Summary
The intestinal tract of mammals is colonized by a large number of microorganisms including trillions of bacteria that are referred to collectively as the gut microbiota. These indigenous microorganisms have co-evolved with the host in a symbiotic relationship. In addition to metabolic benefits, symbiotic bacteria provide the host with several functions that promote immune homeostasis, immune responses and protection against pathogen colonization. The ability of symbiotic bacteria to inhibit pathogen colonization is mediated via several mechanisms including direct killing, competition for limited nutrients and enhancement of immune responses. Pathogens have evolved strategies to promote their replication in the presence of the gut microbiota. Perturbation of the gut microbiota structure by environmental and genetic factors increases the risk of pathogen infection, promotes the overgrowth of harmful pathobionts, and the development of inflammatory disease. Understanding the interaction of the microbiota with pathogens and the immune system will provide critical insight into the pathogenesis of disease and the development of strategies to prevent and treat inflammatory disease.
Keywords: Inflammatory Bowel Disease, Bacterial, Mucosa, Gut microbiota, Colonization resistance
1. INTRODUCTION
The oral cavity and the gastrointestinal tract of vertebrates are colonized by large numbers of microorganisms, including bacteria, fungi, archaea and protozoa, commonly referred to as the microbiota. Microbes colonize mammalian hosts immediately after birth. Many of the resident bacteria are adapted to the intestinal environment and develop complex interactions with other bacteria and host niches to acquire nutrients. The composition of the microbiota is largely defined by nutrient requirements of individual bacteria and highly variable at different locations of the intestinal tract. In neonatal mice, the microbiota is less diverse than that of adult individuals, but as the diet changes from maternal milk to fiber-rich foods it dramatically changes by the acquisition of Clostridiales and Bacteroidales, the dominant taxa found in the adult intestine (1). The small intestine is rich in mono- and di-saccharides as well as amino acids, which support the growth of Proteobacteria and Lactobacillales (2). In contrast, the vast majority of available sugars in the large intestine are diet and host-derived complex carbohydrates which are indigestible by the host. Bacteroidales and Clostridiales harbor enzymes that can break down complex polysaccharides, including fibers and mucins, and use them as an energy source. Consequently, bacteria belonging to the order of Bacteroidales and Clostridiales are the dominant populations within the large intestine.
Millions of years of co-evolution between the host and microbes have led to a mutualistic symbiosis in which the microbiota contributes to many host physiological processes and the host, in turn, provides a nutritious and hospitable environment to the microbes. In addition to metabolic benefits, the microbiota provides the host with several functions that promote the intestinal epithelial barrier, immune homeostasis, optimal immune responses and protection against pathogen colonization. Although the vast majority of intestinal symbionts are mutualistic or commensals (they do not provide a clear benefit to the host), some of the indigenous bacteria can promote disease under certain circumstances and are commonly referred to as pathobionts. In this review, we provide an overview of the current understanding of the role of the microbiota in the regulation of immune responses and host defense against invading pathogens and pathobionts.
2. COLONIZATION RESISTANCE
The normal gut symbionts form a stable community that resists the invasion of non-native bacteria and the expansion of pathobionts. This phenomenon is known as “colonization resistance”, and has been recognized since at least the 1950s (3, 4). An immature bacterial community (such as in infants) or one that is disrupted by antibiotics or diet may lose this protective ability. Colonization resistance actually includes several related aspects: resistance to initial infection, improved tolerance of an established infection, and clearance of the infection. These all arise from the constant competition between normal gut residents (mutualists, commensals, and pathobionts). The mechanisms bacteria use to compete in the gut can be divided into two broad categories: direct and indirect (Figure 1).
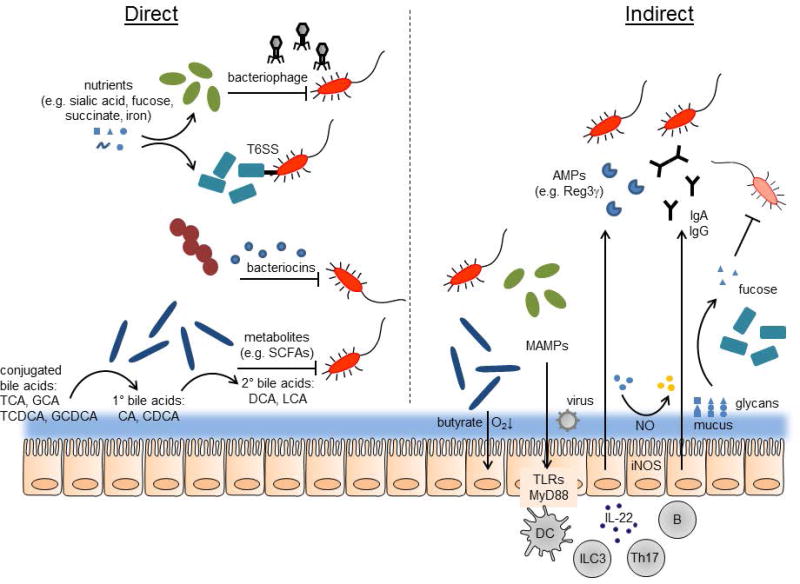
FIGURE 1.
Direct and indirect mechanisms of colonization resistance. Direct mechanisms (left): Symbiotic bacteria scavenge nutrients that would otherwise be available to pathogens (red, with flagella). Bacteriophages, type 6 secretion systems (T6SS), and bacteriocins may target and kill pathogens. Products of bacterial metabolism, such as short-chain fatty acids (SCFAs), can inhibit pathogen growth. Symbiotic bacteria produce enzymes that convert conjugated, primary bile acids to secondary bile acids, which can kill some pathogens. Indirect mechanisms (right): symbionts produce butyrate which can lower oxygen concentration by stimulating host epithelial cell metabolism. Microbe-associate molecular patterns (MAMPs) produced by bacteria and viruses stimulate host innate immunity via TLRs and MyD88, on epithelial cells directly or dendritic cells (DCs). ILC3 and Th17 cells can be activated to produce IL-22, which promotes secretion of AMPs (antimicrobial peptides) such as Reg3g from epithelial cells. B cells produce IgA and IgG antibodies, which can target bacteria in the lumen. Mucus production is stimulated by bacteria, and the mucus is decorated with various glycans. These can be cleaved by bacterial enzymes and the free sugars, such as fucose, can suppress pathogen or pathobiont virulence. The host can also oxidize sugars via reactive nitrogen species produced by inducible nitric oxide synthase (iNOS).
2.1 Direct mechanisms of colonization resistance
The microbiota promotes direct colonization resistance through killing and competition for resources. Bacteria must compete for limited nutrient sources in the gut, as well as physical space. At the same time, they have developed an array of armaments to directly kill competitors. Both of these mechanisms tend to act between more closely-related species: similar bacteria tend to utilize similar nutrients or niches, and have evolved targeted killing mechanisms to compete with their own kind.
2.2 Killing
Evidence suggests that killing or growth suppression can play a dominant role in colonization resistance against some pathogens (5). Bactericidal molecules are found throughout nature; a large portion of these are small polypeptides produced by bacteria, called bacteriocins. These are usually active against closely related bacteria, although some have a wider spectrum of activity. Many have been isolated from gut bacteria of human and animals, lactic acid bacteria found in fermented foods, and common probiotics like the Bifidobacteria (6), so it is reasonable to think that they could be involved in competition in the gut. Indeed, strains of Escherichia coli (E. coli) that could make bacteriocins had improved long-term persistence when introduced into mice compared to non-producers (7). However, these mice were pre-treated with streptomycin, which disrupts the normal community structure. Like E. coli, bacteriocin-producing Enterococcus faecalis (E. faecalis) were better able to colonize mice, in this case without antibiotic pre-treatment. The bacteriocin-positive strain could also inhibit colonization by a different E. faecalis, an opportunistic pathogen, vancomycin-resistant Enterococcus (VRE) (8). A human-derived probiotic Lactobacillus strain protected mice from Listeria monocytogenes infection, and this was dependent on the Lactobacillus bacteriocin (9). The human probiotic E. coli Nissle 1917 may also utilize bacteriocins to compete with and protect mice from Salmonella enterica subsp. Typhimurium (S. Typhimurium) (10). There is also evidence that bacteriocins may contribute to continuous intraspecies competition in the gut (11). Still, it is unclear how big a role bacteriocins play in colonization resistance to relevant enteric pathogens.
Other antibacterial factors are also active in the gut. Bacteriophages (viruses that infect bacteria) can have a profound impact on a population by lysing infected cells, and can also affect bacterial fitness by transferring genetic information. Advances in sequencing technology have revealed abundant and mostly uncharacterized bacteriophages in the human gut; these are present as both viral particles and prophages (bacteriophage DNA that is integrated into a bacterial genome) (12–16). Prophages may be activated during inflammation (17), and the viral community may be different in people with inflammatory bowel disease (12). In mouse experiments, E. faecalis that could produce bacteriophage had a competitive advantage against a related strain (18). Presumably the virus it produced could infect and kill the competing strain. However, other evidence suggests that most of the bacteriophage in the human gut is of the temperate type which does not lyse its targets, and thus bacteriophage “predation” may not play as significant a role in the gut ecosystem as in other environments (16, 19). Like bacteriocins, bacteriophages usually have a very narrow target range. Although they have been used therapeutically for this very reason (20), the extent of their role in colonization resistance is uncertain.
The type VI secretion system (T6SS) is a protein translocation complex found in some gram-negative bacteria, which shares mechanistic similarities to some bacteriophage proteins. It is used by bacteria to transfer effector proteins into other bacterial or eukaryotic cells (21). Recently, a new family of T6SS proteins was found in members of the Bacteroidetes phylum (22), which along with the Firmicutes dominates mammalian guts. The presence of a T6SS and its associated effectors and immunity proteins was shown in several studies to have a major role in the competition between Bacteroides species inhabiting the mouse gut (22–24). Importantly, T6SS-mediated competition is contact-dependent, can involve diverse combinations of effector and immunity proteins, and can have a broader target range than other killing mechanisms.
2.3 Inhibitory metabolites
Metabolic byproducts produced by bacteria can also have an inhibitory effect on other bacteria. Short-chain fatty acids (SCFAs: e.g. acetic, propionic and butyric acid) were identified early on as a key factor in the inhibition of S. Typhimurium growth in the mouse (25) and are also active against pathogenic E. coli (26, 27) and Clostridium difficile (C. difficile) (28). They are produced by anaerobic symbiotic bacteria such as the Bacteroides and Clostridia, which are abundant members of the adult mammalian microbiota. Clostridia species in particular are able to protect mice from S. Typhimurium and C. rodentium, through unknown mechanisms that may include production of inhibitory compounds (29). Importantly, SCFAs require acidic pH for their suppressive activity, a condition which is also maintained by the normal bacteria (30). SCFAs can also affect pathogen virulence: for example, propionate and butyrate can suppress S. Typhimurium virulence factors, while acetate and formate have the opposite effect (31–33). SCFAs can also act on the host, causing it to lower oxygen concentrations and create a less favorable environment for pathogen growth (34).
Bile acids are amphipathic, cholesterol-derived molecules secreted into the small intestine. Their main function is to emulsify fat and fat-soluble vitamins for absorption, but they also have antibacterial properties. Bile acids are usually secreted conjugated to taurine or glycine, which increases their solubility. A variety of gut bacteria produce bile salt hydrolase enzymes that remove the conjugated molecule (35). This may be done to reduce the bile acid’s solubility and hence toxicity, or to obtain the taurine or glycine. The deconjugated primary bile acids can be further converted to secondary bile acids by 7α-dehydroxylation. A much more restricted set of bacteria, mostly Clostridia, have this ability (35). Buffie et al. correlated bacterial species with protection from C. difficile infection in antibiotic-treated mice and humans, and identified Clostridium scindens as a good predictor of resistance (36). C. scindens is capable of creating secondary bile acids by 7α-dehydroxylation that can inhibit C. difficile growth. C. scindens was able to protect mice from C. difficile, as well as restoring secondary bile acid levels. Using metabolomic analysis of resistant or susceptible mice, Theriot et al. similarly found a correlation between the secondary bile acid deoxycholic acid and resistance to C. difficile (37). Interestingly, the primary bile acid chenodeoxycholic acid can also have an indirect protective function by activating innate defenses in the small intestine via its receptor, FXR (38).
2.4 Competition for nutrients and space
Freter proposed the “nutrient niche” hypothesis in 1983, stating that “the populations of most indigenous intestinal bacteria are controlled by one or a few nutritional substrates which a given strain can utilize most efficiently” (39). Subsequent experiments supported the idea that for some bacteria, substrate limitation was indeed an important determinant of their successful colonization of the gut (40, 41). In E. coli, for instance, the ability to utilize one sugar changed the competitive balance between otherwise identical strains in the gut (42). Using an elegant genetic screen, Lee et al. identified a genetic locus, likely related to host glycan utilization, which controls intraspecies competition among gut Bacteroides (43). Maldonado-Gomez et al. examined the metagenomes (sequences of all bacterial genes in the gut) of human subjects to understand why a probiotic strain of Bifidobacteria could sometimes establish itself permanently (instead of transiently colonizing like most probiotics). They found that the availability of an open functional niche involving carbohydrate utilization may be an important factor (44).
Carbohydrate sources are present in ingested food and on host cells and secreted mucus. Gut symbionts, especially the Bacteroides, possess many genes that enable the digestion of complex polysaccharides, which the host and other bacteria can’t access. Ng et al. found that sialic acid and fucose, liberated from host glycans by symbionts like Bacteroides species, were an important sugar source for invading S. Typhimurium and C. difficile (45). Crucially, these sugars only became available to the pathogens when the bacteria that normally consume them were depleted by streptomycin. Similarly, the metabolite succinate, produced by Bacteroides species, became available in the gut after antibiotic treatment, and this promoted C. difficile colonization (46). To effect clearance of Citrobacter rodentium (C. rodentium) from the mouse gut, the microbiota act in concert with the host immune system (47, 48), and competition for sugars appears to play an important role here as well. Once host IgG has targeted virulence factor-expressing C. rodentium and excluded them from the epithelial surface they must compete with symbionts in the gut lumen (48). Here they are outcompeted by a native E. coli species, but not by Bacteroides species, since these presumably prefer other sources of food (such as complex polysaccharides). However, when the diet is changed to one low in polysaccharides but high in simple sugars, Bacteroides now compete with and reduce the numbers of C. rodentium (47). This is an important reminder that diet can have a profound impact on the available nutrients, and the structure, activity and function of the gut microbes (49–51). Microbes can also modify the other major source of sugars in the gut: host glycans. This will be addressed in the following section on indirect mechanisms of resistance.
Iron is another important nutrient for bacteria and it is tightly sequestered by the host, especially during inflammation. Competition for iron might be another explanation for E. coli Nissle’s ability to reduce S. Typhimurium colonization in mice (52).
While the nutrient niche concept has been validated in several cases, untangling the extremely complex metabolic interactions in the gut ecosystem remains a daunting challenge.
2.5 Physical niches
In addition to functional nutrient-based niches, bacteria must compete for physical space. Some species prefer living on the food matter in the lumen, or in the outer mucus layer, or more rarely at the epithelial surface. Close physical contact with the epithelium is an essential part of some pathogens’ lifestyles (e.g. C. rodentium, some pathogenic E. coli, S. Typhimurium), so physical competition for adhesion sites (often glycan structures) could prevent infection or pathology (53). Interestingly, microbes can also change the presence of host adhesion sites indirectly (see below).
2.6 Indirect mechanisms of colonization resistance
Aside from direct competition, microbes can compete with one another indirectly by acting on the host. This usually involves stimulation of the innate or adaptive immune system, but other non-immune defenses can also take part. Microbial stimulation of the host immune system will be explored in detail in sections 3–5, below. Here, we will focus on non-immune factors stimulated by the microbiota that can affect colonization resistance.
2.7 Mucus and glycosylation
The mucus layers in the small intestine, cecum, and colon are a crucial part of the epithelial defenses, and are stimulated by the presence of the microbiota (54). In the cecum and colon, the inner, firmly attached mucus layer effectively excludes bacteria and particles of similar size, but not in GF mice (55). The large intestine mucus’ main component is the glycoprotein Muc2, but it also contains other proteins that can immobilize or kill bacteria (56, 57). Mice lacking Muc2 have higher pathogen loads and more intestinal damage when infected with C. rodentium (58). Likewise, S. Typhimurium growth in the cecum and translocation to the liver is greater in the absence of Muc2 (59).
Muc2 is heavily glycosylated. Glycosylation of the intestinal mucus and epithelium is quite complex and can change in response to microbial colonization (60). This is interesting because host glycans can serve as nutrient sources or adhesion receptors for microbes, including pathogens. One glycosylation modification, α (1,2)fucosylation, is especially intriguing. It is stimulated in the ileal epithelium and mucus of mice by commensal or mutualistic bacteria (61), and may be a useful sugar source for some (62). α (1,2)fucosylation is also robustly activated throughout the small intestine during infection, via MyD88 and IL-22 signaling (63, 64). This supply of fucose during sickness may help pacify dangerous bacteria as well as feeding symbionts: fucose suppresses virulence genes in pathogenic E. coli as well as in members of the normal microbiota (63, 65). Fucosylation also reduces pathology caused by C. rodentium, and prevents an opportunistic pathogen (E. faecalis) from escaping the gut and causing disease (66). Microbes may also affect host modification of free sugars in the gut. Faber et al. found that treating mice with streptomycin led to increased levels of two oxidized sugars, galactarate and glucarate, in the cecal lumen (67). These sugars are formed from galactose and glucose by the host’s inducible nitric oxide synthase enzyme (iNOS), and iNOS expression was increased after streptomycin treatment. Streptomycin may directly trigger iNOS, or it may kill symbionts which normally suppress iNOS and/or consume the oxidized sugars. In either case, the greater availability of these sugars after antibiotic treatment fed S. Typhimurium growth (67).
3. REGULATION OF MYELOID CELLS BY THE GUT MICROBIOTA
The intestinal immune system is immensely shaped by the gut microbiota. Myeloid cells such as neutrophils and macrophages are typically the first immune responders to an infection. Although myelopoiesis occurs in the bone marrow, a broad effect of the gut microbiota on bone marrow hematopoiesis and the functions of intestinal macrophages has been implicated by multiple studies (Figure 2).
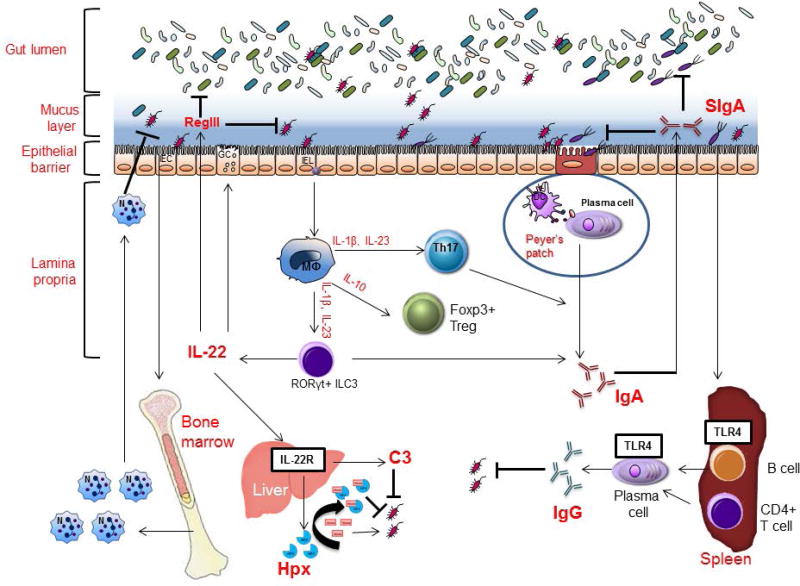
FIGURE 2.
Effects of the gut microbiota on host immune responses. Through TLRs, molecules from gut symbiotic bacteria promote granulopoiesis of neutrophils in the bone marrow and mobilization of neutrophils upon infection. The presence of gut symbiotic bacteria is important for expression of pro-IL-1 in intestinal macrophages, which can be cleaved to mature IL-1 by selective gut Enterobacterial symbionts via NLRP3 inflammation during gut injury to promote proper inflammatory response. IL-1, IL-23 and IL-6 from intestinal macrophages are important to promote mucosal Th17 cell response, and IL-10 from intestinal macrophages as well as microbial short-chain fatty acids (SCFAs) are involved in the development of Tregs in the gut under homeostatic conditions. The presence of the gut microbiota is critical for induction of both T cell-dependent and independent production of IgA antibodies, most of which are specific for gut symbionts and are transferred to intestinal lumen where they target invading bacteria to prevent them from crossing the epithelial barrier. During enteric Citrobacter rodentium or Clostridium difficile infection, IL-22, mostly produced by ILC3 cells, can act systemically to induce hepatocytes to produce hemopexin and complement C3, respectively, to inhibit growth and clearance of systemically translocated bacteria. Under homeostatic conditions, selective members of Gram-negative gut symbionts induce systemic production of IgG antibodies that can recognize bacterial surface antigens, such as murein lipoprotein (MLP) that are expressed on some Gram-negative pathogens, thereby contributing to host defense against systemic infection by gut symbionts or pathogens.
3.1 Neutrophils
Mice treated with broad-spectrum antibiotics were shown to have a decrease in the numbers of stem and progenitors in the bone marrow (68). Khosravi et al reported innate immune defects in GF mice as a result of the absence of the gut microbiota, which leads to impaired early immune responses to pathogens, and recolonization of GF mice with a complex microbiota restores defects in myelopoiesis and resistance to systemic infection with Listeria monocytogenes (69). Mechanistically, microbial molecules such as lipopolysaccharides were shown to sustain steady-state production of neutrophils and priming of neutrophils against bacterial infections through Toll-like receptor signaling (70, 71). In addition, intestinal ILC3 production of IL-17 induced by the gut microbiota leads to G-CSF-mediated granulopoiesis in the bone marrow, which was critical to combat E. coli sepsis in neonatal mice (72). On the other hand, microbiota-mediated priming/activation of neutrophils has been shown to increase the number of activated/aged neutrophils in the circulation, which secrete pro-inflammatory cytokines and granule proteases that damage tissues and exacerbate disease. In mouse models of sickle-cell disease and endotoxin-induced sepsis, depletion of the gut microbiota, significantly reduced the number of circulating aged neutrophils and lessened organ damage (73). We previously reported that during systemic E. coli infection, microbial ligands including those from the microbiota induce NOD1 and TLR4 signaling to mobilize myeloid progenitors to the spleen where these mobilized progenitors mature into neutrophils to combat the infection (74). It is possible that gut microbiota-derived ligands play similar roles in homeostatic mobilization of myeloid progenitors to various organs where they give rise to mature myeloid cells for maintenance of tissue homeostasis. Taken together, microbial molecules from the gut microbiota appear to have a profound and sustained effect on neutrophils, from granulopoiesis of myeloid progenitors in the bone marrow to the response of mature neutrophils to infection and ultimately aging of mobilized neutrophils.
3.2 Intestinal eosinophils
Eosinophils constitute a major population of leukocytes in the gastrointestinal tract under homeostatic conditions, independently of the gut microbiota (75, 76). Although eosinophils are most known as proinflammatory cells that contribute to the pathogenesis of various allergic diseases, more recent studies reveal remarkable functional diversity of eosinophils in different tissues (77). For example, unlike eosinophils from the lung or blood, small intestinal eosinophils display extended lifespans (75, 76). Sugawara et al recently reported that small intestinal eosinophils constitutively secrete high levels of IL-1 receptor antagonist (IL-1Ra), a natural inhibitor of IL-1β, which is promoted by production of GM-CSF by intestinal epithelial cells in a gut microbiota-independent manner. By suppressing the levels of IL-1β in the small intestine, eosinophils suppress Th17 cells, thereby playing a critical role in the maintenance of intestinal homeostasis (76). The function of this major leukocyte population in the gut, however, has not been extensively explored, either in the steady state or in the context of food allergies.
3.2 Intestinal macrophages
There are resident macrophages in every tissue of the body, where they contribute to the maintenance of tissue by acting as the first line of defense against pathogens and by initiating wound repair (78). A recent study demonstrated that yolk sac and fetal liver-derived macrophages are present in the neonatal intestine, which however are diluted by Ly6Chi monocyte-derived macrophages that infiltrate into the intestine around the time of weaning, in a process that is dependent on CCR2 expression of the monocytes and neonatal gut microbiota (79). Consistently, although GF mice exhibit a normal intestinal macrophage compartment at birth, recruitment of Ly6Chi monocytes into the intestine is markedly diminished at 3 weeks of age (79). Intestinal macrophages are hyporesponsive to bacterial Toll-like receptor ligands such as lipopolysaccharides, which is critical to prevent inappropriate activation of inflammatory responses in the intestine (80, 81). Consistently, we have previously demonstrated that a lack of a functional NLRP3 inflammasome in intestinal resident macrophages, which would minimize inappropriate production of IL-1and hence induction of inflammation under steady-state conditions (82). On the other hand, intestinal macrophages constitutively express NLRC4 and pro-IL-1-β, which allows precipitous release of IL-β and initiation of inflammatory response upon infection with enteric pathogens, such as Salmonella. Our studies also revealed that constitutive expression of pro-IL-1-β requires the gut microbiota in a Myd88-dependent manner (82, 83). In addition, in response to intestinal injury, our more recent study showed that gut symbiotic Enterobacteriaceae, and in particular Proteus mirabilis, induce robust NLRP3 inflammasome-mediated IL-1β production in newly recruited Ly6Chi monocytes, which promotes inflammation in the intestine (84). Taken together, our studies and others’ suggest a complex role for the gut microbiota in controlling recruitment of circulating monocytes to replenish resident intestinal macrophages and functional changes of intestinal macrophages in response to injury or infection.
4. REGULATION OF T CELL RESPONSE BY THE GUT MICROBIOTA
A myriad of intestinal immune defects are observed in GF animals, including impaired development of gut-associated lymphoid tissues (GALTs), gut-associated Th17 cells and Tregs, lower numbers of IgA-producing B cells and intraepithelial CD8+ T cells (Figure 2).
4.1 Th17 cells
GF mice have reduced numbers of Th1 and Th17 cells. Th17 cells are a subset of CD4+ effector T cells that make copious amounts of IL-17, which plays important roles in host defense against extracellular pathogens as well as the development of autoimmune diseases (85). Studies have demonstrated that development of Th17 cells in the small intestine can be potently induced by colonization of commensal Clostridia-related segmented filamentous bacteria (SFB) (86–88), and to a lesser extent by colonization of altered Schaedler flora (ASF), a cocktail of 8 defined commensals (89). Two more recent studies elucidated a cascade of events initiated by SFB adhesion to the small intestine epithelium to promote IL-17 expression in RORγt+ CD4+ T cells in the gut. Enhanced SFB adhesion on the epithelial induces the expression and release of serum amyloid A (SAA) from intestinal epithelial cells (IECs), and SAA potentiates the production of IL-1β and IL-23 in CX3CR1+ phagocytes, 2 cytokines that synergistically promote the production of IL-22 in ILC3. IL-22, in turn, reinforces SAA-mediated IL-1β production by phagocytes, which ultimately upregulates IL-17 production in RORgt+ CD4+ T cells (90, 91). It remains unclear what distinct signals from bound SFB are critical for the release of SAA from IECs, and whether this IL-17-inducing mechanism is unique to SFB or all symbiotic bacteria with IEC-adhesive characteristics. Interestingly, the ability of gut symbiotic bacteria to elicit Th17 response could be potentially exploited in the context of anti-tumor immunity. A previous study showed that treatment of cyclophosphamide, a commonly used cancer drug known to stimulate anti-tumor immune response, alters the composition of the gut microbiota in the small intestine and induces translocation of selective Gram-positive bacteria to secondary lymphoid organs. These translocated bacteria stimulate the generation of “anti-tumor” Th17 cells that are important for the therapeutic efficacy of cyclophosphamide (92). Therefore, knowledge of the link between symbiotic bacteria and induction Th17 cells can be harnessed in addressing diseases that involve aberrant Th17 response.
4.2 Tregs
In the absence of the gut microbiota, the number of inducible Foxp3 Helio-Tregs (iTregs) is specifically reduced in colonic lamina propria, but unaffected in the small intestine or mesenteric lymph nodes (93). Treg-inducing activity in mice was found in a cocktail of 46 strains of the Clostridium genus belonging to clusters IV and XIVa (also known as the Clostridium leptum and Clostridium coccoides groups, respectively), and in ASF consisting of 3 Clostridia species belonging to the Clostridium cluster XIV. A cocktail of 17 human Clostridia strains was also identified to possess Treg-inducing activity. Moreover, colonization of Bacteroides fragilis, a human species, was shown to induce Tregs and production of IL-10 by polysaccharide A via TLR2 signaling (94, 95). In addition, bacterial metabolites short-chain fatty acids (SCFAs), which are at reduced levels in the colonic lumen of GF mice, have been shown to regulate the development and function of colonic Tregs (93, 96, 97). SCFAs, particularly butyrate, can directly enhance acetylation of the Foxp3 locus in Tregs (96, 97). In a study of GF mice colonized with 17 human Clostridium strains, luminal levels of SCFAs acetate, propionate, isobutyrate and butyrate were elevated to promote TGFβ production in colonic epithelial cells, which indirectly contributes to the development of colonic Tregs (91). The receptors that are activated by SCFAs, namely several G-protein-coupled receptors (GPRs) such as GPR43, are expressed on both IECs and most hematopoietic cells. Consistently, mice lacking GPR43 have lower numbers of Tregs. It should be noted that the effect of SCFAs on the induction of Tregs appears not restricted to the colon, as studies have shown histone deacetylase (HDAC) inhibitory activity of butyrate that regulates LPS-responsive genes (e.g. Il2, Il6 and RelB) in DCs to promote the differentiation of Tregs. This appears to be a common mechanism that regulates the generation of extrathymic Tregs. Hence, the systemic effect of SCFAs on the induction of Tregs is not consistent with the colon-specific defect in Treg development in GF mice. This suggests that there might be redundant SCFA-independent mechanisms to promote the development of Tregs in most tissues, which however might be lacking in the colon.
4.3 ILC3
Innate lymphoid cells (ILCs) are a population of innate immune cells that differentiate independently of somatic recombination or interactions with cognate antigens presented by MHCII, but exhibit a cytokine profile akin to that of CD4+ T cells. ILCs are found at barrier surfaces of the mammalian body, such as the skin, airway and the gastrointestinal tract. Three distinct groups of ILCs have emerged, based on their cytokine profiles: Tbet+ Group1 ILCs (IFNγ producers), GATA3+ Group2 ILCs (IL-4, IL-13 and IL-9 producers), and RORγt+ ILCs Group 3 (IL-22 and IL-17 producers) (98). There is increasing evidence supporting an important role for ILC3 in intestinal immunity. For example, IL-22 produced by ILC3 promotes production and secretion of RegIIIg, a secretory antimicrobial peptide by intestinal epithelial cells that has been shown to be critical in enteric infection by Citrobacter rodentium (99–101). There have been conflicting reports on whether the development of ILC3 requires gut symbiotic bacteria, with several studies showing normal development of RORγt+ ILCs in both GF mice and antibiotic treated mice, and other studies revealing the lack of RORγt and IL-22 expression in the small intestine of in both GF mice and antibiotic-treated mice. The discrepancy might attribute to diet-derived microbial signals that GF mice in these studies could have been potentially exposed to (98). Nonetheless, gut symbiotic bacteria are believed to influence the function of ILC3, either by directly signaling through pattern-recognition receptors (PRR) on ILC3 or by indirectly regulating intestinal myeloid cells and epithelial cells. Human RORγt+ ILCs were shown to express functional TLR2, which can be stimulated to induce IL-2 that enhances IL-22 production in an autocrine manner (102). The Aryl hydrocarbon receptor (AhR), which can be activated by ligands generated from tryptophan metabolism by gut symbiotic bacteria, is expressed on RORγt+ ILCs and critical for ILC development, IL-22 production, maturation of GALTs, and enteric immunity against Citrobacter rodentium (99–101). In addition, IL-1β and IL-23 produced by intestinal phagocytes can promote IL-22 production RORγt+ ILCs (103). We have previously shown that gut symbiotic bacteria promote the expression of IL-1β in intestinal macrophages under steady-state conditions that likely contribute to the emergence of RORγt+ ILCs (83).
Furthermore, the role of RORγt+ ILCs in regulating the gut microbiota has been unraveled in recent studies. RORγt+ ILC production of lymphotoxin (LTα1β2) promotes the generation of isolated lymphoid follicles (ILFs), which are critical for T cell-independent intestinal IgA production (104, 105). RORγt+ ILCs are also major producers of IL-22, which promotes epithelial cells production of mucins and antimicrobial proteins (RegIIIb, RegIIIg, S100A8, and S100A9), which are integral for compartmentalization of gut bacteria within the gastrointestinal tract (106). The importance of intestinally induced ILC3 in mediating systemic host defense against systemic infection, mainly via IL-22, has been highlighted in our recent studies. We showed that in enteric Clostridium difficile infection (CDI), IL-22, which was shown in a separate study to be produced mainly by ILC3 in CDI (107), can act systemically on hepatocytes that express IL-22R (108). As a result, IL-22 induces production of complement C3 from hepatocytes, which promotes clearance of systemically translocated gut bacteria following CDI and thereby confers protection (108). In addition, our recent study showed that during enteric C. rodentium infection, IL-22 produced by ILC3 systemically induces the production of heme scavenger hemopexin to limit heme availability and thereby suppress the growth of bacteria that have translocated systemically due to damaged gut epithelial barrier (109). Furthermore, Hepworth et al reported that MHCII+ ILC3 cells directly kill CD4+ T cells that are reactive to gut symbiotic bacteria, in a manner reminiscent of how self-reactive T cells are eliminated in the thymus. Notably, MHCII on colonic ILC3 was reduced in pediatric IBD patients, suggestion possible impairment in MHCII+ colonic ILC3 to dampen self-reactive CD4+ T cells in IBD (110). Collectively, these studies support a beneficial role for gut ILC3 in both intestinal and systemic host defense against pathogens or pathobionts as well as dampening otherwise deleterious T cell response to gut symbiotic bacteria, thereby maintaining tissue homeostasis in the intestine.
5. REGULATION OF B CELL IMMUNE RESPONSE BY THE GUT MICROBIOTA
The gut is home to at least 80% of all activated plasmablasts and plasma cells, most of which produce IgA (111). Primary B-cell development, which in humans and mice involves Rag-dependent V (D)J recombination for pre-immune diversification, is thought to occur exclusively in the bone marrow. However, recent evidence suggests a critical role for the gut microbiota during early life in B cell selection and pre-immune immunoglobulin diversification. Using a Rag2-GFP reporter mouse model, Alt and colleagues showed that Rag2+CD19+B220low cells, which are undergoing active V (D)J recombination, are almost undetectable in the first week after birth, emerge and expand rapidly at weaning age (18–24 days), and wane at about 5–6 weeks of age (112). Of note, the gut microbiota expands quickly upon weaning, suggesting a possible role for maternal immunoglobulins in restricting expansion of the gut microbiota prior to weaning before the neonatal immune system is fully developed. Consistently, conventionalization of GF mice increases the levels of pro-B cells systemically and in the gut LP. The transient nature of LP B-cell development upon weaning might be a window for both luminal antigens to shape pre-immune B cell repertoire. The study by Alt and colleagues also revealed similar VH repertoires but markedly different V repertoires of LP and BM Rag2+ B cells, reflecting differential BCR editing within the gut and BM, respectively. BCR editing in the BM is a negative selection to eliminate self-reactive B cells, but it remains unclear whether BCR editing in the gut also serves as a similar tolerance mechanism.
5.1 IgA
IgA is the most abundant isotype of antibodies that can be readily secreted into the lumen of the gut under steady-state conditions. Polymeric IgA binds to the polymeric immunoglobulin receptor (pIgR) expressed on the basolateral surface of the gut epithelium, which transports IgA to the apical surface. IgA is released in the gut lumen upon proteolytic cleavage of the secretory component of pIgR. Mucosal IgA-secreting cells are markedly reduced in GF mice and undetectable in the neonatal gut prior to colonization of symbiotic bacteria, hence suggesting that gut symbiotic bacteria might provide key stimulatory signals to induce production of mucosal IgA. A previous study showed that, during pregnancy in GF female mice, transient inoculation of a mutant E. coli strain (HA107), which was engineered to allow reversible and transient colonization of GF mice, sufficient to induce robust and specific IgA response in the offspring. A potent stimulatory effect on induction of IgA response was observed upon colonization of SFB in GF mice (113). Notably, SFB colonization is associated with induction of Th17 cells, which have been shown to expand and home to the small intestine in mice monocolonized with SFB. In the Peyer’s patches of these mice, Th17 cells acquire a phenotype reminiscent of T follicular helper (TFH) cells to induce the development of IgA-producing germinal center B cells. Consistently, mice lacking Th17 cells are impaired in mounting antigen-specific IgA response after immunization with cholera toxin, thus underscoring an indispensible role for Th17 cells in T cell-dependent generation of antigen-specific IgA antibodies (114). It remains unclear however whether IgA response is induced only by selective members of gut symbiotic bacteria.
The function of IgA in host-microbe symbiosis as well as in host defense has been extensively studied but to an extent remains undefined, given that IgA deficiency is the most common primary immunodeficiency and most affected individuals are asymptomatic (115). It is likely that IgA deficiency can be compensated by other immune mechanisms, such as increased export of SIgM, which is facilitated by pIgR as well. In mice, most intestinal IgA is directed against gut symbiotic bacteria, and coating by SIgA in intestinal lumen preventing invading bacteria from crossing the gut epithelium (116). Studies of mice deficient in pIgR showed impaired export of SIgA and SIgM into the intestinal lumen, elevated serum IgG, and more importantly, increased mucosal penetration of symbiotic bacteria and enhanced systemic antibodies against these translocated bacteria. Macpherson and colleagues reported that IgA restricts penetration of gut symbiotic bacteria to the mesenteric lymph nodes, thereby limiting systemic dissemination of symbiotic bacteria and preserving host-microbe symbiosis (117, 118). Consistently, a recent study demonstrated that intestinal bacteria with high IgA coating confer enhanced susceptibility to DSS-induced colitis in GF mice, suggesting that IgA specifically coats more inflammatory and hence colitogenic intestinal bacteria under homeostatic conditions to confine these harmful bacteria in the intestinal lumen (119). Together these studies highlight an integral role for IgA in compartmentalizing intestinal bacteria, especially those with heightened virulence, in the intestinal lumen to maintain host-microbe symbiosis.
5.2 IgE
IgE antibodies play a critical role in allergies, asthma, and immunity to parasites (120), and studies of human infants and animal models support a role for mucosal microbiota in asthma and atopic disease development (121–123). IgE production appears to be heavily influenced by the gut microbiota as well, albeit by a mechanism seemingly opposite to that underlying gut microbiota-regulated IgA response. Cahenzli et al reported profoundly high levels of IgE are produced in GF mice immediately after weaning, due to active CD4+ T cell-dependent B cell isotype switch to IgE at mucosal sites, particularly Peyer’s patches, in the absence of microbial exposure (124). Colonization of GF mice with diverse microbiota, but not specific symbiotic bacteria, from birth to 4 weeks of age but not thereafter normalizes the IgE levels in adults (124), suggesting that diverse microbial exposure at mucosal sites, while triggering IgA isotype switch, downregulates IgE to baseline levels. The age-specific effect of the gut microbiota to tone down IgE response is intriguing, but the underlying mechanisms remain largely elusive. These studies underscore infancy as a critical window of opportunity for the gut microbiota to educate the host immune system and provide long-term benefits.
5.3 IgG
In contrast to IgA, which is locally induced in GALTs by gut symbiotic bacteria, induction of high-affinity, antigen-specific IgG antibodies is thought to take place in extra-intestinal organs, such as the spleen. However, redundant measures are in place to ensure compartmentalization of symbiotic bacteria in the intestine, including sIgA, patrolling intestinal phagocytes, mucous layer, the epithelium, and lastly the MLNs as a firewall where escaped intestinal bacteria would be killed by phagocytes. For year there was the dogma of systemic ignorance, rather than tolerance, toward intestinal symbiotic bacteria due to complete compartmentalization of intestinal symbiotic bacteria in the intestine. This dogma was supported by a study showing undetectable serum IgG from naïve mice against Enterobacter cloacea, a Gram-positive gut symbiotic bacterium (125). However, higher titers of serum IgG against fecal symbiotic bacteria were frequently reported in patients with either Crohn’s disease or Colitis, which was thought to reflect either prior infections or systemic translocation of gut bacteria in these patients due to “leaky” epithelial barrier (126). In addition, we recently reported that gut symbiotic bacteria promote the adjuvant activity of cholera toxin, an enterotoxin secreted by Vibrio cholera that has been used as a potent mucosal adjuvant, through Nod2-mediated recognition of its microbial agonist by dendritic cells (127).
Evidence from a few recent studies has emerged that the systemic immune system in humans and mice is not ignorant toward gut symbiotic bacteria under homeostatic conditions. We recently reported that in unmanipulated mice and healthy humans, robust levels of serum IgG antibodies were detected that selectively recognized proteins from Gram-negative but not Gram-positive symbiotic bacteria (128) (Figure 2). Consistently, bacterial DNA was detected in the spleens of naïve mice, suggesting some gut symbiotic bacteria likely had made their way to extra-intestinal organs to initiate systemic immune responses, including generation of antigen-specific IgG. This suggests that in the steady state perhaps a small number of Gram-negative Enterobacterial species, which have been shown to possess unique growth advantages that enable them to bloom in an inflamed gut (129), may be uniquely equipped for successful penetration of the gut epithelial barrier, evasion of killing by phagocytes in the mesenteric lymph nodes, and induction of systemic response. Our data also illustrated the requirement for CD4+ T cells and TLR4 on B cells for induction of homeostatic generation of antigen-specific IgG antibodies to gut symbiotic bacteria.
Furthermore, we identified murein lipoprotein (MLP), a secretary outer membrane protein abundantly expressed on Gram-negative Enterobacterial species, as a major bacterial antigen recognized by serum IgG in naïve mice and healthy humans. Bacteremia due to gram-negative symbiotic bacilli, most commonly E. coli, accounts for 25%–50% of all bloodstream infections. Therefore, the potential of homeostatically induced serum anti-MLP IgG, and other still unknown IgG against gut microbiota-derived antigens, should be further investigated for the treatment of Gram-negative sepsis.
Low concentrations of symbiotic bacteria-specific IgG in younger mice from our study, and very likely in human infants and toddlers, might contribute to increased susceptibility of neonates to infection. Maternal IgG antibodies are transported to the infant, through the placenta or breast milk, to provide crucial passive immunity in the developing fetus or infant, respectively. Consistently, a recent study by Koch et al showed that maternal IgG and IgA antibodies in neonatal mice dampen mucosal T follicular helper responses and subsequent germinal center B cell responses following birth (130). Our studies highlight the importance of developing proper homeostatic IgG antibodies against conserved bacterial antigens that may confer critical protection against pathogens later in life. In light of this, excessive use of antibiotics in young children might potentially delay or impair proper development of IgG response and immune memory against symbiotic and pathogenic bacteria and have a profound impact on later life. Therefore, our results underscore the importance of having a balanced microbial community in the intestine in early life, as well as the benefits of maternal antibodies for neonatal health.
6. EFFECTS OF INFLAMMATION ON THE COMMUNITY STRUCTURE OF THE GUT MICROBIOTA
6.1 Selection of symbiotic bacteria that thrive in the inflamed gut
The composition of the human large bowel microbiota exhibits great intersubject variability (131). Nevertheless, there are some conserved features that define a balanced gut microbiota, including a prominent representation of members of the Firmicutes and Bacteroidetes phyla and a lower abundance of members of the Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia phyla (131). Intestinal inflammation can lead to an altered composition of gut microbiota, known as dysbiosis, that is associated with functional changes in the microbial transcriptome, proteome or metabolome (132–134). Growing evidence has shown that perturbation of the gut microbial community may fuel blooms of otherwise low abundance and harmful bacteria which can further exacerbate the intestinal inflammation. Indeed, dysbiosis in the distal gut is often characterized by a decreased in the prevalence of strict anaerobes and an increased relative abundance of facultative anaerobic bacteria (135, 136). In particular, expansion of Enterobacteriaceae is commonly observed in gut dysbiosis in a variety of contexts involving intestinal inflammation (135–141). Enterobacteriaceae, a large family of gram-negative facultative anaerobes that belongs to the class of Gammaproteobacteria and to the phylum Proteobacteria, are usually localized in a close proximity to the gut epithelium due to their relative high tolerance to the oxygen that diffuse from the epithelial barrier. While elevated oxygen levels increase the relative abundance of Enterobacteriaceae, the limited amount of available oxygen in the colon contributes to their low prevalence. Indeed, Enterobacteriaceae represent a small fraction, roughly 0.1%, of the microbial community in the distal gut (131). Interestingly, Enterobacteriaceae are among the most commonly symbionts that overgrow in various settings of gut inflammation, such as IBD, celiac disease, and colon cancer (137, 139, 140). Inflammation-induced environmental and nutritional changes may confer a growth advantage to Enterobacteriaceae. The gut inflammation, that results after an infection by a pathogen, chemically-induce colitis or host immune deficiency, seems to foster the expansion of Enterobacteriaceae by providing a favorable environment for their growth (128, 135, 142).
An increased prevalence of Enterobacteriaceae, including adherent invasive Escherichia coli (AIEC), has been observed in patients with Crohn’s disease (CD) and ulcerative colitis (UC), the two major forms of IBD (139, 143–146). Consistently, the relative luminal abundance of Enterobacteriaceae dramatically increased in either chemical-induced or genetic mouse models of IBD (135, 136, 142). Despite these associations, no single pathogens, including AIEC, has been proven to cause IBD. On the other hand, the expansion of E. coli seems to be a consequence rather than a cause of IBD, most likely due to the ability of Enterobacteriaceae to bloom in an inflamed intestinal environment.
Interestingly, two Enterobacteriaceae species, such as Klebsiella pneumoniae and Proteus mirabilis, isolated from TRUC mice (i.e. T-bet−/− × RAG2−/−) that develop a spontaneous UC-like colitis, can elicit colitis in wild-type mice in the presence of endogenous gut microbiota (142). These observations suggest that inflammation-driven expansion of Enterobacteriaceae could potentially contribute to the pathogenesis of disease development itself.
Antibiotic treatment has been associated with a luminal bloom of E. coli and the pathogen C. rodentium, both members of the family of Enterobacteriaceae, due a disrupted intestinal homeostasis that results in an increased inflammatory tone of the intestinal mucosa in mice (147, 148).
Similarly, evidence showed that a previous antibiotic use may be related to the development of diarrhea-predominant irritable bowel syndrome (149, 150), a clinical condition in humans characterized by an increased abundance of Enterobacteriaceae, Pasteurellaceae and Pseudedomonadaceae families (all members of the phylum of Proteobacteria) (151–154).
Furthermore, several reports confirmed a consistent increase in the relative abundance of Enterobacteriaceae in the stools of infants who developed necrotizing enterocolitis, a fatal disease that is a major cause of mortality in preterm infants (155).
Intestinal inflammation induced by the administration of enteric pathogens or by parasite infection has been associated with a disruption of the intestinal ecosystem that leads to an uncontrolled Enterobacteriaceae expansion within the microbial community inhabiting the distal gut (156–158). Notably, it has been shown that pathogens belonging to the family of Enterobacteriaceae, namely C. rodentium and S. typhimurium utilized virulence factors to promote intestinal inflammation, which in turn conferred a growth advantage for the pathogens in the gut lumen (47, 156).
Finally, expansion of Enterobacteriaceae can negatively affect host defense against bacterial pathogens or injury. For example, clindamycin-induced expansion of Enterobacteriaceae accounted for sustained susceptibility to C. difficile-driven colitis (141). Antibiotic or dextran sulfate sodium (DSS) treatment can lead to the expansion and extraintestinal dissemination of E. coli pathobionts that result in bacteremia, sepsis-like disease and ultimately mouse death (128, 159).
In this context, a recent study has identified an increased fecal abundance of E. coli and Bacteroides species as an important determinant of susceptibility to Campylobacter infection in humans (160).
A study, focused on the modulation of gluten-induced immunopathology by the microbiota, showed that antibiotic treatment, leading to Proteobacteria expansion, further enhanced gluten-induced immunopathology in conventional specific-pathogen-free (SPF) mice. In addition, supplementation of SPF microbiota with E. coli, isolated from a patient with celiac disease, increased the severity of gluten-induced pathology (161). Finally, E. coli and Shigella, isolated from patients with celiac disease, have been shown to increase intestinal permeability in this intestinal disorder, likely due to a reduced expression of tight junction proteins (162). Overall, accumulating evidence suggests that inflammation-inflicted blooms of Enterobacteriaceae are associated with various intestinal diseases and could further exacerbate the intestinal inflammation.
6.2 Selection for the expression of bacterial proteins that sustain microbial survival and perpetuate inflammation
Analyses of bacterial transcriptomes have provided new insights into the effects of inflammation on microbial function and the ability to survive in the inflammatory environment.
AIEC are able to adhere to ileal enterocytes via the common type 1 pili adhesin FimH and recognize the carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), abnormally expressed on CD ileal epithelial cells (163). In this context, FimH DNA sequence analysis recently indicated that AIEC strains predominantly expressed FimH with amino acid mutations of a recent origin. These newly generated mutations in FimH conferred AIEC a significantly higher ability to adhere to CEACAM-expressing intestinal epithelial cells, to persist and to induce gut inflammation in a genetically susceptible host (164).
In an additional study, analyzing the mutational patterns in the FimH gene of mucosa-associated E. coli strains isolated from IBD and non-IBD pediatric patients, different FimH mutational patterns have been found for each E. coli characteristic (i.e. status, phylogroup and adhesion class). E. coli strains from patients with UC showed increased numbers of mutations, while isolates from patients with CD exhibited an enhanced mutational rate but different FimH mutations arose (165). These results suggest that under specific selective pressure the FimH protein undergoes selective amino acid mutations to sustain bacterial survival and imply a distinctive behavior adopted by E. coli to survive under different inflammatory conditions, such as CD and UC.
Thus, it is conceivable that the inflammatory milieu may select for expression of specific microbial proteins that boost bacterial survival and further perpetuate inflammation.
6.3 Mechanisms underlying the bacterial blooms in the inflamed gut
Several mechanisms responsible for Enterobacteriaceae blooms in the inflamed gut have been proposed, including nutritional changes, mucin utilization, production of antimicrobials, anaerobic/aerobic respiration and metal utilization (Figure 3).
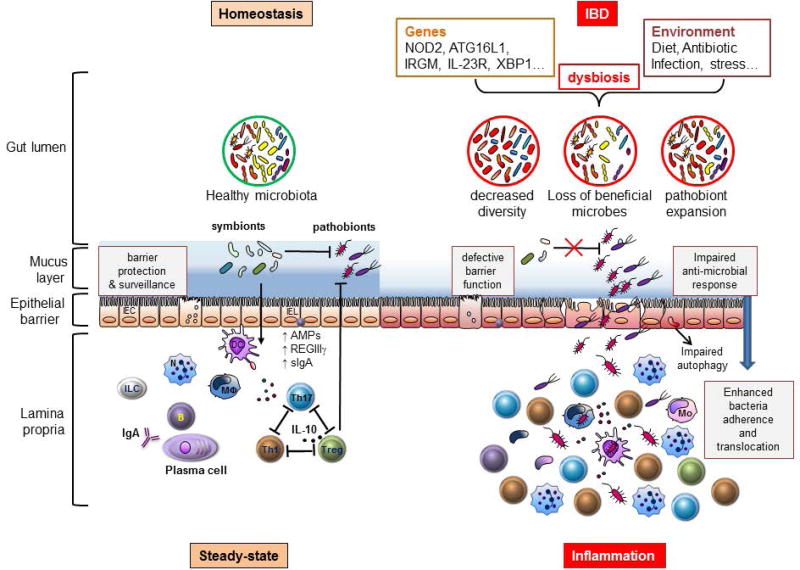
FIGURE 3.
Host-immune interactions play a crucial role in the pathogenesis of Inflammatory Bowel Disease. During homeostasis, gut microbiota critically contribute to the development of host intestinal immunity. Beneficial symbionts usually control the expansion of colitogenic pathobionts through the induction of regulatory immune responses, involving regulatory T (Treg) cells, interleukin-10 (IL-10) and regenerating islet-derived protein 3γ (REGIIIγ). In IBD, confluence of environmental and genetics factors may alter the balance between host immune and gut microbial factors triggering intestinal inflammation. Environmental factors, such as diet and antibiotic treatment, can disrupt the gut microbial community structure. Additionally, variants in NOD2, ATG16L1 and IRGM genes may perturb many aspects of immune homeostasis including reduced muramyl-dipeptide sensing in antigen-presenting cells, impaired anti-microbial responses in Paneth cells, and altered intraepithelial autophagy leading to defective barrier function and/or bacterial killing. These alterations can lead to a reduced overall microbial diversity with loss of beneficial symbionts and/or expansion of pathobionts and ultimately result in an enhanced mucosal adherence and translocation of bacteria leading to the development of chronic inflammation involving expansion of T helper 1 (Th1) and Th17 cells.
IEC, intestinal epithelial cells, IEL, intraepithelial lymphocyte, DC, dendritic cell, MΦ, macrophage, N, neutrophil, Mo, inflammatory monocyte, ILC, innate lymphoid cell, IgA, immunoglobulin A, sIgA, secretory immunoglobulin A, AMPs, anti-microbial peptides.
The microbial communities, inhabiting the distal gut, compete for a limited availability of diet-derived carbohydrates or host mucus-derived glycans (166, 167). Hence, diet plays a crucial role in shaping the composition of gut microbiota and dietary changes can results in perturbation of the gut microbial community structure at species level, but the obligate anaerobic Clostridia and Bacteroidia still maintain their dominance over facultative anaerobic Enterobacteriaceae in the healthy gut (168–171).
Under normal conditions, both Clostridia and Bacteroidia use glycoside hydrolases to break down complex carbohydrates, binding proteins to increase carbohydrate concentration at their surface and finally an active transport system for transporting carbohydrate across the cytoplasmic membrane in Clostridia and across the outer membrane in Bacteroidia (166, 172). On the other hand, Enterobacteriaceae are poorly equipped to degrade complex carbohydrates, due to their paucity of glycoside hydrolases. Enterobacteriaceae can only transport passively oligosaccharides through outer membrane diffusion channels (172). Hence, Enterobacteriaceae are ill-equipped to compete with obligate anaerobes for high-energy nutrients, and this competitive growth disadvantage can explain the dominance of Clostridia and Bacteroidia over Enterobacteriaceae in the healthy distal gut.
During inflammation, the intestinal epithelial damage, resulting in an increased shedding of dead epithelial cells, is associated with an enhanced availability of epithelial cell membrane-derived phospholipids, such as phosphatidylcholine and phosphatidylethanolamine. In particular, ethanolamine can be used as an exclusive source of carbon and/or nitrogen by several bacterial species in the Proteobacteria phylum, as well as pathogenic bacteria, such as Salmonella and Pseudomonas (173). The ability to use ethanolamine could contribute to the pathogenesis of these bacteria by providing a useful source of carbon and/or nitrogen that sustain a successful colonization of the intestine.
An additional mechanism, accounting for the expansion of Enterobacteriaceae in the inflamed gut, includes the mucin utilization. Mucus layer, coating the intestinal epithelium, is composed by two layers; the outer layer is movable, colonized by bacteria and usually limits the colonization of commensals, while the inner layer is firmly attached to the epithelium and largely devoid of bacteria (55). The secreted gel-forming mucin, MUC2, is a major mucin of the colon mucus in both humans and mice. Of note, MUC2-deficient mice showed enhanced bacterial adhesion to the surface epithelium, increased intestinal permeability and increased susceptibility to develop spontaneous or DSS-induced colitis and colorectal cancer (55, 174, 175).
A recent paper underlined the contribution of mucin-derived sialic acid, in promoting the expansion of Enterobacteriaceae during intestinal inflammation induced in mice by DSS treatment (176). Sialic acid, one of the major carbohydrates in mucin, can be taken up by bacteria, unable to de novo synthesize these sugars, such as E. coli, and then incorporated into bacterial capsule and lipooligosaccharides (177).
In addition, S. typhimurium and C. difficile, used a common strategy of catabolizing microbiota-liberated mucosal sugars, such as fucose and sialic acid, during their expansion within the gut (45). Overall, these observations suggest that sialic acid catabolism may confer a growth advantage to Enterobacteriaceae in the inflamed gut.
Enterobacteriaceae can also overcome other bacteria by producing antimicrobial molecules, promoting their blooms in the gut. For example, colicins are bacteriocins, produced by some strains of E. coli, are lethal for phylogenetically close relatives (178). Of note, production of colicin Ib (col1B) conferred a competitive advantage to S. typhimurium over sensitive E. coli strains in the inflamed gut (179). Expression of col1B is positively regulated by low iron availability and the SOS response, conditions that are usually instigated in the inflamed gut by neutrophil recruitment and oxidative stress-induced DNA damage, respectively (180–182). Therefore, the inflammatory milieu in the gut seems to create an advantageous condition that may potentiate the effects of colicins, which act as fitness factors providing a competitive growth advantage for Enterobacteriaceae blooms.
Further mechanisms responsible for the expansion of Enterobacteriaceae in the inflamed distal gut are host-induced changes in the growth conditions in this largely anaerobic environment. During inflammation, gut luminal oxygen levels increased due to elevated blood flow and hemoglobin. Additionally, new respiratory electron acceptors generated during intestinal inflammation may support bacterial growth by anaerobic respiration, including nitrate respiration. For example, it has been shown that nitrate is generated as a by-product of the host inflammatory response. This enrichment in host-derived nitrate can confer a fitness advantage for Enterobacteriaceae, such as E. coli and S. typhimurium, since genes encoding for nitrate reductase are present in the majority of Enterobacteriaceae, but largely absent in obligate anaerobes belonging to Clostridia and Bacteroidia classes (148, 183, 184). In addition, reactive oxygen species (ROS), generated by inflammatory host response, can react with endogenous sulphur compounds (i.e. thiosulfate) to produce a new respiratory electron acceptor, termed tetrathionate. This newly generated electron acceptor provided a selective growth advantage for S. typhimurium over the competing fermenting gut microbes in the inflamed gut (185, 186). These observations indicate that pathogens can utilize host responses to outgrow the intestinal microbiota.
Respiratory flexibility of Enterobacteriaceae allows them to respond to different oxygen availability within the intestine. For example, in the absence of oxygen, E.coli can use nitrate, nitrite, trimethylamine-N-oxide (TMAO), dimethyl sulfoxide (DMSO) and fumarate as electron acceptors (187), while in presence of oxygen E. coli express terminal oxidases that use oxygen as electron acceptor (188). The high levels of oxygen, derived from elevated blood flow and hemoglobin during inflammation, can provide a growth advantage for facultative anaerobes, such as Enterobacteriaceae, over the obligate anaerobes, such as Clostridia and Bacteroidia. For instance, streptomycin treatment led to depletion of butyrate-producing commensal Clostridia in mice, resulting in reduced butyrate levels, elevated epithelial oxygenation, and aerobic bloom of S. typhimurium (34). In addition, C. rodentium used a type III secretion system (T3SS) to promote colonic crypt hyperplasia in mice, which in turn increased oxygenation of the surface epithelium and fueled an aerobic expansion of C. rodentium in the colon (189).
A further mechanism that accounts for Enterobacteriaceae blooms in the intestinal inflammation is the metal acquisition. Iron is a vital nutrient for both the host and the pathogenic bacteria, and it is mostly sequestered in intracellular storages, hence inaccessible to pathogens (190). However, to bypass this iron withholding, many pathogens have evolved high-affinity iron uptake mechanisms that compete against host-mediated restriction. These uptake systems include release of iron-chelating siderophores, heme acquisition systems and transferrin/lactoferrin receptors (190). For example, E. coli was able to produce enterobactin, a catecholate siderophore that acted as a potent inhibitor of neutrophil bacteriocidal myeloperoxidase and conferred a distinct survival advantage to E. coli in the inflamed gut (191). Based on these observations, it is conceivable that siderophores, released from E. coli, serve as dual-purpose molecules, both in iron acquisition and in protection from host-derived oxidative stress.
Analysis of the genomes of phylogenetically diverse AIEC strains, isolated from patients with CD, dogs with granulomatous colitis and mice with ileitis, revealed that AIEC strains overexpressed genes encoding iron acquisition, such as chu operon, as compared to nonpathogenic E. coli. Additionally, AIEC required iron for growing and the presence of chuA (heme iron acquisition) correlated with their ability to persist within macrophages (192). These results were further supported by the observation that siderophore aerobactin enhanced intracellular survival in macrophages and colonization of the mouse intestine by AIEC NRG857c (O83:H1), a clinical isolate of AIEC from the ileum of a CD patient (193).
Overall, these studies highlight the pivotal role of iron acquisition in boosting the expansion of more virulent Enterobacteriaceae in the inflamed gut. Enterobacteriaceae also have evolved strategies to acquire other metals, such as zinc and manganese, to benefit their own growth in the inflamed gut (194–196). Inflammation in the gut commonly fosters the emergence of more virulent species of Enterobacteriaceae that have evolved multiple strategies to evade host immune responses, to outcompete commensal bacteria and to thrive in the inflamed gut.
7. THE MICROBIAL BASIS OF INFLAMMATORY BOWEL DISEASES
7.1 Involvement of microbiota in the pathogenesis of inflammatory bowel diseases
A microbial basis for the development of IBD has long been suspected, but searches for organisms causing IBD have not led to the identification of a specific pathogen. Several experimental and clinical studies have shown that the gut microbiota is an essential factor in driving inflammation in IBD. In humans, the first demonstration of the involvement of intestinal microbiota in the pathogenesis of IBD came from clinical studies in patients with CD, showing that fecal stream diversion prevented recurrence of ileal CD (197) and that exposure of the terminal ileum to the intestinal content triggered postoperative recurrence in patients with CD (198). Furthermore, treatment with antibiotics, such as metronidazole, ciprofloxacin or rifaximin, has been associated with clinical improvement in patients with IBD (199, 200), even though the use of antibiotics as primary or adjuvant therapy for inducing remission in patients with IBD is still controversial.
Animal models of IBD also suggested for a role of gut microbiota in the pathogenesis of these diseases. For instance, SPF mice carrying null mutations in T-cell antigen receptor (TCR) genes developed colitis but the germ-free animals were disease-free. Furthermore, intestinal inflammation was not observed in germ-free TCR-deficient mice colonized with a defined commensal community (201). These observations strongly suggest that intestinal inflammation might be initiated by a specific organism or group of organisms normally present in the gut flora that have yet to be identified.
In addition, HLA-B27-transgenic rats, interleukin (IL)-10- and IL-2-deficient mice raised under conventional conditions spontaneously developed chronic colitis, but the germ-free condition prevented the development of intestinal pathology (202–204).
Additional evidence, arguing for a pathogenic role of the gut microbiota in IBD, derived from an UC-like colitis model that spontaneously occurred in TRUC mice and that was cured by broad spectrum antibiotics (205). The role of microbiota in driving intestinal inflammation has also been confirmed in a model of chronic CD-like ileitis spontaneously developed in SPF, but not germ-free, mice carrying a deletion in the tumor necrosis factor AU-rich elements (206). Taken together, these observations suggest that the luminal microbes provide the stimulus for host immune responses that ultimately lead to mucosal damage in genetically susceptible hosts.
The intestinal epithelial cells and immune cells are constantly in contact with foreign material, including dietary and microbial factors. The mucosal immune system is usually able to protect the host by mounting inflammatory responses against pathogens, but it has also evolved tolerance mechanisms towards not pathogenic bacteria and diet-derived factors.
In IBD, inflammation might arise from abnormal host-microbial interactions that lead to the perturbation of intestinal homeostasis. Notably, patients with IBD showed abnormal mucosal secretion of IgG antibodies, as opposed to a physiological IgA response, against gut commensals (126) as well as higher serum reactivity toward microbial antigens (207) indicating that the tolerance mechanisms towards commensals may be abrogated in patients with IBD.
In addition, T cells isolated from inflamed mucosa of CD patients were hyper-responsive to the stimulation with antigens derived from gut commensals (208). Similarly, intestinal cells isolated from inflamed areas of IBD patients were activated by the exposure to sonicated samples of both autologous and heterologous gut microbiota, while mucosal cells isolated from normal individuals reacted only towards antigens derived from heterologous intestinal microflora (209), suggesting that the immune tolerance to self-intestinal flora is lost in patients with IBD.
It is also conceivable that hyper-reactive T cells can be induced by specific microorganisms. For instance, flagellin is a commensal bacterial protein, known to be a dominant antigen in CD. Flagellin was able to trigger an innate immune response via Toll-like receptor 5 and adoptive transfer of flagellin-specific CD4+ T cells into SCID mice induced severe colitis (210).
IL-17-producing T-helper (Th17) cells are potent mediators of inflammation in different orgarns, including the intestine (211). Colonization of the small intestine of mice with a single commensal, such as segmented filamentous bacterium (SFB), induced the appearance of Th17 cells in the intestinal lamina propria and consistently antibiotic treatment inhibited differentiation of Th17 cells (88). Although SFB contribute to shape the intestinal immune system and their presence has been reported in humans (212), their role in IBD is still unclear and requires further studies. Alike SFB that typically adhere tightly to intestinal epithelium without invading the epithelial cells, other mucus inhabitants, such as Akkermansia species and Mucispirillum genus, have been reported to accumulate in the intestine of mice experiencing DSS-induced recurring colitis (213). Thus, it is conceivable that mucus-dwelling commensals, due to their proximity to the intestinal epithelium, can trigger abnormal host immune responses leading to chronic inflammation and ultimately to mucosal damage in IBD (Figure 3).
The crucial role of intestinal microbes in the pathogenesis of IBD has been further strengthened by the identification of several IBD-susceptibility genes, many involved in mediating host responses to gut microbes. The first identified gene, strongly associated with the risk of developing CD, was the nucleotide-binding oligomerization domain-containing protein 2 (NOD2) encoding for an intracellular receptor for peptidoglycan-derived muramyl dipeptide (MDP) (214, 215). The mechanisms by which NOD2 variants contribute to CD pathogenesis are still unclear, but several hypotheses have been proposed including an impaired MDP sensing in antigen-presenting cells, defective antimicrobial responses in Paneth cells or an altered intraepithelial autophagy (216). However, whether CD-related inflammation is triggered by defective antimicrobial activity within the intestinal crypts and excessive accumulation of pathogenic bacteria and/or pathobionts is still controversial and requires further studies.
Additional IBD-susceptibility genes, such as autophagy related protein 16-like 1 (ATG16L1) and immunity-related GTPase family M (IRGM), are involved in the innate defense mechanism of autophagy (217, 218). Since a functional autophagy is required to limit the replication of intracellular CD-associated AIEC (219, 220), it is conceivable that polymorphisms in these genes could affect the autophagy pathway and promote intracellular proliferation of invasive pathogens leading to chronic inflammation in IBD.
Interestingly, a recent paper has proposed a new gene-environmental etiology for IBD, revealing that polymorphisms in CD-associated genes, NOD2 and ATG16L1, were associated with a defective sensing of protective signals derived from the microbiota (221).
Taken together, the identification of the IBD-susceptibility genes has shed light on essential relationships between genes and gut microbiota, particularly on host immune functions that can affect the assembly of intestinal microbiota.
7.2 Dysbiosis in inflammatory bowel diseases
Advances in culture-independent technologies have provided a more detailed characterization of the gut microbiota in patients with IBD. Analysis of mucosa-associated and luminal bacteria revealed the presence of a dysbiotic microbiota, marked by a reduced overall microbial diversity. Multiple studies have shown less complex profiles of commensal bacteria in IBD patients that seem to be associated with a temporal instability of dominant species when compared to healthy individuals (145, 222–224). An important observation that derived from this evidence is that a loss of total bacterial diversity could have detrimental effects on host health. It has now become evident that distinct members of the gut microbiota exert diverse and non-redundant effects on the host immune system (225), suggesting that a more complex collection of organisms is required for maximal benefits to the host. Given that, depletion of members of the phyla Firmicutes and Bacteroidetes, described in patients with IBD (145), might contribute to the intestinal inflammation seen in these patients. Of note, Bacteriodes fragilis has been shown to protect from the development of experimental IBD in mice through the immunomodulatory activity of its capsular polysaccharide A (PSA) (226). Specific changes, among the phylum of Firmicutes, observed in patients with ileal CD included disappearance of Faecalibacterium prausnitzii, a commensal microbial species that was able to secrete anti-inflammatory metabolites. Interestingly, a lower proportion of Faecalibacterium prausnitzii in ileal mucosa was associated with an enhanced risk of postoperative recurrence of ileal CD (227). In addition, reduced levels of microbes that produce short-chain fatty acids (SCFA) have been observed in patients with IBD. The downside of this loss is further strengthened by the evidence that SCFA-producing bacteria can provide protection from experimental colitis through the differentiation and expansion of regulatory T cells (228).
The loss of beneficial microbes and their immunomodulatory molecules can also provide the opportunity for the expansion of pathogens and/or pathobionts. For instance, a higher number of mucosa-associated Enterobacteriaceae with invasive properties or the presence of intramucosal E. coli has been described in patients with CD (139, 140).
However, there are still questions that remain unanswered. For instance, it is still unclear if intestinal dysbiosis, observed in patients with IBD, is a cause or consequence of gut inflammation. Another key question that remains is whether these diseases are caused by the expansion of pathobionts and/or loss of beneficial symbionts or simply secondary to an aberrant host immune response against gut symbionts.
Two recent cohort studies in pediatric treatment-naïve IBD patients have provided the opportunity to analysis the gut microbiota in patients at the very earliest stage of disease and prior to treatment intervention, showing that intestinal dysbiosis correlated strongly with disease status (229, 230).
Therefore, we can speculate that intestinal inflammation might be initiated by a specific symbiotic bacterium, or group of symbionts normally present at low abundance in the intestine, that may bloom under advantageous conditions, such as mutations in anti-microbial genes. As a result, exaggerated host immune responses may ultimately lead to chronic inflammation that definitively disrupts the intestinal homeostasis in favor of a more extreme dysbiosis. Hence, unraveling the specific microbial contribution to the development of IBD will improve the understanding and management of these diseases.
Concluding Remarks
The integral roles of the gut microbiota in resisting colonization of enteric pathogens, educating and promoting the maturation of the host immune system, and host metabolism, as shown by numerous studies in the past few years, underscore the need to understand mechanisms underlying the myriad of supportive roles of the gut microbiota in human health. Our recent studies on systemic production of complement C3 and hemopexin initiated by intestinally induced IL-22, as well as gut microbiota-induced systemic IgG antibodies that can potentially target pathogens with shared antigens, have unraveled a previously less understood role of the gut microbiota in combating systemic infection. This knowledge can be potentially harnessed in developing treatment for systemic infection that may lead to fatal sepsis. There are likely many others functions of the gut microbiota in equipping the host for systemic infection that are yet to be discovered. There has been increasing evidence linking gut dysbiosis with the manifestation of diseases involving chronic gut inflammation, most notably inflammatory bowel disease. Dampening gut inflammation, utilizing resistant competitors (probiotics), or maneuvering nutritional changes (such as prebiotics and metals) might be potential approaches to restricting blooms of Enterobacterieaceae or disease-causing bacteria in the future.
Acknowledgments
This work was supported by NIH grants T32HL007517 and 5P30DK034933 (to M.Y.Z.), T32DK094775 (to J.M.P), and DK091191 and DK095782 (to G.N.).
References
- 1.Marcobal A, Barboza M, Froehlich JW, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 3.Freter R. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. The Journal of infectious diseases. 1955;97:57–65. doi: 10.1093/infdis/97.1.57. [DOI] [PubMed] [Google Scholar]
- 4.Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1954;86:132–137. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- 5.Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. The Journal of infectious diseases. 2005;191:949–956. doi: 10.1086/428090. [DOI] [PubMed] [Google Scholar]
- 6.Hammami R, Fernandez B, Lacroix C, Fliss I. Anti-infective properties of bacteriocins: an update. Cellular and molecular life sciences : CMLS. 2013;70:2947–2967. doi: 10.1007/s00018-012-1202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillor O, Giladi I, Riley MA. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC microbiology. 2009;9:165. doi: 10.1186/1471-2180-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kommineni S, Bretl DJ, Lam V, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassone-Corsi M, Nuccio SP, Liu H, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428:412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]
- 12.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome research. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. Journal of bacteriology. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern A, Mick E, Tirosh I, Sagy O, Sorek R. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome research. 2012;22:1985–1994. doi: 10.1101/gr.138297.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diard M, Bakkeren E, Cornuault JK, et al. Inflammation boosts bacteriophage transfer between Salmonella spp. Science. 2017;355:1211–1215. doi: 10.1126/science.aaf8451. [DOI] [PubMed] [Google Scholar]
- 18.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinbauer MG. Ecology of prokaryotic viruses. FEMS microbiology reviews. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Sulakvelidze A, Alavidze Z, Morris JG., Jr Bacteriophage therapy. Antimicrobial agents and chemotherapy. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends in microbiology. 2010;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell AB, Wexler AG, Harding BN, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell host & microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wexler AG, Bao Y, Whitney JC, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO reports. 2016;17:1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohnhoff M, Miller CP, Martin WR. Resistance of the Mouse's Intestinal Tract to Experimental Salmonella Infection. I. Factors Which Interfere with the Initiation of Infection by Oral Inoculation. The Journal of experimental medicine. 1964;120:805–816. doi: 10.1084/jem.120.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherrington CA, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. The Journal of applied bacteriology. 1991;70:161–165. doi: 10.1111/j.1365-2672.1991.tb04442.x. [DOI] [PubMed] [Google Scholar]
- 27.Shin R, Suzuki M, Morishita Y. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. Journal of medical microbiology. 2002;51:201–206. doi: 10.1099/0022-1317-51-3-201. [DOI] [PubMed] [Google Scholar]
- 28.Rolfe RD. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infection and immunity. 1984;45:185–191. doi: 10.1128/iai.45.1.185-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YG, Sakamoto K, Seo SU, et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science. 2017;356:315–319. doi: 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohnhoff M, Miller CP, Martin WR. Resistance of the Mouse's Intestinal Tract to Experimental Salmonella Infection. Ii. Factors Responsible for Its Loss Following Streptomycin Treatment. The Journal of experimental medicine. 1964;120:817–828. doi: 10.1084/jem.120.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. Formate acts as a diffusible signal to induce Salmonella invasion. Journal of bacteriology. 2008;190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantois I, Ducatelle R, Pasmans F, et al. Butyrate specifically down-regulates salmonella pathogenicity island 1 gene expression. Applied and environmental microbiology. 2006;72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Molecular microbiology. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 34.Rivera-Chavez F, Zhang LF, Faber F, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell host & microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. Journal of lipid research. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature communications. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inagaki T, Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infection and immunity. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infection and immunity. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guiot HF. Role of competition for substrate in bacterial antagonism in the gut. Infection and immunity. 1982;38:887–892. doi: 10.1128/iai.38.3.887-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweeney NJ, Klemm P, McCormick BA, et al. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infection and immunity. 1996;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldonado-Gomez MX, Martinez I, Bottacini F, et al. Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell host & microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell host & microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamada N, Kim YG, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamada N, Sakamoto K, Seo SU, et al. Humoral Immunity in the Gut Selectively Targets Phenotypically Virulent Attaching-and-Effacing Bacteria for Intraluminal Elimination. Cell host & microbe. 2015;17:617–627. doi: 10.1016/j.chom.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNulty NP, Wu M, Erickson AR, et al. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS biology. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desai MS, Seekatz AM, Koropatkin NM, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353. e1321. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deriu E, Liu JZ, Pezeshki M, et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell host & microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin LZ, Marquardt RR, Zhao X. A strain of Enterococcus faecium (18C23) inhibits adhesion of enterotoxigenic Escherichia coli K88 to porcine small intestine mucus. Applied and environmental microbiology. 2000;66:4200–4204. doi: 10.1128/aem.66.10.4200-4204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johansson ME, Jakobsson HE, Holmen-Larsson J, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell host & microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergstrom JH, Birchenough GM, Katona G, et al. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:13833–13838. doi: 10.1073/pnas.1611400113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergstrom KS, Kissoon-Singh V, Gibson DL, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS pathogens. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zarepour M, Bhullar K, Montero M, et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infection and immunity. 2013;81:3672–3683. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arike L, Holmen-Larsson J, Hansson GC. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology. 2017 doi: 10.1093/glycob/cww134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umesaki Y, Tohyama K, Mutai M. Appearance of fucolipid after conventionalization of germ-free mice. Journal of biochemistry. 1981;90:559–561. doi: 10.1093/oxfordjournals.jbchem.a133506. [DOI] [PubMed] [Google Scholar]
- 62.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pickard JM, Maurice CF, Kinnebrew MA, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goto Y, Obata T, Kunisawa J, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pacheco AR, Curtis MM, Ritchie JM, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pham TA, Clare S, Goulding D, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell host & microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faber F, Tran L, Byndloss MX, et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature. 2016;534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129:729–739. doi: 10.1182/blood-2016-03-708594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khosravi A, Yanez A, Price JG, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fiedler K, Kokai E, Bresch S, Brunner C. MyD88 is involved in myeloid as well as lymphoid hematopoiesis independent of the presence of a pathogen. Am J Blood Res. 2013;3:124–140. [PMC free article] [PubMed] [Google Scholar]
- 71.Balmer ML, Schurch CM, Saito Y, et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol. 2014;193:5273–5283. doi: 10.4049/jimmunol.1400762. [DOI] [PubMed] [Google Scholar]
- 72.Deshmukh HS, Liu Y, Menkiti OR, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang D, Chen G, Manwani D, et al. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525:528–532. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burberry A, Zeng MY, Ding L, et al. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe. 2014;15:779–791. doi: 10.1016/j.chom.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carlens J, Wahl B, Ballmaier M, Bulfone-Paus S, Forster R, Pabst O. Common gamma-chain-dependent signals confer selective survival of eosinophils in the murine small intestine. J Immunol. 2009;183:5600–5607. doi: 10.4049/jimmunol.0801581. [DOI] [PubMed] [Google Scholar]
- 76.Sugawara R, Lee EJ, Jang MS, et al. Small intestinal eosinophils regulate Th17 cells by producing IL-1 receptor antagonist. J Exp Med. 2016;213:555–567. doi: 10.1084/jem.20141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang BG, Seoh JY, Jang MH. Regulatory Eosinophils in Inflammation and Metabolic Disorders. Immune Netw. 2017;17:41–47. doi: 10.4110/in.2017.17.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies LC, Taylor PR. Tissue-resident macrophages: then and now. Immunology. 2015;144:541–548. doi: 10.1111/imm.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bain CC, Bravo-Blas A, Scott CL, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smythies LE, Sellers M, Clements RH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franchi L, Kamada N, Nakamura Y, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seo SU, Kamada N, Munoz-Planillo R, et al. Distinct Commensals Induce Interleukin-1beta via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity. 2015;42:744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hirota K, Ahlfors H, Duarte JH, Stockinger B. Regulation and function of innate and adaptive interleukin-17-producing cells. EMBO Rep. 2012;13:113–120. doi: 10.1038/embor.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ivanov II, Frutos Rde L, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 88.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Geuking MB, Cahenzli J, Lawson MA, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 90.Sano T, Huang W, Hall JA, et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Atarashi K, Tanoue T, Ando M, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 97.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kiss EA, Vonarbourg C, Kopfmann S, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 100.Lee JS, Cella M, McDonald KG, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qiu J, Heller JJ, Guo X, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. 2010;33:752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 103.Hughes T, Becknell B, Freud AG, et al. Interleukin-1beta selectively expands and sustains interleukin-22+ immature human natural killer cells in secondary lymphoid tissue. Immunity. 2010;32:803–814. doi: 10.1016/j.immuni.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cording S, Medvedovic J, Cherrier M, Eberl G. Development and regulation of RORgammat(+) innate lymphoid cells. FEBS Lett. 2014;588:4176–4181. doi: 10.1016/j.febslet.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 105.Tsuji M, Suzuki K, Kitamura H, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 106.Sonnenberg GF, Monticelli LA, Alenghat T, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abt MC, Lewis BB, Caballero S, et al. Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe. 2015;18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hasegawa M, Yada S, Liu MZ, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 2014;41:620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakamoto K, Kim YG, Hara H, et al. IL-22 Controls Iron-Dependent Nutritional Immunity Against Systemic Bacterial Infections. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aai8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hepworth MR, Fung TC, Masur SH, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brandtzaeg P, Halstensen TS, Kett K, et al. Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial lymphocytes. Gastroenterology. 1989;97:1562–1584. doi: 10.1016/0016-5085(89)90406-x. [DOI] [PubMed] [Google Scholar]
- 112.Wesemann DR, Portuguese AJ, Meyers RM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hirota K, Turner JE, Villa M, et al. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yel L. Selective IgA deficiency. J Clin Immunol. 2010;30:10–16. doi: 10.1007/s10875-009-9357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Macpherson AJ, Geuking MB, McCoy KD. Homeland security: IgA immunity at the frontiers of the body. Trends Immunol. 2012;33:160–167. doi: 10.1016/j.it.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 117.Johansen FE, Pekna M, Norderhaug IN, et al. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J Exp Med. 1999;190:915–922. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shimada S, Kawaguchi-Miyashita M, Kushiro A, et al. Generation of polymeric immunoglobulin receptor-deficient mouse with marked reduction of secretory IgA. J Immunol. 1999;163:5367–5373. [PubMed] [Google Scholar]
- 119.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–217. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 121.Herbst T, Sichelstiel A, Schar C, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 122.Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med. 2007;175:561–569. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- 123.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 126.Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–375. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim D, Kim YG, Seo SU, et al. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat Med. 2016;22:524–530. doi: 10.1038/nm.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zeng MY, Cisalpino D, Varadarajan S, et al. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zeng MY, Inohara N, Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koch MA, Reiner GL, Lugo KA, et al. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell host & microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 136.Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS biology. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carvalho FA, Koren O, Goodrich JK, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell host & microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 140.Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 141.Buffie CG, Jarchum I, Equinda M, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infection and immunity. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Garrett WS, Gallini CA, Yatsunenko T, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seksik P, Rigottier-Gois L, Gramet G, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. Journal of clinical microbiology. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. The ISME journal. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 147.Wlodarska M, Willing B, Keeney KM, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infection and immunity. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Spees AM, Wangdi T, Lopez CA, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio. 2013;4 doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maxwell PR, Rink E, Kumar D, Mendall MA. Antibiotics increase functional abdominal symptoms. The American journal of gastroenterology. 2002;97:104–108. doi: 10.1111/j.1572-0241.2002.05428.x. [DOI] [PubMed] [Google Scholar]
- 150.Mendall MA, Kumar D. Antibiotic use, childhood affluence and irritable bowel syndrome (IBS) European journal of gastroenterology & hepatology. 1998;10:59–62. doi: 10.1097/00042737-199801000-00011. [DOI] [PubMed] [Google Scholar]
- 151.Matto J, Maunuksela L, Kajander K, et al. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS immunology and medical microbiology. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 152.Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC gastroenterology. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kerckhoffs AP, Ben-Amor K, Samsom M, et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. Journal of medical microbiology. 2011;60:236–245. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 155.Pammi M, Cope J, Tarr PI, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Barman M, Unold D, Shifley K, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infection and immunity. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Haag LM, Fischer A, Otto B, et al. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PloS one. 2012;7:e35988. doi: 10.1371/journal.pone.0035988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raetz M, Hwang SH, Wilhelm CL, et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nature immunology. 2013;14:136–142. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nature medicine. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dicksved J, Ellstrom P, Engstrand L, Rautelin H. Susceptibility to Campylobacter infection is associated with the species composition of the human fecal microbiota. mBio. 2014;5:e01212–01214. doi: 10.1128/mBio.01212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Galipeau HJ, McCarville JL, Huebener S, et al. Intestinal microbiota modulates gluten-induced immunopathology in humanized mice. The American journal of pathology. 2015;185:2969–2982. doi: 10.1016/j.ajpath.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cinova J, De Palma G, Stepankova R, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PloS one. 2011;6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Barnich N, Carvalho FA, Glasser AL, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. The Journal of clinical investigation. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dreux N, Denizot J, Martinez-Medina M, et al. Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS pathogens. 2013;9:e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Iebba V, Conte MP, Lepanto MS, et al. Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease. Infection and immunity. 2012;80:1408–1417. doi: 10.1128/IAI.06181-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature reviews Microbiology. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sonnenburg ED, Zheng H, Joglekar P, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. The ISME journal. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO reports. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nature reviews Microbiology. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 175.Van der Sluis M, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 176.Huang YL, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nature communications. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bouchet V, Hood DW, Li J, et al. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cascales E, Buchanan SK, Duche D, et al. Colicin biology. Microbiology and molecular biology reviews : MMBR. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nedialkova LP, Denzler R, Koeppel MB, et al. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS pathogens. 2014;10:e1003844. doi: 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ward PP, Uribe-Luna S, Conneely OM. Lactoferrin and host defense. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2002;80:95–102. doi: 10.1139/o01-214. [DOI] [PubMed] [Google Scholar]
- 181.Bachman MA, Miller VL, Weiser JN. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS pathogens. 2009;5:e1000622. doi: 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Butala M, Zgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cellular and molecular life sciences : CMLS. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Winter SE, Winter MG, Xavier MN, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rivera-Chavez F, Winter SE, Lopez CA, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS pathogens. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Winter SE, Thiennimitr P, Winter MG, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lopez CA, Winter SE, Rivera-Chavez F, et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3 doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochimica et biophysica acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 188.Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, Haase VH. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. The Journal of clinical investigation. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lopez CA, Miller BM, Rivera-Chavez F, et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353:1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS pathogens. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Singh V, Yeoh BS, Xiao X, et al. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nature communications. 2015;6:7113. doi: 10.1038/ncomms8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dogan B, Suzuki H, Herlekar D, et al. Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflammatory bowel diseases. 2014;20:1919–1932. doi: 10.1097/MIB.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 193.Nash JH, Villegas A, Kropinski AM, et al. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC genomics. 2010;11:667. doi: 10.1186/1471-2164-11-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kehl-Fie TE, Chitayat S, Hood MI, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell host & microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu JZ, Jellbauer S, Poe AJ, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell host & microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Diaz-Ochoa VE, Lam D, Lee CS, et al. Salmonella Mitigates Oxidative Stress and Thrives in the Inflamed Gut by Evading Calprotectin-Mediated Manganese Sequestration. Cell host & microbe. 2016;19:814–825. doi: 10.1016/j.chom.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rutgeerts P, Goboes K, Peeters M, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 198.D'Haens GR, Geboes K, Peeters M, Baert F, Penninckx F, Rutgeerts P. Early lesions of recurrent Crohn's disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–267. doi: 10.1016/s0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- 199.Thia KT, Mahadevan U, Feagan BG, et al. Ciprofloxacin or metronidazole for the treatment of perianal fistulas in patients with Crohn's disease: a randomized, double-blind, placebo-controlled pilot study. Inflammatory bowel diseases. 2009;15:17–24. doi: 10.1002/ibd.20608. [DOI] [PubMed] [Google Scholar]
- 200.Prantera C, Lochs H, Grimaldi M, et al. Rifaximin-extended intestinal release induces remission in patients with moderately active Crohn's disease. Gastroenterology. 2012;142:473–481. e474. doi: 10.1053/j.gastro.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 201.Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. The American journal of pathology. 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 202.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 203.Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infection and immunity. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Taurog JD, Richardson JA, Croft JT, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. The Journal of experimental medicine. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schaubeck M, Clavel T, Calasan J, et al. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut. 2016;65:225–237. doi: 10.1136/gutjnl-2015-309333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mow WS, Vasiliauskas EA, Lin YC, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn's disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 208.Pirzer U, Schonhaar A, Fleischer B, Hermann E, Meyer zum Buschenfelde KH. Reactivity of infiltrating T lymphocytes with microbial antigens in Crohn's disease. Lancet. 1991;338:1238–1239. doi: 10.1016/0140-6736(91)92104-a. [DOI] [PubMed] [Google Scholar]
- 209.Duchmann R, Neurath MF, Meyer zum Buschenfelde KH. Responses to self and non-self intestinal microflora in health and inflammatory bowel disease. Research in immunology. 1997;148:589–594. doi: 10.1016/s0923-2494(98)80154-5. [DOI] [PubMed] [Google Scholar]
- 210.Lodes MJ, Cong Y, Elson CO, et al. Bacterial flagellin is a dominant antigen in Crohn disease. The Journal of clinical investigation. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Laboratory animals. 1993;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- 213.Berry D, Kuzyk O, Rauch I, et al. Intestinal Microbiota Signatures Associated with Inflammation History in Mice Experiencing Recurring Colitis. Frontiers in microbiology. 2015;6:1408. doi: 10.3389/fmicb.2015.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 215.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 216.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature genetics. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cellular microbiology. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nature genetics. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 221.Chu H, Khosravi A, Kusumawardhani IP, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 223.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. Journal of clinical microbiology. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Science translational medicine. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 227.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 229.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shaw KA, Bertha M, Hofmekler T, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome medicine. 2016;8:75. doi: 10.1186/s13073-016-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]