Summary
Nuclear receptors recruit multiple coactivators sequentially to activate transcription. This “ordered” recruitment allows different coactivator activities to engage the nuclear receptor complex at different steps of transcription. Estrogen receptor (ER) recruits SRC-3 primary coactivator and secondary coactivators p300/CBP and CARM1. CARM1 recruitment lags behind the binding of SRC-3 and p300 to ER. Combining cryo-EM structure analysis and biochemical approaches, we demonstrate that there is a close crosstalk between early and late recruited coactivators. The sequential recruitment of CARM1 not only adds a protein arginine methyltransferase activity to the ER-coactivator complex but also alters the structural organization of the pre-existing ERE/ERα/SRC-3/p300 complex. It induces a p300 conformational change and significantly increases p300 HAT activity on histone H3K18 residues, which in turn promotes CARM1 methylation activity on H3R17 residues to enhance transcriptional activity. This study reveals a structural role for a coactivator sequential recruitment and biochemical process in ER-mediated transcription.
eTOC Blurb
Nuclear receptors sequentially recruit different coactivators to regulate transcription. Yi et al. combined cryo-EM structure analysis and biochemical approaches to demonstrate that late-recruited coactivators impact the structure and specific functions of early-recruited nuclear receptor/coactivator complexes to enhance transcription.
Introduction
Estrogen receptor (ER) belongs to a superfamily of nuclear receptors that act as ligand-dependent transcriptional factors. ER is activated upon binding of steroid estrogen hormones. Moreover, it recruits a variety of coregulators to the target gene promoter/enhancer region to regulate gene transcription. In animal cells, there are over 400 transcriptional coregulators identified to date and a number of these interact with ER (Foulds et al., 2013; Lonard et al., 2007; Lonard and O’Malley, 2012). Coregulators are not recruited to an ER responsive element (ERE) DNA at the same time but proceed through a sequential recruitment mechanism to form different ER-coactivator complexes that can play different roles in transcription activation (Burakov et al., 2002; Dilworth and Chambon, 2001; Metivier et al., 2003; Shang et al., 2000). These coactivators can either be bridging factors to bring other coactivators or transcription machinery to the ER dimer, or they can have their own enzymatic activity, such as histone acetyltransferases and methyltransferases. The latter activities promote histone modifications, chromatin remodeling, transcription initiation and elongation. Kinetic chromatin immunoprecipitation (ChIP) has been employed to study the “ordered recruitment” of coactivators (Burakov et al., 2002; Metivier et al., 2003; Shang et al., 2000; Sharma and Fondell, 2002). However, the precise structural role that this temporal recruitment process plays in regulating transcription initiation is unclear.
Steroid receptor coactivator family members (SRC-1/2/3) are primary coactivators that are recruited to ER upon estrogen stimulation. Serving as scaffolding proteins, SRCs then recruit secondary coactivators to the ER complex including p300/CBP, a histone acetyltransferase, and CARM1 (Chen et al., 1999a), a protein methyltransferase. SRCs and p300 are among the first coactivators recruited by ER (Metivier et al., 2003; Shang et al., 2000). Here we demonstrate biochemically and structurally how CARM1 can be recruited to the ER complex at a slightly later time point than SRC. CARM1 is a type I protein arginine methyltransferase (PRMT), which catalyzes an asymmetric dimethylation on histone H3 Arg-17, Arg-26 and Arg-42 (Casadio et al., 2013; Schurter et al., 2001), as well as a variety of other proteins including coactivators p300/CBP (Chevillard-Briet et al., 2002; Xu et al., 2001) and SRCs (Feng et al., 2006; Naeem et al., 2007). CARM1 was the first protein methyltransferase identified as a nuclear receptor coactivator (Chen et al., 1999a). It synergizes with p300/CBP to enhance ER target gene transcription (Chen et al., 2000; Lee et al., 2002). This function requires the methyltransferase activity of CARM1 and it is dependent on SRCs (Chen et al., 2000; Ma et al., 2001). Loss of CARM1 or an enzyme-dead CARM1 mutant knock-in in mouse reveals significantly reduced estrogen regulated gene transcription (Kim et al., 2010; Yadav et al., 2003).
Using CARM1 as an example of a later recruited coactivator, we investigated the impact of its recruitment on the structural organization and the function of the ER/SRC-3/p300 complex. In our recent study of the structure of the ERE-bound ER/SRC-3/p300 core complex (Yi et al., 2015), we found that two SRC-3 proteins independently bind to the ER dimer, and participate in recruiting a single p300 protein to the ER complex (Yi et al., 2015). Here, using a combination of structural and biochemical approaches, we demonstrate that CARM1 is not simply an add-on to a preexisting ER/SRC-3/p300 complex. It triggers significant structural and functional changes to the estrogen receptor-coactivator complex. Our study presents a mechanism for cooperative interaction between sequentially recruited coactivators in the activation of nuclear receptor-mediated transcription.
Results
Biochemical evidence for sequential recruitment of ER co-activators
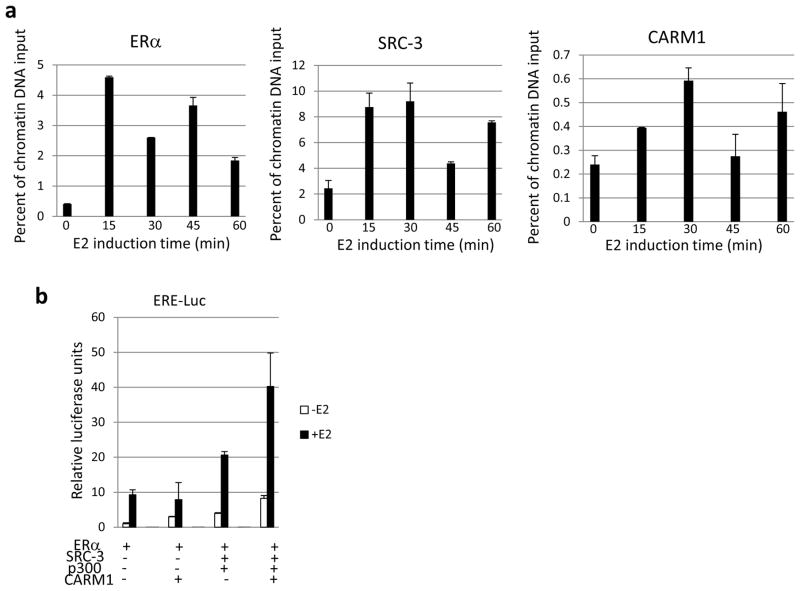
Of the three SRCs, SRC-3 has been documented as the primary recruiter for p300/CBP to ER, and it plays an essential role in ER-mediated transcription (Foulds et al., 2013; List et al., 2001; Shao et al., 2004). In addition to p300/CBP, SRC-3 also recruits other secondary coactivators, such as CARM1, through a different activation domain at its C-terminus. To determine the chronological sequence of binding of SRC-3 and CARM1 to ER, we performed a ChIP experiment in MCF-7 breast cancer cells at different time points after estrogen stimulation (0, 15, 30, 45 and 60 minutes). Shown in Figure 1a is the recruitment of ER, SRC-3 and CARM1 to an ER-targeted gene TFF1 (pS2) promoter. Although this type of assay is difficult to quantify in a precise temporal fashion, the following was noted. ER recruitment peaked ~15 min after estrogen induction. The peak of SRC-3 recruitment was also observed at ~15 min. Recruitment of CARM1, however, reached a maximal level only at 30 min. This suggested that CARM1 recruitment followed after the binding of SRC-3 to ER in cells. We also observed that the recruitment of ER, SRC-3 and CARM1 decreased after it reached a peak and then increased again. This cyclic recruitment of nuclear receptor and coactivators is consistent with previous reports (Burakov et al., 2002; Metivier et al., 2003; Shang et al., 2000). To determine whether this sequential coactivator recruitment also occurs in other ER distal enhancer and proximal promoter binding sites, we examined the binding of ER, SRC-3 and CARM1 to ERE binding sites at GREB1 distal enhancer (−35.4 kb from isoform a TSS), GREB1 proximal promoter (ERE1, −1.6 kb from isoform a TSS), and TFF1 distal enhancer (ERE3, −9.9 kb from the TSS). Similar results were observed (Figure S1a–c). Recruitment of CARM1 synergistically activated estrogen-dependent ERE-driven luciferase activity when transiently overexpressed with SRC-3 and p300 in HeLa cells (Figure 1b), similar to previous reports on SRC-2 (GRIP1) (Chen et al., 2000; Lee et al., 2002).
Figure 1.
ER sequentially recruits different coactivators. (a) CARM1 recruitment to the ER complex followed behind the recruitment of SRC-3. Shown are the ChIP results of ERα, SRC-3 and CARM1 in MCF-7 cells stimulated with 10nM E2 at different time points. (b) CARM1 synergized with SRC-3 and p300 to activate ER-mediated transcription. The ER-mediated transcription activity was measured through an ERE-driven luciferase reporter in the absence or presence of 10nM 17-β estradiol (E2). Data are represented as mean ± SEM.
CryoEM evidence of CARM1 recruitment to the ER complex
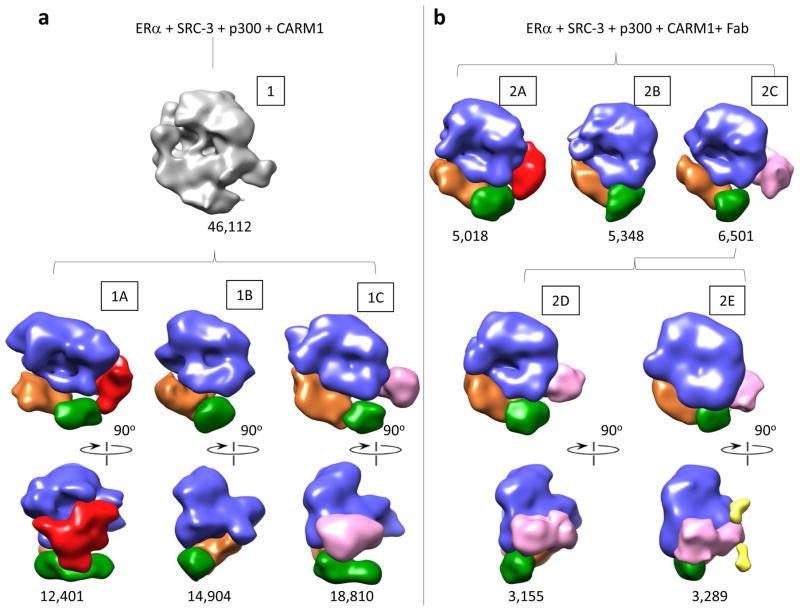
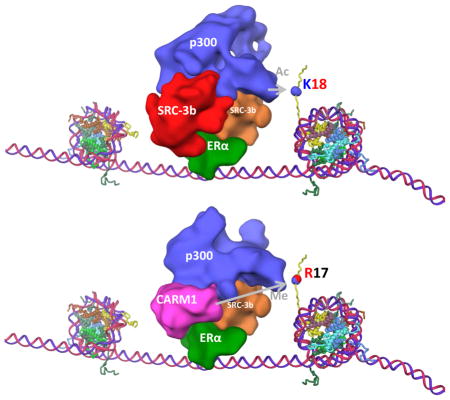
We previously reported a structure for the ERE/ERα/SRC-3a/SRC-3b/p300 complex (Yi et al., 2015). To understand the structural basis for the synergistic transcriptional activation upon binding of CARM1, we investigated whether CARM1 binding alters the structure of the DNA-bound ERα/SRC-3a/SRC-3b/p300 complex. We incubated purified CARM1 protein with ERα, SRC-3, p300 proteins and a biotinylated ERE DNA in the presence of 1μM 17β-estradiol. The assembled protein complex was purified using magnetic streptavidin beads and then prepared for cryo-EM imaging. A SDS-PAGE gel of this assembled complex followed by silver staining is shown in Figure S2a. A raw image of the complex is shown in Figure S2b. With five interacting components and the possibility of sequential recruitment of CARM1 (shown in Figure 1a) significant compositional variability was expected. Thus we classified the image dataset into three groups using the EMAN2 reconstruction software (Ludtke, 2016)(Figure 2a maps 1A,1B and 1C). Each of these maps was interpreted by segmenting the density following the same protocol as described previously for the ERE/ERα/SRC-3a/SRC-3b/p300 complex (Yi et al., 2015). Striking similarities emerged. One of the groups (Figure 2a map 1A and Figure S4a) is similar to the map of ERE/ERα/SRC-3a/SRC-3b/p300 (EMDB:EMDB-6259) (Yi et al., 2015), representing complexes that contain two SRC-3 molecules, but do not have any additional density (i.e. CARM1) bound. The most variable region between this group and the other two groups occurred at the SRC-3b binding site of the ERE/ERα/SRC-3a/SRC-3b/p300 complex (Figure 2a and Supplementary Movie 1). The SRC-3b density was missing in map 1B (Figure 2a and Figure S4b), while a significantly smaller density was detected in map 1C (Figure 2a). This variation is likely the result of CARM1 binding. To our surprise, we did not observe a map similar to map 1A, but with extra density attributable to CARM1. We thus interpret that map 1B represents a state in which SRC-3b is dissociated from the complex, and map 1C represents a complex in which CARM1 replaces SRC-3b.
Figure 2.
Workflow for stepwise 3D classification of ERα + SRC-3 + p300 + CARM1 (a) and of ERα + SRC-3 + p300 + CARM1 + Fab (b) datasets using new multirefine and e2refinemulti in EMAN2. Each subset was refined separately with specified particle number. Each map was segmented to annotate different proteins with p300 in Blue, ER in Green, SRC-3a/b in Orange and Red, respectively, CARM1 in Pink and CARM1 Fab in Yellow.
To test our hypothesis, we added Fab generated from CARM1-specific monoclonal antibody to the assembled protein complex sample and applied the same classification process to this new dataset (Figure 2b and Figure S4c–d). We initially obtained three groups. Maps 2A and 2B (Figure 2b) are similar to maps 1A and 1B (Figure 2a), respectively. The third group was further classified into two groups, which showed the absence (map 2D) and presence (the yellow density in map 2E, Figure S4d, and Supplementary Movie 2) of density in the position close to the putative CARM1. We interpret these additional densities in map 2E to be CARM1-specific monoclonal Fabs. The Fab density indeed binds to the pink region, substantiating that the pink density is CARM1. Comparison between the complexes without and with CARM1 (Figure 2a) thus indicates that CARM1 binding displaces SRC-3b from the ER complex.
Effects of CARM1 on the ER complex formation
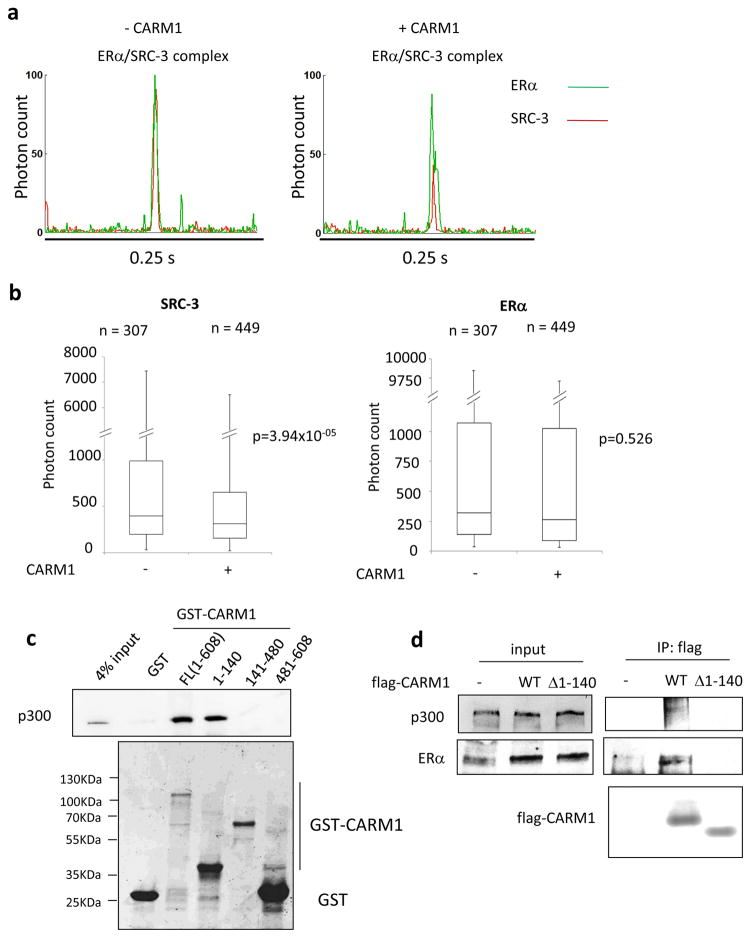
In an attempt to confirm that the dissociation of one SRC-3 from the ER complex upon CARM1 binding also occurs in cells, we utilized an advanced Flow Proteometric system (Chou et al., 2014; Chou et al., 2016) to analyze the stoichiometry of SRC-3 and ER within a single ER complex in the absence or presence of CARM1. HeLa cells were transiently transfected with ERα with or without CARM1 co-transfection and stimulated with estrogen before harvesting. ERα and endogenous SRC-3 were then labeled with green (Atto488) and red (Alexa647) fluorescence probes, respectively. Coincidence of green and red fluorescence signals were detected when individual ER/SRC-3 complexes flew through the detection spot in a microfluidic based flow-proteometric system (Figure 3a). Interestingly, overexpression of CARM1 significantly reduced the SRC-3 fluorescence intensity in the complex (Figure 3 a and b). In contrast, the ERα fluorescence intensity did not change significantly (Figure 3b). These results indicated that CARM1 overexpression can decrease the ratio of SRC-3:ER in the ER complex in living cells, and is consistent with our structural result whereby CARM1 replaces one of the SRC-3 binding molecule. In the ChIP-qPCR analysis (Figure 1a and Figure S1), not all ER binding sites we assessed have reduced SRC-3 occupancy upon CARM1 recruitment. This is likely due to cell heterogeneity, so the presence of SRC-3 complexed with coactivators other than CARM1 in a particular gene locus, may mask the reduction of SRC-3 complexed with CARM1.
Figure 3.
CARM1 overexpression reduced SRC-3:ER ratio and its N-terminal domain mediated the interaction with p300. (a) Raw Flow Proteometric analysis data for single ER/SRC-3 complex in HeLa cells without (left panel) or with (right panel) CARM1 overexpression. (b) Box Whisker Plot of SRC-3 and ERα fluorescence intensities detected in single ER/SRC-3 complexes without or with CARM1 expression. (c) The N-terminal domain of CARM1 (1-140aa) mediated the interaction between CARM1 and p300 in vitro. Shown is the GST pull-down experiment using GST-fused CARM1 full length and different fragments to pull down purified p300 protein. (d) CARM1 N-terminal domain deletion significantly reduced its ability to interact with p300 and ER in cells. Shown is a co-IP experiment.
N terminal domain of CARM1 interacts with p300
CARM1 has 608 amino acids and contains three domains. The central domain (141-480 aa) contains methyltransferase activity and SRC binding activity (Teyssier et al., 2002). However, both N- and C-terminal domains are indispensable for its synergistic transcriptional activation with p300 along with the methyltransferase activity (Teyssier et al., 2002). When fused to a Gal4-DNA binding domain, the N-terminal domain does not activate reporter gene transcription (Teyssier et al., 2002). Thus, it is not clear how the N-terminal domain of CARM1 contributes to its coactivator function. In our cryoEM structure, the CARM1 Fab used to label CARM1 recognizes the N-terminal region. Interestingly, it binds to the junction region between CARM1 and p300, suggesting that the N-terminal region of CARM1 is involved in the binding with p300. In order to validate this interaction, we generated three different fragments of CARM1 fused with GST constructs and then performed GST pull-down experiments using purified p300 protein. As shown in Figure 3c, only the full-length CARM1 and the N-terminal region of CARM1 (1-140 aa) were able to pull-down p300 successfully. To test further whether the N-terminal domain is responsible for the interaction between CARM1 and p300 in cells, an additional co-IP experiment was carried out in cells transiently transfected with ERα and flag tagged CARM1 wild type (WT) or with the 1-140 aa deletion mutant (Δ1-140). As shown in Figure 3d, WT CARM1 had a significant interaction with endogenous p300 and ER compared to the Δ1-140 mutant. These results indicate the importance of the N-terminal domain for CARM1’s interaction with p300.
Structure and functional changes of p300 upon binding to CARM1
Comparing the complex with two SRC-3s to the complex with SRC-3a and CARM1, as expected, the most significant changes are observed in the vicinity of SRC-3b (Figure 2a). There are also some changes in the structure of p300 (Figure 4a and b). This suggested that p300 undergoes a conformational change when CARM1 binds to the complex. As a multi-domain protein, p300 contains NRID (nuclear receptor interacting domain), CH1/TAZ1 (Cys/His rich region 1/Transcription Adaptor putative zinc finger 1), KIX (CREB interaction region) BRMD (bromodomain), CH2/HAT (histone acetyltransferase domain), CH3/TAZ2 and SRCID (SRC interaction domain) regions. Four of these domains (NRID, CH1/TAZ1, KIX and SRCID, shown in red lines in Figure S5a), interact with the two SRC-3s in the ERα/SRC-3/p300 complex (Yi et al., 2015). Since CARM1 appears to occupy the same location as SRC-3b in the complex while inducing a p300 conformational change, we then asked whether CARM1 and SRC-3 contact the same surface on p300. A co-IP experiment was performed in cells transiently transfected with each of the seven different domains of p300 and HA tagged CARM1. As shown in Figure S5a, only BRMD and CH3/TAZ2 domains of p300 (shown in blue lines) interacted with HA-CARM1, different from the regions interacting with SRC-3 (shown in red lines) (Yi et al., 2015). This change in binding could explain the conformational change of p300 upon CARM1 recruitment.
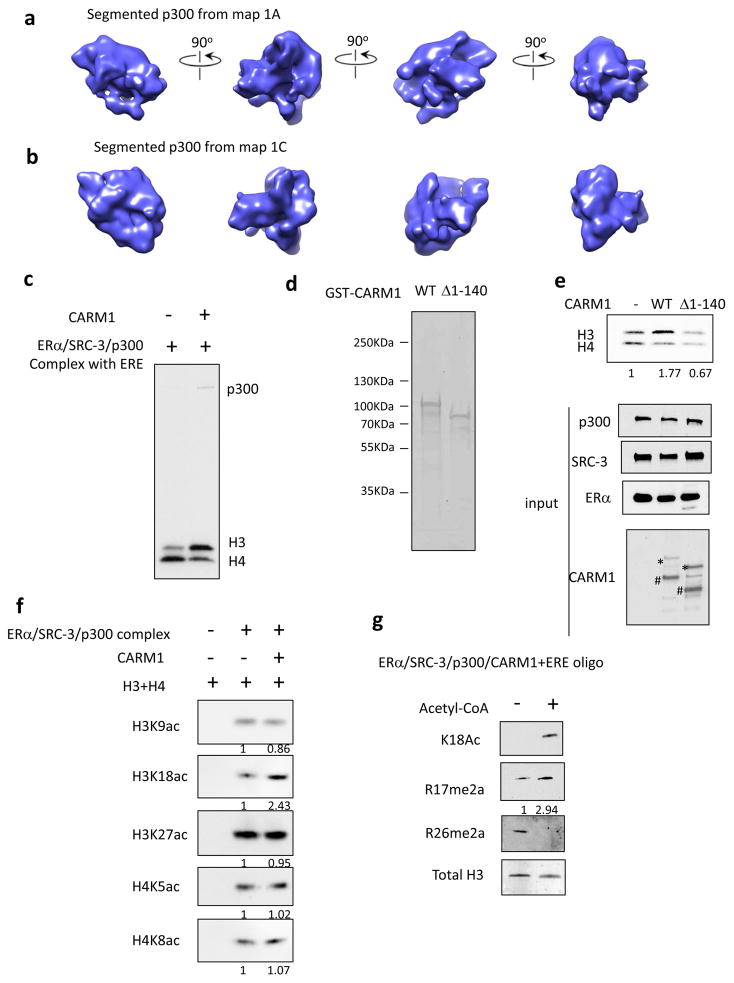
Figure 4.
Communication between p300 and CARM1 within the ER complex regulates HAT and HMT activity. (a) Segmented p300 from map 1A. (b) Segmented p300 from map 1C. (c) CARM1 increased p300 autoacetylation and its HAT activity on histone H3 in vitro. Shown is the autoradiograph of 3H-labeled acetylation. (d) Coomassie blue staining of purified GST-CARM1 WT and Δ1-140 mutant from bacteria. (e) Deletion of the N-terminal domain abolished the ability of CARM1 to regulate p300 HAT activity in vitro. Top panel, the autoradiograph of 3H-labeled acetylation. Bottom panels, Western blot analysis on the levels of ER, SRC-3, p300 and CARM1 WT or Δ1-140 mutant in the input. * represents the uncleaved GST-CARM1. # represents GST-removed CARM1 through thrombin cleavage. (f) CARM1 selectively increased p300 HAT activity on H3K18. Shown is the Western blot analysis of the levels of histone acetylation in the in vitro HAT assay using different acetylation antibodies. (g) p300-mediated acetylation significantly increased CARM1-mediated H3R17 methylation. Shown is the autoradiograph of 3H-labeled methylation in vitro.
Since the binding of CARM1 changes the quaternary structure of the ER complex and induces a conformational change in p300 (Figure 4a and b), we next investigated whether these structural changes have a functional impact. p300 has a histone acetyltransferase activity critical for promoting chromatin remodeling. The ERE/ERα/SRC-3/p300 complex is able to acetylate both histones H3 and H4 in an in vitro histone acetylation assay (Figure 4c). Addition of CARM1 to the complex significantly increased the levels of p300 autoacetylation and H3 acetylation but not H4 acetylation (Figure 4c). This suggests that introduction of CARM1 to the complex selectively enhanced p300 HAT activity on histone H3.
Since the N-terminal domain of CARM1 is important for interacting with p300, our next experiment was to test whether deletion of the N-terminal domain could affect the ability of CARM1 to regulate p300 HAT activity. We utilized a GST fused CARM1Δ1-140 and WT in a bacterial expression vector and then purified the proteins from bacteria (Figure 4d). The N-terminal GST was then removed through thrombin cleavage in order to avoid any potential steric hindrance for CARM1 to interact with p300. We found that the bacterial expressed CARM1 WT increased p300 HAT activity on H3 but not on H4 although the intensity is lower compared to the CARM1 purified from human cells (Figure 4e). This might be due to decreased post-translational modifications in the bacterial-expressed protein, which could lower the efficiency of CARM1 incorporation into the complex. Most importantly, the N-terminal deletion mutant of CARM1 lost the ability to regulate p300 HAT activity. Our results suggest that the ability of CARM1 to interact with p300 through its N-terminus is essential for regulating p300 activity.
Effects of CARM1-p300 communication on histone modification
Multiple lysine residues in H3 can be acetylated and they are usually linked to transcription activation. To determine which lysine acetylation was affected by CARM1, we performed an in vitro histone acetylation assay and used several different H3 and H4 lysine acetylation-specific antibodies to detect the level of lysine acetylation in the absence or presence of CARM1. As shown in Figure 4f, only H3K18 acetylation was significantly increased with the addition of CARM1. It has previously been reported that H3K18 acetylation levels at ER targeted gene promoters rapidly increases upon estrogen stimulation (Daujat et al., 2002); p300/CBP is the major HAT mediating H3K18 acetylation in cells (Jin et al., 2011). It plays an essential role in ligand-induced RNA Pol II recruitment and in nuclear receptor-targeted gene transcription activation (Jin et al., 2011). We also observed decreased H3K18 acetylation of ER binding sites at both distal enhancer (Figure S5b) and proximal promoter regions (Figure S5c) when CARM1 was knocked-down in estradiol-stimulated MCF-7 cells, suggesting that CARM1 indeed affects p300-mediated H3K18 acetylation in cells. Crosstalk between p300/CBP-mediated histone acetylation and CARM1-mediated histone methylation has been reported previously (Daujat et al., 2002; Yue et al., 2007). Acetylation at the H3K18 site dramatically promotes CARM1-mediated H3R17 methylation (Daujat et al., 2002; Yue et al., 2007). Since CARM1 binding increased p300-mediated H3K18ac in vitro, we next determined whether this increase could in turn regulate CARM1 activity on H3R17 methylation using an in vitro histone methylation assay. We first incubated CARM1 with estradiol, ER, SRC-3, p300, ERE oligo and H3 for 30 min in the absence or presence of acetyl CoA, the acetylation donor, to initiate an in vitro acetylation reaction. The methylation donor SAM then was added to the mixture to start the methylation reaction. As shown in Figure 4g, the addition of acetyl CoA induced H3K18 acetylation. The level of H3R17 methylation significantly increased with the addition of acetyl CoA. In contrast, CARM1-mediated H3R26 methylation was inhibited. These results suggest that p300-mediated histone acetylation selectively increases CARM1-mediated H3R17 methylation.
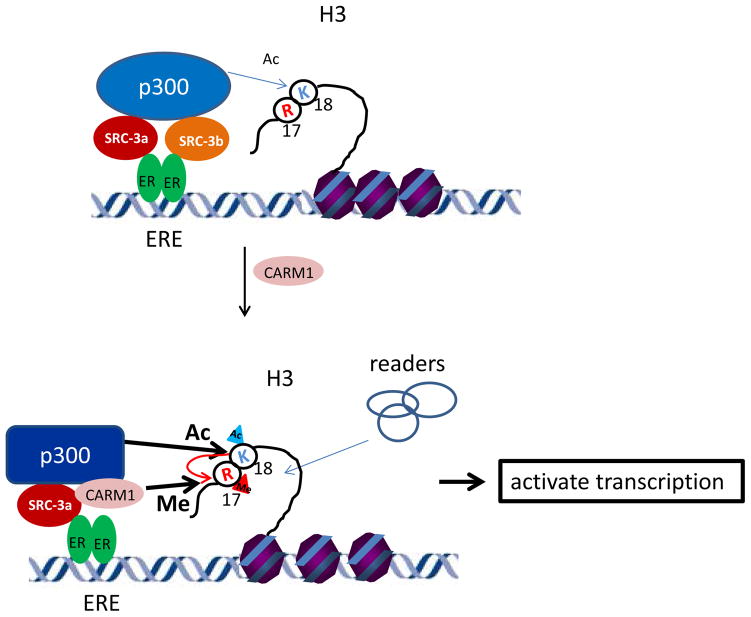
Model of sequential recruitment of ER-mediated coactivators and its impact on transcription activation
Combining all of our structural and biochemical analyses, we propose the following model for ER-mediated coactivator sequential recruitment and transcription activation (Figure 5 and Supplementary Movie 3). Upon estrogen stimulation, an ER dimer binds to ERE in target gene promoter/enhancer regions. Each ER monomer recruits one SRC-3 and the two SRC-3s work together to securely lock one p300 into the ER complex, which then initiates histone H3 acetylation. Subsequent CARM1 binding causes the release of one SRC-3 from the complex and induces a p300 conformational change, resulting in increased p300 HAT activity on H3K18, which in turn enhances CARM1 on the adjacent residue R17 methylation. These estrogen induced histone modifications then bring in specific reader proteins to promote activation of transcription.
Figure 5.
A working model of the impact of coactivator sequential binding on ER-mediated transcriptional activation. Upon estrogen stimulation, an ER dimer binds two SRC-3 molecules which then recruit p300. The binding of p300 promotes basal level of histone H3K18 acetylation at the ER binding sites. The recruitment of CARM1 displaces one SRC-3 molecule from the complex and induces a p300 conformational change to enhance p300-mediated H3K18 acetylation. Increased H3K18ac further promotes CARM1-mediated H3R18 dimethylation. These histone epigenetic modifications then bring in reader proteins to promote transcription.
Discussion
Synergistic action is often seen with nuclear receptor coactivators, and it is generally believed that different coactivators cooperate to promote this synergism. Yet it is not clear exactly how these coactivators communicate with each other, especially for sequentially recruited coactivators, to activate transcription synergistically. Here we present structural and biochemical evidence to demonstrate that the sequential process of coactivator recruitment plays a role in this cooperative interaction (Supplementary movie 3).
We reported previously that two SRC-3s are needed to form an initial stable ERE/ERα/SRC-3a/SRC-3b/p300 core complex (Yi et al., 2015). This is a critical step to establish the core ER-coactivator complex and to bring in p300 protein to ER genomic binding sites. Based on our map classification analysis (Figure 2a), the ER complex can exist with either one SRC-3 or two SRC-3s. Interestingly, our cryoEM analysis shows that CARM1 is located only at one of the SRC-3 binding sites (SRC-3b) (Figure 2a Map 1C) and is confirmed by our Fab complex structure (Figure 2b). Previously, we showed that this SRC-3 binding site has a weak interaction with ER and p300 (Yi et al., 2015). However, the CARM1 binding has a profound impact on the function of the new ER-coactivator complex. First, CARM1 binds to the complex where it can easily access methylation of its substrates-SRC-3, p300 and histones. SRC-3b is located in proximity to ER, SRC-3a and p300. It is likely that SRC-3b is no longer needed once p300 is recruited to the complex (Figure 2a Map 1B). Upon CARM1 binding, the absence of SRC-3b avoids a steric hindrance that would have limited the ability of CARM1 to efficiently interact with other components in the complex. Second, CARM1 binding at the SRC-3b location induces a further p300 conformational change since it contacts different regions in p300 compared to SRC-3 (Figure 4a and b). This structural alteration induced by sequential coactivator recruitment both further alters p300 HAT activity and also promotes CARM1 HMT activity on histone H3. It was reported previously that p300-mediated histone acetylation regulates CARM1 activity (Daujat et al., 2002; Yue et al., 2007). Here we present evidence that the crosstalk between the two proteins is bi-directional. CARM1 binding also influences p300 to create an optimal environment for CARM1-mediated histone methylation. Ultimately, the cooperative interaction between CARM1 and p300 enhances both histone H3K18 acetylation and H3R17 methylation, contributing to the synergistic activation of target gene transcription (Supplement Movie 3).
A link between CARM1 or p300-mediated enzymatic reactions on cofactors and ER-coactivator complex dissociation has been reported previously (Chen et al., 1999b; Feng et al., 2006; Lee et al., 2005). CARM1 methylates p300 at the C-terminal SRC-interacting domain, which inhibits the interaction between SRC and p300 (Lee et al., 2005). SRC-3 methylation can destabilize the SRC-3/CARM1 complex (Feng et al., 2006). p300-mediated SRC acetylation also dissociates SRC from ER (Chen et al., 1999b). The purified p300 and CARM1 proteins we used likely still have bound cofactors (acetyl-coA or ado-Met), which allow the acetylation or methylation reactions to occur. The recruited CARM1 together with p300 promotes p300 and SRC-3 methylations as well as SRC-3 acetylation, which then disrupts the association of the weaker-binding SRC-3b with p300, CARM1 and ER, leading to its dissociation from the complex (Supplementary movie 3).
We found herein that H3R17 methylation but not H3R26me2a significantly increased when H3 is acetylated by p300 (Figure 4g). Both H3R17me2a and H3R26me2a are generated by CARM1 (Schurter et al., 2001). They all have an adjacent lysine residue that can be acetylated, K18 and K27, respectively. Elevated H3R17me2a levels were found in nuclear receptor targeted gene promoters upon hormone stimulation (Bauer et al., 2002; Denis et al., 2009; Ma et al., 2001; Metivier et al., 2003), which correlates with CARM1 recruitment (Metivier et al., 2003). It is linked to gene activation (Bauer et al., 2002). Several reports have identified “readers” for H3R17me2a, including Tudor domain containing protein TDRD3 and a transcription elongation associated PAF1 complex (PAF1c) (Wu and Xu, 2012; Yang et al., 2010; Yang et al., 2014). TDRD3 recruits topoisomerase IIIB (TOP3B), which plays a subsequent role in transcription elongation by resolving a transcription-mediated R-loop (Yang et al., 2014). These reader proteins are believed to be effector molecules mediating CARM1 and H3R17me2a-dependent transcription activity.
In contrast to the well-known activation role of H3R17me2a in transcription, little is known about H3R26me2a. We recently found that H3R26 methylation represses transcription through inhibiting the binding of the Super Elongation Complex (SEC) with H3K27ac, an active transcription mark (Zhang et al., 2017). Thus, the two marks generated by CARM1 can be active or repressive depending on relative levels of each modification. Our study demonstrates that recruitment of CARM1 to the ER complex selectively increases CARM1 activity on the active histone mark through a cooperative interaction with p300. This could be one of the main mechanisms by which CARM1 exerts a positive effect in regulating transcription as a nuclear receptor coactivator. Interestingly, the R17me2a reader proteins also appear to be involved in transcription elongation. This implies that CARM1 recruitment at a later stage after estrogen stimulation prepares the ER complex to functionally participate in transcription elongation after the initial activation reaction for transcription. The observed inhibition of H3R26 methylation could be influenced by squelching since H3R17 methylation is significantly increased.
Although CARM1 shares a highly conserved “PRMT core” region that possesses arginine methyltransferase activity along with other PRMT family members (Zhang et al., 2000), it has unique substrate specificity (Di Lorenzo and Bedford, 2011; Frankel et al., 2002). CARM1 knockout mice are neonatal lethal (Yadav et al., 2003), indicating a non-redundant function of CARM1 in vivo. The N-terminal domain is the most variable region among PRMTs (Zhang et al., 2000). It is required for CARM1 coactivator function but does not contribute to its autonomous transcription activation activity (Teyssier et al., 2002). The crystal structure of CARM1 N-terminal domain (28-140 aa) reveals that it has a Pleckstrin Homology (PH) domain-like fold structure (Troffer-Charlier et al., 2007). The PH domain is found in a variety of proteins and it frequently serves as a protein-protein interaction platform (Scheffzek and Welti, 2012). The protein partner interacting with the N-terminal PH domain of CARM1 has not been identified previously. Our structural (Figure 2) and biochemical studies (Figure 3c) now demonstrate that this domain mediates the interaction between CARM1 and p300. This could explain a previous observation that deletion of this region abolishes the synergistic activation between CARM1 and p300 (Teyssier et al., 2002). Interestingly, the electron density of residues 28-146 is not visible in the crystal structure of CARM1 (28-507aa), indicating that the position of this PH domain is very flexible (Troffer-Charlier et al., 2007). Clearly, it is immobilized upon binding to the ER complex (Figure 2). This “unordered” feature of the N-terminal domain may be important for CARM1 to associate with a variety of transcription factor complexes and to function as a coactivator (An et al., 2004; Chen et al., 2002; Covic et al., 2005; Koh et al., 2002; Zika et al., 2005).
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bert W. O’Malley (berto@bcm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Culture Conditions
MCF-7, 293T and HeLa cells were cultured in DMEM (high glucose) medium supplemented with 10% Fetal bovine serum, 100 units/ml of penicillin and 100 μg/ml of streptomycin. For ChIP and luciferase reporter assays, cells were maintained in charcoal-stripped serum containing medium for 3 days before estrogen stimulation. All cells were incubated at 37°C incubator with 5% CO2.
METHOD DETAILS
Chromatin Immunoprecipitation
MCF-7 cells were cultured in 10% charcoal-stripped serum containing medium for 3 days and then treated with 10 nM or 100 nM estradiol for 0, 15, 30, 45 and 60 minutes. Cells were then fixed in 1% formaldehyde and subjected to Chromatin immunoprecipitation using anti-ERα, anti-SRC-3, anti-CARM1 (Invitrogen), anti-H3K18ac (Active Motif) specific antibodies following the manufacturer’s protocol (Active motif). Binding of these proteins to TFF1 gene promoter was assessed through real time PCR using the following primers. Forward: 5′-GGATTAAGGTCAGGTTGGAGG; Reverse: 5′-CATGGTCAAGCTACATGGAAGG. TFF1 ERE3 (−9.9 kb from the TSS); Forward: 5′-GTCGTTGCCAGCGTTTCC Reverse: 5′-CTTCTCCACGCCCTGTAAATTT; GREB1 ERE1 (−1.6 kb from isoform a TSS): Forward: 5′-GTGGCAACTGGGTCATTCTGA; Reverse: 5′-CGACCCACAGAAATGAAAAGG. GREB1 distal enhancer (−35.4 kb from isoform a TSS) Forward: 5′-CAGGGGCTGACAACTGAAAT; Reverse: 5′-GAGAGGGTGGTGACACTTGG.
Flow Proteometric analysis
Individual ERα/SRC-3 complexes were analyzed by the flow-proteometry system (Chou et al., 2014; Chou et al., 2016). In brief, ER (0.8 μg) plasmid DNA were transfected into HeLa cells (1×105) with or without CARM1 plasmid DNA (1.6 μg) using Lipofectamin 3000 (Life Technologies). After 48 hours of incubation for protein expression, cells were fixed with 1% paraformaldehyde in PBS for 8 min in room temperature. The fixation was then stopped using 125mM glycine. The cells were then incubated with 0.25% Triton X-100 in PBS for 10 min in room temperature to increase the permeability. After PBS washing, the cells were incubated with blocking reagent (6% BSA in PBS) for 1hr in room temperature followed by incubation with anti-SRC-3 and anti-ERα (H-184, Santa Cruz) primary antibodies (diluted in 6% BSA) in 4°C for overnight. The cells were then washed with PBS to remove unbound antibodies and then incubated with fluorescence conjugated secondary antibodies (Alexa 647 conjugated secondary antibody, Life Technologies, Atto488 conjugated secondary antibody, Sigma). After PBS washing, a secondary crosslinking process (1% paraformaldehyde for 5 min in room temperature) was performed. The cells were then lysed in lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% (v/v) Nonidet P-40, 1 mM EDTA) and the protein complexes were broken-down under sonication into a single complex level. The lysates were then centrifuged 1 min at 16,000 × g and the supernatants were collected and further diluted in detection buffer (20 mM HEPES-KOH, pH 7.9, 0.1 mM KCl, 2 mM MgCl2, 15 mM NaCl, 0.2 mM EDTA, 10% (v/v) glycerol) for flow-proteometric analysis. The sample lysates were flowed in a microchannel device (Chou et al., 2014) and each ER/SRC-3 complex (SRC-3 labeled with Alexa 647 and ER labeled with Atto488) was detected by the flow-proteometric device (Chou et al., 2016). The fluorescence intensities of SRC-3 and ER in individual ER/SRC-3 complex were analyzed. Each individual photon burst represents a single target protein.
GST pull-down
CARM1 full-length and different fragments were fused to GST in a pGEX-4T1 vector and the proteins were expressed in E. coli. Bacterial lysates containing expressed different GST-CARM1 proteins were incubated with 6 μl of glutathione sepharose 4B beads (GE Healthcare Life Sciences) for 1 hour and then washed 3 times with wash buffer (20mM HEPES pH 7.6, 150mM KCl, 1mM DTT, 0.1% NP40, 8% glycerol and protease inhibitor cocktail). The beads were then incubated with 40ng purified recombinant p300 from baculovirus for 1.5 hours. After wash, the beads were boiled in 2x SDS sample buffer and then loaded to a 4–15% SDS-PAGE.
In vitro HAT and HMT assay
For in vitro HAT assay using human cell expressed CARM1 protein (EMD Millipore), 0.4 μg of p300 protein, SRC-3, ERα (Invitrogen) proteins and 10ng ERE oligo (Santa Cruz), 1 μM 17-β estradiol (Sigma) were incubated with or without 0.5 μg of CARM1 for 1 hour before the addition of recombinant histone H3.1 and H4 (1 mg each per reaction, NEB) in a 20 μl reaction mixture containing 20 mM Tris-HCl (pH 8.0), 4 mM EDTA, 1 mM PMSF, 0.5 mM DTT and 1 μl [3H] acetyl coenzyme (3.69 Ci/mmol; Perkin Elmer) for 1 hour at 30°C. Reactions were stopped by the addition of 6x SDS sample buffer, and proteins were separated in a 4–15% SDS-PAGE. Following fixation with buffer containing 50% methanol and 20% acetic acid, gels were treated with autoradiography amplify reagent (Amersham Biosciences) for 20 min, dried, and exposed to x-ray films.
For in vitro HMT assay, the procedure is essentially the same as in vitro HAT assay except that the reaction was incubated with or without cold acetyl coenzyme. After 1 hour incubation of the HAT assay, 1 μl [3H] Ado-Met was added into the reaction for another 1 hour at 30°C. The reaction was then stopped by the addition of 6x sample buffer.
For in vitro HAT assay using bacterial expressed CARM1, the complexes were assembled on a biotinylated GREB1-ERE DNA using the same procedure as described for cryo-EM sample preparation. The DNA-bound ER-coactivator complexes were then separated from unbound proteins using Dynabeads M-280 Streptavidin (Invitrogen). 1 μg of H3.1, H4 and 1 ml [3H] acetyl-CoA were then added to the beads to initiate the HAT assay.
Cryo-EM specimen preparation and data collection
0.6 μg of human ERα, SRC-3, p300 and CARM1 proteins were incubated with 1 μg of biotinylated ERE containing DNA derived from GREB1 enhancer sequence (Foulds et al., 2013) and 1 μM 17-β estradiol (Sigma) on ice for 30 min. The reaction was then incubated with 20 μl of Dynabeads M-280 Streptavidin (Invitrogen) for 15 min followed by EcoRI restriction enzyme cleavage. For CARM1 Fab labeling, Fab was generated from CARM1 monoclonal antibody (Thermo Fisher) using Pierce Fab preparation kit (Thermo Fisher) following the manufacturer’s instruction. Fab was then incubated with the assembled complex for 30 minutes on ice. The complex was kept on ice before vitrification on the grid. A 1.5 μl aliquot of the ERα + SRC-3 + p300 + CARM1 samples was applied onto a continuous carbon film supported by a 200-mesh R1.2/1.3 Quantifoil grid (Quantifoil). The grid was previously washed and glow discharged. After applying the sample, the grid was blotted and rapidly frozen in liquid ethane using a Vitrobot IV (FEI), with constant temperature and humidity during the process of blotting. The grid was stored in liquid nitrogen before imaging. 1,876 images of frozen-hydrated ERα + SRC-3 + p300 + CARM1 complex particles were acquired on a Technai G2 Polara electron microscope (FEI) operated at 300 kV using a K2 Summit direct electron detector camera (Gatan). Images were collected automatically by SerialEM (Mastronarde, 2005) in dose fractionation super-resolution counting mode at a nominal magnification of ×23,000, corresponding to a calibrated physical pixel size of 1.62 Å and super-resolution pixel size of 0.81 Å with a dose rate on the camera set to ~10 electrons pixel−1s−1. 1,686 images of complex with Fab were collected semi-automatically automatically by SerialEM (Mastronarde, 2005) with a defocus range of −1.5 to −3.5 μm using JEM3200FSC with an in-column energy filter (20 ev width) and a K2 Summit camera in super resolution counting mode at magnification of 20k x. The total exposure time for both datasets was 6 s, leading to a total accumulated dose of 22 electrons Å−2 on the specimen. Each image stack was fractionated into 30 subframes, each with an accumulation time of 0.2 s per frame. 22 images of ERα/SRC-3/p300/CARM1 complex for tilt pair validation were collected on a JEM2010F operated at 200 KV using a CCD camera at a nominal magnification of ×20,000, corresponding to a calibrated pixel size of 3.62 Å.
Image processing, 3D reconstruction and classification
Dose-fractionated super-resolution raw image stacks of ice-embedded ERα/SRC-3/p300/CARM1 complex were binned 2 × 2 by Fourier cropping resulting in a pixel size of 1.62 Å for further image processing. Each image stack was subjected to motion correction using ‘dosefgpu_driftcorr’ (Li et al., 2013), and a sum of subframes 1–29 in each image stack was used for further image processing (Figure S2b).
Following the same procedure as described previously (Yi et al., 2015), using EMAN2 program e2boxer.py we manually boxed out 46,527 particle images from ERα + SRC-3 + p300 + CARM1 dataset excluding particles that have DNA wrapping or interfere with neighboring particles. Defocus of the particles in each frame average was automatically determined by both EMAN2 program e2ctf.py and ctffind3. Particles were then phase flipped to form stack files for further processing. 2D reference free class averages were computed by e2refine2d.py (Figure S2d). Initial models for every reconstruction were generated from scratch by the e2initialmodel.py program using selected unsupervised 2D averages of good quality based on visual comparison without applying any symmetry (Figure S3a). The same strategy was used to process the dataset of CARM1-binding Fab with 16,883 particle images. The fact that the structures of the various complexes are so similar despite completely independent processing, including generation of new initial models, is additional evidence for the reliability of the maps with such a compositionally variable data set.
As an additional validation, we performed tilt-pair analysis (Rosenthal and Henderson, 2003) (Figure S3c). The distribution of particle orientations was also sufficient to have no missing data in Fourier space with near-isotropic resolution (Figure S2e).
3D classification of the raw particle images
After the initial map was obtained and validated, we subjected the original data to classification employing recent revisions to the EMAN2 e2refinemulti program, which has been specially designed to classify heterogeneous particles by randomly removing map fragments (Ludtke, 2016). Following the particle classification, we carried out another round of refinement for each subset of particle data and computed gold standard FSCs (Figure S3b). Similar to EMAN2’s resolution testing strategy where initial models are phase-randomized, this multi-reference refinement strategy used significantly perturbed initial models, so the agreement among the final maps is a strong statement that these maps are data-derived, and not due to bias.
Map segmentation
The classified cryo-EM maps of ERα/SRC-3/p300/CARM1 and ERα/SRC-3/p300/CARM1/Fab complexes were segmented using Segger (Pintilie et al., 2010), with a similar procedure as described previously for the map of ERE/ERα/SRC-3a/SRC-3b/p300 complex (Yi et al., 2015). Briefly, watershed regions were first computed in both maps; these regions correspond to local areas of high densities (peaks), with boundaries that follow lower-density voxels (valleys). A total of 4 steps of smoothing and grouping were then applied, which groups the initial small regions into larger regions. Comparison to the previous segmentation of ERα/SRC-3/p300/CARM1 showed that 3 of these larger regions roughly correspond to the ER, SRC-3a and p300 components. A few small interactive ungrouping/regrouping adjustments were made, as the automated grouping approach can sometimes mis-attribute regions to components, especially when there are size differences between components as in this study. In both maps, a region was further identified as CARM1, which occupies the same region as the SRC-3b in the former segmentation. In the map for ERE/ERα/SRC-3a/p300/CARM1/Fab, two new small regions at the boundary between p300 and CARM1 were also identified and labeled as Fab1 and Fab2.
QUANTIFICATION AND STATISTICAL ANALYSIS
All error bars shown in the Figures represent mean ± standard error the mean.
DATA AND SOFTWARE AVAILABILITY
All the cryo-EM maps have been deposited to EMDB under accession code EMD-8830, 8831 and 8832. Original imaging data have been deposited to Mendeley Data and are available at http://dx.doi.org/10.17632/2hxs654ntj.1
Supplementary Material
Supplementary movie 1: A side by side comparison between complex with two SRC-3s (Map 1A) and the complex with one SRC-3 and CARM1 in dataset of ERα+SRC-3+p300+CARM1 (Map 1C), related to Figure 2.
Supplementary movie 2: A side by side comparison between CARM1 complex with Fab (Map 2E) and without Fab (Map 1C), related to Figure 2.
Supplementary movie 3: Sequential process of ER coactivator SRC-3/p300/CARM1 recruitment, using a crystallographic model of the histone-DNA complex (Davey et al., 2002) (PDB 1kx5), segmented maps 1A and 1C, related to Figure 5.
Highlights.
ER sequentially recruits SRC-3 and CARM1 coactivators to ER binding sites
CARM1 recruitment replaces one of two SRC-3 proteins from the complex
CARM1 recruitment induces p300 conformational change and promotes H3K18ac
Increased histone H3K18 acetylation in turn enhanced CARM1-mediated H3R17 methylation
Acknowledgments
This work is supported by Komen Foundation (5PG12221410) and DOD R038318-I to B.W.O; DOD W81XWH-15-1-0536 to P.Y.; NIH grants (HD8818 and NIDDK59820 to B.W.O.; P41GM103832 and R01GM079429 to W.C.; CNIHR and R21AI122418 to Q.F; R01GMGM072804 to I.I.S.); CPRIT grants (RP150648 to B.W.O. and DP150052 to M.C.H.); and NCI Cancer Center Support Grant P30CA125123 (BCM Monoclonal Antibody/recombinant Protein Expression Core Facility).
Footnotes
Author Contributions
P.Y., Z.W., M.F.S, W.C. and B.W.O conceived and designed the experiments. P.Y., Q.F. S.H. and C.E.F. performed the biochemical experiments. C-K. C. performed the Flow Proteometric analysis. Z.W., G.D.P., G. Z., I.S. and S.J.L. did cryo-EM imaging and computational analysis. P.Y., Z.W., G.D.P., M.F.S., M-C. H., W.C. and B.W.O. interpreted the data and wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov D, Crofts LA, Chang CP, Freedman LP. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem. 2002;277:14359–14362. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- Casadio F, Lu X, Pollock SB, LeRoy G, Garcia BA, Muir TW, Roeder RG, Allis CD. H3R42me2a is a histone modification with positive transcriptional effects. Proc Natl Acad Sci U S A. 2013;110:14894–14899. doi: 10.1073/pnas.1312925110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang SM, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999a;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999b;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. Embo J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CK, Lee HH, Tsou PH, Chen CT, Hsu JM, Yamaguchi H, Wang YN, Lee HJ, Hsu JL, Lee JF, et al. mMAPS: a flow-proteometric technique to analyze protein-protein interactions in individual signaling complexes. Sci Signal. 2014;7:rs1. doi: 10.1126/scisignal.2004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CK, Tsou PH, Hsu JL, Lee HH, Wang YN, Kameoka J, Hung MC. Analysis of Individual Signaling Complexes by mMAPS, a Flow-Proteometric System. Curr Protoc Mol Biol. 2016;114:20 11 21–20 11 22. doi: 10.1002/0471142727.mb2011s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. Embo J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F. Functional connection between deimination and deacetylation of histones. Mol Cell Biol. 2009;29:4982–4993. doi: 10.1128/MCB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, Hamilton RA, Gates LA, Zhang Z, Li C, et al. Proteomic Analysis of Coregulators Bound to ERalpha on DNA and Nucleosomes Reveals Coregulator Dynamics. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. Embo J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, Bedford MT. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem. 2010;285:1147–1152. doi: 10.1074/jbc.M109.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J Biol Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List HJ, Lauritsen KJ, Reiter R, Powers C, Wellstein A, Riegel AT. Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells. J Biol Chem. 2001;276:23763–23768. doi: 10.1074/jbc.M102397200. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ. Single-Particle Refinement and Variability Analysis in EMAN2.1. Methods Enzymol. 2016;579:159–189. doi: 10.1016/bs.mie.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Welti S. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 2012;586:2662–2673. doi: 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shao W, Keeton EK, McDonnell DP, Brown M. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc Natl Acad Sci U S A. 2004;101:11599–11604. doi: 10.1073/pnas.0402997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci U S A. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C, Chen D, Stallcup MR. Requirement for multiple domains of the protein arginine methyltransferase CARM1 in its transcriptional coactivator function. J Biol Chem. 2002;277:46066–46072. doi: 10.1074/jbc.M207623200. [DOI] [PubMed] [Google Scholar]
- Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. Embo J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xu W. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc Natl Acad Sci U S A. 2012;109:5675–5680. doi: 10.1073/pnas.1114905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014;53:484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wang Z, Feng Q, Pintilie GD, Foulds CE, Lanz RB, Ludtke SJ, Schmid MF, Chiu W, O’Malley BW. Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol Cell. 2015;57:1047–1058. doi: 10.1016/j.molcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. Embo J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. Embo J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Nikolai BC, Gates LA, Jung SY, Siwak EB, He B, Rice AP, O’Malley BW, Feng Q. Crosstalk between histone modifications indicates that inhibition of arginine methyltransferase CARM1 activity reverses HIV latency. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zika E, Fauquier L, Vandel L, Ting JP. Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-gamma-inducible MHC-II gene expression. Proc Natl Acad Sci U S A. 2005;102:16321–16326. doi: 10.1073/pnas.0505045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary movie 1: A side by side comparison between complex with two SRC-3s (Map 1A) and the complex with one SRC-3 and CARM1 in dataset of ERα+SRC-3+p300+CARM1 (Map 1C), related to Figure 2.
Supplementary movie 2: A side by side comparison between CARM1 complex with Fab (Map 2E) and without Fab (Map 1C), related to Figure 2.
Supplementary movie 3: Sequential process of ER coactivator SRC-3/p300/CARM1 recruitment, using a crystallographic model of the histone-DNA complex (Davey et al., 2002) (PDB 1kx5), segmented maps 1A and 1C, related to Figure 5.