(a) Scheme of refinement and classification procedures for the cognate dataset. (b) FSC curves for the cognate Structures. (c) Scheme of refinement and classification procedures for the near-cognate dataset. (d) FSC curves for the near-cognate Structures.
Extended Data Figure 2. Cryo-EM densities for ternary complex in each Structure.
(a) Cryo-EM density for ternary complex and codon in Structure I is shown at 3 σ after applying a B-factor of −36 Å2. (b) Cryo-EM density for cognate tRNA and codon in Structure I is shown as in (a). (c) Cryo-EM density for the anticodon and codon, which are not base paired, in Structure I is shown at 4 σ after applying a B-factor of −36 Å2. (d) Cryo-EM density for ternary complex and codon in Structure II is shown at 3 σ after applying a B-factor of −50 Å2. (e) Cryo-EM density for cognate tRNA and codon in Structure II is shown as in (d). (f) Cryo-EM density for the anticodon and codon, which are base paired, in Structure II is shown at 4.5 σ after applying a B-factor of −50 Å2. (g) Cryo-EM density for ternary complex and codon in Structure III is shown at 4 σ after applying a B-factor of −100 Å2. (h) Cryo-EM density for cognate tRNA and codon in Structure II is shown as in (g). (i) Cryo-EM density for the anticodon and codon, which are base paired, in Structure III is shown at 5 σ after applying a B-factor of −150 Å2. (j) Cryo-EM density for ternary complex and codon in Structure I-nc is shown at 3 σ. (k) Cryo-EM density for near-cognate tRNA and codon in Structure I-nc is shown as in (j). (l) Cryo-EM density for the anticodon and codon, which are not base paired, in Structure I-nc is shown at 3.5 σ for T tRNA and 16s rRNA or 4 σ for mRNA. (m) Cryo-EM density for ternary complex and codon in Structure II-nc is shown at 3 σ after applying a B-factor of −25 Å2. (n) Cryo-EM density for near-cognate A*/T tRNA and codon in Structure II-nc is shown as in (m). (o) Cryo-EM density for the anticodon and codon, which are interacting in Structure II-nc is shown at 4.5 σ after applying a B-factor of −25 Å2. (p) Cryo-EM density for ternary complex and codon in Structure III-nc is shown at 4 σ after applying a B-factor of −50 Å2. (q) Cryo-EM density for near-cognate tRNA and codon in Structure II-nc is shown as in (p). (r) Cryo-EM density for the anticodon and codon, which are base paired, in Structure III-nc is shown at 5.2 σ after applying a B-factor of −60 Å2.
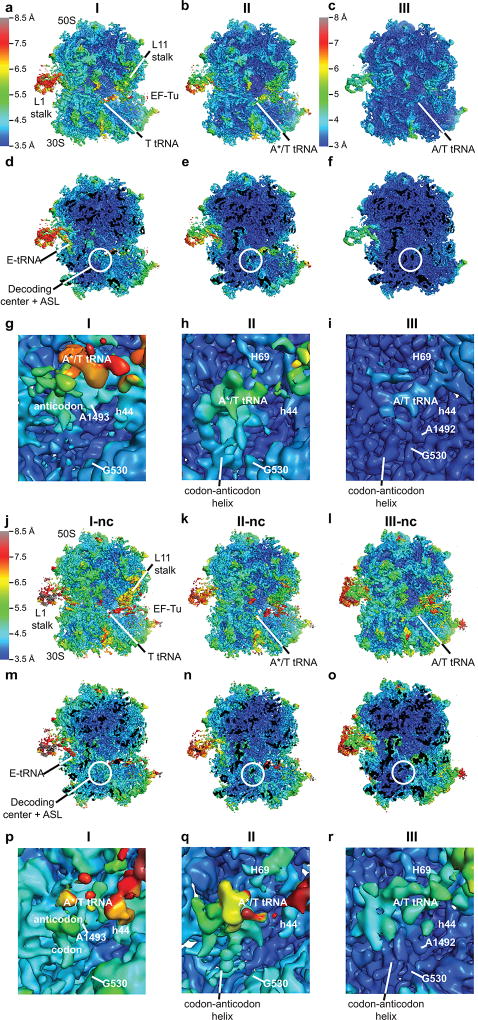
Extended Data Figure 3. Local resolution of cryo-EM maps of the cognate and near-cognate complexes.
Local resolution of each cryo-EM map was determined using Blocres. (a) An overview of the Structure I map. The unsharpened map is shown at 5 σ colored using scale ranging from 3.5–8.5 Å shown on left. (b) An overview of the Structure II map shown as in (a). (c) An overview of the Structure III map. The unsharpened map is shown at 5 σ colored using scale ranging from 3.0–8.0 Å shown on left. (d–f) Slab views at the ribosome interior in maps corresponding to Structure I (d), Structure II (e) and Structure III (f) prepared and colored as in panels a–c, respectively. (g) Close-up view of DC of Structure I. The map was sharpened by applying a B-factor of −36 Å2 and is shown at 4.5 σ colored as in (a). (h) Close-up view of DC of Structure II. The map was sharpened by applying a B-factor of −50 Å2 and is shown at 5 σ colored as in (a). (i) Close-up view of DC of Structure III. The map was sharpened by applying a B-factor of −100 Å2 and is shown at 4 σ colored as in (c). (j) An overview of the Structure I-nc map. The unsharpened map is shown at 5 σ and is colored using scale ranging from 3.5–8.5 Å shown on left. (k) An overview of the Structure II-nc map as in (j). (l) An overview of the Structure III-nc map as in (j). (m–o) Slab views at the ribosome interior in maps corresponding to Structure I-nc (m), Structure II-nc (n) and Structure III-nc (o) prepared and colored as in panel j. (p) Close-up view of DC of Structure I-nc. The unsharpened map is shown at 4.5 σ colored as in (j). (q) Close-up view of DC of Structure II-nc. The map was sharpened by applying a B-factor of −25 Å2 and is shown at 5 σ colored as in (j). (r) Close-up view of DC of Structure III-nc. The map was sharpened by applying a B-factor of −50 Å2 and is shown at 5 σ colored as in (j).
Extended Data Figure 4. 30S domain closure and aa-tRNA conformations in cognate and near-cognate complexes.
(a) Comparison of the 30S conformations among Structures I (magenta), II (gray) and III (multi-colored). Superposition was achieved by structural alignment of 23s rRNA. (b) Superposition of Structure II (gray) and III (multi-colored) highlighting the movement of the shoulder including the 530 loop toward the 30S body including h44. (c) Different conformations of aa-tRNA in Structures I and II: T tRNA (Structure I) is relaxed, whereas A*/T tRNA (Structure II) is kinked to base-pair with mRNA. (d) Interaction of T tRNA in Structure I with the DC is shown in surface representation. All atoms within 15 Å of residues 30–38 of T tRNA are shown except for 16S residues 950–964 and 984–985, which were omitted for clarity. (e) Interaction of A*/T tRNA in Structure II with the DC is shown in surface representation as in (d). (f) Cognate tRNA anticodon samples positions between those in Structure I and Structure II. Additional focused classification into four classes revealed intermediate classes with A-site tRNA density midway between the T tRNA and A*/T tRNA conformations. The cryo-EM density, within 15 Å of residues 30–38 of T or A*/T tRNA, is shown with exceptions as in (d), at 3 σ after applying a B-factor of +200 Å2. (g) Near-cognate tRNA anticodon samples positions between those in Structure I-nc and Structure II-nc. Additional focused classification into four classes revealed intermediate classes with A site tRNA density midway between the T tRNA and A*/T tRNA conformations. The cryo-EM density is shown as in (f).
Extended Data Figure 5. Sliding of tRNA elbow along the L11 stalk from Structure I to III towards the P tRNA agrees with distance changes inferred from single-molecule FRET studies of tRNA decoding8,76.
(a) Overview of Structure III with box highlighting the location of tRNA elbow and L11 stalk. (b) tRNA elbow residues G19 and C57 slide along L11 stalk residues 1095 and 1067 from Structure II (gray) to Structure III (green). Superposition was achieved by aligning on residues 1095 and 1067 of L11 stalk. (c) The elbow of T tRNA (green) and L11 stalk in Structure I. (d) The elbow of A*/T tRNA (green) and L11 stalk in Structure II. (e) The elbow of A/T tRNA (green) and L11 stalk in Structure III. (f) The distance between nucleotide 47 of T tRNA (magenta) and nucleotide 8 of P tRNA (orange) is shown. These locations were used in smFRET studies of tRNA decoding8,76. (g) The distance between nucleotide 47 of A*/T tRNA (gray) and nucleotide 8 of P tRNA (orange) is shown. (h) The distance between nucleotide 47 of A/T tRNA (green) and nucleotide 8 of P tRNA (orange) is shown. The distance changes between T or A*/T tRNA to A/T tRNA are consistent with the change from a low FRET value of 0.35 in the early tRNA decoding states to a mid-FRET value of 0.5 in the GTP activated tRNA decoding state, as described in Methods.
Extended Data Figure 6. Conformational differences in the decoding centers of cognate and near-cognate Structures I–III.
(a) Cryo-EM density (shown as mesh) of the DC in Structure I. The map was sharpened by applying a B-factor of −36 Å2 and density is shown at 3.5 σ for mRNA and anticodon of of T tRNA, 5.5 σ for G530, 4.0 σ for A1492, A1493 and A1913. (b) Cryo-EM density for the G530 region in Structure I is shown with two conformations of G530 syn (top) and anti (bottom). Both conformations fit with similar local cross-correlation coefficients (Methods). The map was sharpened by applying a B-factor of −75 Å2 and density is shown at 5 σ. (c) Cryo-EM density for the DC in Structure II. The map was sharpened by applying a B-factor of −75 Å2 and density is shown at 5.5 σ for G530 or at 4 σ for the mRNA and the anticodon of A*/T tRNA, A1492, A1493 and A1913. Density for residue 1492 (shown in gray) is compatible with two conformations, in and out of h44. (d) Cryo-EM density for the G530 region in Structure II. The map was sharpened by applying a B-factor of −100 Å2 and density is shown at 5 σ. (e) Cryo-EM density for the DC in Structure III. The map was sharpened by applying a B-factor of −150 Å2 and density is shown at 4.5 σ for the mRNA and the anticodon of A/T tRNA, G530, A1492, A1493, and A1913. (f) Cryo-EM density for the G530 region in Structure III. The map was sharpened by applying a B-factor of −150 Å2 and density is shown at 5 σ. (g) Cryo-EM density (gray mesh) showing the anti conformation of G530 (yellow model) in Structure II. The map was sharpened by applying a B-factor of −100 Å2 and density is shown at 5 σ. (h–i) Cryo-EM density showing the anti conformation of G530 in Structure III. The map was sharpened by applying a B-factor of −150 Å2 and density is shown at 5 σ.
(j) Cryo-EM density of the DC in Structure I-nc. The map was not B-factor sharpened and density is shown at 3.75 σ for mRNA, the anticodon of T tRNA, A1492, A1493, and A1913, or at 5 σ for G530. (k) Cryo-EM density for the DC in Structure II-nc. The map was sharpened by applying a B-factor of −25 Å2 and density is shown at 4.3 σ for mRNA, the anticodon of tRNA, A1492, A1493, and A1913, or at 5.5 σ for G530. (l) Cryo-EM density of Structure II-nc for the 30S shoulder including G530. The map was sharpened by applying a B-factor of −120 Å2 and density is shown at 3.5 σ. (m) Cryo-EM density for the DC in Structure III-nc. The map was sharpened by applying a B-factor of −50 Å2 and density is shown at 4.75 σ for mRNA and the anticodon of tRNA, G530, A1492, A1493, and at 4.5 for A1913. (n) Cryo-EM density of Structure III-nc for the 30S shoulder including G530. The map was sharpened by applying a B-factor of −100 Å2 and density is shown at 4.5 σ. (o) Cryo-EM density showing the syn conformation of G530 in Structure II-nc. The map was sharpened by applying a B-factor of −120 Å2 and density is shown at 3.7 σ. (p–q) Cryo-EM density showing the anti conformation of G530 in Structure III-nc. The map was sharpened by applying a B-factor of −120 Å2 and density is shown at 3.3 σ. (r) Nucleotide 34 of the anticodon stacks on C1054 in the cognate Structure II. (s) Cryo-EM density (gray mesh) for the cognate Structure II. The map was sharpened by applying a B-factor of −100 Å2 and density is shown at 5 σ. (t) Cryo-EM density for the cognate Structure III. The map was sharpened by applying a B-factor of −150 Å2 and density is shown at 5.5 σ. (u) Nucleotide 34 of the near-cognate anticodon in Structure II-nc is shifted by ~2 Å from C1054, relative to its position in the cognate complex (shown in panel r). (v) Cryo-EM density for the near-cognate Structure II-nc. The map was sharpened by applying a B-factor of −120 Å2 and density is shown at 3.5 σ. (w) Cryo-EM density for the near-cognate Structure III-nc. The map was sharpened by applying a B-factor of −50 Å2 and density is shown at 5.5 σ. Modification of U34 of tRNALys to 5-methylaminomethyl-2-thiouridine (mnm5s2U34) is shown in panels (u–w).
Extended Data Figure 7. Anchoring of EF-Tu to 30S shoulder in Structures I, II and III and to SRL in Structures III and III-nc.
(a) Overview of Structure III with boxes highlighting locations of EF-Tu contacts to 30S shoulder (dashed box) and to SRL (solid box). (b) The contacts of EF-Tu with the 30S shoulder are similar among Structures I (purple), II (gray) and III (red). (c) Cryo-EM density for EF-Tu (red) and 16S rRNA (pale yellow) in Structure I. The map was sharpened by applying a B-factor of −36 Å2 and is shown at 3 σ. (d) Cryo-EM density for EF-Tu and 16S rRNA in Structure II. The map was sharpened by applying a B-factor of −75 Å2 and is shown at 3.5 σ. (e) Cryo-EM density for EF-Tu and 16S rRNA in Structure III. The map was sharpened by applying a B-factor of −100 Å2 and is shown at 4.5 σ. (f) Cryo-EM density for EF-Tu (red) and the sarcin-ricin loop (SRL) of 23S rRNA (pale cyan) in Structure III. The map was sharpened by applying a B-factor of −150 Å2 and is shown at 4.5 σ. (g) Cryo-EM density for EF-Tu and SRL of 23S rRNA in Structure III-nc. The map was sharpened by applying a B-factor of −50 Å2 and is shown at 4.5 σ.
Extended Data Figure 8. Modifications of A37 in tRNAPhe and tRNALys, and magnesium ion coordination near G530.
(a) Cryo-EM density for the codon-anticodon helix in Structure II shows that the 2-methylthio moiety of 2-methylthio-N6-(2-isopentenyl)-adenosine at position 37 of tRNAPhe (ms2i6A37) stacks on U1 of the A-site codon. The map was sharpened by applying a B-factor of −75 Å2 and density is shown at 4.8 σ. (b) Cryo-EM density for the codon-anticodon helix in Structure III shows that ms2i6A37 of tRNAPhe stacks on U1 similarly to that in Structure II. The map was sharpened by applying a B-factor of −150 Å2 and density is shown at 6 σ. (c) Cryo-EM density for the codon-anticodon helix in Structure III-nc shows that 6-threonylcarbamoyl adenosine at position 37 of tRNALys (t6A37) stacks on A1. The map was sharpened by applying a B-factor of −120 Å2 and density is shown at 4.5 σ. (d) Cryo-EM density for Structure II shows the N6 modification of ms2i6A37 of tRNAPhe in close proximity to U33. The map was sharpened by applying a B-factor of −75 Å2 and density is shown at 4 σ. (e) Cryo-EM density for Structure III shows the N6 modification of ms2i6A37 of tRNAPhe in close proximity to U33. The map was sharpened by applying a B-factor of −150 Å2 and density is shown at 4 σ. (f) Cryo-EM density for Structure III-nc shows the N6 modification of t6A37 of tRNALys. The map was sharpened by applying a B-factor of −120 Å2 and density is shown at 3.5 σ. (g) In Structure II, three magnesium ions (magenta) are coordinated (dotted lines) by G530 and codon-anticodon helix (in some instances, the coordination likely occurs via water molecules). Density for magnesium ions (mesh) was sharpened by applying a B-factor of −75 Å2, shown at 4 σ. (h) In Structure III, the magnesium ions shift with G530. Density was sharpened by applying a B-factor of −150 Å2, shown at 4 σ. (i) In Structure III-nc, three magnesium ions are seen at equivalent position to those in Structure III. Density was sharpened by applying a B-factor of −120 Å2, shown at 3 σ.
Extended Data Table 1.
Refinement statistics for all structures
| Structure I | Structure II | Structure III | Structure I-nc | Structure II- nc |
Structure III- nc |
|
|---|---|---|---|---|---|---|
| PDB code | 5UYK | 5UYL | 5UYM | 5UYN | 5UYP | 5UYQ |
| EMDB code | 8615 | 8616 | 8617 | 8618 | 8619 | 8620 |
|
|
||||||
| Data collection | ||||||
| EM equipment | FEI Titan Krios | FEI Titan Krios | FEI Titan Krios | FEI Titan Krios | FEI Titan Krios | FEI Titan Krios |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Detector | K2 summit | K2 summit | K2 summit | K2 summit | K2 summit | K2 summit |
| Pixel size (Å) | 1.64 | 1.64 | 1.64 | 1.64 | 1.64 | 1.64 |
| Electron dose (e−/Å2) | 50 | 50 | 50 | 30 | 30 | 30 |
| Defocus range (µm) | 0.4 – 5.0 | 0.4 – 5.0 | 0.4 – 5.0 | 0.5 – 5.0 | 0.5 – 5.0 | 0.5 – 5.0 |
|
|
||||||
| Reconstruction | ||||||
| Software | Frealign v9.10–9.11 | Frealign v9.10–9.11 | Frealign v9.10–9.11 | Frealign v9.10–9.11 | Frealign v9.10–9.11 | Frealign v9.10–9.11 |
| Number of particles in final map | 6,726 | 10,431 | 153,597 | 4,629 | 6,910 | 5,758 |
| Final resolution (Å) | 3.9 | 3.6 | 3.2 | 4 | 3.9 | 3.8 |
| Map-sharpening B factor (Å2) | −36 | −50 | −100 | 0 | −25 | −25 |
| Model fitting | ||||||
| Software | Chimera & Pymol | Chimera & Pymol | Chimera & Pymol | Chimera & Pymol | Chimera & Pymol | Chimera & Pymol |
|
|
||||||
| Model composition | ||||||
| Non-hydrogen atoms | 154,413 | 153,755 | 154,140 | 153,718 | 153,781 | 153,760 |
| Protein residues | 6,563 | 6,476 | 6,476 | 6,474 | 6,476 | 6,476 |
| RNA bases | 4,812 | 4,810 | 4,810 | 4,809 | 4,811 | 4,810 |
| Ligands (Zn2+/Mg2+) | 0, 0 | 0, 0 | 2, 383 | 0, 0 | 0, 0 | 0, 1 |
| Ligands/Modifications (GDPCP, fMet, Phe, Lys) | 1 (GDPCP) | 3 (GDPCP, fMet, Phe) | 3 (GDPCP, fMet, Phe) | 1 (GDPCP) | 3 (GDPCP, fMet, Lys) | 3 (GDPCP, fMet, Lys) |
| Refinement | ||||||
| Software | RSRef & Phenix | RSRef & Phenix | RSRef & Phenix | RSRef & Phenix | RSRef & Phenix | RSRef & Phenix |
| Correlation Coefficient* | 0.84 | 0.84 | 0.88 | 0.79 | 0.80 | 0.79 |
| Real space R-factor† | 0.19 | 0.19 | 0.20 | 0.20 | 0.20 | 0.20 |
|
|
||||||
| Validation (proteins) | ||||||
| Molprobity Score‡ | 2.22 | 2.31 | 2.31 | 2.4 | 2.5 | 2.4 |
| Clash score, all atoms‡ | 12.0 | 13.1 | 13.0 | 13.6 | 13.3 | 14.5 |
| Poor rotamers (%)‡ | 0.9 | 1.1 | 1.2 | 1.1 | 1.6 | 1.3 |
| Favored rotamers (%)‡ | 94.9 | 93.8 | 94.6 | 93.6 | 92.8 | 92.9 |
| Ramachandran-plot statistics | ||||||
| Outlier (%)‡ | 1.7 | 2.2 | 2.0 | 2.8 | 3.1 | 2.7 |
| Favored (%)‡ | 86.4 | 85.6 | 86.5 | 83.5 | 82.6 | 84.5 |
| R.m.s. deviations†,§ | ||||||
| Bond length (Å) | 0.007 | 0.008 | 0.006 | 0.005 | 0.005 | 0.005 |
| Bond angle (°) | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 |
|
|
||||||
| Validation (RNA) | ||||||
| Good sugar puckers (%)‡ | 99.6 | 99.5 | 99.5 | 99.6 | 99.6 | 99.4 |
| Good backbone conformation (%)‡ | 88.4 | 88.2 | 88.4 | 88.4 | 88.3 | 87.1 |
Extended Data Table 2.
Distances among cognate Structures I to III and near-cognate Structures I-nc to III-nc, reflecting movements of the 30S shoulder domain relative to the head and body of the 30S subunit.
| Region | Distance* (RMSD, all-atom), Å |
Distance* (RMSD, all-atom), Å |
Distance* (RMSD, all-atom), Å |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| I to II | I to III | II to III | I-nc to II-nc |
II-nc to III-nc |
I-nc to III-nc |
I to I-nc |
II to II-nc |
III to III-nc |
|
| 50S subunit, used to align 70S ribosomes (23S rRNA, excluding L1 and L11 stalks) | 0.46 | 0.51 | 0.47 | 0.52 | 0.71 | 0.71 | 0.57 | 0.56 | 0.66 |
| Body, 30S central region (nt 580–920 of 16S rRNA) | 0.49 | 1.4 | 1.4 | 0.52 | 1.1 | 1.1 | 0.61 | 0.58 | 0.75 |
| Head (nt 960–1400 of 16S rRNA) | 0.56 | 1.6 | 1.7 | 0.60 | 1.2 | 1.2 | 0.71 | 0.75 | 0.86 |
| Shoulder, near 30S center (h18, nt 510–540, 16S rRNA) | 0.90 | 3.2 | 3.2 | 0.78 | 2.8 | 2.7 | 0.89 | 0.93 | 0.85 |
| Shoulder, periphery (h16, nt 400–440) | 0.54 | 4.6 | 4.6 | 0.58 | 4.3 | 4.1 | 0.65 | 0.67 | 0.88 |
Root-mean-square differences (RMSD) that are more or equal to 3× RMSD for the 23S rRNA, are shown in bold blue font. Superposition of Structures relative to each other was obtained by structural alignment of 23S rRNA excluding the L1 and L11 stalks.
Supplementary Material
Acknowledgments
We thank Alexander Park for help with preparing ribosome complexes; Chen Xu and Mike Rigney for help with preparing and screening cryo-EM grids at the cryo-EM facility at Brandeis University; Zhiheng Yu, Chuan Hong, Alexis Rohou and Ruben Diaz-Avalos for data collection at Janelia Research Campus; Dmitri Ermolenko for sharing an EF-Tu-overexpression plasmid and helpful comments on the manuscript; Alexei Korennykh, Darryl Conte Jr. and members of the Korostelev laboratories for helpful comments on the manuscript. This study was supported by NIH Grants R01 GM106105 and GM107465 (to A.A.K.) and P01 GM62580 (to N.G.). A.B.L. performed this work as a Howard Hughes Medical Institute Fellow of the Helen Hay Whitney Foundation.
Footnotes
Author Contributions
ABL, NG, and AAK designed the project; ABL prepared ribosome complexes, collected and analyzed cryo-EM data; GD assisted with protein purification and prepared ribosomes; NG and AAK oversaw cryo-EM data processing; ABL and AAK built and refined structural models; ABL and AAK wrote the manuscript; all authors contributed to manuscript finalization.
Competing interests
The authors declare that there are no conflicts of interest.
Literature Cited
- 1.Ogle JM, Murphy FV, IV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form. Cell. 2002;111:721–732. doi: 10.1016/S0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina MV, Pape T, Fricke R, Kuhn L, Wintermeyer W. Initial binding of the elongation factor Tu.GTP.aminoacyl-tRNA complex preceding codon recognition on the ribosome. The Journal of Biological Chemistry. 1996;271:646–652. doi: 10.1074/jbc.271.2.646. [DOI] [PubMed] [Google Scholar]
- 3.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nature Structural & Molecular Biology. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 4.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. The EMBO Journal. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gromadski KB, Rodnina MV. Kinetic Determinants of High-Fidelity tRNA Discrimination on the Ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/S1097-2765(04)00005-X. [DOI] [PubMed] [Google Scholar]
- 6.Gromadski KB, Daviter T, Rodnina MV. A Uniform Response to Mismatches in Codon-Anticodon Complexes Ensures Ribosomal Fidelity. Mol. Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Ieong K-W, Johansson M, Ehrenberg M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. PNAS. 2015;112:9602–9607. doi: 10.1073/pnas.1506823112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nature Structural & Molecular Biology. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 9.Ieong K-W, Uzun Ü, Selmer M, Ehrenberg M. Two proofreading steps amplify the accuracy of genetic code translation. PNAS. 2016;113:13744–13749. doi: 10.1073/pnas.1610917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voorhees RM, Ramakrishnan V. Structural Basis of the Translational Elongation Cycle. Annu. Rev. Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 11.Maracci C, Rodnina MV. Review: Translational GTPases. Biopolymers. 2016;105:463–475. doi: 10.1002/bip.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E.coli ribosome. The EMBO Journal. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. The EMBO journal. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeing TM, et al. The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The Mechanism for Activation of GTP Hydrolysis on the Ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hausner TP, Atmadja J, Nierhaus KH. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 17.Bilgin N, Ehrenberg M. Mutations in 23 S Ribosomal RNA Perturb Transfer RNA Selection and can Lead to Streptomycin Dependence. Journal of Molecular Biology. 1994;235:813–824. doi: 10.1006/jmbi.1994.1041. [DOI] [PubMed] [Google Scholar]
- 18.Daviter T, Wieden H-J, Rodnina MV. Essential Role of Histidine 84 in Elongation Factor Tu for the Chemical Step of GTP Hydrolysis on the Ribosome. Journal of Molecular Biology. 2003;332:689–699. doi: 10.1016/S0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- 19.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 20.Ogle JM, et al. Recognition of Cognate Transfer RNA by the 30S Ribosomal Subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 21.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nature Structural & Molecular Biology. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 22.Selmer M, et al. Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 23.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 24.Zeng X, Chugh J, Casiano-Negroni A, Al-Hashimi HM, Brooks CL. Flipping of the ribosomal A-site adenines provides a basis for tRNA selection. Journal of Molecular Biology. 2014;426:3201–3213. doi: 10.1016/j.jmb.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrode P, Huter P, Clementi N, Erlacher M. Atomic mutagenesis at the ribosomal decoding site. RNA biology. 2016:0. doi: 10.1080/15476286.2016.1256535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khade PK, Shi X, Joseph S. Steric complementarity in the decoding center is important for tRNA selection by the ribosome. Journal of Molecular Biology. 2013;425:3778–3789. doi: 10.1016/j.jmb.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grigorieff N. ed BT - Methods in Enzymology. Academic Press; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loveland AB, et al. Ribosome•RelA structures reveal the mechanism of stringent response activation. eLife. 2016;5:e17029. doi: 10.7554/eLife.17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 31.Schuwirth BS, et al. Structures of the Bacterial Ribosome at 3.5 Å Resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 32.Nissen P, et al. Crystal Structure of the Ternary Complex of Phe-tRNAPhe, EF-Tu, and a GTP Analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 33.Fischer N, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 34.Valle M. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. The EMBO Journal. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark H, et al. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- 36.Ledoux S, Uhlenbeck OC. Different aa-tRNAs Are Selected Uniformly on the Ribosome. Mol. Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fahlman RP, Dale T, Uhlenbeck OC. Uniform Binding of Aminoacylated Transfer RNAs to the Ribosomal A and P Sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Ieong K-W, Mellenius H, Ehrenberg M. Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs. RNA. 2016;22:896–904. doi: 10.1261/rna.055632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers T, Noller HF. Evidence for functional interaction between elongation factor Tu and 16S ribosomal RNA. PNAS. 1993;90:1364–1368. doi: 10.1073/pnas.90.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nature Structural & Molecular Biology. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- 41.Abdi NM, Fredrick K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santer M, et al. Functional effects of a G to U base change at position 530 in a highly conserved loop of Escherichia coli 16S RNA. Biochemistry. 1993;32:5539–5547. doi: 10.1021/bi00072a007. [DOI] [PubMed] [Google Scholar]
- 43.Taliaferro DL, Farabaugh PJ. Testing constraints on rRNA bases that make nonsequence-specific contacts with the codon•anticodon complex in the ribosomal A site. RNA. 2007;13:1279–1286. doi: 10.1261/rna.552007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westhof E. Isostericity and tautomerism of base pairs in nucleic acids. FEBS letters. 2014;588:2464–2469. doi: 10.1016/j.febslet.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 45.Fischer N, et al. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature. 2016;540:80–85. doi: 10.1038/nature20560. [DOI] [PubMed] [Google Scholar]
- 46.Budkevich TV, et al. Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement. Cell. 2014;158:121–131. doi: 10.1016/j.cell.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. PNAS. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 49.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 50.Traub P, Mizushima S, Lowry CV, Nomura M. In: Vol. 20 Part C. Nucleic Acids and Protein Synthesis. BT - Methods in Enzymology, editor. Academic Press; 1971. pp. 391–407. [Google Scholar]
- 51.Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Kremer JR, Mastronarde DN, McIntosh JR. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 54.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J. Struct. Biol. 2015 doi: 10.1016/j.jsb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 57.Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. J. Struct. Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 58.Gabashvili IS, et al. Solution Structure of the E. coli 70S Ribosome at 11.5 Å Resolution. Cell. 2000;100:537–549. doi: 10.1016/S0092-8674(00)80690-X. [DOI] [PubMed] [Google Scholar]
- 59.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Cardone G, Heymann JB, Steven AC. One number does not fit all: Mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rozov A, Westhof E, Yusupov M, Yusupova G. The ribosome prohibits the G•U wobble geometry at the first position of the codon–anticodon helix. Nucleic Acids Research. 2016;44:6434–6441. doi: 10.1093/nar/gkw431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bairoch A, Boeckmann B, Ferro S, Gasteiger E. Swiss-Prot: Juggling between evolution and stability. Briefings in Bioinformatics. 2004;5:39–55. doi: 10.1093/bib/5.1.39. [DOI] [PubMed] [Google Scholar]
- 64.Tishchenko S, et al. High-resolution crystal structure of the isolated ribosomal L1 stalk. Acta Crystallographica. Section D, Biological Crystallography. 2012;68:1051–1057. doi: 10.1107/S0907444912020136. [DOI] [PubMed] [Google Scholar]
- 65.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Korostelev A, Bertram R, Chapman MS. Simulated-annealing real-space refinement as a tool in model building. Acta Crystallogr D Biol Crystallogr. 2002;58:761–767. doi: 10.1107/s0907444902003402. [DOI] [PubMed] [Google Scholar]
- 67.Chapman MS. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron-density function. Acta Crystallographica Section A. 1995;51:69–80. doi: 10.1107/S0108767394007130. [DOI] [Google Scholar]
- 68.Gonen T, et al. Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou G, Wang J, Blanc E, Chapman MS. Determination of the relative precision of atoms in a macromolecular structure. Acta Crystallographica. Section D, Biological Crystallography. 1998;54:391–399. doi: 10.1107/s0907444997011530. [DOI] [PubMed] [Google Scholar]
- 70.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 71.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods (San Diego, Calif.) 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature Protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 73.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D. Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- 75.Tinoco I, Gonzalez RL. Biological mechanisms, one molecule at a time. Genes & Development. 2011;25:1205–1231. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalez RL, Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. RNA. 2007;13:2091–2097. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The models generated and analyzed during the current study are available from the RCSB Protein Data Bank: 5UYK (Structure I), 5UYL (Structure II), 5UYM (Structure IIII), 5UYN (Structure I-nc), 5UYP (Structure II-nc) and 5UYQ (Structure III-nc). The cryo-EM maps used to generate models are available from the Electron Microscopy Database: EMBD-8615 (Structure I), EMBD-8616 (Structure II), EMBD-8617 (Structure IIII), EMBD-8618 (Structure I-nc), EMBD-8619 (Structure II-nc) and EMBD-8620 (Structure III-nc).