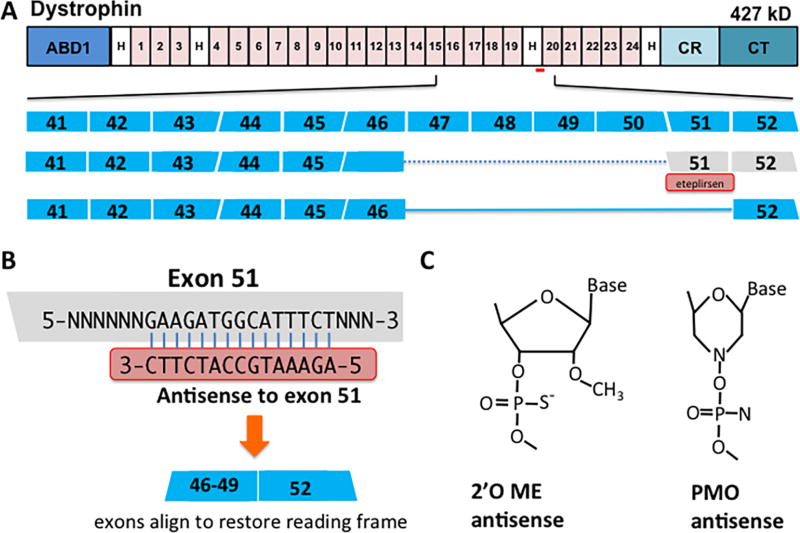
Duchenne Muscular Dystrophy (DMD) arises from mutations in the dystrophin gene. The dystrophin gene is composed of 79 exons, and the majority of mutations in DMD are deletions, often spanning multiple exons 1. In 2016, the FDA granted accelerated approval for eteplirsen (Exondys51™), an antisense oligonucleotide compound designed to block exon 51 of dystrophin in order to restore the reading frame in DMD patients with specific mutations (Figure 1A) 2. This treatment is directed at approximately 10–15% of DMD patients (~1500 treatment eligible individuals). Eteplirsen’s approval is game changing for the field of molecular gene correction. However, eteplirsen’s approval was viewed as controversial due to unconventional clinical trial data and limited efficacy.
Figure.
Eteplirsen (Exondys51) mediated readframe correction of DMD mutations. A. Top row: schematic of the dystrophin protein and its domains. ABD1, actin binding domain; H, hinge regions; CR, cysteine rich domain; CT, C-terminal domain; Numbered are the spectrin-like repeats. Red bar, indicates the approximate exon 51 targeting region. Middle rows: Schematic showing eteplirsen-mediated reading frame correction of a DMD frameshift mutation. The normal dystrophin locus from exons 41 to 52 is shown indicating the reading frame of each exon. Many DMD patients have variable sized deletions spanning exons 47 to 50, disrupting the reading frame (dashed blue line). Eteplirsen (red box) is an antisense oligonucleotide that binds to exon splicing enhancer sequences in exon 51 causing its exclusion from mRNA. By excluding 51, exons 46 (or 47, 48 and 49) joins to exon 52 (solid blue line) to restore an open reading frame. B. Antisense oligonucleotides have complementary sequences to those within an exon, in this case Exon 51. Chemical modifications to the antisense oligonucleotides permit the double stranded hybrid to evade nucleases that would normally target the hybrid for destruction. C. Two antisense chemical modifications are shown that were tested as drugs for treating DMD. The first, 2’-O-methyl-phosphorothioate (2’OE Me) is a charged moiety that was tested in drisapersen. The second, phophorodiamidate morpholino (PMO) was used in eteplirsen.
The strategy of exon skipping to treat DMD arose from seeing “revertent” fibers in dystrophic muscle, which vary depending on the specific mutation 3. In DMD muscle, dystrophin is absent from the myofiber sarcolemma, and revertent fibers are sporadic dystrophin-positive fibers, typically less than 1% of the total. Revertent fibers represent intrinsic splicing events that ‘skip’ around the mutation and restore the reading frame, leading to an internally-deleted, but functional, dystrophin protein. Dystrophin’s protein structure with its internally repeating structure is ideal for exon skipping. Removing a portion of the internal repeat structures leaves intact dystrophin’s ability to stabilize the sarcolemma. DMD mutations that create internal deletions, yet maintain the reading frame, cause the milder Becker muscular dystrophy (BMD). Furthermore, the X-linked nature of dystrophin also supports exon skipping as only one allele per genome requiring correction.
Antisense oligonucleotides are short RNA or DNA oligomers, chemically modified to avoid exo- and endonuclease degradation. Antisense oligonucleotides can engage sequences found within exons by targeting exon splicing enhancer sequences (called ESEs). ESEs normally direct inclusion of an exon into the mature mRNA. In DMD, blocking ESEs results in exclusion of specific exons. By excluding specific sequences, a restored reading frame can be created. Antisense compounds represent the reverse and complementary sequences to mRNAs, and thus result in a double stranded hybrid with the mRNA (Figure 1B). Normally, this double stranded structure is destroyed by nucleases within cells, and it is this mechanism that can exploit antisense approaches to destroy a target mRNA. However, in DMD the goal is not to destroy the mRNA, but rather to skip selective exons to re-establish an open reading frame. Chemical modifications to antisense oligonucleotides improve the stability of RNA hybrids by avoiding nuclease-mediated degradation (Figure 1C). Drisapersen, a compound tested in DMD, used a specific chemistry called 2’-O-methyl-phosphorothioate-modified nucleotides to target exon 51. Drisapersen, which was delivered subcutaneously, was tested in 3 placebo controlled trials (> 300 DMD subjects) but failed to meet pre-specified clinical endpoints of prolonged 6 minute walk distance 2. Furthermore, renal toxicity, seen as proteinuria, and infusion site reactions were reported with the drisapersen studies, and the FDA did not approve the drug resulting in a halt in further development. Analysis of outcomes in DMD boys treated for 36 months has suggested potential benefit 4. The negatively charged residues found on 2’-O-methyl-phosphorothioates are thought to contribute to dose limiting toxicity.
Eteplirsen, which is delivered intravenously, uses an alternative chemistry referred to as phophorodiamidate morpholino (PMO). PMOs are neutral compounds and therefore can be dosed higher with fewer side effects (Figure 1B). In a phase 2 study, eteplirsen was tested in a small, short-term placebo controlled trial using a surrogate endpoint of dystrophin protein expression. The cohort of 12 subjects included three arms: two different doses administered weekly, and a placebo. At the end of 24 weeks, no functional benefit was seen using 6 min walk distance, but analysis of dystrophin expression in muscle biopsies suggested an increase of dystrophin expression in those who received eteplirsen. Based on these data, an open label extension phase ensued with all 12 subjects. After 48 weeks, there was a modest improvement in walk distance (67m) in patients who received eteplirsen from the outset as compared to placebo/delayed controls. After 36 months of open label extension, 83% of the 12 patients who received eteplirsen remained ambulatory while at this same age, based on historical data, a genotype-matched control group was half nonambulatory. In April 2016, these data were considered by an advisory board to the FDA at a public hearing, after which the majority of the advisory board did not find sufficient evidence for efficacy, although the advisory board was not uniform in its opinions. After internal debate, the FDA ultimately chose to approve eteplirsen and requested follow up data from an ongoing Phase III trial.
A major source of controversy surrounding eteplirsen effect was the degree of dystrophin protein produced, which was monitored from muscle biopsies. A re-analysis of the data by an FDA pathologist did not confirm the same high level of expression as originally reported, although did confirm an increase in dystrophin expression with treatment. Whether dystrophin exhibits a threshold effect, under which dystrophin content is nonfunctional is not known. Genetic studies suggest that even small amounts of dystrophin confer functional benefit since patients with “skippable” DMD mutations have longer ambulation. For example, mutations encompassing exon 44 produce a range of dystrophin protein expression, accompanied by a parallel range of clinical phenotype 5. Moreover, longer ambulation tends to correlate with improved cardiopulmonary function and preserved upper limb strength. Thus, even a modest increase of dystrophin expression is likely to be clinically meaningful. However, the cost of eteplirsen (estimated at $300,000 annually) and the cost-benefit equation appeared to factor into the regulators’ analysis.
In DMD, cardiomyocytes are adversely affected by loss of dystrophin. In the absence of dystrophin, cardiomyocytes, like skeletal myofibers, become unusually fragile and subject to breakdown. In DMD, the heart develops a progressive dilated cardiomyopathy. In principle exon skipping should work mechanistically the same in the heart as in muscle. However, current antisense chemistries are thought to have low tissue penetration into the heart, as compared to skeletal muscle, so eteplirsen may have little direct efficacy in the hearts of treated DMD patients. Indirect benefit, from improved respiratory muscle function may be gained from eteplirsen treatment. Newer chemistries and modifications for antisense technology are in development, with the intent of improving tissue distribution, reducing toxicity, and restricting conformation to adapt more favorable binding to target sequences. These next generation antisense chemistries are expected to allow lower and perhaps less frequent dosing and permit multi-exon skipping more readily and could have better cardiac penetration.
Ongoing studies are now aimed at testing additional antisense sequences, targeting the next most skippable exons in the DMD gene, and preclinical data exists to support development for a variety of other disorders. Recently, the neuromuscular field also welcomed FDA approval of nusinersen, an antisense treatment for Spinal Muscular Atrophy. Ultimately, many of these same targets for exon skipping may be suitable for more permanent gene editing strategies, where correction targets the DNA. There has been considerable preclinical progress in adapting CRISPr/Cas9 methods to in vivo use, and gene editing has an advantage of requiring perhaps only a single treatment. This first small step of eteplirsen approval paves the path for more efficient exon skipping and ultimately more permanent means of molecular correction.
Acknowledgments
Funding Sources
EMM and EW are partially supported by NIH HL61322.
Footnotes
Conflicts of Interests Disclosure
EMM and EW are co-inventors of a patent related to exon skipping.
References
- 1.Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, Dawkins H, Lamont L, Roy AJ, Chamova T, Guergueltcheva V, Chan S, Korngut L, Campbell C, Dai Y, Wang J, Barisic N, Brabec P, Lahdetie J, Walter MC, Schreiber-Katz O, Karcagi V, Garami M, Viswanathan V, Bayat F, Buccella F, Kimura E, Koeks Z, van den Bergen JC, Rodrigues M, Roxburgh R, Lusakowska A, Kostera-Pruszczyk A, Zimowski J, Santos R, Neagu E, Artemieva S, Rasic VM, Vojinovic D, Posada M, Bloetzer C, Jeannet PY, Joncourt F, Diaz-Manera J, Gallardo E, Karaduman AA, Topaloglu H, El Sherif R, Stringer A, Shatillo AV, Martin AS, Peay HL, Bellgard MI, Kirschner J, Flanigan KM, Straub V, Bushby K, Verschuuren J, Aartsma-Rus A, Beroud C, Lochmuller H. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Human Mutation. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aartsma-Rus A, Krieg AM. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Therapeutics. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony K, Arechavala-Gomeza V, Ricotti V, Torelli S, Feng L, Janghra N, Tasca G, Guglieri M, Barresi R, Armaroli A, Ferlini A, Bushby K, Straub V, Ricci E, Sewry C, Morgan J, Muntoni F. Biochemical characterization of patients with in-frame or out-of-frame DMD deletions pertinent to exon 44 or 45 skipping. JAMA Neurology. 2014;71:32–40. doi: 10.1001/jamaneurol.2013.4908. [DOI] [PubMed] [Google Scholar]
- 4.Goemans NM, Tulinius M, van den Hauwe M, Kroksmark AK, Buyse G, Wilson RJ, van Deutekom JC, de Kimpe SJ, Lourbakos A, Campion G. Long-Term Efficacy, Safety, and Pharmacokinetics of Drisapersen in Duchenne Muscular Dystrophy: Results from an Open-Label Extension Study. PloS One. 2016;11:e0161955. doi: 10.1371/journal.pone.0161955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bello L, Morgenroth LP, Gordish-Dressman H, Hoffman EP, McDonald CM, Cirak S. DMD genotypes and loss of ambulation in the CINRG Duchenne Natural History Study. Neurology. 2016;87:401–409. doi: 10.1212/WNL.0000000000002891. [DOI] [PMC free article] [PubMed] [Google Scholar]