Abstract
NOTCH1 is frequently mutated in adenoid cystic carcinoma (ACC). To test the idea that immunohistochemical staining (IHC) can identify ACCs with NOTCH1 mutations, we performed IHC for activated NOTCH1 (NICD1) in 197 cases diagnosed as ACC from 173 patients. NICD1 staining was positive in 194 cases (98%) in two major patterns: subset positivity, which correlated with tubular/cribriform histology; and diffuse positivity, which correlated with a solid histology. To determine the relationship between NICD1 staining and NOTCH1 mutational status, targeted exome sequencing data was obtained on 14 diffusely NICD1-positive ACC specimens from 11 patients and 15 subset NICD1-positive ACC specimens from 15 patients. This revealed NOTCH1 gain-of-function mutations in 11 of 14 diffusely NICD1-positive ACC specimens, whereas all subset-positive tumors had wild type NOTCH1 alleles. Notably, tumors with diffuse NICD1 positivity were associated with significantly worse outcomes (p=0.003). To determine if NOTCH1 activation is unique among tumors included in the differential diagnosis with ACC, we performed NICD1 IHC on a cohort of diverse salivary gland and head and neck tumors. High fractions of each of these tumor types were positive for NICD1 in a subset of cells, particularly in basaloid squamous cell carcinomas; however, sequencing of basaloid squamous cell carcinomas failed to identify NOTCH1 mutations. These findings indicate that diffuse NICD1 positivity in ACC correlates with solid growth pattern, the presence of NOTCH1 gain of function mutations, and unfavorable outcome, and suggest that staining for NICD1 can be helpful in distinguishing ACC with solid growth patterns from other salivary gland and head and neck tumors.
Keywords: Notch signaling, Notch mutations, adenoid cystic carcinoma, next generation sequencing
Introduction
Adenoid cystic carcinoma (ACC) is an uncommon cancer that usually arises in the salivary glands or other secretory glands of the head and neck, upper airways, and breast (for recent review, see (1)). Though most cases are slow growing, ACC has a propensity for local spread due to perineural invasion and often ultimately metastasizes, most commonly to the lung, bone, and liver. Local or distal recurrence after surgical resection, with or without radiation therapy, is common, and for these patients the prognosis is poor, even with aggressive chemotherapy regimens.
ACC has several growth patterns, including tubular, cribriform, and solid. Tumors with tubular/cribriform growth patterns are composed of mixtures of p63-positive myoepithelial cells and KIT-positive epithelial cells, which form luminal or gland-like structures. By contrast, tumors with solid growth patterns are comprised mainly of KIT-positive epithelial cells and have been associated with worse outcomes (2–5). Many ACCs show a mixture of growth patterns, and some studies suggest that even a minor tumor component showing a solid growth pattern imparts a less favorable prognosis (5).
Notch receptors participate in a conserved signaling pathway that regulates many cell fate decisions and has diverse cell lineage-dependent effects on cellular behavior (for recent review, see (6)). Notch signaling depends of successive ligand-induced cleavages. The first is carried out by ADAM metalloproteases, which cleave at a site within the juxtamembrane negative regulatory region of Notch. The sets the stage for a second cleavage carried out by gamma-secretase, which allows the intracellular portion of Notch to translocate to the nucleus and turn on Notch target genes. Genes encoding Notch receptor and other components of the Notch pathway are recurrently mutated in many cancers, including ACC (7–11). We recently noted in a small series of primary ACCs and patient-derived xenograft (PDX) models that epithelial differentiation in ACC is associated with activation of NOTCH1 (12–14), as assessed by immunohistochemical staining with a rabbit monoclonal antibody specific for a neoepitope located at the N-terminus of the NOTCH1 intracellular domain (NICD1) that is created when NOTCH1 is activated by gamma-secretase cleavage (15). These findings suggested that IHC staining for NICD1 might provide a simple, rapid, and inexpensive means of identifying NOTCH1-mutated ACCs with solid growth patterns, a subtype that can be diagnostically challenging, particularly in small biopsies with limited tumor sampling.
In this study, we tested these ideas by performing NICD1 IHC on a large cohort of ACC specimens as well as a panel of epithelial salivary gland and head and neck tumors. We find that NICD1 staining is a common feature among the diverse tumors studied, but that diffuse NICD1 positivity, which correlates with the presence of NOTCH1 gain-of-function mutations, is restricted to ACCs with solid growth patterns. Notably, we also find that ACCs with diffuse NICD1 positivity in our cohort are associated with a worse outcome. These studies suggest that IHC for NICD1 is useful in identifying ACCs with pathogenic mutations in NOTCH1 and has value as a prognostic marker.
Materials and Methods
Cases
Formalin-fixed paraffin-embedded (FFPE) samples were obtained from the tissue archives of Massachusetts General Hospital and Brigham and Women’s Hospital with Institutional Review Board approval. FFPE samples were cut at 4-micron thickness, transferred to Superfrost Plus charged slides, and baked for 60 min at 60°C prior to IHC. Tumors were characterized for growth pattern as tubular/cribriform, solid, mixed tubular/cribriform and solid, or too scant to reliably assess growth pattern. Case characteristics are summarized in Table 1.
Table 1.
Characteristics of adenoid cystic carcinoma cases selected for study.
| Number of patients | Sex | Age (years) | Number of specimens | Tumor origin | Specimens Subjected to IHC staining | Growth Pattern |
|---|---|---|---|---|---|---|
| 173 | Female (111) Male (62) |
Range: 12–89 Median: 56 |
197 | Head and neck (major and minor salivary gland) (108); upper airway (60); breast (2); prostate (2); thyroid (1) | Head and neck (primary or local recurrence, 99) upper airway (primary or local recurrence, 54); thyroid (primary, 1); breast (primary, 2); metastatic lesions (41) | Tubular / cribriform (145); solid (16); mixed (16); scant, cannot assess (20) |
Immunohistochemistry
To explore the association between NOTCH1 activation and histological features in ACC, we performed immunohistochemistry on 197 FFPE cases diagnosed as ACC from 173 patients using a rabbit monoclonal antibody specific for a neoepitope in NICD1 that is created when NOTCH1 is cleaved by gamma-secretase (15). IHC was performed on a Leica Bond III automated immunostainer following Bond Epitope Retrieval 2 for 40 min. NICD1 staining was performed using rabbit monoclonal antibody clone D3B8 (Cell Signaling Technologies, Beverly, MA) at 1:50 for 60 min. Staining with diaminobenzidine (DAB) was developed using the Bond Polymer Refine Detection Kit (Leica). Slides were counterstained with hematoxylin. Scoring of IHC results (negative, subset positive, or diffusely positive) was done independently by J.C.A. and by W.F. and D.P.S. Diffuse positivity was defined as staining in >90% of cells within a high power (40x) field in an immunoreactive area of the tumor (judged by positive NICD1 staining in endothelial cells, which constitute an internal positive control (15)). Subset positivity was defined as staining in <90% of cells within a high power (40x) field within immunoreactive areas of the tumor, while negative was defined as the absence of staining in tumor cells within immunoreactive areas of the tumor.
Next Generation Targeted Exome Sequencing
Next generation sequencing was performed on all ACCs with diffuse NICD1 staining for which adequate viable FFPE samples were available (n = 15), as well as 15 ACCs with subset NICD1 staining, which were selected at random based on tissue availability. Targeted exome sequencing was performed using an NGS assay (“OncoPanel”) that detects somatic mutations, copy number variations, and structural variants in tumor DNA extracted from FFPE samples (16, 17). The assay covers the exonic sequences of 300 cancer genes and also surveys 113 introns across 35 genes for genomic rearrangements. Areas of tumor in unstained slides were manually removed with a razor blade and used for DNA isolation. DNA (200ng) was enriched for sequences of interest using a solution phase Agilent SureSelect hybrid capture kit and then used for library preparation. Libraries were sequenced on an Illumina HiSeq 2500 sequencer. Sequence reads were aligned to human reference genome GRCh37 (hg19) with the Burrows–Wheeler Alignment tool (18). Aligned data were sorted, duplicate marked, and indexed with Picard tools. Base-quality score recalibration and local realignment around insertions and deletions was achieved with the Genome Analysis Toolkit (19, 20). Single nucleotide variants were called with MuTect (21). Copy number alterations were determined by comparing normalized sample depth of coverage against a median from a panel of normal samples using an internally developed algorithm (RobustCNV). Structural variants were detected from aligned sequence data using BreaKmer (17).
Nanostring Analysis
To detect truncated NOTCH1 mRNA transcripts in ACC biopsies, a antisense probe set was designed and synthesized by Nanostring that included probes specific for individual exons or spliced exon pairs spanning exons 2–34 of NOTCH1. RNA was isolated from paraffin sections containing ACC using an RNeasy FFPE kit (Qiagen). RNA quantified on a NanoDrop spectrophotometer was hybridized to probe sets, captured on an nCounter cartridge, and quantified on a Nanostring Digital Analyzer. nSolver software was used to normalize the data, which was then exported to Excel for further analysis.
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) was performed according to established methods using a laboratory developed, break-apart probe set for detection of MYB rearrangement. Bacterial artificial chromosomes (BACs) for probe construction were selected using the University of California Santa Cruz Biotechnology Genome Browser and Database (http://genome.ucsc.edu, genome assembly hg38) and obtained from Invitrogen (Carlsbad, CA). DNA was isolated from bacterial cultures using the Plasmid Maxi Kit (QIAGEN, Valencia, CA) and fluorescently labeled via nick translation (Abbott Molecular, Des Plaines, IL). Clone specificity was verified by PCR and by FISH to metaphase chromosome spreads prepared from normal male blood specimens.
Survival Analysis
Patient information was obtained from the electronic medical record of Partners Healthcare under a protocol approved by the Institutional Review Board. Patient vitality was determined from the electronic medical record and from publicly available information (e.g., obituaries). Kaplan-Meier curves were generated with GraphPad Prism 7 software. The significance of differences in survival between groups was assessed using the Log-rank (Mantel-Cox) test.
Results
Relationship between growth pattern and NOTCH1 activation in adenoid cystic carcinoma
Virtually all tumors diagnosed as ACC, regardless of growth pattern, stained positively for NICD1 (Table 2); representative images of NICD1 staining are shown in Figure 1. All but two tumors with a tubular/cribriform growth pattern were positive for NICD1 in at least a subset of cells, often in a suprabasilar pattern (Figure 1A), consistent with prior work showing that p63-positive myoepithelial cells are NICD1-negative (14). By contrast, 13 of 16 ACCs with a solid growth pattern were diffusely positive for NICD1 (81%) (Figure 1B), while tumors with mixed tubular/cribriform and solid growth patterns showed a mixture of NICD1 staining patterns (Table 2), with diffuse NICD1 positivity typically correlating with areas with a solid growth pattern. We also noted, in the process of staining ACCs of salivary gland origin, that normal salivary gland epithelium was uniformly negative for NICD1 (Figure 1A). Three tumors diagnosed as ACC were completely negative for NICD1 staining. One was a tumor with classic tubular/cribriform ACC morphology. A second NICD1-negative tumor arose in the middle turbinate and exhibited an unusual ribbon-like growth pattern (Figure 1C). The third NICD1-negative tumor was a lung metastasis from a sinonasal mass that showed basaloid features, nuclear pleomorphism, and focal gland-like microcystic structures (Figure 1D). Notably, positive staining was observed in internal positive control cells (endothelial cells and/or adjacent benign squamous mucosa) in all three of these cases, suggesting that each of these tumors is a true negative.
Table 2.
Relationship between growth pattern and NICD1 staining pattern in ACC
| Growth Pattern | NICD1 Staining Pattern |
|---|---|
| Tubular / cribriform (145) | Subset positive (141, 97.2%) Diffusely positive (1) Subset and focally diffusely positive (1) Negative (2) |
| Solid (16) | Diffusely positive (13, 81.3%) Subset positive (2) Negative (1) |
| Tubular / cribriform and solid (16) | Subset positive (6) Subset and focally diffusely positive (4) Diffusely positive (6) |
Figure 1.
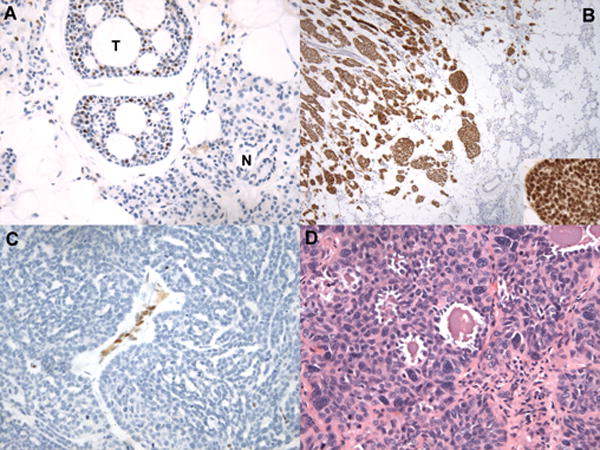
Histologic correlates of NICD1 staining in selected cases diagnosed as ACC. A) NICD1 staining in a tubular/cribriform ACC (hematoxylin counterstain). The field contains several nests of tubular/cribriform ACC showing typical subset NICD1 staining (T) adjacent to normal parotid gland (N), which is NICD1 negative. B) Diffuse NICD1 positivity in a solid ACC. Low-power view demonstrates infiltration of normal salivary gland by an ACC with a solid growth pattern and strong, diffuse NICD1 positivity; note nuclear staining in the high-power inset image. C) Absence of NICD1 staining in a tumor diagnosed as ACC involving the middle turbinate showing a ribbon-like or trabecular to solid growth pattern. Other areas in this tumor showed a tubular/cribriform growth pattern reminiscent of ACC. Note NICD1 staining in endothelial cells, which provide an internal control for immunoreactivity. D) H&E-stained section of a NICD1-negative tumor diagnosed as ACC showing squamoid differentiation and microcystic areas.
Diffuse NICD1 staining correlates with NOTCH1 mutation status in adenoid cystic carcinoma
We hypothesized that ACCs with diffuse NICD1 positivity would be enriched for cases with NOTCH1 gain-of-function mutations. To test this idea, targeted exome sequencing was performed successfully on DNA isolated from 29 FFPE specimens: 14 with diffuse NICD1 positivity and either solid (n=13) or tubular/cribriform (n=1) growth patterns; and 15 with subset NICD1 positivity and either tubular/cribriform growth patterns (n = 14) or tubular/cribriform and focally solid growth patterns (n =1) (Table 3). Diffuse NICD1 positive cases selected for study included three different biopsies from one patient and two different biopsies from a second patient. Of the 14 tumors with diffuse NICD1 positivity, we found that 11 harbored sequence variants within NOTCH1 mutational hotspots (exons 25-28, exon 34) that are predicted to produce NOTCH1 gain-of-function. The observed frequency of NOTCH1 mutations in ACCs from different patients that are diffusely positive for NICD1 (8 of 11 tumors, or 73%) is substantially higher than the frequency of NOTCH1 mutations that has been observed in NGS studies on unselected ACCs (7–11). By contrast, none of the 15 tumors with subset positivity for NICD1 had NOTCH1 mutations, a frequency significantly lower than that observed in ACCs with diffuse NICD1 positivity (p < 0.001, Fisher exact test).
Table 3.
Tumor characteristics and targeted exome sequencing results in ACC.
| Tumor Number | NICD1 Staining Pattern | Tumor Location / Type | Growth Pattern | NOTCH1 Mutations | Additional Mutations / Aberrations (Affected gene or chromosomal region) |
|---|---|---|---|---|---|
| 1 | Diffuse | Lung (metastasis) | Tubular/cribriform | None | PTEN |
| 2 | Diffuse | Thyroid (primary) | Solid | NOTCH1 c.7507C>T (p.Q2503*), exon 34 - in 57% of 38 reads | PIK3CA |
| 3 | Diffuse | Parotid gland (primary) | Solid | NOTCH1 c.7400_7400C>GTC (p.S2467fs), exon 34 - in 24% of 41 reads | PIK3CA |
| 4 | Diffuse | Trachea (primary) | Solid | NOTCH1 c.4744_4747CCGG>G (p.P1582del), exon 26 - in 36% of 131 reads | None |
| 5 | Diffuse | Trachea (primary) | Solid | NOTCH1 c.5162T>G (p.V1721G), exon 27 - in 38% of 113 reads; NOTCH1 c.7400C>A (p.S2467*), exon 34 - in 45% of 57 reads | None |
| 6 | Diffuse | Oropharynx | Solid | None | None |
| 7–9 | Diffuse (x3) | Trachea x 3 (primary + 2 recurrences) | Solid (x3) | One copy loss of NOTCH1 exons 1–27 (x3) | 6q loss except MYB exons 1-15 |
| 10 | Diffuse | Maxillary sinus (primary) | Solid | NOTCH1 c.4631A>C (p.H1544P), exon 26 - in 60% of 81 reads; NOTCH1 c7398_7399GT>T (p.2466fs), exon 34 – in 55% of 58 reads | ARID1A, CREBBP, 6q abnormality involving MYB |
| 11 | Diffuse | Liver (metastasis) | Solid | NOTCH1 c.4680C>A (p.D1560E), exon 26 - in 57% of 49 reads | ARID1A, KDM6A |
| 12 | Diffuse | Nasopharynx (primary) | Solid | None | None |
| 13-14 | Diffuse (x2) | Trachea (primary) + liver (metastasis) | Solid (x2) | NOTCH1 exon 27 duplication (primary), NOTCH1 exon 28 duplication (liver metastasis) | ARID1A (trachea); PMS2 (both biopsies) |
| 15 | Subset | Maxillary sinus (recurrence) | Tubular/cribriform | None | Low copy number gain of 9q34.3 |
| 16 | Subset (focally diffuse) | Submandibular gland (primary) | Tubular/cribriform and solid (focal) | None | Single copy number loss of 9q34.3 |
| 17 | Subset | Maxillary sinus, orbit (recurrence) | Tubular/cribriform | None | Numerous copy number changes |
| 18 | Subset | Submandibular gland (primary) | Tubular/cribriform | None | None |
| 19 | Subset | Pleura (metastasis) | Tubular/cribriform | None | None |
| 20 | Subset | Palate (primary) | Tubular/cribriform | None | NFIB-MYB fusion |
| 21 | Subset | Lung (metastasis) | Tubular/cribriform | None | Numerous copy number variants |
| 22 | Subset | Lung (metastasis) | Tubular/cribriform | None | MYB-NFIB fusion; del 22q11.21 |
| 23 | Subset | Liver (metastasis) | Tubular/cribriform | None | CREBP c.4557C>A (p.Y1519*), exon 27 - in 19% of 296 reads; low copy number gain of 9q34.3 and 6q23.3 (MYB) |
| 24 | Subset | Parotid (primary) | Tubular/cribriform | None | TP53, numerous copy number variants |
| 25 | Subset | Trachea (primary) | Tubular/cribriform | None | MUTYH |
| 26 | Subset | Lung (metastasis) | Tubular/cribriform | None | PIK3CA, RUNX1 |
| 27 | Subset | Lung (metastasis) | Tubular/cribriform | None | ARID2 |
| 28 | Subset | Lung (metastasis) | Tubular/cribriform | None | NRAS, ARID1A, KDM6A |
| 29 | Subset | Parotid (primary) | Solid | None | None |
Average read coverage, NOTCH1: exon 25, 106 (range 35–230); exon 26, 98 (range 28–216); exon 27, 146 (range 50–291); exon 28, 94 (range 27–242); exon 34, 94 (range 31–213)
Different NOTCH1 mutations (n = 11) in this ACC cohort include a range of examples of the two major types of gain-of-function mutations previously identified in human cancers: mutations in exons 25–28 that lead to ligand-independent NOTCH1 activation (22); and mutations in exon 34 that result in deletion of the C-terminal NOTCH1 PEST domain. Identified exon 25–28 mutations include two mutations in the Lin12-Notch repeats (LNRs) and several point substitutions/deletions in the core of the Notch negative regulatory region (Figure 2A), as well as two in-frame insertions stemming from small duplications (Figure 2B). In addition, three different tumor biopsies from another patient showed loss of NOTCH1 exons 1–27 in one allele. This pattern of copy number loss is consistent with the presence of a rearranged, truncated NOTCH1 allele expressing short NOTCH1 transcripts containing only exons 28-34, a type of activating mutation described previously in triple negative breast cancer (13, 23). To evaluate this possibility, we performed Nanostring analysis of tumor RNA using a probe set spanning the NOTCH1 locus (Figure 2C). This revealed excess signal for NOTCH1 exons 28–34 relative to exons 1–27 in all 3 tumor samples with a 5′ NOTCH1 copy number loss (p < 0.0001, two-sided t-test) as well as in a control triple negative breast cancer cell line, MB-157, that contains only truncated NOTCH1 alleles (13). By contrast, samples from ACCs without NOTCH1 genomic copy number variation lacked NOTCH1 RNA 5′/3′ exon imbalances.
Figure 2.
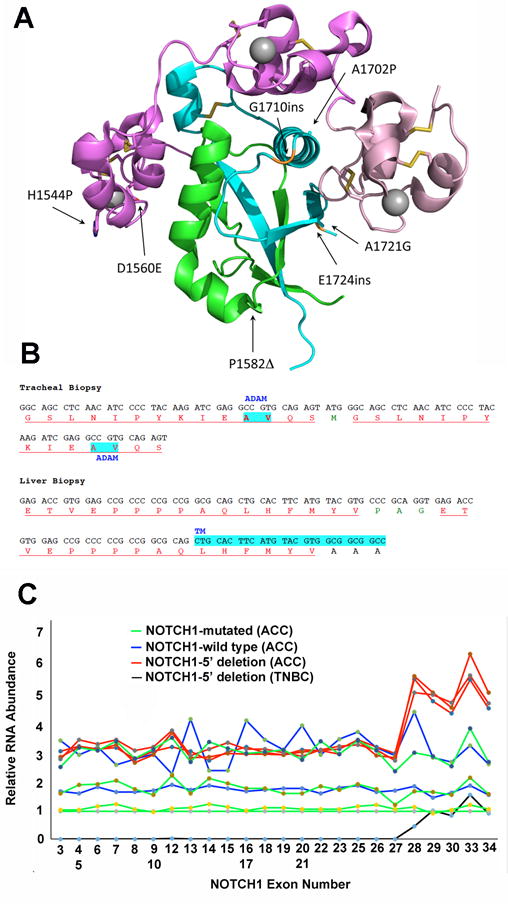
NOTCH1 mutations in ACC. A) Positions of point substitutions and in-frame indels in the NOTCH1 extracellular negative regulatory region (NRR). NOTCH1 structure is modeled based on X-ray crystallographic studies of Gordon et al. (34). B) Sequence of juxtamembrane insertional mutations caused by small NOTCH1 duplications in biopsies of ACC involving the trachea and liver in the same ACC patient. Residues in red correspond to duplicated amino acid residues; residues in green correspond to unique intervening amino acid residues; and residues in black correspond to normal, unduplicated amino acid residues. Highlighted AV residues in the sequence from the tracheal biopsy corresponds to the normal site of ADAM cleavage in NOTCH1, while highlighted residues in the sequence from the liver biopsy corresponds to the NOTCH1 transmembrane domain (TM). C) Nanostring analysis of RNA isolated from FFPE ACC tissue. RNA was isolated from two specimens containing ACCs with wild type NOTCH1 alleles; four ACC specimens with NOTCH1 alleles with point substitutions or small indels (NOTCH1 mutated); three ACC specimens with loss of copy number spanning NOTCH1 exons 1-27; and the triple negative breast cancer cell line MB-157, which has homozygous or hemizygous NOTCH1 rearrangements and lacks NOTCH1 exons 1–27 (13). RNA abundance expressed as Nanostring signal strength (Y-axis) for each exon covered by probes spanning the NOTCH1 locus (X-axis) was first normalized using average signal strength internal control housekeeping genes and then was expressed relative to signal strength for a NOTCH1-mutated ACC with no evidence of allelic imbalance, for which signal strength for each exon was arbitrarily set at 1. Note that excess signals for NOTCH1 exons 28-34, which encode the NOTCH1 transmembrane domain and intracellular domain, is seen in the control MB-157 triple negative breast cancer cell line (black) and in the 3 samples (red) prepared from specimens with copy number loss involving exons 1-27.
Diffusely NICD1 positive ACC is associated with MYB rearrangements
ACC is often associated with rearrangements involving the MYB gene on chromosome 6p that lead to MYB overexpression (7, 14). To confirm that the subset of ACCs with diffuse NICD1 positivity is associated with MYB gene rearrangements, we performed fluorescence in situ hybridization (FISH) with “split apart” probes flanking MYB on 9 diffusely NICD1 positive cases, using subset NICD1 positive ACC cases as controls (n = 6). All 9 diffusely NICD1 positive cases produced abnormal hybridization signals (Table 4). In 8 of these cases, the signals were consistent with the presence of a MYB rearrangement, while in 1 case one of two MYB signals were lost. However, NGS on this case revealed that all of MYB’s 15 exons were retained on both alleles (Table 3), indicating that this tumor also contains a MYB rearrangement rather than a MYB deletion. Thus, MYB gene rearrangements appear to be common in ACCs with diffuse NICD1 positivity.
Table 4.
Summary of MYB split-apart FISH results
| NICD1 staining pattern | MYB FISH Pattern | ||
|---|---|---|---|
| Normal | Rearranged | Copy Number Loss | |
| Diffusely positive (n = 9) | 0 | 8 | 1 |
| Subset positive (n = 6) | 3 | 3 | 0 |
Diffuse NICD1 staining correlates with outcome in adenoid cystic carcinoma
To determine if NICD1 staining pattern correlates with outcome, we obtained vitality data for all patients whose tumors were subjected to IHC and NOTCH1 mutational analysis. Kaplan-Meier analysis revealed that ACCs with diffuse NICD1 positivity were associated with significantly worse outcome than NICD1 subset positive tumors (median survival of 56.5 versus 140 months, p = 0.003) (Figure 3).
Figure 3.
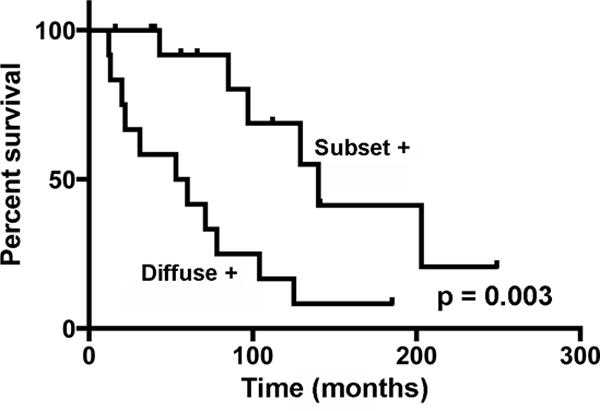
Kaplan-Meier survival curves for patients with NICD1 diffusely positive (n=12) or NICD1 subset positive (n=15) ACC.
NICD1 staining and Notch mutations in non-ACCs
The differential diagnosis of ACC includes other benign and malignant basaloid epithelial tumors of salivary gland and other regions of the head and neck. Few of these tumors have been subjected to NGS to date, and we therefore used NICD1 IHC to screen a diverse group of non-ACCs for tumors that might harbor NOTCH1 gain of function mutations. As shown in Table 5, a high fraction of non-ACCs also showed subset NICD1 positivity. Clear-cut diffuse NICD1 positivity was not observed, but we noted that basaloid squamous cell carcinoma (bSCC) often was strongly NICD1-positive in a subset of cells (Figure 4).
Table 5.
NICD1 staining in non-ACC tumors
| Diagnosis | Number of cases | Positive NICD1 staining |
|---|---|---|
| Pleomorphic adenoma | 22 | 12/22 (subset) |
| Epithelioma / myoepithelioma | 9 | 9/9 (subset) |
| Basal cell adenoma | 22 | 19/22 (subset) |
| Basal cell adenocarcinoma | 3 | 2/3 (subset) |
| Polymorphous low-grade adenocarcinoma | 17 | 7/17 (subset) |
| Basaloid squamous cell carcinoma | 13 | 13/13 (often strong staining, major subset of cells) |
Figure 4.
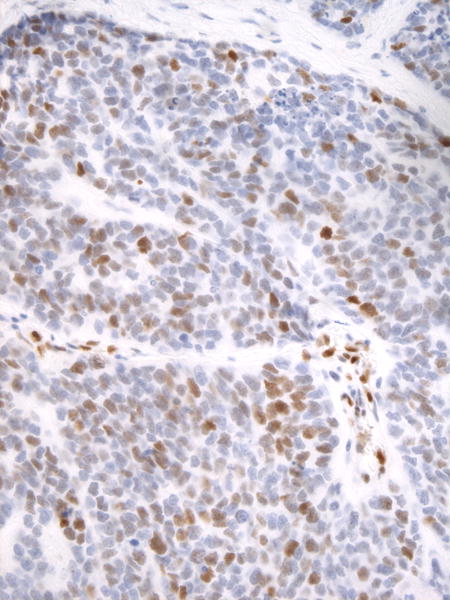
Immunohistochemical staining for NICD1 in a representative case of basaloid squamous cell carcinoma
To exclude the occurrence of NOTCH1 gain-of-function mutations in bSCC, a tumor that has not been previously characterized by genomic sequencing, NGS was performed on all 13 cases (Table 6). This revealed that bSCC is commonly associated with TP53 mutations (8 of 13 cases) and with genomic instability marked by segmental gains and losses in numerous chromosomes (all cases), features that are less frequently seen in ACC (Table 3). As a further point of comparison with ACC, FISH analysis also was performed, which identified frequent changes in copy number involving MYB but only identified one tumor with a MYB gene rearrangement (Table 3). Three mutations involving Notch genes were identified. One mutation, P2415del, is an in-frame deletion of a proline residue within an unstructured C-terminal NOTCH1 region of unknown functional significance. The other two mutations, R1608* and H2107Gfs*4, disrupt regions of NOTCH1 that are required for function and therefore correspond to loss-of-function mutations. Thus, despite frequent evidence of NOTCH1 activation in bSCC, mutational data point to selective pressure for loss rather than gain of Notch function in this tumor.
Table 6.
Basaloid squamous cell carcinoma NGS results
| Case # | Pathogenic Mutations | CNVs |
MYB FISH Results |
|---|---|---|---|
| 1 |
FBXW7 c.1513C>A (p.R505S) - in 23% of 327 reads KIT c.1540G>T (p.E514*) - in 19% of 197 reads TET2 c.4354C>T (p.R1452*) - in 22% of 283 reads |
3p11.1-3p22.1 loss 3q12.3-3q27.3 loss 11q14.2-11q23.3 loss 13q12.2-13q33.1 loss |
Normal |
| 2 |
TP53 c.568C>A (p.P190T) - in 65% of 136 reads EED c.322_323insGA (p.D109Efs*4) - in 51% of 446 reads ARID1B c.956G>T (p.G319V) - in 62% of 27 reads FGFR1 c.1615G>A (p.G539R) - in 48% of 286 reads IKZF3 c.244G>A (p.E82K) - in 51% of 346 reads KDM6B c.485C>T (p.S162F) - in 15% of 106 reads |
Complex gains and losses involving chromosomes 1, 2, 3, 4, 6, 8, 9, 10, 11, 12, 13, 14, 16, and 17 | Polysomy |
| 3 |
TP53 c.404G>A (p.C135Y) - in 91% of 275 reads GNAQ c.524C>T (p.T175M) - in 47% of 270 reads KDM5C c.1879C>T (p.R627C) - in 94% of 100 reads PIK3CA c.652G>C (p.E218Q) - in 30% of 725 reads STAG1 c.3595G>A (p.E1199K) - in 19% of 575 reads |
Complex gains and losses involving chromosomes 2, 3, 5, 7, 8, 9, 11, 13, 14, 15, 16, 17, 18, 20, and X | Polysomy |
| 4 | TP53 c.482C>A (p.A161D) - in 67% of 369 reads | Complex gains and losses involving chromosomes 2, 3, 4, 5, 7, 8, 9, 11, 12, 14, 15, 17, 19, 20, and X | Polysomy |
| 5 | None | Complex gains and losses involving chromosomes 1, 2, 3, 4, 5, 9, 10, 11, 12, 13, and 18 | Normal |
| 6 |
TP53 c.503_525delACATGACGGAGGTTGTGAGGCGC (p.H168Lfs*5) - in 22% of 413 reads CDKN2A c.172C>T (p.R58*) - in 33% of 105 reads CDKN2A c.238C>T (p.R80*) - in 25% of 185 reads SMARCA4 c.3403C>G (p.R1135G) - in 26% of 230 reads CYLD c.1609_1610delAA (p.K537Efs*5) - in 30% of 383 reads |
Complex gains and losses involving chromosomes 1, 3, 4, 5, 6, 8, 9, 11, 14, 16, 19, and 20 | Polysomy |
| 7 | None | Complex gains and losses involving chromosomes 4, 6, 9, 10, 11, 12, 14, 16, 18, and 20 | Normal |
| 8 |
TP53 c.216delC (p.V73Wfs*50) - in 64% of 139 reads NOTCH2 c.4822A>T (p.R1608*) - in 39% of 307 reads |
Complex gains and losses involving chromosomes 1, 2, 3, 4, 6, 7, 8, 9, 11, 12, 13, 14, 15, 16, 17, 18, 19, and 20 | Polysomy |
| 9 |
FBXW7 c.1394G>A (p.R465H) - in 33% of 254 reads FGFR3 c.746C>G (p.S249C) - in 34% of 246 reads KMT2D c.166C>T (p.Q56*) - in 31% of 360 reads |
3q12.3-3q27.3 gain 4q31.3 gain 6p21 loss 6q15-6q26 loss 13q13.1-33.1 loss 20q11.21-13.32 gain |
Monosomy |
| 10 |
TP53 c.783-20_795del TCTTTTCCTATCCTGAGTAGTGGTAATCTACTG - in 11% of 102 reads NOTCH1 c.7244_7246delCAC (p.P2415del) - in 6% of 131 reads PIK3CA c.1633G>A (p.E545K) - in 17% of 680 reads SMAD4 c.1333C>T (p.R445*) - in 57% of 157 reads |
Complex gains and losses involving chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 21, and X | Normal |
| 11 | NOTCH1 c.6321delC (p.H2107Qfs*4) - in 19% of 338 reads | Complex gains and losses involving chromosomes 1, 2, 3, 5, 6, 9, 12, 13, 16, 17, 19, 20, 22, and X | Polysomy |
| 12 |
TP53 c.1010G>T (p.R337L) - in 85% of 204 reads ARID1A c.465_471delCCGGAGC (p.S157Lfs*73) - in 36% of 226 reads KMT2D c.8401C>T (p.R2801*) - in 41% of 360 reads |
Complex gains and losses involving chromosomes 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17, 18, 19, 20, and X | Polysomy |
| 13 | TCF7L1 c.1071G>A (p.M357I) - in 25% of 329 reads | Low copy number gains and losses involving chromosomes 1, 3, 4, 16, 20, 22, and X | Rearranged (66% of nuclei) |
Average read coverage, NOTCH1: exon 25, 230 (range 133–422); exon 26, 196 (range 123–361); exon 27, 282 (range 181–505); exon 28, 222 (range 120–376); exon 34, 182 (range 113–311)
Discussion
This study shows that that diffuse NICD1 staining is common in ACCs with solid growth patterns, is highly associated with NOTCH1 gain-of-function mutations, and is associated with worse outcomes. Among tumors of salivary gland and head and neck origin, activation of NOTCH1 is a common feature, but only ACCs show diffuse NICD1 positivity and NOTCH1 mutations in a subset of cases, suggesting that the selective pressure for Notch gain-of-function mutations is specific for ACC among this group of tumors. Prior NGS studies of ACC have identified NOTCH1 gain-of-function mutations in a subset of cases, varying from 0% to 19% (overall 12/172 cases, or 7%)(7–10). By contrast, we detected NOTCH1 mutations in 8 of 11 ACCs (73%) with diffuse NICD1 positivity, suggesting that NICD1 staining is effective at identifying ACCs with dysregulated NOTCH1 signaling.
Our findings add to the weight of evidence implicating Notch signaling in the pathogenesis of ACC and have several translational implications. Among other epithelial cancers, only triple negative breast cancer has been reported to be associated with NOTCH1 gain-of-function mutations (13, 23). The association of ACCs with a solid growth pattern and diffuse NICD1 positivity suggests that NICD1 staining may be of diagnostic utility in this subset of ACCs, particularly in small biopsies, albeit with an important caveat. In reviewing our ACC cohort, we noted that NICD1 staining intensity often varied from area to area and that patchy negative areas were not uncommon. The NICD1 epitope is in an unstructured region of the intracellular domain of NOTCH1 (24), which may make it susceptible to degradation post-tumor resection. This limitation is partially mitigated by the existence of internal positive control cells, such as vascular endothelial cells, in virtually all tissue biopsies. Conversely, lack of NICD1 staining in several of our cases may stem from misdiagnosis. One NICD1-negative neoplasm arising in the middle turbinate had an unusual ribbon-like growth pattern, while a second NICD1-negative tumor, a lung metastasis from a sinonasal mass, had a morphology that was reminiscent of HPV-related carcinoma with adenoid cystic-like features (25). Further work is needed to determine the value of NICD1 staining in the differential diagnosis of epithelial neoplasms. The association of diffuse NICD1 staining and worse outcome also suggests that NICD1 staining is a useful prognostic marker for ACC, an idea that is reinforced by a recent report showing that NOTCH1 mutations are associated with worse outcome in ACC patients (26). Finally, clinical trials of several Notch pathway inhibitors are underway in patients with relapsed/refractory ACC. Prior work has shown that high levels of NICD1 in T-ALL and triple negative breast cancer cell lines correlate with sensitivity to gamma-secretase inhibitors (13, 27), and T-ALLs that have shown the greatest response to gamma-secretase inhibitors in the context of clinical trials have been NOTCH1-mutated (28, 29). Thus, it is possible that diffuse NICD1 positivity may be predictive of tumor response to Notch pathway inhibitors. We acknowledge, however, that while solid growth pattern, NOTCH1 mutation, and diffuse NICD1 staining are associated, these correlations are not uniform in all cases; thus, further work will be necessary to determine which of these markers, alone or in combination, is most informative.
Several of the NOTCH1 mutations we identified in ACC are unusual. Two in-frame insertions due to sequence duplications occurred in a primary tracheal tumor and a liver metastasis from the same patient, suggesting that the genetic background in this tumor predisposed to this rare type of NOTCH1 mutation. A pathogenic mutation also was identified in the mismatch repair gene PMS2 in both specimens, but typical indel mutations indicative of a mismatch repair defect were not seen in the NGS data sets and the basis for recurrent insertional NOTCH1 mutations in this tumor is unclear. We also noted that 3 of 4 PEST domain mutations fell in codon 2466 or 2467, a site that is mutated at low frequency in other tumors, such as T-ALL (27). Of note, of 9 NOTCH1 exon 34 mutations reported in ACC by other groups, 5 affected codon 2467 (9, 10), suggesting that this is a mutational hotspot in ACC. Enrichment for specific PEST domain mutations is not a prominent feature in T-ALL, but is characteristic of chronic lymphocytic leukemia, in which approximately 90% of mutations are a del(CT) involving codon 2514 (30, 31); the same del(CT) mutation also appears to be mutational hotspot in mantle cell lymphoma (32). Whether these tumor-specific mutational patterns reflect selective pressure for loss of different C-terminal regions of NOTCH1 in different cell contexts or are a manifestation of differing mutational mechanisms in various tumors remains to be determined.
Our mutational data also sound a note of clinical caution. Three of the identified NOTCH1 gain-of-function mutations, a deletion spanning exons 1–27 of NOTCH1 and two insertional mutations that reduplicate juxtamembrane sequences, create mutated alleles encoding receptors that are resistant to NOTCH1 inhibitory antibodies (33) that are being tested in clinical trials, including in patients with ACC (NCT02662608); thus, our findings suggest that if inhibitory antibody therapy is to be given, NOTCH1 mutational testing should be carried out to ensure that the tumor is expressing a “targetable” form of NOTCH1. By contrast, because the antibody used to stain NICD1 recognizes a neoepitope created by gamma-secretase cleavage, mutated NOTCH1 receptors in tumors with diffuse NICD1 positivity should “targetable” by gamma-secretase inhibitors, regardless of the mechanism of NOTCH1 activation.
Acknowledgments
This work was supported in part by a grant from the Adenoid Cystic Carcinoma Research Foundation. Technical work was performed with the support of the Specialized Histopathology Core Laboratory, a component of the Dana Farber/Harvard Cancer Center.
The work was supported by a grant from the Adenoid Cystic Carcinoma Research Foundation to J.C.A.
Footnotes
Disclosures/Conflict of Interest
The authors have nothing to disclose and declare no conflicts of interest.
References
- 1.Dillon PM, Chakraborty S, Moskaluk CA, Joshi PJ, Thomas CY. Adenoid cystic carcinoma: A review of recent advances, molecular targets, and clinical trials. Head Neck. 2016;38(4):620–7. doi: 10.1002/hed.23925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nascimento AG, Amaral AL, Prado LA, Kligerman J, Silveira TR. Adenoid cystic carcinoma of salivary glands. A study of 61 cases with clinicopathologic correlation. Cancer. 1986;57(2):312–9. doi: 10.1002/1097-0142(19860115)57:2<312::aid-cncr2820570220>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 3.da Cruz Perez DE, de Abreu Alves F, Nobuko Nishimoto I, de Almeida OP, Kowalski LP. Prognostic factors in head and neck adenoid cystic carcinoma. Oral Oncol. 2006;42(2):139–46. doi: 10.1016/j.oraloncology.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Bjorndal K, Krogdahl A, Therkildsen MH, Charabi B, Kristensen CA, Andersen E, et al. Salivary adenoid cystic carcinoma in Denmark 1990–2005: Outcome and independent prognostic factors including the benefit of radiotherapy. Results of the Danish Head and Neck Cancer Group (DAHANCA) Oral Oncol. 2015;51(12):1138–42. doi: 10.1016/j.oraloncology.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 5.van Weert S, van der Waal I, Witte BI, Leemans CR, Bloemena E. Histopathological grading of adenoid cystic carcinoma of the head and neck: analysis of currently used grading systems and proposal for a simplified grading scheme. Oral Oncol. 2015;51(1):71–6. doi: 10.1016/j.oraloncology.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722–35. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 7.Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45(7):791–8. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest. 2013;123(7):2965–8. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross JS, Wang K, Rand JV, Sheehan CE, Jennings TA, Al-Rohil RN, et al. Comprehensive genomic profiling of relapsed and metastatic adenoid cystic carcinomas by next-generation sequencing reveals potential new routes to targeted therapies. Am J Surg Pathol. 2014;38(2):235–8. doi: 10.1097/PAS.0000000000000102. [DOI] [PubMed] [Google Scholar]
- 10.Morris LG, Chandramohan R, West L, Zehir A, Chakravarty D, Pfister DG, et al. The Molecular Landscape of Recurrent and Metastatic Head and Neck Cancers: Insights From a Precision Oncology Sequencing Platform. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rettig EM, Talbot CC, Jr, Sausen M, Jones S, Bishop JA, Wood LD, et al. Whole-Genome Sequencing of Salivary Gland Adenoid Cystic Carcinoma. Cancer Prev Res (Phila) 2016;9(4):265–74. doi: 10.1158/1940-6207.CAPR-15-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106(44):18740–4. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoeck A, Lejnine S, Truong A, Pan L, Wang H, Zang C, et al. Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov. 2014;4(10):1154–67. doi: 10.1158/2159-8290.CD-13-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48(3):265–72. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluk MJ, Ashworth T, Wang H, Knoechel B, Mason EF, Morgan EA, et al. Gauging NOTCH1 Activation in Cancer Using Immunohistochemistry. PLoS One. 2013;8(6):e67306. doi: 10.1371/journal.pone.0067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abo RP, Ducar M, Garcia EP, Thorner AR, Rojas-Rudilla V, Lin L, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43(3):e19. doi: 10.1093/nar/gku1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26(12):4642–51. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17(12):1646–51. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson SE, Ilagan MX, Kopan R, Barrick D. Thermodynamic analysis of the CSL x Notch interaction: distribution of binding energy of the Notch RAM region to the CSL beta-trefoil domain and the mode of competition with the viral transactivator EBNA2. J Biol Chem. 2010;285(9):6681–92. doi: 10.1074/jbc.M109.019968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop JA, Ogawa T, Stelow EB, Moskaluk CA, Koch WM, Pai SI, et al. Human papillomavirus-related carcinoma with adenoid cystic-like features: a peculiar variant of head and neck cancer restricted to the sinonasal tract. Am J Surg Pathol. 2013;37(6):836–44. doi: 10.1097/PAS.0b013e31827b1cd6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrarotto R, Mitani Y, Diao L, Guijarro I, Wang J, Zweidler-McKay P, et al. Activating NOTCH1 Mutations Define a Distinct Subgroup of Patients With Adenoid Cystic Carcinoma Who Have Poor Prognosis, Propensity to Bone and Liver Metastasis, and Potential Responsiveness to Notch1 Inhibitors. J Clin Oncol. 2017;35(3):352–60. doi: 10.1200/JCO.2016.67.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 28.Papayannidis C, DeAngelo DJ, Stock W, Huang B, Shaik MN, Cesari R, et al. A Phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J. 2015;5:e350. doi: 10.1038/bcj.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoechel B, Bhatt A, Pan L, Pedamallu CS, Severson E, Gutierrez A, et al. Complete hematologic response of early T-cell progenitor acute lymphoblastic leukemia to the γ-secretase inhibitor BMS-906024: genetic and epigenetic findings in an outlier case. Cold Spring Harb Mol Case Stud. 2015;1(1):a000539. doi: 10.1101/mcs.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 31.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–71. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- 33.Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, et al. Characterization of notch1 antibodies that inhibit signaling of both normal and mutated notch1 receptors. PLoS One. 2010;5(2):e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113(18):4381–90. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]


