Abstract
Microfluidics is a vibrant and expanding field that has the potential for solving many analytical challenges. Microfluidics show promise to provide rapid, inexpensive, efficient, and portable diagnostic solutions that can be used in resource-limited settings. Researchers have recently reported various microfluidic platforms for biomarker analysis applications. Sample preparation processes like purification, preconcentration and labeling have been characterized on-chip. Additionally, improvements in microfluidic separation techniques have been reported for cellular and molecular biomarkers. This review critically evaluates microfluidic sample preparation platforms and separation methods for biomarker analysis reported in the last two years. Key advances in device operation and ability to process different sample matrices in a variety of device materials are highlighted. Finally, current needs and potential future directions for microfluidic device development to realize its full diagnostic potential are discussed.
Keywords: Biomarkers, Capillary electrophoresis, Solid-phase extraction, Microfluidics
Graphical Abstract
1. Introduction
Disease diagnostics are important for improving human health, and the effective treatment of many life-threatening conditions is dependent upon the accuracy and speed of the diagnosis, which can result in improved human life expectancy. Technologies currently used in healthcare diagnostics often require expensive instrumentation or a modern testing laboratory, neither of which are feasible in many developing nations or in remote locations. Hence, low-cost, rapid, portable, and easy to use tools are desirable to advance clinical diagnostics, especially in developing countries or remote areas that lack appropriate infrastructure.
Analyses of biomarkers, biomolecular indicators of medical conditions, hold excellent potential for clinical diagnosis of various diseases. These biomarkers are frequently found in complex biological matrices or bodily fluids, which almost always require sample preparation prior to analysis. Sample preparation steps often need large volumes (> mL) and experienced personnel, which further increase the analysis time and cost. Thus, fast and effective sample preparation techniques are necessary to facilitate early diagnosis.
Microfluidic systems offering advantages like low cost per device, rapid analysis, and small sample requirements [1] have potential to transform diagnostics, especially in developing countries or remote locations due to amenability to point-of-care testing. Biomarker analysis has been one of the most actively pursued applications in miniaturization of chemical analyzers. A number of microfluidic systems have been reported recently that can perform sample preparation steps like purification, preconcentration and labeling on a chip prior to quantitation [1–3]. Separation techniques have also advanced for the analysis of molecular biomarkers in a microfluidic setup.
This manuscript critically reviews microfluidic techniques reported for biomarker sample preparation and separation over the last two years. Developments in microfluidics for biomarker analysis prior to 2015 have been reviewed previously [4, 5]. Herein, we specifically focus on microfluidic sample preparation methods, such as biomarker purification from biological matrices and preconcentration of trace components, and approaches that are used for biomarker separation. Furthermore, novel aspects of device design and analysis methods are highlighted.
2. On-chip sample preparation methods
Microfluidics can be used to miniaturize and integrate sample preparation processes on a microchip platform. Typically, building these systems requires innovations in device design and manufacturing; fluid transport, automation and control; preparation of samples before analysis; separation; multiplexing; and detection [6]. Often sample specimens are limited in volume, contain matrix-related interferences, require multiplex analysis and have low target analyte concentrations [7]; therefore, sample preparation is a key part of analysis. Commonly used sample preparation processes include analyte purification, enrichment and labeling. In this section we focus on select techniques for microfluidic sample preparation.
2.1 Molecular affinity extraction
On-chip sample preparation can be used to selectively extract, preconcentrate and label selected analytes in an automated fashion. The ability to extract trace amounts of desired analytes from a complex sample matrix such as blood significantly simplifies the analysis [8]. Such on-chip sample preparation could replace laborious benchtop processes, and thus decrease analysis time and potentially allow point-of-care usage. Affinity approaches using antibodies or aptamers on a solid support can purify target species in blood from undesired matrix components that complicate analysis. A summary of key information related to molecular affinity extraction work discussed in this section is given in Table I.
Table I.
Microfluidic devices for affinity-based analysis. Additional abbreviations used: Carcinoembryonic antigen (CEA), recombinant avian influenza A virus (rH7N9), cancer antigen-125 (CA-125), cluster of differentiation 24 (CD24).
| Affinity method |
Biomarkers | Disease/condition | Immobilization | Detection | Concentration | Reference |
|---|---|---|---|---|---|---|
| Antibody | EpCAM | Epithelial cancer | Channel surface | Fluorescence | 2–2000 pg/mL | 18 |
| TNFα | Inflammation | Channel surface | Electrochemical | 4–50 ng/mL | 19 | |
| ErbB2 | Breast cancer | Covalent to TiO2 nanofibers | Electrochemical | 1 fM – 0.1 µM | 20 | |
| CEA, rH7N9 | Influenza A | in-solution and in-gel | Colorimetric | 1–100 pg/mL | 22 | |
| Influenza virus epitope | Influenza | in-solution | Colorimetric | 1–100 µg/mL | 23 | |
| Bcl-2 | Apoptosis | Covalent to tin oxide | Mass spectrometry | 140 nM | 24 | |
| Gastric cancer biomarker panel | Gastric cancer | Channel surface | Electrochemical | pg/mL-ng/mL | 25 | |
| E. coli | E. coli infection | Channel surface | Fluorescence | 105–107 cfu/mL | 28 | |
| PGA | Anthrax | Magnetic beads | SERS | 100 pg/mL-100 µg/mL | 29 | |
| Apolipoprotein A1 | Bladder cancer | Magnetic beads | Semiconductor based ion-sensitivity | 12.5–1000 ng/mL | 30 | |
| CA-125, EpCAM, CD24 | Ovarian cancer | Magnetic beads | Fluorescence | 7.5×105 – 1×107 particles/mL | 31 | |
| Aβ peptides | Alzheimer’s disease | Magnetic beads | Fluorescence | 25–100 ng | 32 | |
| E. coli | Pathogens | Nanoparticle clusters | UV-Vis | 100–105 cfu/mL | 33 | |
| Aptamer | PSA | Prostate cancer | Channel surface | Chemiluminescence | 3–50 ng/mL | 36 |
| Hemoglobin | Diabetes | Magnetic beads | Fluorescence | 0.7–14.8 g/dL | 37 | |
| VEGF | Cervical cancer | Channel surface | UV-Vis, Fluorescence | 2–40 ng/mL | 38 | |
| Creatine kinase | Cardiac damage | Covalent to gold electrode | Electrochemical | 10 pg/mL-100 ng/mL | 39 |
2.1.1 Antibody-based extraction
An antibody (Ab) offers high selectivity and specificity towards its target antigen, and can be used in microfluidic systems for the selective capture of desired molecules [9, 10]. Antibodies can be placed in a microfluidic setup by device surface modification [11, 12] or through a solid support like porous polymer monoliths [13], beads [14, 15] or nanoparticles [16, 17] introduced into microchannels.
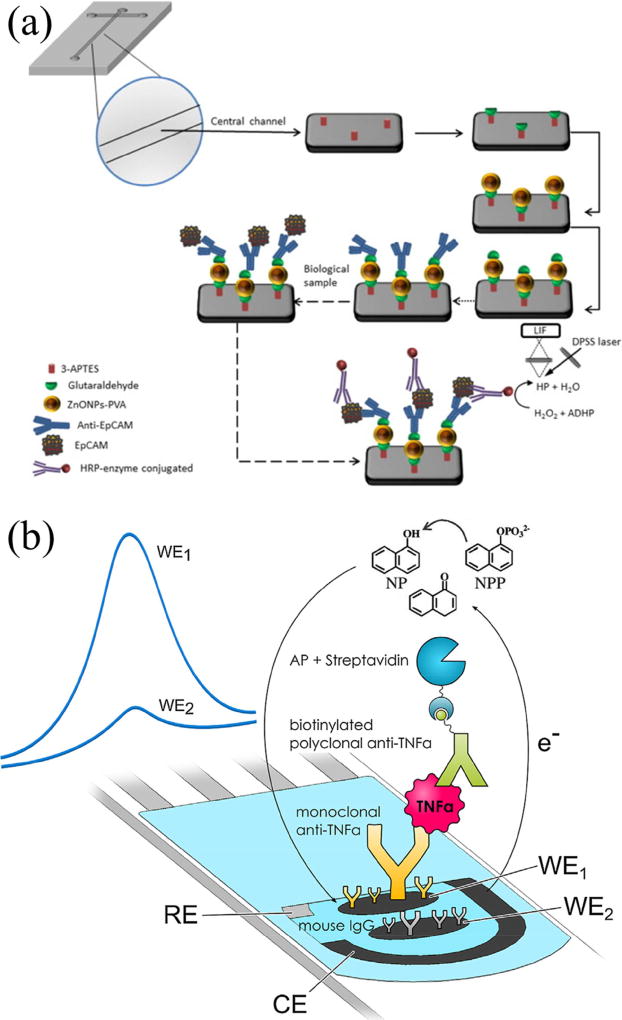
An immunosensor was developed on a PDMS treated glass microfluidic device using Ab-conjugated polyvinyl alcohol covered zinc oxide nanoparticles for the extraction of epithelial cell adhesion molecule (EpCAM), a biomarker for epithelial cancers [18]. Whole blood was centrifuged and lysed off-chip to prepare the supernatant that was introduced into the microfluidic devices. Bound EpCAM interacted with anti-EpCAM conjugated to horseradish peroxidase, which catalyzed the oxidation of non-fluorescent 10-acetyl-3,7-dihydroxyphenoxazine to fluorescent resorufin, as shown in Fig. 1a. Extraction results from blood samples obtained from cancer patients and healthy volunteers were compared to a commercially available test, and a linear correlation was obtained from 2–2000 pg mL−1 EpCAM with a detection limit of 1.3 pg mL−1. Future work in correlation of measured EpCAM levels with cancer incidence with minimum off-chip sample preparation would be impactful.
Figure 1.
Immunocapture for biomarker detection. (a) Surface modification scheme for immobilization of anti-EpCAM for detection of EpCAM. Adapted with permission from Fernández-Baldo et al. Microchem. J., 128 (2016) 18–25. (b) Sandwich immunoassay for electrochemical detection of TNF-α. Adapted with permission from Eletxigerra et al. Sens. Actuators, B, 221 (2015) 1406–1411.
In a different study a paper microfluidic device was reported that utilized an antibody-based sandwich assay for detection of tumor necrosis factor alpha (TNFα), an inflammation biomarker [19]. Carbon electrodes were printed on the paper device and anti-TNFα immobilization through both covalent binding and physical adsorption was tested for immunocapture and electrochemical detection, as shown in Fig. 1b. The limit of detection for TNFα was 4 ng mL−1, and diluted human serum samples spiked with TNFα were analyzed down to 20 ng mL−1, although further improvements in detection will be needed to analyze TNFα at native levels in blood. With further work in miniaturizing the electrochemical detection instrumentation, such disposable devices may show promise for detection of diseases, potentially in a point-of-care setting in developing countries. Ali et al. [20] also reported a microfluidic device for electrochemical detection of a breast cancer biomarker, epidermal growth factor receptor 2 protein family (ErbB2). Using carbodiimide linkage methods Anti-ErbB2 was covalently attached to graphene foam modified with titanium dioxide nanofibers, which served as an immuno-electrode inside a polydimethylsiloxane (PDMS)-glass microfluidic device. Detection by differential pulse voltammetry and electrochemical impedance spectroscopy worked for ErbB2 from 100 nM to 1 fM in the presence of interfering antigens. Experimental results were obtained in buffer solutions, so studies with cell lysate or serum samples as a next step would provide a greater impact.
With the advent of smartphones, diagnosis based on ubiquitous camera capabilities offers great potential for point-of-care [21] and microfluidics applications. One recent example used an on-chip complement fixation test for the detection of carcinoembryonic antigen and recombinant avian influenza A virus using a PDMS-glass microchip [22]. Fluidic-based and agar gel-based complement fixation tests were developed to indicate the presence of a specific antibody or antigen. Colorimetric changes for concentrations in the range of 1–100 pg/mL were easily imaged and analyzed using a smartphone, indicating strong potential for point-of-care application. Another study reported a smartphone-operated microfluidic colorimetric immunoassay for detection of influenza infection [23]. The PDMS microfluidic device contained nitrocellulose paper with influenza virus epitope spots to capture the corresponding primary antibodies present in the diluted sample. These primary antibodies were then detected using alkaline phosphatase conjugated secondary antibodies that induced a color change for a concentration range of 1–100 µg/mL in diluted serum. The device was battery operated and portable, and the total analysis time was 18 min. The results in both these studies involved colorimetric changes that were imaged and analyzed using a smartphone, which as shown previously [21] can be advantageous for point-of-care applications. Improved microfluidic devices that can effectively detect clinically relevant concentration of biomarkers in typical biological matrices with little to no sample preparation are needed to further advance smartphone-enabled diagnostics.
Yang et al. [24] reported a PDMS microfluidic device containing wells separated by pneumatic valves for affinity capture, tryptic digestion and isotopic labeling leading to mass spectrometric analysis of an apoptosis-related protein, Bcl-2. Anti-Bcl-2 antibodies were covalently linked to indium tin oxide on the bottom of the wells, and sequence coverage of 50% was reported for mass spectrometric analysis after tryptic digestion and iTRAQ labeling of captured Bcl-2. Going forward, cell studies and multiplex biomarker capture are important capabilities that should be addressed for these systems to have greater impact in proteomics.
In a different study an electrochemical microfluidic chip for the multiplexed detection of gastric cancer biomarkers was reported [25]. Gold working electrodes were fabricated on glass, antibodies were covalently bound and the device was formed by bonding a PDMS mold cover. Six biomarkers for gastric cancer (carcinoembryonic antigen, carbohydrate antigen 19–9, H. pylori CagA protein, P53 oncoprotein, and pepsinogen I and II) were electrochemically detected from serum samples using these immunosensor chips with a linear correlation from clinically relevant pg/mL to ng/mL concentrations. Future work in developing a concentration-based assay would eventually allow early diagnosis and monitoring of gastric cancer.
PDMS has been widely used for making microfluidic devices but it has disadvantages like hydrophobicity and poor fabrication scalability [26, 27]. Thus, a thiol-acrylate resin that exhibits low background fluorescence was reported for making microfluidic devices at room temperature [28]. Simple electrostatic interaction between the channel walls and an Ab to E. coli was used for immobilization. Using these devices, 105 cfu mL−1 of fluorescently labeled E. coli were detected. Although this resin shows promise as a material for microfluidic devices, improvement in the detection limit, immobilization method and ability to detect unlabeled bacteria will be needed for utility in disease diagnosis.
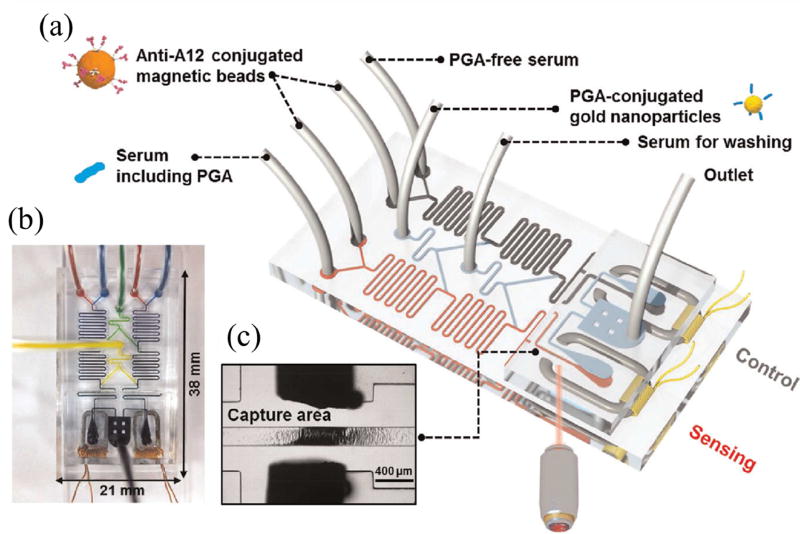
Beads are routine solid supports for antibodies and can be easily manipulated within fluidic networks [14]; thus, microfluidic systems that utilize beads for immuno-capture and detection of disease biomarkers are being developed [15]. Gao et al. [29] used antibody-conjugated magnetic beads for the detection of an anthrax biomarker, poly-γ-D-glutamic acid (PGA), in human serum using surface-enhanced Raman scattering (SERS) in a microfluidic setup, shown in Fig. 2. Detection was done using competition between PGA and PGA-conjugated gold nanoparticles for anti-PGA linked magnetic beads in a PDMS microdevice. A linear decrease in SERS signal was observed for clinically relevant PGA concentrations ranging from 100 pg/mL to 100 µg/mL in human serum. Future work should focus on extension of the approach to additional biomarkers and developing multiplexing methods.
Figure 2.
Microfluidic SERS immunoassay for detection of PGA. (a) Assay overview, (b) image of device and (c) image showing captured magnetic beads. Adapted with permission from Gao et al. Biosens. Bioelectron. 72 (2015) 230–236.
Another study reported a magnetic bead-based immunoassay for urinary protein biomarker detection in a microfluidic device [30]. Magnetic beads with epoxy groups were conjugated with antibodies to capture apolipoprotein A1, a bladder cancer biomarker. Addition of negatively charged DNA on the antigen-antibody complex increased charge, resulting in signal enhancement on the semiconductor sensor platform within the microdevice. Apolipoprotein A1 was measured in urine samples within 20% error compared to established methods, with a limit of detection of 10 ng/mL. Although this microfluidic device shows promising results for quantitation of clinically useful levels of a urine biomarker, the error in concentration measurement has room for improvement.
Zhao et al. [31] also used magnetic beads to capture intact exosomes from human serum for the detection of three ovarian cancer biomarkers (see Table I). A microchip containing a serpentine channel with Y-shaped inlets was fabricated in PDMS for mixing beads and capturing exosomes. These beads were then collected in a microchamber using a magnet, incubated with fluorescent antibodies for three ovarian cancer exosomal markers and detected by multi-color fluorescence imaging. Comparable results to a conventional assay for cancer vs. healthy samples were obtained using the microchip in a 40 min analysis time. Future efforts to correlate detected biomarker concentrations with occurrence of cancer will be needed for diagnostic applications.
Another group developed a PDMS microfluidic device that used antibody-coated magnetic beads for selective capture of Aβ peptides, biomarkers of Alzheimer’s disease [32]. The device had a nanoporous hydrogel membrane for peptide preconcentration, and a microchip electrophoresis (µCE) channel for separation of Aβ peptides. Using this integrated device 25 ng of Aβ peptide spiked in 100 µL cerebrospinal fluid (CSF) was detected. To further improve this method, mixing of Ab-coated magnetic beads with CSF could be done on-chip and analysis of patient CSF samples instead of spiked ones should be done. Lee et al. [33] used antibody-conjugated magnetic nanoparticle clusters in a 3D printed helical channel device to capture E. coli in milk. The concentration of captured bacteria was determined by UV-Vis absorption, and the limit of detection was 100 cfu/mL in milk. This study indicates the potential of 3D printed fluidic devices for biomarker analysis; however, the channel dimensions are larger than traditional microfluidic dimensions.
2.1.2 Aptamer-based extraction
Aptamers are short oligonucleotides that have high affinity for their target molecule; they have been used for capture and extraction in microfluidics. Compared to antibodies, aptamers offer advantages like easy synthesis, high stability and low cross-reactivity [34, 35], but aptamer research has not been pursued as deeply as the research of antibodies.
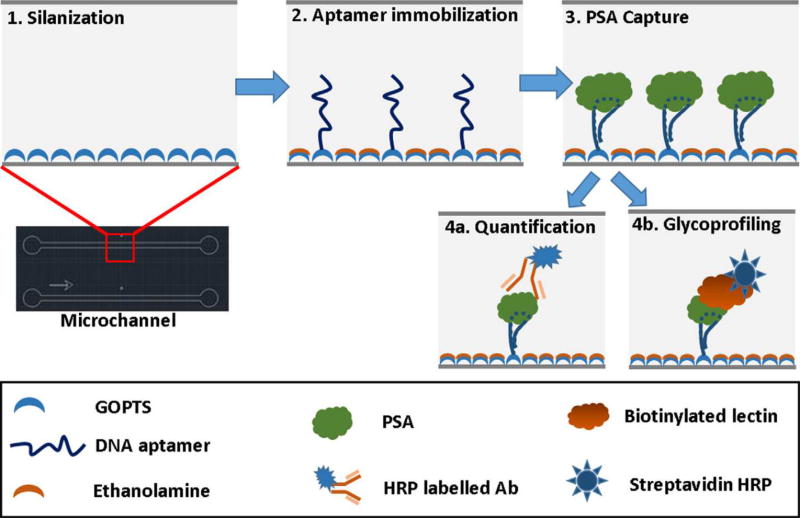
Jolly et al. [36] reported aptamer-based microfluidic immunoassays for prostate cancer biomarker measurement. Amine-linked aptamers covalently attached to PDMS channels derivatized with (3-glycidyloxypropyl) trimethoxysilane were used to capture prostate specific antigen (PSA) from buffer solution, as shown in Fig. 3. In one immunoassay, free PSA was measured by introducing secondary antibodies, followed by chemiluminescence detection with a limit of detection of 0.5 ng/mL. Additionally, detection of glycosylated PSA was done using a biotinylated lectin after aptamer capture, with a limit of detection of 3 ng/mL. This device shows a novel aptamer-based assay that can detect clinical levels of free PSA; however, multiplexed experiments using serum samples still need to be evaluated for greater impact. In a different report aptamers were used for sandwich immunoassays in a PDMS microfluidic device for potential diagnosis of diabetes [37]. Two parallel assays were conducted for quantitation of glycated hemoglobin and total hemoglobin. Analytes were captured from pretreated blood samples by incubating with aptamer-coated magnetic beads. After capture, a second labeled aptamer was added for fluorescence detection. This assay showed good correlation in the quantitation of typical human blood hemoglobin levels compared with benchtop HPLC results, in a shorter analysis time. Going forward, hemoglobin concentrations should be assayed on control vs. diabetic blood samples.
Figure 3.
Channel surface modification scheme for aptamer-based detection of PSA. Adapted from Jolly et al. Biosens. Bioelectron. 79 (2016) 313–319.
Lin et al. [38] reported a PDMS microfluidic device for cell culture and used aptamer-functionalized microchannels for analysis of vascular endothelial growth factor (VEGF). Amine-modified aptamers were attached to the carboxy-silane derivatized surface via carbodiimide coupling. Cell-cell communication was studied under different low oxygen conditions and for various distances between cell cultures. The results showed faster cell migration under oxidative stress, and captured VEGF (indicating tumor development) was detected using fluorescence and UV-Vis absorption. Another group developed an aptamer-based electrochemical microfluidic biosensor for the detection of creatine kinase, a cardiac biomarker [39]. A gold electrode surface was coated with a carboxy-terminated thiol, which was then functionalized with amine-linked aptamers via carbodiimide coupling. Impedance signal for creatine kinase was linear from 10 pg/mL to 100 ng/mL (relevant to clinical concentrations) in both buffer and culture media samples. A heart-on-chip cardiac bioreactor was integrated with this device, and doxorubicin-induced cardiac damage was assessed through changes in creatine kinase concentration. Both these studies demonstrate microfluidic cell culture devices that detect secreted proteins; future work enabling detection of multiple biomarkers for diagnostic purposes would be impactful.
To summarize, a variety of microfluidic platforms have been reported recently for affinity capture of targeted biomarkers using antibodies and aptamers. A key area of improvement needed for many of these systems is the ability to maintain good detection limits with biological matrices. Multiplexing is another pursuit that could improve disease diagnosis in these microfluidic devices. Finally, efforts are needed to design diagnostic assays that are well suited for point-of-care applications.
2.2 Sample preconcentration
To achieve rapid analysis in a point-of-care setup, a system must measure analytes from blood or other specimens with minimal sample preparation. Since biomarkers are often present in trace amounts, a preconcentration step can be desirable to improve detection [40]. Sample preconcentration on a microfluidic platform can be achieved through electrokinetic means, filtration or chromatographic interactions, as summarized in Table II.
Table II.
On-chip sample preconcentration.
| Mechanism | Medium | Device material |
Analytes | Enrichment | References |
|---|---|---|---|---|---|
| Electrokinetic | Liquid phase | PDMS | DNA | 500 | 41 |
| PDMS | Angiotensin | 450 | 42 | ||
| Nanoporous membrane | Paper | Bovine serum albumin | 800 | 43 | |
| PDMS | Lipid vesicles | 160 | 44 | ||
| SPE | Size selective membrane | PDMS | Albumin | 500 | 45 |
| PDMS-COC | Ferritin | 100 | 46 | ||
| Porous polymer monolith | COC | Preterm birth peptide biomarker Ferritin | 50 | 47 |
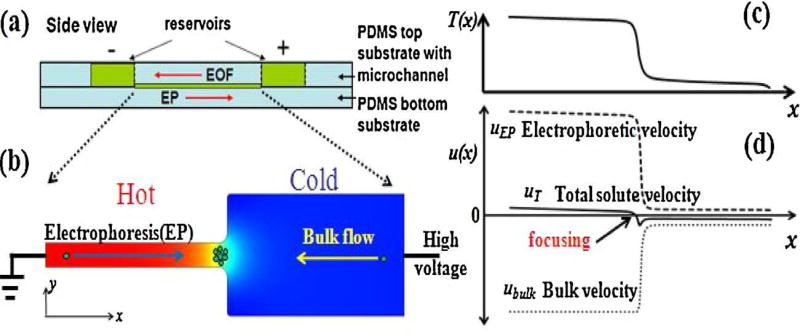
Ge et al. [41] developed an approach to concentrate DNA in a PDMS microchannel, where temperature gradient focusing (TGF) occurred at the interface of a channel that expanded rapidly (see Fig. 4), supplementing electrokinetic concentration. A combination of high-frequency AC with DC voltage reduced the backpressure due to electroosmosis and improved efficiency. DNA preconcentration of 500-fold was achieved in 40 s of operation. This device showed a good preconcentration efficiency for model DNA; however, device performance with real samples still needs to be evaluated.
Figure 4.
Working principle of Joule heating induced temperature gradient focusing. (a) Device schematic. (b) Numerical simulation showing temperature profile within the device. (c) Temperature along the microfluidic channel. (d) Velocity profiles along the microchannel under the combined AC and DC fields. Adapted with permission from Ge et al. Anal. Chim. Acta, 858 (2015) 91–97.
Combining hydrodynamic control with electrokinetic methods can be beneficial. Indeed, Cong et al. [42] developed an electrokinetic sample preconcentration microfluidic device with electrokinetic or hydrodynamic injection, a pneumatic valve for preconcentration and µCE separation. With the valve closed during electrokinetic injection, current flowed but bulk flow was blocked, allowing ion concentration polarization for sample preconcentration at the closed valve interface. A preconcentrated sample was hydrodynamically injected for µCE, showing 450-fold enrichment. This initial demonstration with model analytes in buffer solution could be improved upon with analysis of biomarkers in a complex sample matrix.
Paper microfluidics can provide inexpensive but often low performance devices. Paper microfluidics for electrokinetic preconcentration of model analytes were demonstrated by forming a cation selective Nafion membrane to induce ion concentration polarization [43]. Preconcentration was further enhanced by decreasing channel depth through a two-sided wax-printing process, resulting in an 800-fold increase in the concentration of bovine serum albumin. In future efforts this method should be tested on disease-related biomarkers in blood or other sample matrices.
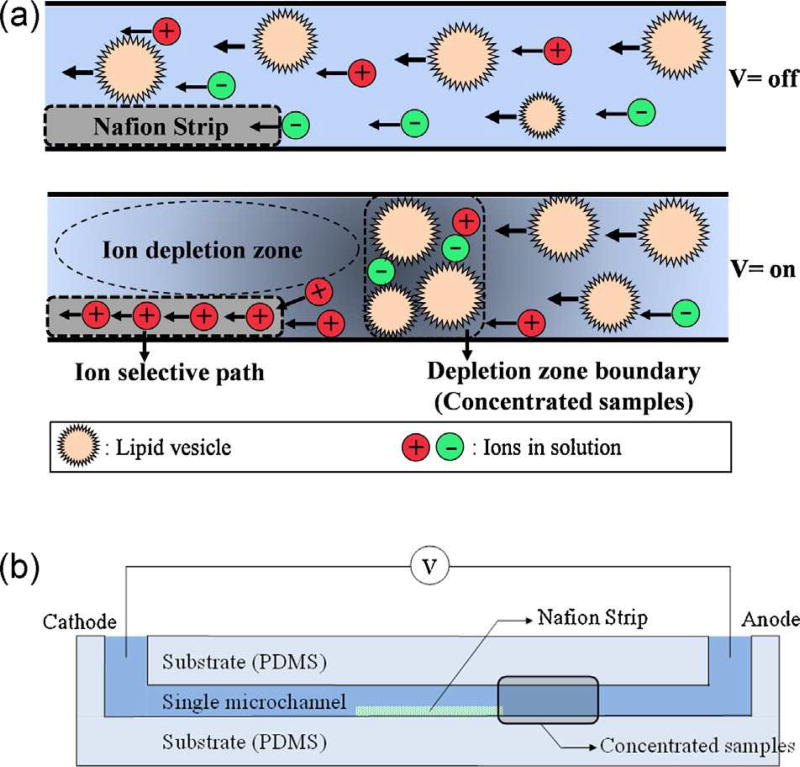
Similarly, Lee et al. [44] fabricated an ion-selective Nafion membrane inside PDMS microfluidic channels to electrokinetically concentrate lipid vesicles, as illustrated in Fig. 5. Under an applied voltage ion concentration polarization occurred at the interface of the microchannel with the Nafion, causing a 160-fold lipid vesicle enrichment. Further development is needed to achieve greater preconcentration as well as for application to specific biomarkers.
Figure 5.
Nafion-based enrichment of lipid vesicles in a microfluidic device. (a) Working mechanism for the lipid vesicle preconcentrators. (b) Side schematic view of the microfluidic device. Adapted with permission from Lee et al. Sens. Actuators, B, 229 (2016) 276–280.
To provide analyte selectivity a PDMS microfluidic device was developed with integrated polycarbonate track etched membranes having different sized nanopores [45]. This double-membrane microfluidic device processed a urine sample, with the 100 nm pore membrane excluding particles and cells in human urine, but passing proteins, small molecules and ions. A second 10 nm pore membrane passed small molecules and ions but excluded albumin, which was then analyzed by µCE. This approach took 2 min to process a urine sample, and yielded a 6–100 µg/mL linear range and 1.5 µg/mL limit of detection for albumin. This work shows promise in analyzing proteins in a real sample matrix; future efforts to detect additional biomarkers in more complex matrices would be desirable.
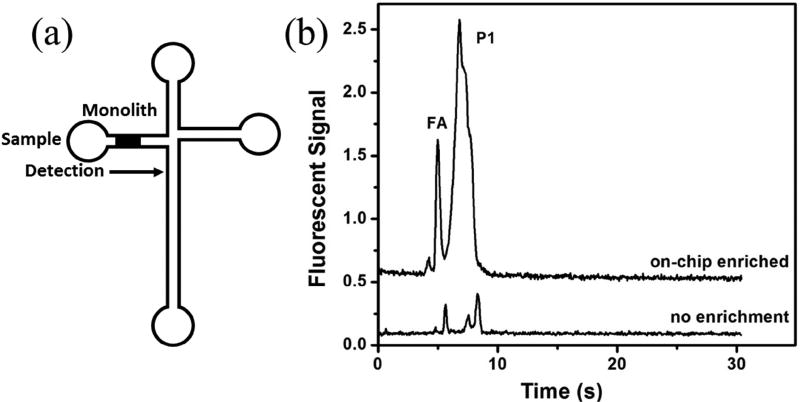
On-chip pneumatic pumps and valves offer advantages such as reproducibility and precise control of fluids. Our group developed a microfluidic device integrating solid phase extraction (SPE) and µCE to analyze for a preterm birth biomarker [46]. In this device, SPE was performed in a reversed-phase porous polymer monolith made inside cyclic olefin copolymer (COC) material, and the hydrodynamic controls were formed in PDMS. An integrated SPE-µCE analysis was performed on a preterm birth protein biomarker, ferritin, and a 100-fold enrichment factor was achieved relative to µCE without on-chip SPE, which makes this device amenable for detection of serum levels of ferritin. Although a real sample matrix was not tested, this work lays a foundation for the development of pneumatically operated integrated microfluidic systems for biomarker analysis, potentially integrating additional sample preparation steps like analyte labeling. Very recently, Sonker et al. [47] reported an integrated SPE device for electrokinetic preconcentration of preterm birth biomarkers. A reversed-phase monolith inside a COC microchannel was used to perform pH-based SPE of clinically relevant low nanomolar concentrations of preterm birth biomarkers with 50-fold sample preconcentration. Additionally, this SPE method was integrated with µCE for combined enrichment (15-fold) and separation of a peptide preterm birth biomarker, as shown in Fig. 6. This electrokinetically operated microfluidic device for biomarker analysis in diluted serum could be further enhanced with future integration of on-chip analyte labeling and immunoaffinity extraction steps.
Figure 6.
Microfluidic device for pH-mediated SPE and µCE of preterm birth biomarkers. (a) Integrated device layout. (b) Electropherogram of an on-chip preconcentrated preterm birth biomarker peptide (P1) and a model peptide (FA) compared to µCE without preconcentration on a monolith. Adapted with permission from Sonker et al. Electrophoresis, DOI: 10.1002/elps.201700054 (2017).
2.3 Sample labeling
In addition to preconcentration and purification, analyte labeling is an important sample preparation step that can be performed on a chip to save time. On-chip labeling requires loading, reacting and purifying, and typically uses a support inside the microchannels; fluorescent labeling is the most common approach being explored.
Herzog et al. [48] developed an integrated microfluidic device on a glass substrate for electrokinetic labeling and separation of peptides and proteins. The integrated device had a reactor for fluorescent labeling with Atto 425, a separation compartment for free flow isoelectric focusing, and a pH sensor layer to calibrate pI values. This device allowed the analysis of proteins and peptides in 5 min, offering process integration and speed; however, the resolution of the separated mixture still needs to be improved.
Our group used reversed-phase porous polymer monolith SPE for the preconcentration and fluorescent labeling of model proteins with fluorescein isothiocyanate (FITC) and Alexa Fluor 488 [49]. Different reversed-phase monolith recipes and eluent compositions were optimized for selective elution of fluorescent dye and labeling of bovine serum albumin and heat shock protein 90. Recently, we advanced this on-chip SPE and fluorescent labeling technique to process preterm birth biomarkers [50]. Octyl methacrylate reversed-phase monoliths were polymerized inside channels in a COC device as the sample preconcentration platform. Successful FITC labeling of three preterm birth biomarkers was accomplished using this device. Both of these studies indicate the potential of microfluidic devices to integrate preconcentration and labeling to minimize off-chip sample preparation time and effort. Future efforts should focus on lowering biomarker concentrations detected, subsequent integration with separation by µCE and on-chip processing of samples in complex matrices.
3. Molecular separation techniques
Multiple biomarkers are often analyzed simultaneously in clinical diagnostic applications; thus, separation is an integral part of biomarker panel analysis. Various separation techniques have been developed and explored in microfluidic systems for analysis of biomarkers [3, 4]. Molecular separation techniques in microfluidics discussed in this section are summarized in Table III.
Table III.
Molecular separation methods for biomarker analysis.
| Technique | Analytes | Device material |
Concentration | Reference |
|---|---|---|---|---|
| Gel electrophoresis | Bovine serum albumin, ovalbumin, trypsin inhibitor, parvalbumin | PDMS-glass | 0.05 mg/mL | 51 |
| DNA fragments | PMMA | 10–20 ng/µL | 52 | |
| Electrophoresis | mAbs | Glass | 0.5–1 mg/mL | 53 |
| D-amino acids | Glass | 0.3–0.5 mM | 54 | |
| TK1 | PMMA | 2–25 µg/mL | 55 | |
| Ferritin, preterm birth peptide biomarker | PDMS-COC | 100–500 nM | 56 | |
| Myoglobin, carbonic anhydrase, catalase | Glass | 0.1–0.3 µg/µL | 57 |
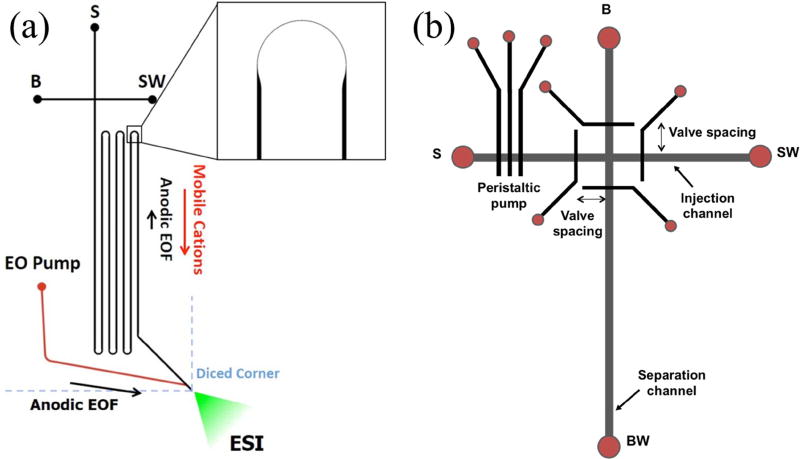
Shameli and Ren [51] developed a PDMS-glass microfluidic chip for two-dimensional separation of proteins by combining TGF with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Initially a mixture of bovine serum albumin, ovalbumin, trypsin, and parvalbumin was injected and separated by TGF in the first dimension; subsequently, the TGF peaks were analyzed by denaturing gel electrophoresis in a different channel. In an 8 min analysis time a 70% improvement in the peak resolution of a model protein sample was achieved compared to a separation with only TGF. However, further improvement in separation efficiency to achieve baseline resolution is desirable for quantitative applications. Another group reported a gel-based microchip for preconcentration, separation and extraction of DNA fragments [52]. The device was fabricated by thermally bonding 4 PMMA layers, agarose gel electrophoresis was used to separate DNA and a cellulose ester membrane was used for extraction of DNA fragments. Sample was preconcentrated prior to isotachophoresis, and two parallel channels were used for separation with one channel containing a reference DNA ladder. PCR products were analyzed and an extraction efficiency of 50% was reported. With further work in biological matrices, this device could be used for separation of nucleic acid biomarkers in the future. Redman et al. [53] reported an integrated µCE-electrospray ionization device for separation of monoclonal antibodies (mAbs). A T-shaped device had a separation channel with tapered turns and which extended to the edge of the device to produce an electrospray, as shown in Fig. 7a. This microchip was further combined with mass spectrometry for identification of separated mAbs, showing potential for rapid mass spectrometric analysis of biomarkers using microfluidics, provided more complex sample matrices can be analyzed.
Figure 7.
Innovations in µCE device designs. (a) A µCE-electrospray ionization device. Adapted with permission from Redman et al. Anal. Chem. 87 (2015) 2264–2272. (b) A pressure-actuated device for µCE of preterm birth biomarkers. Adapted with permission from Sahore et al. Anal. Bioanal. Chem., 408 (2016) 599–607.
In a different study a T-shaped electrophoresis device for separation of D-amino acids (biomarkers of Vibrio cholerae infection) followed by electrochemical detection was reported [54]. A graphene electrode at the end of the separation channel allowed the amperometric detection of liberated H2O2 when D-amino acids reacted with D-amino acid oxidase; in contrast L-amino acids showed no amperometric signal. Although this work utilized a novel signal generating mechanism, further improvements in analyte concentration are needed for potential clinical applications. Pagaduan et al. [55] reported a µCE device for determination of thymidine kinase 1 (TK1), a cancer biomarker. A microchip immunoaffinity assay was reported for measuring Ab-TK1 complex after separating it from the unbound mAb. Although this study reported separation of purified TK1 in buffer incubated with mAb off-chip, there is potential for translation to clinical application if the assay can be performed with adequate detection limits in a relevant matrix like blood.
Sahore et al. [56] reported a pressure-actuated microfluidic device for separation of biomarkers associated with preterm birth. A three-layer PDMS device (see Fig. 7b) was fabricated with integrated valves and a peristaltic pump for pressure-actuated injection for µCE of these biomarkers. Injection was optimized for valve spacing and actuation rate, eliminating bias and yielding an increase in signal, resolution and number of theoretical plates compared to electrokinetic injection. Although this device was used to separate off-chip-labeled biomarkers in buffer, further integration with on-chip sample preparation should enable analysis in more complex matrices. Another study reported a two-dimensional electrophoresis microdevice for separation of proteins using pH gradient isoelectric focusing and zone electrophoresis [57]. The device was constructed with glass and used acidic and basic buffers driven through a voltage difference to create a multilayer pH gradient for isoelectric focusing of proteins. Using this setup, a mixture of proteins (myoglobin, carbonic anhydrase, and catalase) was separated at concentrations ranging from 0.1 to 0.3 µg/µL. Although not shown in this work, native proteins could be utilized for separation and studying protein interactions in the future.
4. Conclusions and future trends
Important innovations and developments continue to be made in sample preparation and separation of biomarkers in microfluidics. Significant advances have been achieved in affinity capture, preconcentration and sample labeling processes on-chip, as well as integrating these together. Antibody- or aptamer-conjugated channels, monoliths and particles have been employed for extraction of biomarkers. Additionally, analyte preconcentration and labeling have been achieved on-chip using various solid support and stacking approaches. Some progress has been seen in combining multiple sample preparation steps within the same platform to provide integrated analysis. Clearly, further research that leads to improvements in limits of detection, sample purification, multiplexing and ability to rapidly analyze samples directly from biological matrices is still needed.
Separation is a key microfluidic technology that continues to advance; for example, through innovative strategies for µCE of biomarkers. Integration of sample preparation with separation and other analyses has moved forward, but at a much slower pace, and advances in such integration would be highly beneficial for development of sample-to-answer diagnostic devices. One area of focus would be to perform whole blood processing on chip to fractionate cells from serum [58, 59], followed by other integrated microfluidic sample preparation and analysis steps like affinity extraction, preconcentration and separation to provide automated quantitation of target biomarkers. An attractive characteristic of microfluidics is portability, which is amenable to point-of-care applications, and substantial ongoing efforts focus on developing portable diagnostic microfluidic devices with easily accessible detection like smartphone enabled sensing [60]. Additionally, the supporting instrumentation for microfluidics including electrical, optical and data analysis systems should be integrated within the device or miniaturized to facilitate point-of-care diagnosis [61–63].
Microfluidics has made progress on potential biomarker analysis applications in the last few years. However, microfluidic platforms still require significant improvements in analysis time, clinically relevant limits of detection, accuracy and cost. Further work in transitioning academic “chip in the lab” research assays toward allowing minimally instrumented microfluidic biomarker diagnostics is required to make point-of-care applications a reality. One promising future direction is the development of 3D printing to create microfluidic devices. Many papers claim to have 3D printed microfluidic devices [64], but most have fluidic features at the mm or sub-mm scale, while very few publications have achieved the ~100 µm size range of 3D printed fluidic features [65–67]. 3D printing could also increase the rate at which new microfluidic designs are tested, which should hasten innovation. However, to achieve this promising potential, diagnostic applications for biomarker detection using 3D printed microfluidics [68] will need to be expanded and improved upon significantly in the future. We believe that with these developments, microfluidics will continue to have a major impact on biomarker analysis and point-of-care diagnostics in future years.
Highlights.
We critically review recent advances in using microfluidic devices for biomarker sample preparation and separation.
We discuss improvements in on-chip sample preparation techniques, including affinity extraction, preconcentration and labeling.
We highlight developments in molecular biomarker separations.
We provide a critical evaluation of papers, detailing promising directions for additional work.
We discuss possible future trends that will help to increase the impact of microfluidics in improving human health.
Acknowledgments
We thank the National Institutes of Health (R01 EB006124) and a Roland K. Robins graduate research fellowship granted to Mukul Sonker from the Department of Chemistry and Biochemistry at Brigham Young University for financial support of this work. We are also grateful to Yao-Kuang Lee and Lauren Stolworthy for assistance in preparing the graphical abstract.
Biographies

Mukul Sonker is currently a Biochemistry PhD candidate in the Department of Chemistry and Biochemistry at Brigham Young University. As a part of Professor Adam Woolley’s research group, Mukul’s research work focuses on the development of an integrated microfluidic sample-to-answer platform for diagnosis of preterm births. Mukul was born in New Delhi, India and also received a B.S. degree in Pharmacy from University of Delhi, India.

Adam T. Woolley is University Professor in the Department of Chemistry and Biochemistry at Brigham Young University in Provo, Utah, USA. He is recipient of a Presidential Early Career Award for Scientists and Engineers (2006) and the ACS Division of Analytical Chemistry Award for Young Investigators in Separation Science (2007). His current research focuses on three general topics: the creation of novel and sophisticated integrated microfluidic systems for enhanced biomarker analysis, biotemplated nanofabrication, and the design of simple miniaturized biomolecular assays.

Vishal Sahore is a postdoctoral fellow in the Department of Chemistry and Biochemistry at Brigham Young University in Provo, Utah, USA. Currently, he is developing an integrated microfluidic technology for the assessment of preterm birth risk. He did his Ph.D. at the University of Arkansas, Fayetteville, developing redox-magnetohydrodynamics microfluidics technology for lab-on-a-chip applications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nge PN, Rogers CI, Woolley AT. Advances in Microfluidic Materials, Functions, Integration, and Applications. Chem. Rev. 2013;113:2550–2583. doi: 10.1021/cr300337x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 3.Tetala KKR, Vijayalakshmi MA. A review on recent developments for biomolecule separation at analytical scale using microfluidic devices. Anal. Chim. Acta. 2016;906:7–21. doi: 10.1016/j.aca.2015.11.037. [DOI] [PubMed] [Google Scholar]
- 4.Pagaduan JV, Sahore V, Woolley AT. Applications of microfluidics and microchip electrophoresis for potential clinical biomarker analysis. Anal. Bioanal. Chem. 2015;407:6911–6922. doi: 10.1007/s00216-015-8622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nahavandi S, Baratchi S, Soffe R, Tang S-Y, Nahavandi S, Mitchell A, Khoshmanesh K. Microfluidic platforms for biomarker analysis. Lab Chip. 2014;14:1496–1514. doi: 10.1039/c3lc51124c. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, He Z, Chen Q, Lin J-M. Biochemical analysis on microfluidic chips. TrAC Trends Anal. Chem. 2016;80:213–231. [Google Scholar]
- 7.Karle M, Vashist SK, Zengerle R, von Stetten F. Microfluidic solutions enabling continuous processing and monitoring of biological samples: A review. Anal. Chim. Acta. 2016;929:1–22. doi: 10.1016/j.aca.2016.04.055. [DOI] [PubMed] [Google Scholar]
- 8.Cui F, Rhee M, Singh A, Tripathi A. Microfluidic Sample Preparation for Medical Diagnostics. Annu. Rev. Biomed. Eng. 2015;17:267–286. doi: 10.1146/annurev-bioeng-071114-040538. [DOI] [PubMed] [Google Scholar]
- 9.Mairhofer J, Roppert K, Ertl P. Microfluidic Systems for Pathogen Sensing: A Review. Sensors. 2009;9:4804–4823. doi: 10.3390/s90604804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wujcik EK, Wei H, Zhang X, Guo J, Yan X, Sutrave N, Wei S, Guo Z. Antibody nanosensors: a detailed review. RSC Adv. 2014;4:43725–43745. [Google Scholar]
- 11.Makamba H, Kim JH, Lim K, Park N, Hahn JH. Surface modification of poly(dimethylsiloxane) microchannels. Electrophoresis. 2003;24:3607–3619. doi: 10.1002/elps.200305627. [DOI] [PubMed] [Google Scholar]
- 12.Bai Y, Koh CG, Boreman M, Juang Y-J, Tang I-C, Lee LJ, Yang S-T. Surface Modification for Enhancing Antibody Binding on Polymer-Based Microfluidic Device for Enzyme-Linked Immunosorbent Assay. Langmuir. 2006;22:9458–9467. doi: 10.1021/la061123l. [DOI] [PubMed] [Google Scholar]
- 13.Knob R, Sahore V, Sonker M, Woolley AT. Advances in monoliths and related porous materials for microfluidics. Biomicrofluidics. 2016;10:032901. doi: 10.1063/1.4948507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gijs MAM. Magnetic bead handling on-chip: new opportunities for analytical applications. Microfluid. Nanofluid. 2004;1:22–40. [Google Scholar]
- 15.Reverté L, Prieto-Simón B, Campàs M. New advances in electrochemical biosensors for the detection of toxins: Nanomaterials, magnetic beads and microfluidics systems. A review. Anal. Chim. Acta. 2016;908:8–21. doi: 10.1016/j.aca.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 16.Cretich M, Daaboul GG, Sola L, Ünlü MS, Chiari M. Digital detection of biomarkers assisted by nanoparticles: application to diagnostics. Trends Biotechnol. 2015;33:343–351. doi: 10.1016/j.tibtech.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Ng E, Chen K, Hang A, Syed A, Zhang JXJ. Multi-Dimensional Nanostructures for Microfluidic Screening of Biomarkers: From Molecular Separation to Cancer Cell Detection. Ann. Biomed. Eng. 2016;44:847–862. doi: 10.1007/s10439-015-1521-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-Baldo MA, Ortega FG, Pereira SV, Bertolino FA, Serrano MJ, Lorente JA, Raba J, Messina GA. Nanostructured platform integrated into a microfluidic immunosensor coupled to laser-induced fluorescence for the epithelial cancer biomarker determination. Microchem. J. 2016;128:1825. [Google Scholar]
- 19.Eletxigerra U, Martinez-Perdiguero J, Merino S. Disposable microfluidic immuno-biochip for rapid electrochemical detection of tumor necrosis factor alpha biomarker. Sens. Actuators, B. 2015;221:1406–1411. [Google Scholar]
- 20.Ali MA, Mondal K, Jiao Y, Oren S, Xu Z, Sharma A, Dong L. Microfluidic Immuno-Biochip for Detection of Breast Cancer Biomarkers Using Hierarchical Composite of Porous Graphene and Titanium Dioxide Nanofibers. ACS Appl. Mater. Interfaces. 2016;8:20570–20582. doi: 10.1021/acsami.6b05648. [DOI] [PubMed] [Google Scholar]
- 21.Martinez AW, Phillips ST, Carrilho E, Thomas SW, III, Sindi H, Whitesides GM. Simple Telemedicine for Developing Regions: Camera Phones Paper-Based Microfluidic Devices for Real-Time, Off-Site Diagnosis. Anal. Chem. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Shi Z, Fang C, Gao A, Li CM, Yu L. Versatile microfluidic complement fixation test for disease biomarker detection. Anal. Chim. Acta. 2016;916:67–76. doi: 10.1016/j.aca.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 23.Garg N, Vallejo D, Boyle D, Nanayakkara I, Teng A, Pablo J, Liang X, Camerini D, Lee AP, Felgner P. Integrated On-Chip Microfluidic Immunoassay for Rapid Biomarker Detection. Procedia Eng. 2016;159:53–57. [Google Scholar]
- 24.Yang M, Nelson R, Ros A. Toward Analysis of Proteins in Single Cells: A Quantitative Approach Employing Isobaric Tags with MALDI Mass Spectrometry Realized with a Microfluidic Platform. Anal. Chem. 2016;88:6672–6679. doi: 10.1021/acs.analchem.5b03419. [DOI] [PubMed] [Google Scholar]
- 25.Xie Y, Zhi X, Su H, Wang K, Yan Z, He N, Zhang J, Chen D, Cui D. A Novel Electrochemical Microfluidic Chip Combined with Multiple Biomarkers for Early Diagnosis of Gastric Cancer. Nanoscale Res. Lett. 2015;10:477. doi: 10.1186/s11671-015-1153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald JC, Whitesides GM. Poly(dimethylsiloxane) as a Material for Fabricating Microfluidic Devices. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Ellis AV, Voelcker NH. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis. 2010;31:2–16. doi: 10.1002/elps.200900475. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Tullier MP, Patel K, Carranza A, Pojman JA, Radadia AD. Microfluidics using a thiolacrylate resin for fluorescence-based pathogen detection assays. Lab Chip. 2015;15:4227–4231. doi: 10.1039/c5lc00971e. [DOI] [PubMed] [Google Scholar]
- 29.Gao R, Ko J, Cha K, Jeon JH, Rhie G-e, Choi J, deMello AJ, Choo J. Fast and sensitive detection of an anthrax biomarker using SERS-based solenoid microfluidic sensor. Biosens. Bioelectron. 2015;72:230–236. doi: 10.1016/j.bios.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y-H, Peng P-Y. Semiconductor sensor embedded microfluidic chip for protein biomarker detection using a bead-based immunoassay combined with deoxyribonucleic acid strand labeling. Anal. Chim. Acta. 2015;869:34–42. doi: 10.1016/j.aca.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16:489–496. doi: 10.1039/c5lc01117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohamadi RM, Svobodova Z, Bilkova Z, Otto M, Taverna M, Descroix S, Viovy J-L. An integrated microfluidic chip for immunocapture, preconcentration and separation of β-amyloid peptides. Biomicrofluidics. 2015;9:054117. doi: 10.1063/1.4931394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee W, Kwon D, Choi W, Jung GY, Au AK, Folch A, Jeon S. 3D-Printed Microfluidic Device for the Detection of Pathogenic Bacteria Using Size-based Separation in Helical Channel with Trapezoid Cross-Section. Sci. Rep. 2015;5:7717. doi: 10.1038/srep07717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox JC, Hayhurst A, Hesselberth J, Bayer TS, Georgiou G, Ellington AD. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res. 2002;30:e108. doi: 10.1093/nar/gnf107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunka DHJ, Stockley PG. Aptamers come of age – at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 36.Jolly P, Damborsky P, Madaboosi N, Soares RRG, Chu V, Conde JP, Katrlik J, Estrela P. DNA aptamer-based sandwich microfluidic assays for dual quantification and multi-glycan profiling of cancer biomarkers. Biosens. Bioelectron. 2016;79:313–319. doi: 10.1016/j.bios.2015.12.058. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Chang K-W, Wang C-H, Yang C-H, Shiesh S-C, Lee G-B. On-chip, aptamer-based sandwich assay for detection of glycated hemoglobins via magnetic beads. Biosens. Bioelectron. 2016;79:887–893. doi: 10.1016/j.bios.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Chen Q, Liu W, Zhang J, Wang S, Lin Z, Lin J-M. Oxygen-induced cell migration and on-line monitoring biomarkers modulation of cervical cancers on a microfluidic system. Sci. Rep. 2015;5:9643. doi: 10.1038/srep09643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin SR, Zhang YS, Kim D-J, Manbohi A, Avci H, Silvestri A, Aleman J, Hu N, Kilic T, Keung W, Righi M, Assawes P, Alhadrami HA, Li RA, Dokmeci MR, Khademhosseini A. Aptamer-Based Microfluidic Electrochemical Biosensor for Monitoring Cell-Secreted Trace Cardiac Biomarkers. Anal. Chem. 2016;88:10019–10027. doi: 10.1021/acs.analchem.6b02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giri B, Pandey B, Neupane B, Ligler FS. Signal amplification strategies for microfluidic immunoassays. TrAC Trends Anal. Chem. 2016;79:326–334. [Google Scholar]
- 41.Ge Z, Wang W, Yang C. Rapid concentration of deoxyribonucleic acid via Joule heating induced temperature gradient focusing in poly-dimethylsiloxane microfluidic channel. Anal. Chim. Acta. 2015;858:91–97. doi: 10.1016/j.aca.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 42.Cong Y, Katipamula S, Geng T, Prost SA, Tang K, Kelly RT. Electrokinetic sample preconcentration and hydrodynamic sample injection for microchip electrophoresis using a pneumatic microvalve. Electrophoresis. 2016;37:455–462. doi: 10.1002/elps.201500286. [DOI] [PubMed] [Google Scholar]
- 43.Yeh S-H, Chou K-H, Yang R-J. Sample pre-concentration with high enrichment factors at a fixed location in paper-based microfluidic devices. Lab Chip. 2016;16:925–931. doi: 10.1039/c5lc01365h. [DOI] [PubMed] [Google Scholar]
- 44.Lee SJ, Rhee H, Jeon T-J, Kim D. Preconcentration of lipid vesicles using concentration polarization in a microfluidic chip. Sens. Actuators, B. 2016;229:276–280. [Google Scholar]
- 45.Li F, Guijt RM, Breadmore MC. Nanoporous Membranes for Microfluidic Concentration Prior to Electrophoretic Separation of Proteins in Urine. Anal. Chem. 2016;88:8257–8263. doi: 10.1021/acs.analchem.6b02096. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S, Sahore V, Rogers CI, Woolley AT. Development of an integrated microfluidic solid-phase extraction and electrophoresis device. Analyst. 2016;141:1660–1668. doi: 10.1039/c5an02352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonker M, Knob R, Sahore V, Woolley AT. Integrated electrokinetically driven microfluidic devices with pH-mediated solid-phase extraction coupled to microchip electrophoresis for preterm birth biomarkers. Electrophoresis. 2017;38:1743–1754. doi: 10.1002/elps.201700054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herzog C, Poehler E, Peretzki AJ, Borisov SM, Aigner D, Mayr T, Nagl S. Continuous on-chip fluorescence labelling, free-flow isoelectric focusing and marker-free isoelectric point determination of proteins and peptides. Lab Chip. 2016;16:1565–1572. doi: 10.1039/c6lc00055j. [DOI] [PubMed] [Google Scholar]
- 49.Yang R, Pagaduan JV, Yu M, Woolley AT. On chip preconcentration fluorescence labeling of model proteins by use of monolithic columns: device fabrication, optimization, and automation. Anal. Bioanal. Chem. 2015;407:737–747. doi: 10.1007/s00216-014-7988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonker M, Yang R, Sahore V, Kumar S, Woolley AT. On-chip fluorescent labeling using reversed-phase monoliths and microchip electrophoretic separations of selected preterm birth biomarkers. Anal. Methods. 2016;8:7739–7746. doi: 10.1039/C6AY01803C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shameli SM, Ren CL. Microfluidic Two-Dimensional Separation of Proteins Combining Temperature Gradient Focusing and Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis. Anal. Chem. 2015;87:3593–3597. doi: 10.1021/acs.analchem.5b00380. [DOI] [PubMed] [Google Scholar]
- 52.Wu R, Seah YP, Wang Z. Microfluidic chip for stacking, separation and extraction of multiple DNA fragments. J. Chromatogr. A. 2016;1437:219–225. doi: 10.1016/j.chroma.2016.01.076. [DOI] [PubMed] [Google Scholar]
- 53.Redman EA, Batz NG, Mellors JS, Ramsey JM. Integrated Microfluidic Capillary Electrophoresis-Electrospray Ionization Devices with Online MS Detection for the Separation and Characterization of Intact Monoclonal Antibody Variants. Anal. Chem. 2015;87:2264–2272. doi: 10.1021/ac503964j. [DOI] [PubMed] [Google Scholar]
- 54.Batalla P, Martín A, López MÁ, González MC, Escarpa A. Enzyme-Based Microfluidic Chip Coupled to Graphene Electrodes for the Detection of D-Amino Acid Enantiomer-Biomarkers. Anal. Chem. 2015;87:5074–5078. doi: 10.1021/acs.analchem.5b00979. [DOI] [PubMed] [Google Scholar]
- 55.Pagaduan JV, Ramsden M, O’Neill K, Woolley AT. Microchip immunoaffinity electrophoresis of antibody-thymidine kinase 1 complex. Electrophoresis. 2015;36:813–817. doi: 10.1002/elps.201400436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahore V, Kumar S, Rogers CI, Jensen JK, Sonker M, Woolley AT. Pressure-actuated microfluidic devices for electrophoretic separation of pre-term birth biomarkers. Anal. Bioanal. Chem. 2016;408:599–607. doi: 10.1007/s00216-015-9141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin F, Yu S, Gu L, Zhu X, Wang J, Zhu H, Lu Y, Wang Y, Deng Y, Geng L. In situ photoimmobilised pH gradient isoelectric focusing and zone electrophoresis integrated two-dimensional microfluidic chip electrophoresis for protein separation. Microchim. Acta. 2015;182:2321–2328. [Google Scholar]
- 58.Shields IV CW, Reyes CD, López GP. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip. 2015;15:1230–1249. doi: 10.1039/c4lc01246a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohammadi M, Madadi H, Casals-Terré J, Sellarès J. Hydrodynamic and direct-current insulator-based dielectrophoresis (H-DC-iDEP) microfluidic blood plasma separation. Anal. Bioanal. Chem. 2015;407:4733–4744. doi: 10.1007/s00216-015-8678-2. [DOI] [PubMed] [Google Scholar]
- 60.Yang K, Peretz-Soroka H, Liu Y, Lin F. Novel developments in mobile sensing based on the integration of microfluidic devices and smartphones. Lab Chip. 2016;16:943–958. doi: 10.1039/c5lc01524c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyd-Moss M, Baratchi S, Di Venere M, Khoshmanesh K. Self-contained microfluidic systems: a review. Lab Chip. 2016;16:3177–3192. doi: 10.1039/c6lc00712k. [DOI] [PubMed] [Google Scholar]
- 62.Samiei E, Tabrizian M, Hoorfar M. A review of digital microfluidics as portable platforms for lab-on-a-chip applications. Lab Chip. 2016;16:2376–2396. doi: 10.1039/c6lc00387g. [DOI] [PubMed] [Google Scholar]
- 63.Patino T, Mestre R, Sanchez S. Miniaturized soft bio-hybrid robotics: a step forward into healthcare applications. Lab Chip. 2016;16:3626–3630. doi: 10.1039/c6lc90088g. [DOI] [PubMed] [Google Scholar]
- 64.Bhattacharjee N, Urrios A, Kang S, Folch A. The upcoming 3D-printing revolution in microfluidics. Lab Chip. 2016;16:1720–1742. doi: 10.1039/c6lc00163g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong H, Beauchamp M, Perry S, Woolley AT, Nordin GP. Optical approach to resin formulation for 3D printed microfluidics. RSC Adv. 2015;5:106621–106632. doi: 10.1039/C5RA23855B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong H, Woolley AT, Nordin GP. High density 3D printed microfluidic valves, pumps, and multiplexers. Lab Chip. 2016;16:2450–2458. doi: 10.1039/c6lc00565a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beauchamp MJ, Nordin GP, Woolley AT. Moving from millifluidic to truly microfluidic sub-100-µm cross-section 3D printed devices. Anal. Bioanal. Chem. 2017;409:4311–4319. doi: 10.1007/s00216-017-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waheed S, Cabot JM, Macdonald NP, Lewis T, Guijt RM, Paull B, Breadmore MC. 3D printed microfluidic devices: enablers and barriers. Lab Chip. 2016;16:1993–2013. doi: 10.1039/c6lc00284f. [DOI] [PubMed] [Google Scholar]