Abstract
Yatakemycin (YTM) is an extraordinarily toxic DNA alkylating agent with potent antimicrobial and antitumor properties and the most recent addition to the CC-1065 and duocarmycin family of natural products. While bulky DNA lesions the size of those produced by YTM are normally removed from the genome by the nucleotide excision repair (NER) pathway, YTM adducts are also a substrate for the bacterial DNA glycosylases AlkD and YtkR2, unexpectedly implicating base excision repair (BER) in their elimination. The reason for the extreme toxicity of these lesions and the molecular basis for how they are eliminated by BER have been unclear. Here, we describe the structural and biochemical properties of YTM adducts responsible for their toxicity, and define the mechanism by which they are excised by AlkD. These findings delineate an alternative strategy for repair of bulky DNA damage and establish the cellular utility of this pathway relative to that of NER.
INTRODUCTION
A large number of environmental toxins, chemotherapeutic drugs, and cellular metabolites, including those used by microbes as defense mechanisms, are alkylating agents that generate a chemically diverse spectrum of modifications on both the phosphodeoxyribose backbone and the nucleobases of DNA1. These lesions are both mutagenic and cytotoxic as a result of their ability to mistemplate or block nucleotide incorporation during transcription and replication. The majority of alkylation damage is eliminated from the genome by two excision repair pathways. Bulky and/or helix-distorting lesions are removed through the nucleotide excision repair (NER) pathway2,3. In prokaryotes, NER is initiated by the UvrABC incision complex, which isolates the damage within a 12-nucleotide segment that is subsequently replaced with undamaged DNA by helicase, polymerase, and ligase activities. Conversely, small nucleobase modifications are removed by the base excision repair (BER) pathway4,5, which is initiated by lesion-specific DNA glycosylases that hydrolyze the N-glycosidic bond linking the damaged nucleobase to the backbone. The resulting apurinic/apyrimidinic (AP) site is processed by endonuclease, exonuclease, polymerase, and ligase activities. Unlike the UvrABC complex, which acts on a broad spectrum of bulky lesions, DNA glycosylases are each specific for a limited number of small lesions6.
The size of the lesions repaired by BER is limited by the DNA glycosylases that initiate the pathway. These enzymes typically employ a base-flipping mechanism to extrude the modified nucleobase from the DNA duplex in order to capture it inside a nucleobase binding pocket on the surface of the protein6–8. This binding pocket allows for damage recognition through shape and charge complementary with the modified nucleobase, while also providing a scaffold for residues that catalyze hydrolysis of the N-glycosidic bond. Importantly, the nucleobase binding pocket also imposes a steric limit on the size of the lesions that can be excised by DNA glycosylases.
We recently discovered that the DNA glycosylase AlkD from Bacillus cereus excises cationic N3- and N7-alkylpurine lesions exclusively through contacts with the phosphodeoxyribose backbone, while leaving the modified nucleobase stacked in the duplex9. This unique mechanism eliminates the steric constraints of a nucleobase binding pocket and allows AlkD to excise bulky alkylpurine adducts that are refractory to removal by base-flipping DNA glycosylases9,10. Furthermore, we showed that this ability extends to adducts produced by yatakemycin (YTM), a 680-Da alkylating agent belonging to the spirocyclopropylcyclohexadienone family of natural products, which includes CC-1065, duocarmycin A, and duocarmycin SA11–14 (Fig. 1). This discovery suggested that excision of 3-yatakemycinyl-2′-deoxyadenosine (YTMA) lesions is a general feature of DNA glycosylases in the AlkD family, contradicting the original report that YtkR2, an ortholog of AlkD (26% identity and 48% similarity) from the YTM-producer Streptomyces sp. TP-A0356, had uniquely evolved to provide self-resistance to YTM15.
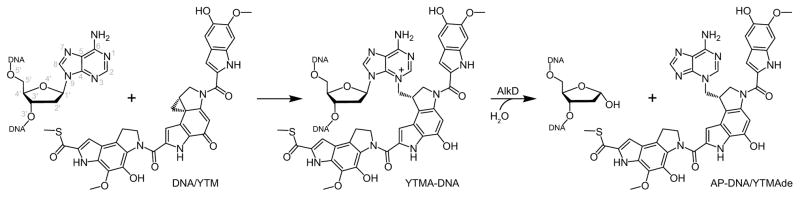
Figure 1.
DNA modification by YTM and YTMA excision by AlkD.
YTM, CC-1065, and the duocarmycins are extraordinarily potent antimicrobial and antitumor drugs12–14,16. These bulky alkylating agents preferentially bind in the deeper, narrower minor groove of AT-rich regions of DNA and undergo DNA binding-induced activation of the spirocyclopropylcyclohexadienone moiety to form covalent adducts at N3 of adenine17–21. The resultant lesions provide a striking degree of stabilization to the DNA duplex, and have been shown to severely hinder transcription and replication22. Moreover, the stability that these bulky adducts impart has been postulated to account for the ineffectiveness with which they are removed by the NER pathway22,23. However, the cause of this increased duplex stability has not been convincingly explained.
We now provide a molecular basis for the efficacy of YTM and related drugs through an examination of the YTMA lesion and its thermodynamic and structural effects on DNA, as well as its recognition and repair by AlkD. YTM adducts impart a remarkable stabilization to the DNA duplex through a network of CH/π interactions between the YTM moiety and the deoxyribose groups of the surrounding nucleotides. This stabilization is likely responsible for both the slow spontaneous depurination of YTMA lesions and the apparent difficulty with which these lesions are repaired by the NER pathway. Remodeling of YTMA-DNA by AlkD, however, disrupts the CH/π interactions involving the modified strand and provides a water nucleophile access to the N-glycosidic bond. The catalytic power of AlkD is underscored by rapid excision of YTMA lesions under single-turnover conditions and substantial retention of enzyme activity for a substrate analog that abrogates the activity of other DNA glycosylases. Surprisingly, the combined activities of AlkD-mediated BER and NER provide only weak protection against YTM toxicity in cells. We attribute this low level of resistance to the extreme stability of the YTMA-DNA substrate and a deleterious persistence of the AlkD/AP-DNA product complex. We discuss structural differences between YtkR2 and AlkD that would alleviate product inhibition by YtkR2, providing the producing strain with effective self-resistance against YTM toxicity.
RESULTS
BER and NER provide limited resistance to YTM
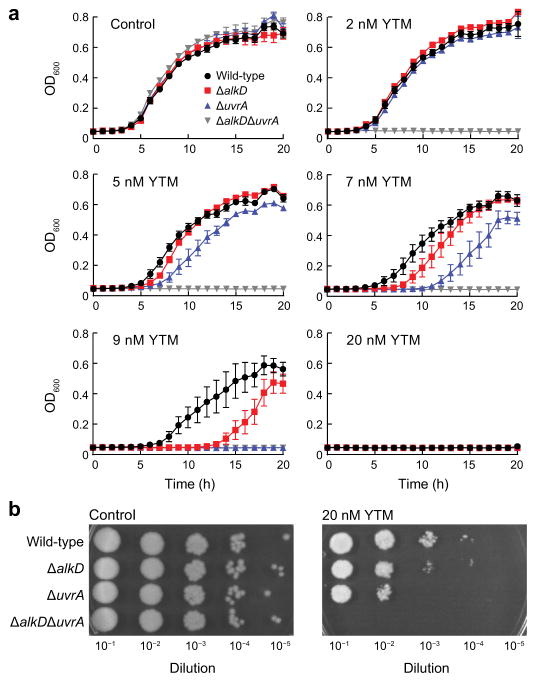
We previously showed that AlkD provides Bacillus anthracis cells with a small degree of protection from YTM toxicity9. Because NER has been shown to provide resistance to CC-1065 and analogs of the duocarmycins24–26, we wanted to compare the relative efficiencies of AlkD-mediated BER and UvrA-mediated NER in YTM resistance. Using a series of B. anthracis deletion strains that lacked either alkD, uvrA, or both, we monitored growth in the presence of increasing concentrations of YTM. In the absence of YTM, none of the deletion strains displayed a growth defect (Fig. 2a,b). However, in the presence of YTM, all deletion strains exhibited varying degrees of increased sensitivity (Fig. 2a,b). The ΔuvrAΔalkD strain was the most sensitive, and showed a pronounced growth defect at 2 nM YTM, a concentration at which the other deletion strains showed normal growth. At 5–9 nM YTM, growth of the ΔuvrA and ΔalkD strains was inhibited beyond that of the wild-type strain, with ΔuvrA cells showing greater sensitivity than ΔalkD cells. Strikingly, all strains, including wild-type B. anthracis, were markedly inhibited at 20 nM YTM. These results indicate that NER and AlkD-mediated BER provide similarly weak levels of protection against YTM toxicity.
Figure 2. Determination of YTM resistance.
(a) Growth of B. anthracis deletion strains in liquid medium. Error bars represent s.e.m. from four replicate experiments. (b) Growth of B. anthracis deletion strains on solid medium. YTM was omitted from all control experiments.
YTMA lesions dramatically stabilize duplex DNA
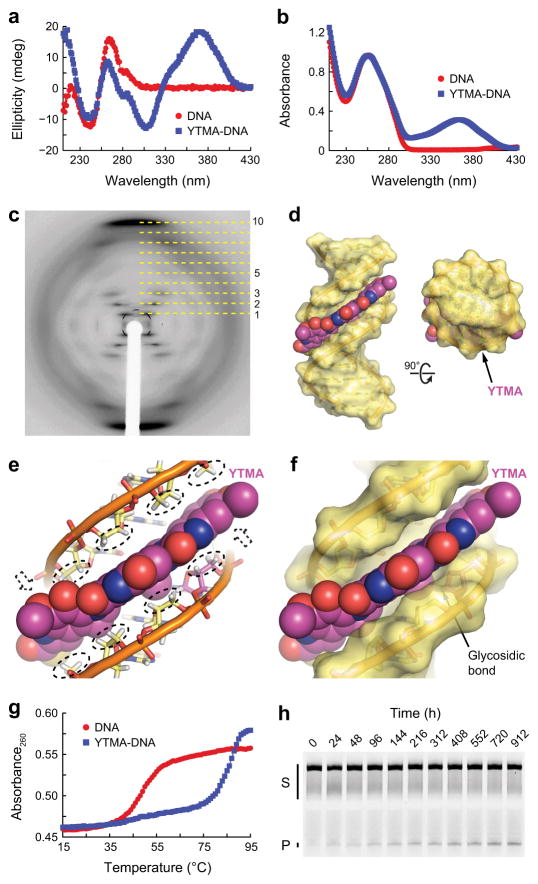
To understand the molecular basis for the extreme toxicity of YTM and the resistance of YTMA lesions to repair by both BER and NER in cells, we characterized yatakemycinylated DNA using a combination of crystallographic and biophysical methods. The circular dichroism spectrum of YTMA-DNA showed negative and positive bands at ~240 and 280 nm, respectively, characteristic of B-form DNA27 (Fig. 3a). The two additional bands present above 300 nm in the spectrum of YTMA-DNA are likely caused by dichroism of the YTM moiety itself, which produces a broad peak centered at ~360 nm in the YTMA-DNA absorbance spectrum (Fig. 3b). However, we cannot eliminate the possibility that the spectral differences between yatakemycinylated and unmodified DNA, at least in part, reflect structural changes in the DNA induced by the YTMA lesion. Efforts to determine the structure of YTMA-DNA by X-ray crystallography generated crystals that produced smeared reflections, indicative of the partial lateral and angular disorder present in fibers (Fig. 3c). The fiber diffraction pattern from YTMA-DNA is unmistakably representative of B-form DNA, which can be identified by a repeating diamond pattern and layer lines that form a distinct central cross (lines 1–3 and 5) and intense meridian arcs (line 10)28–30. Thus, the circular dichroism spectrum and the fiber diffraction pattern of YTMA-DNA are both consistent with a B-DNA structure.
Figure 3. Characterization of YTMA-DNA.
(a,b) Circular dichroism (a) and absorbance (b) spectra of unmodified and yatakemycinylated DNA collected at 15°C. (c) Fiber diffraction pattern from a paracrystalline YTMA-DNA dodecamer. Layer lines are indicated with dashed lines. (d) Hypothetical model of YTMA-DNA. (e) CH/π interactions between YTMA and DNA. Hydrogen atoms that contact the YTM moiety are indicated with dashed ovals or dashed arrows. (f) Close-up of the YTMA-DNA model. (g) Thermal melting profiles of unmodified and yatakemycinylated DNA measured at 260 nm. Melting temperatures [Tm,DNA = 49°C and Tm,YTMA-DNA = 85°C] were calculated by fitting the data to a polynomial function and determining the temperature at which the second-order derivative was equal to zero. (h) Spontaneous depurination of YTMA at 25°C. Full-length YTMA-DNA substrate (S) and cleaved AP-DNA product (P) were separated by denaturing gel electrophoresis (Supplementary Fig. 2). Complete denaturation of the YTMA-DNA duplex was prevented by the high melting temperature of the substrate. Double-stranded YTMA-DNA, single-stranded YTMA-DNA, and YTMA-DNA that denatured during electrophoresis are collectively indicated with a black bar.
Based on our crystallographic and biophysical data, we constructed a model of YTMA-DNA by manually docking the YTM moiety of yatakemycinyladenine (YTMAde) nucleobase from a crystallographic AlkD/AP-DNA product complex (described below) into ideal B-form DNA (Fig. 3d–f). Despite the lesion and the DNA being maintained as rigid bodies, the conformation of the YTM moiety seemed to be well suited to fit into the minor groove, leaving only the hydrophilic edge exposed to solvent. In this predominantly buried arrangement, the π-faces of all three YTM subunits form an extensive network of CH/π interactions with deoxyribose groups in both DNA strands (Fig. 3e). Individually, CH/π interactions are often fairly weak (~1 kcal mol−1)31. However, the combined strength of the 24 CH/π interactions present in our YTMA-DNA model would create a significant energetic barrier to base-pair opening or thermal denaturation. Consistent with this prediction, we found that a single YTMA lesion in a GC-rich dodecamer increased the melting temperature (Tm) of the duplex by 36°C (Fig. 3g). Our YTMA-DNA model also predicts that without the unlikely dissociation of YTMAde, C1′ will be sterically occluded from nucleophilic attack by a water molecule, rendering YTMA resistant to hydrolysis (Fig. 3f). Indeed, we found that at 25°C the spontaneous rate of YTMA depurination [knon = (1.8 ± 0.1) × 10−8 s−1] (Fig. 3h and Supplementary Results, Supplementary Fig. 1) is 61-fold slower than that of 3-methyl-2′-deoxyadenosine (3mA) [knon = 1.1 × 10−6 s−1] (interpolated from published rates at 22 and 39°C)32. This difference corresponds to a half-life of 1.1 weeks for 3mA versus 1.2 years for YTMA. Taken together, these data suggest that YTMA lesions substantially increase the energetic barrier to duplex denaturation and YTMA depurination by preventing separation of the DNA strands surrounding the adduct. Moreover, previous characterization of DNA modified by CC-1065 and the duocarmycins17,18,22 suggests these properties may extend to lesions produced by all members of this family of natural products.
AlkD excises YTMA lesions by prying open the minor groove
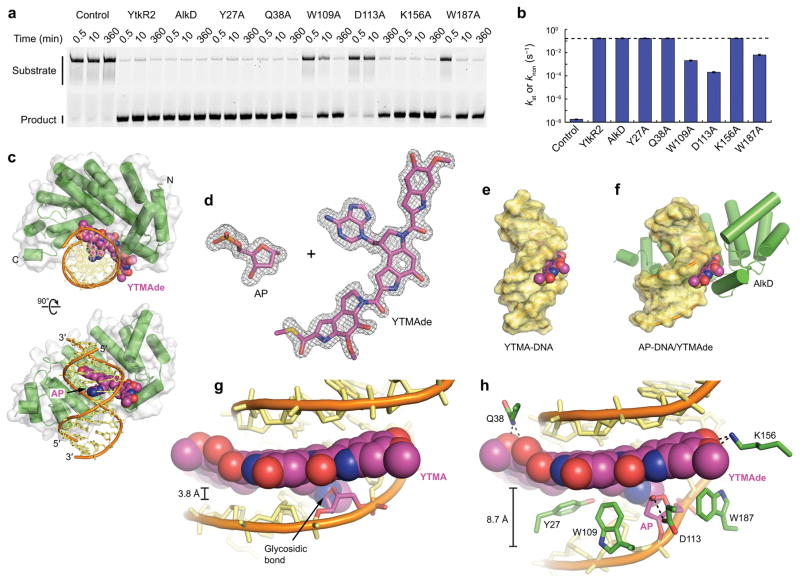
We previously showed that AlkD is capable of excising YTMA lesions in vitro9. In order to assess the enzymatic rate enhancement and the relative activity of AlkD for YTMA versus other cationic alkylpurine lesions, we measured the rate of YTMA excision under single-turnover conditions. With a 50-fold molar excess of enzyme to DNA, both AlkD and YtkR2 converted all substrate to product within 30 s (Fig. 4a and Supplementary Fig. 1), the first time point in the assay, corresponding to the maximum measureable rate of excision under these conditions. As such, the catalytic rate constant determined for excision of YTMA [kst = (1.6 ± 0.1) × 10−1 s−1] is a lower limit. Despite being underestimated, this rate equates to a 107-fold enhancement over the rate of non-enzymatic YTMA depurination under the same conditions (Fig. 4b and Supplementary Table 1). By contrast, the rate of excision of 7-methyl-2′-deoxyguanosine (7mG) [kcat = 1.3 × 10−2 s−1] by AlkD corresponds to a rate enhancement of only 104-fold33.
Figure 4. Excision of YTMA by AlkD.
(a) Separation of full-length YTMA-DNA substrate from cleaved AP-DNA product by denaturing gel electrophoresis (Supplementary Fig. 2). Reaction mixtures contained 0 (control) or 5 μM enzyme and 100 nM YTMA-DNA. (b) Rate constants for enzymatic YTMA excision and non-enzymatic YTMA depurination (control). The dashed line indicates the maximum rate that could be measured under the assay conditions. Rate constants were calculated by fitting the fraction product to a single-exponential or linear (control) model (Supplementary Fig. 1). Error bars represent s.d. from three replicate experiments. (c) Ternary product complex between AlkD, AP-DNA, and YTMAde. (d) AP site and YTMAde nucleobase extracted from the product complex. For clarity, relative positions are altered. The annealed omit mFo−DFc electron density map is contoured to 3.5σ and carved around the omitted atoms with a 2-Å radius. (e) Hypothetical model of YTMA-DNA. (f) AP-DNA/YTMAde remodeled by AlkD. (g) Blocked minor groove in the hypothetical YTMA-DNA substrate. (h) Opened minor groove in the AlkD/AP-DNA/YTMAde product complex. Hydrogen-bonding interactions are indicated with dashed lines.
To ascertain the structural basis for the preferential and apparently facile excision of YTMA lesions by AlkD, we determined a crystal structure of AlkD in the presence of YTMA-DNA. X-ray diffraction data extending to 1.6 Å revealed a ternary AlkD/AP-DNA/YTMAde product complex in which the glycosidic bond of YTMA had been cleaved (Fig. 4c,d and Supplementary Table 2). Overall, the structure is remarkably similar to those of previously determined product complexes containing much smaller 3-methyladenine (3mAde) and 3-deaza-3-methyladenine (3d3mAde) nucleobases9 (Supplementary Figs. 3 and 4). In all structures, a common set of DNA binding residues remodels the duplex, bending the helical axis by ~30° away from the protein and widening the minor groove by 4–5 Å around the lesion (Fig. 4e–h). In stark contrast to the products of base-flipping DNA glycosylases6–8, the excised nucleobases in these complexes are stacked between flanking nucleotides and paired with an opposing thymidine. In the complex containing YTMAde, the YTM moiety is bound in a cleft that follows the minor groove. Contacts between AlkD and this bulky group significantly extend the protein–ligand binding interface, providing an additional 43% of buried surface area compared to the complex containing 3d3mAde (Supplementary Fig. 3). Interestingly, the only hydrogen bonds between AlkD and YTMAde are provided by Gln38 and Lys156, which contact the four hydroxyl and methoxy substituents on the left and right subunits (Fig. 4h). All other hydrogen bonds with YTMAde are mediated by water (Supplementary Fig. 5). The most prominent binding contacts with the YTMAde nucleobase occur through CH/π interactions provided by Tyr27, Trp109, and Trp187 (Fig. 4h). Together, these residues form an aromatic shelf below the YTM group, with Tyr27 acting as a CH/π acceptor for the pyrrolidine moiety in the left subunit, and Trp109 and Trp187 serving as CH/π donors for the indole moieties in the middle and right subunits.
DNA remodeling of this kind by DNA glycosylases is generally associated with inducing strain that allows for detection of DNA damage and reducing the energetic barrier to base-pair opening, which facilitates base flipping34–36. DNA remodeling by AlkD, however, likely plays a different, or additional, role. By asymmetrically widening the minor groove around the YTMAde nucleobase, AlkD leaves intact all CH/π interactions between the YTM moiety and the unmodified strand, while prying apart the YTM moiety and the modified strand (Fig. 4g,h). This separation allows Trp109, Asp113, and Trp187 to form the same contacts with the AP site in the complex containing YTMAde as in the complex containing 3d3mAde (Supplementary Figs. 3 and 4). In both structures, Asp113 forms a hydrogen-bonding interaction with O1′, while Trp109 and Trp187 form CH/π interactions with C2′, C4′, and C5′ (Fig. 4h). For methylpurine lesions, these residues have been shown to catalyze excision by stabilizing the development of positive charge on the deoxyribose ring in the transition state9,33,37. Asp113 has also been proposed to catalyze excision by positioning and deprotonating the nucleophilic water.
We mutated the three putative catalytic residues that contact the AP site, as well as three residues that contact the YTMAde nucleobase, to assess the role that each plays in YTMA excision. As noted above, the rate of excision measured for wild-type AlkD is a lower limit. The same maximum measurable rate was observed with the AlkD-Y27A, -Q38A, and -K156A mutants, masking any possible differences above this threshold (Fig. 4a,b and Supplementary Table 1). However, the AlkD-W109A, -D113A, and -W187A mutants were substantially impaired, exhibiting at least 76-fold, 760-fold, and 25-fold reductions, respectively (Fig. 4a,b and Supplementary Table 1). These reduced rates were not significantly altered by increasing the concentration of the enzymes [kst,W109A = (2.4 ± 0.4) × 10−3 s−1, kst,D113A = (1.9 ± 0.1) × 10−4 s−1, and kst,W187A = (7.2 ± 2.0) × 10−3 s−1] (Supplementary Fig. 1), indicating the mutants were present at a saturating concentration but have a diminished ability to stabilize the cationic transition state. These rates are nonetheless at least 104-fold greater than the rate of spontaneous depurination (Fig. 4a,b and Supplementary Table 1). Such high residual activity is consistent with the redundant roles of Trp109, Asp113, and Trp187 in stabilizing the development of positive charge on the deoxyribose ring, but may also be indicative of the importance of widening the minor groove around the YTMA lesion. Even with diminished stabilization of the transition state, eliminating the steric hindrance that prevents a water nucleophile from accessing the glycosidic bond may significantly enhance the rate of hydrolysis.
Fluorinated YTMA is not refractory to excision by AlkD
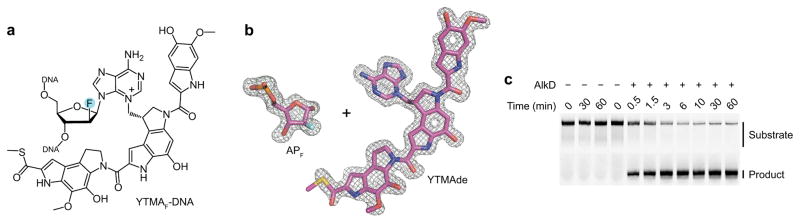
The high excision activity retained by the AlkD mutants hindered our efforts to crystallize an AlkD/YTMA-DNA substrate complex. Alternatively, we created a fluorinated YTMA (YTMAF)-DNA substrate analog by yatakemycinylating DNA containing 2′-fluoroarabinoadenosine (Fig. 5a). Addition of electron-withdrawing substituents (e.g., hydroxyl and fluoro groups) to C2′ is a common strategy employed to stabilize the glycosidic bond of deoxynucleotides. In all reported cases, 2′-fluoro groups have inhibited base excision and enabled crystallization of enzyme–substrate complexes38–40. However, our attempts to crystallize an AlkD/YTMAF-DNA complex were unsuccessful. While we were able to grow crystals that diffracted beyond 1.6 Å, the corresponding electron density maps revealed that the “stable” substrate analog had been converted to a fluorinated AP (APF) site and a YTMAde nucleobase (Fig. 5b and Supplementary Table 2). Relative to a product complex containing the same AP-DNA construct without the 2′-fluoro substituent, the overall structures are virtually identical [r.m.s.d. = 0.47 Å for all non-hydrogen atoms], including all catalytic contacts with the AP and APF sites (Supplementary Fig. 6 and Supplementary Table 2).
Figure 5. Excision of a fluorinated YTMA analog by AlkD.
(a) YTMAF. (b) APF site and YTMAde nucleobase extracted from the AlkD/APF-DNA/YTMAde product complex. For clarity, relative positions are altered. The annealed omit mFo−DFc electron density map is contoured to 3.5σ and carved around the omitted atoms with a 2-Å radius. The fluorine atom is colored cyan. (c) Separation of full-length YTMAF-DNA substrate from cleaved APF-DNA product by denaturing gel electrophoresis (Supplementary Fig. 2). Reaction mixtures contained 0 or 5 μM AlkD and 100 nM YTMAF-DNA.
Our crystallographic finding was corroborated by the observation that AlkD excises YTMAF lesions in solution. Under the single-turnover conditions used with the non-fluorinated YTMA-DNA substrate, AlkD fully removed YTMAF in less than 10 min [kst = (2.4 ± 0.1) × 10−2 s−1] (Fig. 5c). To our knowledge, only uracil DNA glycosylase has been observed to excise a fluorinated lesion 41, albeit at a rate 103-fold slower than that catalyzed by AlkD. The addition of electron-withdrawing substituents to C2′ increases the activation barrier to depurination by destabilizing the cationic transition state42. Seemingly, this heightened barrier is insurmountable for most DNA glycosylases that use a traditional base-flipping mechanism and rely upon a single catalytic residue to stabilize the development of positive charge on the deoxyribose ring. Conversely, the non-base-flipping mechanism employed by AlkD allows for charge stabilization by three residues—Trp109, Asp113, and Trp187—which seem to provide sufficient electron donation to largely offset the effects of electron withdrawal by the 2′-fluoro group.
Persistence of the product complex impedes repair
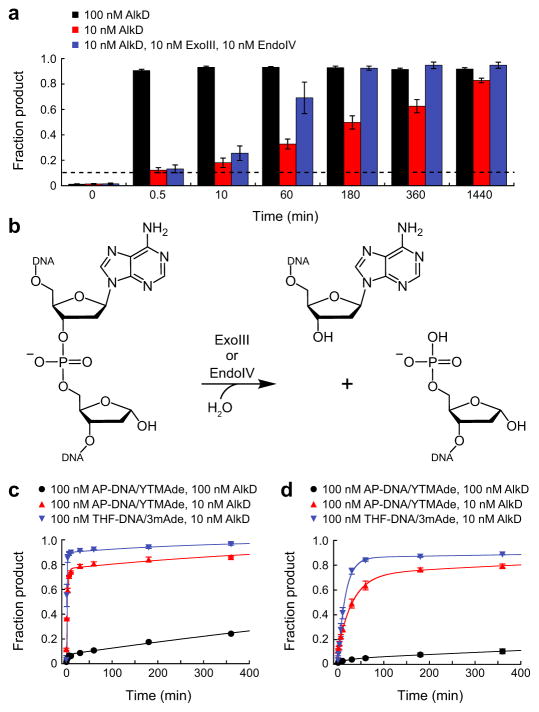
The proficiency with which AlkD excised YTMA and YTMAF lesions in vitro seemed at odds with the weak protection that AlkD provided to cells challenged with YTM. We examined two possible explanations for this paradox. First, we determined if AlkD could efficiently excise YTMA lesions under multiple-turnover conditions, which may be more relevant to repair in vivo. With 0.1 molar equivalents of AlkD, the first enzymatic turnover of YTMA-DNA was complete within 30 s (Fig. 6a and Supplementary Fig. 7). However, subsequent turnovers required far more time, and the time required for each successive turnover progressively increased as the fraction product increased. Both observations are consistent with product inhibition. Moreover, the addition of Exonuclease III (ExoIII) and Endonuclease IV (EndoIV), the two bacterial AP endonucleases that perform the next step in the BER pathway43 (Fig. 6b), only modestly increased the rate of YTMA excision (Fig. 6a and Supplementary Fig. 7). Second, we ascertained if ExoIII and EndoIV could readily incise DNA at AP sites produced from YTMA, or if AlkD and/or YTMAde nucleobase would hinder incision. Using AP-DNA and YTMAde generated with 0.1 and 1 molar equivalents of AlkD, we observed two rates of incision in each reaction (Fig. 6c,d, Supplementary Fig. 7, and Supplementary Table 3). We attributed the first, faster rate to incision of unbound DNA, and the second, much slower rate to incision of DNA bound by AlkD. For each AP endonuclease, the first rates of incision were similar at both concentrations of AlkD, as were the second rates (Supplementary Table 3). However, the fraction product formed at each rate differed substantially, presumably as a function of the proportion of unbound to bound AP-DNA (Fig. 6c,d). To determine if inhibition of AP endonuclease activity resulted from the presence of AlkD, YTMAde, or both, we repeated the incision assays with 0.1 molar equivalents of AlkD but replaced YTMAde and AP-DNA with 3mAde and tetrahydrofuran (THF)-DNA. Again, we observed two rates, both of which were comparable to the rates of incision of AP-DNA with YTMAde (Fig. 6c,d, Supplementary Fig. 7, and Supplementary Table 3). Together, these data indicate that AlkD tightly and preferentially binds abasic sites, either with or without YTMAde. This is consistent with our current and previous crystal structures of AlkD bound to abasic DNA9,10, and with our previous biochemical data showing that AlkD binds THF-DNA with 104-fold higher affinity [Kd = 1.5 nM] than unmodified DNA44. Tight, preferential binding of AP sites by AlkD would inhibit both multiple turnover of YTMA lesions and further repair by the BER pathway, explaining the weak level of protection that AlkD provides against YTM toxicity in vivo.
Figure 6. Inhibition of AP-DNA incision.
(a) Single- and multiple-turnover excision of YTMA by AlkD. All reactions contained 100 nM YTMA-DNA. The dashed line indicates the fraction product expected with 10 nM AlkD after one enzymatic turnover. (b) AP-DNA incision by ExoIII or EndoIV. (c,d) Incision of AP-DNA/YTMAde and THF-DNA/3mAde by 1 nM ExoIII (c) or EndoIV (d). Error bars in a, c, and d represent s.d. from three replicate experiments.
DISCUSSION
This work delineates the molecular basis for an alternative repair pathway for YTMA lesions. The adducts formed by YTM, CC-1065, and the duocarmycins are too large to be excised by traditional DNA glycosylases and too stabilizing to the DNA duplex to be efficiently recognized by the UvrABC complex. Consequently, these compounds have extraordinarily high levels of toxicity, not unlike those of bifunctional alkylating agents that produce DNA interstrand crosslinks (ICLs). Given the dramatic increase in duplex stability provided by YTMA and related lesions, owing to the extensive network of CH/π interactions that they form between opposing DNA strands, these lesions should arguably be regarded as non-covalent ICLs. In prokaryotes, ICLs are repaired through the combined activities of NER and either homologous recombination or translesion synthesis45. Since the adducts generated by YTM, CC-1065, and the duocarmycins neither destabilize nor significantly distort DNA17–19,22, reliance on the UvrABC incision complex will likely hinder ICL repair in the same manner that it hinders NER. As such, none of the traditional repair pathways are well suited for these bulky and highly duplex-stabilizing lesions.
YtkR2 presumably evolved to eliminate this repair deficiency in Streptomyces sp. TP-A0356 by specifically excising YTMA lesions, and thereby providing self-resistance to YTM. While orthologs of YtkR2, including AlkD, are prevalent in non-YTM producing bacteria10,46, they likely have not undergone the same selective pressure. Despite YtkR2 and AlkD sharing thirteen of sixteen DNA binding residues, they share only four of twelve YTMAde binding residues (Supplementary Fig. 8). Notably, these differences seem to create a larger binding cleft in YtkR2, with fewer contacts with YTMAde. While initially surprising, this may serve to destabilize the ternary product complex and promote dissociation of YtkR2, thereby avoiding the inhibition of AP endonuclease activity observed with AlkD. There is also the possibility that YtkR2 functions within a more specialized BER pathway. The gene cluster that encodes YtkR2 and the enzymes necessary for biosynthesis of YTM also encodes proteins with low sequence similarity to Escherichia coli EndoIV and TatD, the latter of which is a 3′→5′ exonuclease specific for single-stranded DNA15,47. Since orthologs of EndoIV and TatD with high sequence similarity to the E. coli enzymes exist elsewhere in the genome of Streptomyces sp. TP-A035648, the divergent proteins in the YTM producer may have evolved to specifically process AP sites generated from YTMA.
The ability of AlkD to excise YTMA lesions, despite its divergent binding surface, as well as 7-pyridyloxobutyl-2′-deoxyguansine and O2-pyridyloxobutyl-2′-deoxycytidine lesions10 suggests that AlkD-mediated BER of bulky lesions may be more general than previously thought. Seemingly the only strict requirement is that the substrate be positively charged, and therefore activated for excision9. There are no protein contacts in the major groove, which could possibly block repair of major groove adducts, and the contacts in the minor groove seem to be largely non-specific. Only two residues—Gln38 and Lys156—form hydrogen-bonding interactions with YTMAde (Supplementary Fig. 8), and the most prominent binding contacts, the CH/π interactions formed by Tyr27, Trp109, and Trp187, select only for aromatic groups. Furthermore, since most interactions with the hydrophilic edge of YTMAde are mediated by water (Supplementary Fig. 5), the partially solvent-filled cavity in which the YTM moiety is bound would be able to accommodate a variety of chemically and structurally diverse adducts. Taken together, these characteristics suggest that AlkD is capable of removing a broad spectrum of bulky lesions, both within and outside the spirocyclopropylcyclohexadienone family.
Extension of the BER pathway beyond repair of small modifications is not limited to orthologs of AlkD. Two unrelated DNA glycosylases capable of unhooking covalent ICLs have been identified recently in bacteria and vertebrates49,50. Similar to ytkR2, the Streptomyces sahachiroi azi36/orf1 gene provides self-resistance to the natural product azinomycin B, a potent bifunctional alkylating agent, by encoding a DNA glycosylase specific for azinomycin B-derived ICLs. While the cellular utility of BER-mediated removal of bulky and crosslinked adducts remains to be fully elucidated, these studies show that the spectrum of lesions eliminated from the genome by BER is primarily dependent on the DNA glycosylases that initiate the pathway. Moreover, the continued identification of novel DNA glycosylases with non-traditional substrates is likely to further expand the known role of BER in genomic maintenance, including providing self-resistance to genotoxic natural products.
ONLINE METHODS
Protein purification
Streptomyces sp. TP-A0356 YtkR29 and Bacillus cereus AlkD51 were purified as previously described. AlkD mutants were generated by site-directed mutagenesis and purified in the same manner as wild-type AlkD. However, cells overproducing AlkD-D113A and AlkD-K156A required induction at 16°C and lysis by gentle sonication to improve the yield of soluble protein. Escherichia coli ExoIII and EndoIV were purchased from New England Biolabs.
YTMA-DNA preparation
Oligodeoxynucleotides were purchased from Integrated DNA Technologies or ChemGenes and used without further purification. Complementary strands were annealed in annealing buffer (10 mM MES, pH 6.5, and 40 mM NaCl) by rapidly heating to 85°C and then slowly cooling to room temperature (~22°C). Duplex DNA was yatakemycinylated by incubating 10 μM DNA with 150 μM YTM in annealing buffer supplemented with 10% (v/v) DMSO at room temperature for 1–10 days. Reactions longer than one day were only necessary for DNA containing 2′-fluoroarabinoadenosine at the site of yatakemycinylation. Free YTM was then removed by passing the reaction mixture through a Sephadex G25 column equilibrated in annealing buffer. YTMA-DNA intended for crystallographic or biophysical experiments was frozen, lyophilized to dryness, and resuspended in a minimal volume of annealing buffer or melting buffer (10 mM sodium phosphate, pH 7.0, and 10 mM NaCl), respectively. Complete buffer exchange was ensured by subsequent diafiltration (3,000 NMWL). Yatakemycinylation of the desired 2′-deoxyadenosine nucleotide was verified by denaturing gel electrophoresis of the products generated by enzymatic excision of YTMA and alkaline cleavage of the resultant AP-DNA (described below). The presence of a single product band of the expected length indicated modification of only the desired nucleotide. Yatakemycinylation of a different nucleotide would create a cleaved product of a different length.
YTMA-DNA characterization
Absorbance and circular dichroism measurements were collected from yatakemycinylated and unmodified oligodeoxynucleotide duplexes [d(CCCCAXAGCCCG)/d(CGGGCTTTGGGG); X = YTMA or A] on a Jasco-810 spectropolarimeter equipped with a Peltier temperature controller. Solutions containing 5 μM duplex DNA resuspended in melting buffer were equilibrated at 15°C for 3 min and then scanned from 210 to 430 nm in 1-nm steps (1-nm bandwidth). Reported spectra represent the average values from three scans. Thermal melting profiles were collected from solutions containing 2.5 μM duplex DNA by equilibrating the samples at 15°C for 3 min and then measuring changes in absorbance at 260 nm (1-nm bandwidth) as the solutions were heated at 1°C per minute to 95°C. Melting temperatures were calculated by fitting the data to a polynomial function and determining the temperature at which the second-order derivative was equal to zero.
YTMA-DNA crystallization
Yatakemycinylated DNA [d(CGCGAXTTCGCG)/d(CGCGAATTCGCG); X = YTMA] was crystallized using the hanging-drop vapor-diffusion method. Drops were prepared from 1 μL of DNA stock [1.1 mM YTMA-DNA], 1 μL of crystallization buffer [40 mM sodium cacodylate, pH 6.0, 12 mM spermine tetrahydrochloride, 80 mM NaCl, and 10% (v/v) MPD], and 1 μL of seed solution [submicroscopic crystals of unmodified DNA], and equilibrated against 1 mL of reservoir solution [35% (v/v) MPD] at 16°C. After several weeks, yellow crystals were harvested and flash-cooled in liquid nitrogen.
AlkD/AP-DNA/YTMAde crystallization
Ternary product complexes were formed by incubating equal volumes of 0.45 mM AlkD and 0.54 mM 12-mer YTMA-DNA [d(CCCCAXAGCCCG)/d(CGGGCTTTGGGG); X = YTMA], 0.54 mM 9-mer YTMA-DNA [d(AGGCAXAGC)/d(TGCTTTGCC); X = YTMA], or 1.1 mM 9-mer YTMAF-DNA [d(AGGCAXAGC)/d(TGCTTTGCC); X = YTMAF] at 4°C for 30 min. Complexes originating from 12-mer YTMA-DNA were crystallized using the hanging-drop vapor-diffusion technique. Drops were assembled from 2 μL of protein-DNA solution [0.22 mM AlkD and 0.27 mM DNA], 2 μL of reservoir solution [19% (w/v) PEG 4,000, 42 mM sodium acetate, pH 4.6, 85 mM ammonium acetate, and 5% (v/v) glycerol], and 1 μL of seed solution [submicroscopic crystals of AlkD and DNA containing a 1mA lesion], and equilibrated against an additional 500 μL of reservoir solution at 21°C. Crystals were harvested after several days, briefly soaked in reservoir solution supplemented with 15% (v/v) glycerol, and flash-cooled in liquid nitrogen. Complexes originating from 9-mer YTMA-DNA were crystallized using the sitting-drop vapor-diffusion method. Drops were prepared from 2 μL of protein-DNA solution [0.22 mM AlkD and 0.27 mM DNA], 2 μL of reservoir solution [25% (w/v) PEG 8,000, 50 mM HEPES, pH 7.0, and 50 mM CaCl2], and 1 μL of additive solution [5% (w/v) benzamidine hydrochloride], and equilibrated against an additional 500 μL of reservoir solution at 21°C. Crystals were harvested after several days, briefly soaked in reservoir solution supplemented with 15% (v/v) glycerol, and flash-cooled in liquid nitrogen. Complexes originating from 9-mer YTMAF-DNA were also crystallized using the sitting-drop vapor-diffusion method. Drops were prepared from 2 μL of protein-DNA solution [0.22 mM AlkD and 0.54 mM DNA], 1 μL of reservoir solution [21% (w/v) PEG 8,000, 50 mM HEPES, pH 7.0, and 50 mM CaCl2], 1 μL of seed solution [submicroscopic crystals of AlkD and DNA containing an A•C mismatch], and 1 μL of additive solution [5% (w/v) benzamidine hydrochloride], and equilibrated against an additional 500 μL of reservoir solution at 21°C. Crystals were harvested after 20 h, briefly soaked in reservoir solution supplemented with 15% (v/v) glycerol, and flash-cooled in liquid nitrogen.
X-ray data collection and refinement
X-ray diffraction data were collected on beamlines 21-ID-F (λ = 0.97872 Å) and 21-ID-G (λ = 0.97857 Å) at the Advanced Photon Source. Each dataset was collected from a single cryopreserved crystal at −173°C and processed using HKL200052. Data collection statistics are provided in Supplementary Table 2. Initial phases were determined by molecular replacement from a model of AlkD (PDB accession 3BVS)51 positioned using Phaser53. DNA was then manually built in Coot54. The entireties of the 9-mer and 12-mer oligodeoxynucleotide duplexes were readily apparent in all product complexes, as was excised YTMAde nucleobase. AlkD residues 1–225 were clearly defined in both 9-mer complexes. Similarly, AlkD residues 1–226 were well-ordered in the 12-mer structure. The last 11 residues (227–237) at the C-terminus of AlkD could not be reliably modeled in any structure. Atomic coordinates, anisotropic temperature factors (TLS), and fractional occupancies were refined for all non-hydrogen atoms using PHENIX55. Hydrogen atoms were placed in riding positions and were not refined against the X-ray data. The final models were validated using MolProbity56 and contained 97.4–97.8% of residues in the favored regions of the Ramachandran plot, 2.2–2.6% of residues in the allowed regions, and no residues in the disallowed regions. Additional refinement and validation statistics are included in Supplementary Table 2. Figures were prepared in PyMOL (https://www.pymol.org). mFo−DFc omit maps were generated with PHENIX after removing the AP site and the YTMAde nucleobase and performing simulated annealing on the remaining AlkD/DNA complex.
YTMA-DNA modeling
The YTM moiety of YTMAde from the 12-mer product complex was manually placed in the minor groove of ideal B-form DNA [d(CCCCAAAGCCCG)/d(CGGGCTTTGGGG)] that was generated using Coot. Both the modification and the DNA were maintained as rigid bodies. Placement was dictated by the geometric requirements of a covalent bond between the YTM moiety and the deoxyadenosine nucleotide and the avoidance of steric clashes with the remainder of the DNA duplex.
Single-turnover YTMA excision
Base excision was monitored by alkaline cleavage of the abasic products produced from yatakemycinylated DNA [FAM-d(CGGGCGGCGGCAXAGGGCGCGGGCC)/d(GGCCCGCGCCCTTTGCCGCCGCCCG); X = YTMA or YTMAF]. Reaction mixtures containing 5 or 50 μM enzyme, 100 nM YTMA-DNA or YTMAF-DNA, 50 mM Bis-Tris propane, pH 6.5, 100 mM NaCl, 5% (v/v) glycerol, 2 mM DTT, and 0.1 mM EDTA were incubated at 25°C. Aliquots were periodically removed and quenched by incubating with 0.2 M NaOH at 70°C for 30 min. An equal volume of loading buffer [80% (w/v) formamide, 10 mM EDTA, pH 8.0, 1 mg mL−1 bromophenol blue, and 1 mg mL−1 xylene cyanol] was then added before samples were heated at 70°C for an additional 5 min. Cleaved product was then separated from full-length substrate by denaturing PAGE57, allowing the fraction product to be determined from the fluorescence intensities of the corresponding bands. Rate constants were calculated by fitting the data to a single-exponential model. No-enzyme controls were conducted in parallel, but data were fit to a linear model. Experiments were performed in triplicate.
Multiple-turnover YTMA excision
YTMA excision was monitored by alkaline cleavage of the abasic product generated from yatakemycinylated DNA [FAM-d(CGGGCGGCGGCAXAGGGCGCGGGCCTTTTT)/d(GGCCCGCGCCCTTTGCCGCCGCCCGTTTTT); X = YTMA]. Reaction mixtures containing 10 or 100 nM AlkD, 0 or 10 nM ExoIII, 0 or 10 nM EndoIV, 100 nM YTMA-DNA, 25 mM Bis-Tris propane, pH 6.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mg mL−1 BSA were incubated at 25°C. Aliquots were periodically removed and quenched by incubating with 0.2 M NaOH at 70°C for 30 min. After addition of an equal volume of loading buffer, samples were incubated at 70°C for an additional 5 min and analyzed as described above. Experiments were performed in triplicate.
AP-DNA incision
AP-DNA/YTMAde substrate [FAM-d(CGGGCGGCGGCAXAGGGCGCGGGCCTTTTT)/d(GGCCCGCGCCCTTTGCCGCCGCCCGTTTTT); X = AP] was generated by incubating 500 nM YTMA-DNA with 50 or 500 nM AlkD in 25 mM Bis-Tris propane, pH 6.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mg mL−1 BSA at 25°C for 24 h. THF-DNA/3mAde substrate [FAM-d(CGGGCGGCGGCAXAGGGCGCGGGCCTTTTT)/d(GGCCCGCGCCCTTTGCCGCCGCCCGTTTTT); X = THF] was prepared by mixing THF-DNA and 3mAde in an equal molar ratio. Reaction mixtures were prepared with 10 or 100 nM AlkD, 1 nM ExoIII or EndoIV, 100 nM AP-DNA/YTMAde or THF-DNA/3mAde, 25 mM Bis-Tris propane, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mg mL−1 BSA and incubated at 25°C. Aliquots were periodically removed and quenched by adding an equal volume of loading buffer supplemented with 0.1 M EDTA, pH 8.0, and incubating at 70°C for 5 min. Samples were analyzed as described above. Rate constants were calculated by fitting the data to a double-exponential model. Experiments were performed in triplicate.
YTM resistance
Bacillus anthracis Sterne deletion strains were generated by established methods9. Growth experiments and spot assays were performed as previously described9, except LB plates used in the spot assays were incubated at room temperature. All experiments were performed in triplicate or quadruplicate.
Data availability
Atomic coordinates and structure factors were deposited in the Protein Data Bank under accession codes 5UUF (12-mer AlkD/AP-DNA/YTMAde complex), 5UUG (9-mer AlkD/AP-DNA/YTMAde complex), and 5UUH (9-mer AlkD/APF-DNA/YTMAde complex).
Supplementary Material
Acknowledgments
We thank Yasuhiro Igarashi for providing yatakemycin. This work was funded by the National Science Foundation (MCB-1517695) and the National Institutes of Health (R01 ES019625). Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Argonne National Laboratory, was supported by the U.S. Department of Energy (DE-AC02-06CH11357). Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (085P1000817). E.A.M. was partially supported by the Vanderbilt Training Program in Environmental Toxicology (T32 ES007028).
Footnotes
Author contributions
E.A.M. and B.F.E. conceived the project; E.A.M., R.S., and B.F.E. designed experiments; E.A.M. performed biochemical, biophysical, and structural experiments; R.S. performed cellular experiments; E.A.M., R.S., and B.F.E. analyzed data; E.A.M. and B.F.E. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. ASM Press; 2006. [Google Scholar]
- 2.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: The UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 3.Nouspikel T. Nucleotide excision repair: Variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 5.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slupphaug G, et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 8.Stivers JT. Site-specific DNA damage recognition by enzyme-induced base flipping. Prog Nucleic Acid Res Mol Biol. 2004;77:37–65. doi: 10.1016/S0079-6603(04)77002-6. [DOI] [PubMed] [Google Scholar]
- 9.Mullins EA, et al. The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature. 2015;527:254–258. doi: 10.1038/nature15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanka LJ, Dietz A, Gerpheide SA, Kuentzel SL, Martin DG. CC-1065 (NSC-298223), a new antitumor antibiotic. Production, in vitro biological activity, microbiological assays and taxonomy of the producing microorganism. J Antibiot. 1978;31:1211–1217. doi: 10.7164/antibiotics.31.1211. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi I, et al. Duocarmycin A, a new antitumor antibiotic from Streptomyces. J Antibiot. 1988;41:1915–1917. doi: 10.7164/antibiotics.41.1915. [DOI] [PubMed] [Google Scholar]
- 13.Ichimura M, et al. Duocarmycin SA, a new antitumor antibiotic from Streptomyces sp. J Antibiot. 1990;43:1037–1038. doi: 10.7164/antibiotics.43.1037. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi Y, et al. Yatakemycin, a novel antifungal antibiotic produced by Streptomyces sp TP-A0356. J Antibiot. 2003;56:107–113. doi: 10.7164/antibiotics.56.107. [DOI] [PubMed] [Google Scholar]
- 15.Xu H, et al. Self-resistance to an antitumor antibiotic: A DNA glycosylase triggers the base-excision repair system in yatakemycin biosynthesis. Angew Chem. 2012;51:10532–10536. doi: 10.1002/anie.201204109. [DOI] [PubMed] [Google Scholar]
- 16.Martin DG, et al. CC-1065 (NSC 298223), a potent new antitumor agent. Improved production and isolation, characterization and antitumor activity. J Antibiot. 1981;34:1119–1125. doi: 10.7164/antibiotics.34.1119. [DOI] [PubMed] [Google Scholar]
- 17.Lin CH, Patel DJ. Solution structure of the covalent duocarmycin A-DNA duplex complex. J Mol Biol. 1995;248:162–179. doi: 10.1006/jmbi.1995.0209. [DOI] [PubMed] [Google Scholar]
- 18.Eis PS, et al. High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA. J Mol Biol. 1997;272:237–252. doi: 10.1006/jmbi.1997.1223. [DOI] [PubMed] [Google Scholar]
- 19.Schnell JR, Ketchem RR, Boger DL, Chazin WJ. Binding-induced activation of DNA alkylation by duocarmycin SA: Insights from the structure of an indole derivative-DNA adduct. J Am Chem Soc. 1999;121:5645–5652. [Google Scholar]
- 20.Parrish JP, Kastrinsky DB, Wolkenberg SE, Igarashi Y, Boger DL. DNA alkylation properties of yatakemycin. J Am Chem Soc. 2003;125:10971–10976. doi: 10.1021/ja035984h. [DOI] [PubMed] [Google Scholar]
- 21.Tichenor MS, et al. Systematic exploration of the structural features of yatakemycin impacting DNA alkylation and biological activity. J Am Chem Soc. 2007;129:10858–10869. doi: 10.1021/ja072777z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swenson DH, et al. Mechanism of interaction of CC-1065 (NSC 298223) with DNA. Cancer Res. 1982;42:2821–2828. [PubMed] [Google Scholar]
- 23.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 24.Selby CP, Sancar A. ABC excinuclease incises both 5′ and 3′ to the CC-1065-DNA adduct and its incision activity is stimulated by DNA helicase II and DNA polymerase I. Biochemistry. 1988;27:7184–7188. doi: 10.1021/bi00419a004. [DOI] [PubMed] [Google Scholar]
- 25.Jin SG, et al. Excision repair of adozelesin-N3 adenine adduct by 3-methyladenine-DNA glycosylases and UvrABC nuclease. Mol Cells. 2001;11:41–47. [PubMed] [Google Scholar]
- 26.Kiakos K, et al. DNA sequence selective adenine alkylation, mechanism of adduct repair, and in vivo antitumor activity of the novel achiral seco-amino-cyclopropylbenz[e]indolone analogue of duocarmycin AS-I-145. Mol Cancer Ther. 2007;6:2708–2718. doi: 10.1158/1535-7163.MCT-07-0294. [DOI] [PubMed] [Google Scholar]
- 27.Gray DM, Ratliff RL, Vaughan MR. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 28.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 29.Lucas AA, Lambin P, Mairesse R, Mathot M. Revealing the backbone structure of B-DNA from laser optical simulations of its X-ray diffraction diagram. J Chem Ed. 1999;76:378–383. [Google Scholar]
- 30.Lucas AA. A-DNA and B-DNA: Comparing their historical X-ray fiber diffraction images. J Chem Ed. 2008;85:737–743. [Google Scholar]
- 31.Shibasaki K, Fujii A, Mikami N, Tsuzuki S. Magnitude of the CH/π interaction in the gas phase: Experimental and theoretical determination of the accurate interaction energy in benzene-methane. J Phys Chem A. 2006;110:4397–4404. doi: 10.1021/jp0605909. [DOI] [PubMed] [Google Scholar]
- 32.Osborne MR, Phillips DH. Preparation of a methylated DNA standard, and its stability on storage. Chem Res Toxicol. 2000;13:257–261. doi: 10.1021/tx990182e. [DOI] [PubMed] [Google Scholar]
- 33.Parsons ZD, Bland JM, Mullins EA, Eichman BF. A catalytic role for C-H/π interactions in base excision repair by Bacillus cereus DNA glycosylase AlkD. J Am Chem Soc. 2016;138:11485–11488. doi: 10.1021/jacs.6b07399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramstein J, Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci USA. 1988;85:7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair. 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Gold B, Stone MP, Marky LA. Looking for Waldo: A potential thermodynamic signature to DNA damage. Acc Chem Res. 2014;47:1446–1454. doi: 10.1021/ar500061p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullins EA, Rubinson EH, Eichman BF. The substrate binding interface of alkylpurine DNA glycosylase AlkD. DNA Repair. 2014;13:50–54. doi: 10.1016/j.dnarep.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett TE, et al. Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J Am Chem Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pidugu LS, et al. Structural basis for excision of 5-formylcytosine by thymine DNA glycosylase. Biochemistry. 2016;55:6205–6208. doi: 10.1021/acs.biochem.6b00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 42.Scharer OD, Kawate T, Gallinari P, Jiricny J, Verdine GL. Investigation of the mechanisms of DNA binding of the human G/T glycosylase using designed inhibitors. Proc Natl Acad Sci USA. 1997;94:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daley JM, Zakaria C, Ramotar D. The endonuclease IV family of apurinic/apyrimidinic endonucleases. Mutat Res. 2010;705:217–227. doi: 10.1016/j.mrrev.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Mullins EA, Shi R, Kotsch LA, Eichman BF. A new family of HEAT-like repeat proteins lacking a critical substrate recognition motif present in related DNA glycosylases. PLoS One. 2015;10:e0127733. doi: 10.1371/journal.pone.0127733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alseth I, et al. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Mol Microbiol. 2006;59:1602–1609. doi: 10.1111/j.1365-2958.2006.05044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YC, Li CL, Hsiao YY, Duh Y, Yuan HS. Structure and function of TatD exonuclease in DNA repair. Nucleic Acids Res. 2014;42:10776–10785. doi: 10.1093/nar/gku732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komaki H, Ichikawa N, Hosoyama A, Fujita N, Igarashi Y. Draft genome sequence of Streptomyces sp TP-A0356, a producer of yatakemycin. Genome Announc. 2015;3:e01446–15. doi: 10.1128/genomeA.01446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, et al. Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B induced DNA damage. Nucleic Acids Res. 2016;44:187–197. doi: 10.1093/nar/gkv949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell. 2016;167:498–511. doi: 10.1016/j.cell.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubinson EH, Metz AH, O’Quin J, Eichman BF. A new protein architecture for processing alkylation damaged DNA: The crystal structure of DNA glycosylase AlkD. J Mol Biol. 2008;381:13–23. doi: 10.1016/j.jmb.2008.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullins EA, et al. An HPLC-tandem mass spectrometry method for simultaneous detection of alkylated base excision repair products. Methods. 2013;64:59–66. doi: 10.1016/j.ymeth.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors were deposited in the Protein Data Bank under accession codes 5UUF (12-mer AlkD/AP-DNA/YTMAde complex), 5UUG (9-mer AlkD/AP-DNA/YTMAde complex), and 5UUH (9-mer AlkD/APF-DNA/YTMAde complex).