Summary
The appearance of ten high-resolution cryo-EM maps of proteins, ribosomes, and viruses was compared to the experimentally phased crystallographic electron density maps of four proteins. We found that maps calculated at a similar resolution by the two techniques are quite comparable in their appearance, although cryo-EM maps, even when sharpened, seem to be a little less detailed. An analysis of models fitted to the cryo-EM maps indicated the presence of significant problems in almost all of them, including incorrect geometry, clashes between atoms, and discrepancies between the map density and the fitted models. In particular, the treatment of the atomic displacement (B) factors was meaningless in almost all analyzed cryo-EM models. Stricter cryo-EM structure deposition standards and their better enforcement are needed.
Keywords: electron density, cryo electron microscopy, resolution, structure quality
eTOC blurb
Wlodawer et al. analyzed the maps and models corresponding to the highest-resolution cryo-EM structures and found that whereas the resolution claims are mostly valid, the quality of the final models is frequently not fully acceptable and stronger deposition standards are needed.
Introduction
The three principal methods used to construct atomic models of macromolecules are crystallography (X-ray, neutron, and electron diffraction), nuclear magnetic resonance (NMR), and, most recently, cryo-electron microscopy (Cryo-EM). As of May 8, 2017 the Protein Data Bank (PDB) (Berman et al., 2000) contained almost 130,000 individual deposits, 116,200 obtained by X-ray crystallography (additional 113 by neutron diffraction and 65 by electron diffraction), 11,836 obtained by solution and solid state NMR, and 1510 by cryo-EM. The original three-dimensional electron density map for myoglobin, the first protein to have its structure determined, was computed using X-ray diffraction data extending to only 6 Å resolution (Kendrew et al., 1958). That map allowed general description of the structure and placement of the helices, but did not yield an atomic model. It required the subsequently computed 2 Å map (Kendrew et al., 1960) to successfully place the amino acid residues and provide a model that included all protein atoms. Of course, the technique of crystallography has undergone major changes and improvements in more than half a century since those pioneering efforts were first described, but it is still not possible to assign de-novo atomic coordinates to maps calculated at very low resolution, although it is possible to dock known atomic structures to such maps. The resolution limit that allows de novo building is difficult to define precisely, but an estimate might be provided by the successful tracing of the proteins at a resolution as low as 3.3–3.5 Å (Liu et al., 2017; Nozawa et al., 2017; Schulte et al., 2017), or at even much lower resolution of ~4 Å for a cryo-EM reconstruction of the flagellar-like filament of the archaeon Ignicoccus hospitalis (Braun et al., 2016).
Whereas NMR does not directly address the question of resolution (although NMR structures are often described in the literature as “high resolution”) (Wlodawer and Dauter, 2017), the concept of resolution is inherently relevant to both crystallography and cryo-EM. For crystallographic analyses, resolution refers to the extent of observable diffraction data, although the question where to terminate the data sets (and whether to use extremely weak high-resolution structure amplitudes) has not been completely settled (Karplus and Diederichs, 2012). Thus the limit of resolution may depend on the size and location of the detector, or indeed on statistical properties of the data in the highest resolution shell, described by factors such as Rmerge, Rmeas, CC1/2, or <I>/<σI>. An electron density map based on very strong data truncated at a particular resolution because of the detector limits may look better than a map at the same nominal resolution computed with data that were barely observable. Thus, although map resolution may provide some guidance to the expected accuracy of the model that was built into it, this indicator is not a perfect predictor of model quality.
The situation is even more complicated in the field of cryo-EM, a technique that is very quickly gaining prominence for large macromolecular structures (the current minimum size is on the order of 100,000 daltons (Merk et al., 2016), or may become even lower) and for structures of molecules that may concurrently assume several conformations. Whereas in the past electron microscopy was able to provide only crude images of the macromolecules, the arrival of new, very fast detectors capable of counting electrons directly, as well as of vastly improved and accelerated computations, allowed very significant improvement of the accuracy of structures determined by cryo-EM (Subramaniam et al., 2016). Thus about 150 sets of coordinates found in the PDB in May 2017 have already been determined at the claimed resolution of 3.5 Å or higher, 35 of which report resolution better than or equal to 3 Å.
However, a question needs to be raised about how the limits of resolution are derived for cryo-EM structures and whether such limits are defined in a way that is compatible with their utilization in crystallography. The most common way of estimating the resolution limits in cryo-EM is based on comparison of two independently computed maps, each based on half of the available images, and calculating their correlation (Rosenthal and Henderson, 2003; van Heel and Schatz, 2005). An objective criterion for the assessment of resolution involves Fourier transformation of two such maps and calculating shell correlations (FSC – Fourier shell correlation). Based on theoretical considerations, (Rosenthal and Henderson, 2003) proposed that FSC of 0.143 in the outermost shell should correspond to the claimed resolution limit, and this parameter has been generally accepted in subsequent publications.
The approach taken by us here is practical rather than based on theory – we have computed a series of experimental electron density maps based on diffraction data sets collected at different resolution from crystals of different proteins, and compared them with the deposited cryo-EM maps. Thus we do not attempt to provide a statistically meaningful comparison between the levels of details visible in these maps, but rely only on our subjective impression. Additionally, we discuss some other problems that appear to be common in structures determined by cryo-EM.
Results
The electron density maps for four crystal structures determined in our laboratories using single-wavelength anomalous diffraction (SAD) were used for assessing the dependence of the appearance of the maps on resolution of diffraction data. Data extending to the resolution between 1.8 and 0.94 Å were originally utilized in structure determination. The location of the anomalous scatterers was determined using the program SHELX independently of any previous work, and the electron density maps were improved by cycles of density modification. It must be stressed that these maps were based strictly on the experimental data without the utilization of any atomic coordinates for phasing, in contrast to the 2Fo-Fc maps that are used during crystal structure refinement and that do utilize model information in phase calculation. In that way, these experimentally derived maps are more equivalent to the cryo-EM maps, since both types of maps are not influenced by any previous knowledge about the macromolecular models that were fitted to them, other than the shape of molecular envelopes.
Fig. 1 shows selected regions of ferredoxin (Fig. 1A–C), insulin (Fig. 1D–F), sedolisin (Fig. 1G–I), and thioesterase (Fig. 1H–L) and the corresponding maps calculated with data extending to 1.8, 2.2, and 2.8 Å. All maps were calculated using SAD phases derived at the maximum resolution of data (see STAR Methods) and were contoured at 1σ level. Although the nominal resolution of the maps is the same in each column, the maps for ferredoxin and insulin appear to be more detailed than the maps for sedolisin and thioesterase, most likely due to better phases that resulted from complete data extending to higher resolution.
Figure 1.
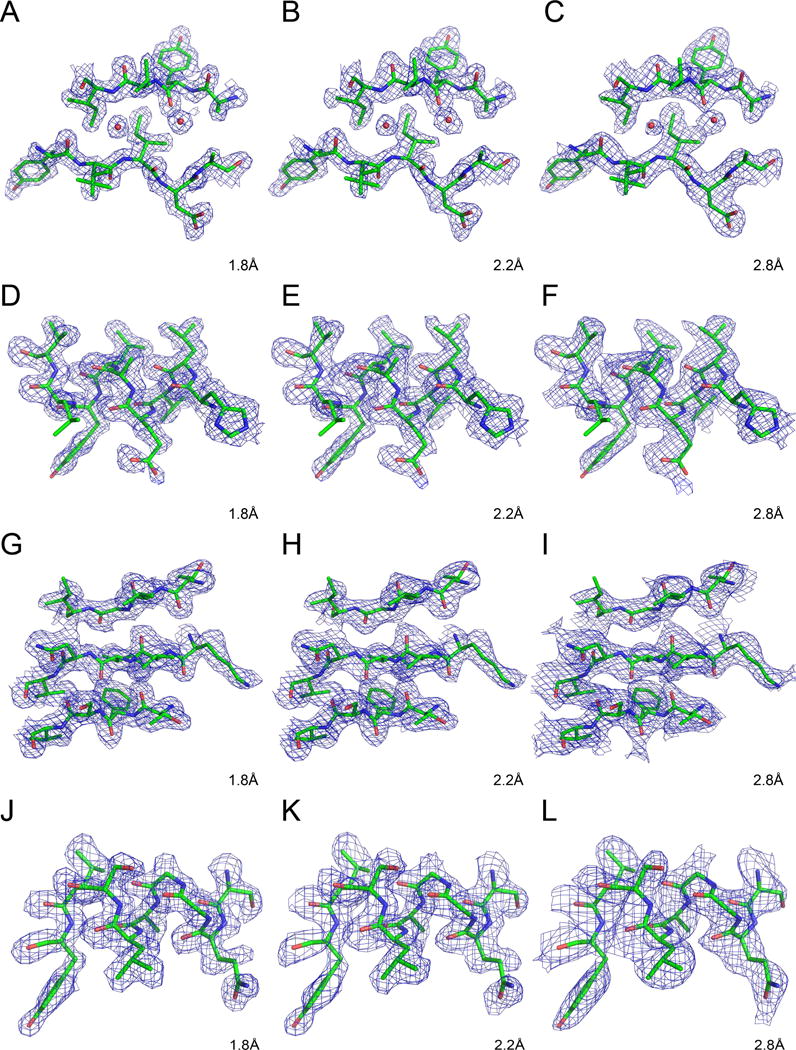
Experimentally derived electron density maps for ferredoxin (A–C), insulin (D–F), sedolisin (G–I), and thioesterase (J–L). All maps were calculated using identical grids and were contoured at 1σ level. Whereas data extending to the full resolution of the available data sets were used for phase calculation (see STAR Methods), they were truncated to the resolution shown in the respective panels for calculation of the electron density.
Ten high-resolution cryo-EM structures available in the PDB were selected for detailed analysis (Table 1). Nine of them were among the data sets that claimed the highest resolution (1.8–2.9 Å), whereas the tenth one was determined at a lower resolution (3.5 Å) and was included for comparison. The three highest resolution structures were solved in a single laboratory, whereas the other structures were determined in a number of different venues. Literature references are available for nine out of the ten structures listed in Table 1.
Table 1.
High-resolution cryo-EM structures selected from the PDB and used for the present assessment.
| Protein name | PDB ID | Resolution | Reference |
|---|---|---|---|
| Glutamate dehydrogenase | 5k12 | 1.8 Å | (Merk et al., 2016) |
| β - gal actosidase | 5a1a | 2.2 Å | (Bartesaghi et al., 2015) |
| P97 with inhibitor | 5ftj | 2.3 Å | (Banerjee et al., 2016) |
| T. cruzi 60S ribosomal subunit | 5t5h | 2.54 Å | (Liu et al., 2016b) |
| Rotavirus VP6 | 3j9s | 2.6 Å | (Grant and Grigorieff, 2015) |
| Human rhino virus C | 5k0u | 2.79 Å | (Liu et al., 2016a) |
| Leishmania large ribosomal subunit | 3jcs | 2.8 Å | (Shalev-Benami et al., 2016) |
| Grapevine fanleaf virus with nanobody | 5foj | 2.8 Å | — |
| Anthrax toxin protective antigen pore | 3j9c | 2.9 Å | (Jiang et al., 2015) |
| Human 20S proteasome core | 5a0q | 3.5 Å | (da Fonseca and Morris, 2015) |
The highest resolution cryo-EM structure available in the PDB in May 2017 was the 1.8 Å model of glutamate dehydrogenase (PDB ID 5k12)(Merk et al., 2016). The protein forms a hexamer (chains A–F) with both the coordinates and atomic displacement parameters (B factors) strictly constrained among the six molecules. The map available in the PDB (EMD-8194) is not sharpened, whereas both a sharpened (by the application of B = −90.0 Å2) and unsharpened maps were deposited in the EMDB. The figures in the publication (Merk et al., 2016) display the sharpened map. Whereas no map contour level is explicitly specified in the published figures, the contour level of 3σ recommended in the EMDB reproduces most published images. However, it appears that the images shown in Fig. 3 of (Merk et al., 2016) were contoured at a higher contour level (~3.5σ) to emphasize the holes in aromatic rings. The raw map (Fig. 2A) calculated for residues A192–A199, A381–A391, and waters A636 and A648 found in the core of the enzyme, contoured at 3σ level, shows clear density for the side chains but only rather indistinct bulges for the carbonyls, whereas the latter are much clearer in the sharpened map contoured at the same level (Fig. 2B). A puzzling aspect of the published coordinate set is the range of B factors, spread from 0.0 to over 165 Å2. Such large differences are seen even for bonded atoms, for example 1.47 and 165 Å2 for Cβ and Sγ of CysA89, respectively. Many atoms have a fixed B of 20.00 Å2 and the B factors for all water molecules are either 25.31 Å2 or 30.00 Å2, indicating that the values of the atomic displacement parameters included in this model are largely meaningless.
Figure 2.
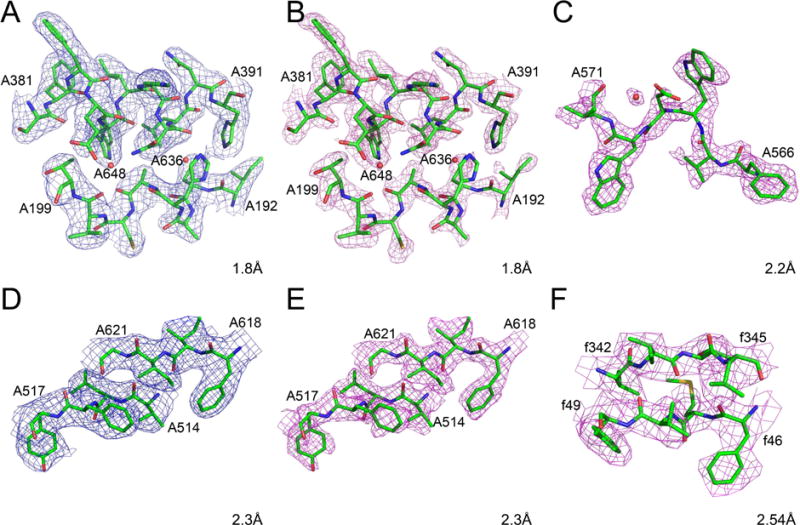
Cryo-EM maps and models for fragments of selected high-resolution protein structures. The raw maps are colored blue and the sharpened maps magenta. A–B) Glutamate dehydrogenase, residues A192-A199, A381-A391, and waters A636 and A648. C) β-galactosidase, residues A566-A571 and water A5035. D–E) P97 with inhibitor, residues A514-A517 and A618-A621. F) T. cruzi 60S ribosomal subunit, residues f46-f49 and f342-f345. The grid for all maps is as deposited, and the contour levels are 3σ, with the exception of panel F, contoured at 1.5σ.
The structure of β-galactosidase complexed with phenylethyl β-D-thiogalactopyranoside, a cell-permeable inhibitor, was determined by cryo-EM at the resolution of 2.2 Å (PDB ID 5a1a) (Bartesaghi et al., 2015). The protein is a homotetramer, with each protomer containing just over 1000 amino acid residues, and the structure was refined with strict inter-protomer constraints. It appears that only the sharpened map, EMD-2984 (B −75.0 Å2), is available in both the PDB and EMDB. The contour level recommended by the authors is 3σ, although the map contoured at this level appears to be too skinny. An example showing residues A566-A571 and water A5035 and the corresponding map is shown in Fig. 2C. Whereas a majority of the B factors in the deposited model are in the range of 10–40 Å2, a significant number of atoms have B factors of exactly 20.0, 30.0, and 100.0, often next to bonded atoms with very different B’s. Thus also in this case the B factors do not appear to have much physical meaning. A particularly interesting aspect of this structure is the presence of an inhibitor bound to the enzyme. However, when the map is contoured at the recommended level, only scattered density covers some of its atomic positions, and the fit of the inhibitor model to density is not fully convincing even when contoured at 1.5σ. A significant number of residues appear to have been fitted with incorrect side chain rotamers, the presence of several cis peptides is very unlikely, and the coordination of most metal ions is not optimal.
The structure of p97, a hexameric AAA+ ATPase (PDB ID 5ftj), has been determined at the resolution of 2.3 Å in complex with inhibitor UPCDC30245 (Banerjee et al., 2016). The sharpened map is available in the PDB (EMD-3295), but both that map and the uncorrected one are found in the EMDB. The recommended contour level of 3.4σ appears to be too high in the areas outside of the very center of each molecule. The model and the original map are shown in Fig. 2D, whereas the sharpened map covering the same region is shown in Fig. 2E. There appears to be quite significant discrepancy between the model and the density near the periphery of the molecule (for example, residues 609–616 do not fit the clearly visible density). Some cis peptides (for example, Ser462) are most likely wrong. As shown in Fig. 2 of the original publication, a large part of the inhibitor resides in very clear density, although the fit of the last visible aromatic ring to the density is not optimal. Also, the “3.2 Å” hydrogen bond between N10 of the inhibitor and the carboxylate of Glu498, marked in Fig. 2D of the original publication, must be in error since the actual distance is 3.99 Å and the alignment of the relevant atoms precludes the possibility of hydrogen bonding.
The structure of the Trypanosoma cruzi 60S ribosomal subunit was determined at 2.54 Å resolution (PDB ID 5t5h (Liu et al., 2016b)). Only one map, presumably the sharpened one (EMD-8361), is present in both the PDB and EMDB. The author-recommended contour level of 1.3σ appears to be correct. The fragment consisting of residues f46–f49 and f342–f345 is shown in Fig. 2F. Some side chains (for example, ArgY57, Metf47) are modeled with incorrect conformers, even though the density for them is quite clear. The B factors range from 0.0 to 80.00 A2, with a significant number set to either 30.00 or 20.00 A2. Mg ions have B factors of either 30.00 or 30.64 A2, thus the assignment of the atomic displacement factors is clearly incomplete.
Single-particle cryo-EM reconstruction of rotavirus VP6 (PDB ID 3j9s) was performed at 2.6 Å resolution with the aim of establishing the optimal exposure that would maximize the signal-to-noise ratio in the images (Grant and Grigorieff, 2015). The sharpened map (B −175 Å2) obtained by 13-fold averaging of the density for a VP6 trimer was submitted to the EMDB (EMD-6272) and PDB, but no original map is available. The recommended contour level is 3σ, although displaying the density at the 2σ level appears to agree better with the figures shown in the original publication. A map showing the fragment A167–A177 is shown in Fig. 3A. Since neither the residues nor the contour level are identified in the legend of Fig. 2 of (Grant and Grigorieff, 2015), it is not possible to directly reproduce them in order to verify how these images were created. No attempt to assign B factors was made and B values were set at 20.00 for all atoms. Several ions (Zn, Ca, Cl) and about a hundred water molecules were also modeled. Surprisingly, many water molecules do not seem to be residing in any density, even when contoured at extremely low levels, despite being shown in Fig. 2 of (Grant and Grigorieff, 2015).
Figure 3.
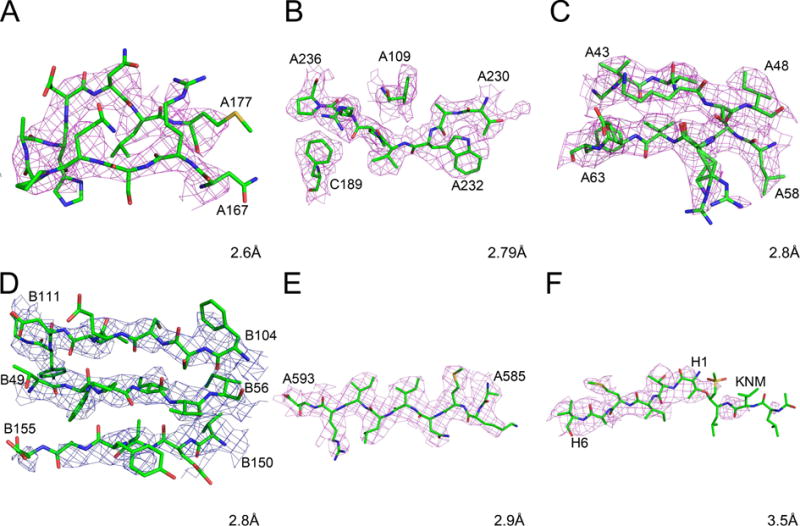
Cryo-EM maps and models for fragments of selected high-resolution protein structures. The raw maps are colored blue and the sharpened maps magenta. A) Rotavirus VP6, residues A167-A177. B) Human rhinovirus C, residues A230-A236, A109, and C189. C) Leishmania large ribosomal subunit, residues A43-A48 and A58-A63. D) Grapevine fanleaf virus complexed with a nanobody, residues B49-B54, B104-B111, and B150-B155. E) Anthrax toxin protective antigen pore, residues 585–593. F) Human 20S proteasome core, residues H1-H6 and the inhibitor KNM.
A 2.79 Å resolution map of rhinovirus C was reconstructed using 8973 icosahedrally averaged particles and sharpened using a B factor of −108.6 Å2 (Liu et al., 2016a). The PDB ID of the structure is 5k0u and only the sharpened map is present in the PDB and EMDB as EMD-8189. The recommended map contour level is 2.95σ, although the map in the area of the spike, shown in Fig. 1C of the original publication, was most likely contoured lower. The quality of the map is generally very high, due in part to extensive symmetry averaging. For example, the density in the area near TrpA232 is very clear (with a visible bump for the carbonyl), but its appearance suggests that both the main chain torsion angles and the rotamer of the side chain are incorrect (Fig. 3B). The four polypeptide chains correspond to the proteins VP1, VP2, VP3, and VP4. The range of B factors is from ~7 to over 100 Å2, with some well-defined side chains (for example, TrpC71) assigned B values of 30.00 Å2 for all atoms. The B factors for ~60 modeled water molecules (some of which are located in marginal density or too close to the protein) range from 13.50 to 57.28 Å2. In this model the values of B factors, excluding the unrefined ones, seem to represent the fit of the atoms to the density in a meaningful way.
The model of the large subunit of the Leishmania ribosome, determined at 2.8 Å resolution, consists of over 117,000 atoms (Shalev-Benami et al., 2016). Its PDB ID is 3jcs and only a sharpened map is available (EMD-6583). The recommended contour level is 3.3σ. The structure was refined in real space with Phenix (Adams et al., 2010), including group B refinement in which separate values were used for the main chain and the side chains of the proteins, as well as for the backbone and bases of RNA. The mean B was 39.2 Å2, with the range of ~5 Å2 to over 150 Å2. Surprisingly, double conformations of some residues (for example, ArgA60) were modeled (Fig. 3C), even though not supported by the density (and unlikely to be observable at this resolution).
The so far unpublished 2.8 Å resolution structure of a complex of grapevine fanleaf virus with nanobody was deposited in the PDB (ID 5foj) and a single map is found in the PDB and EMDB (EMD-3246). It is not clear whether this map is sharpened; its recommended contour level is 2.67σ (Fig. 3D). The model is characterized by a significant number of stereochemical violations, including unlikely cis peptide bonds, improper side chain rotamers, and a high clashscore. No solvent has been modeled. Density is particularly poor for the nanobody molecule (A). B factors are all less than 1.0 for molecule A (with the exception of the N-terminal residue, for which the main chain B is 30.00 Å2, and the side chain B 20.00 Å2), whereas B values for molecule B range from ~40 Å2 to ~130 Å2. These very different ranges of B factors do not make any physical sense.
The structure of the anthrax protective antigen pore was determined at the resolution of 2.9 Å (Jiang et al., 2015) and deposited in the PDB (ID 3j9c) and EMDB (EMD-6224). The seven-fold averaged map was sharpened with B of −95 Å2 and the recommended contour level is 2.8σ. This unusually looking protein consists of a globular head and a long tail consisting of a twisted β hairpin in each protomer, which forms a 14-stranded β-barrel. No solvent has been modeled and only two octahedrally coordinated Ca ions were included. The B factors range from 3.2 to ~440 Å2, with the highest values present between the middle and the bottom of the barrel. More than 30 residues forming the bottom part of the barrel are not in any density, even contoured at a very low level. However, the density is quite clear for the globular part of the molecule, although many side chains extend outside of the map (Fig. 3E).
The structure of a complex of the human 20S proteasome with a substrate analog (PDB ID 5a0q) was reported at a lower resolution of 3.5 Å (da Fonseca and Morris, 2015) and is included here for comparison with the higher resolution structures discussed above. The map (EMD-2981) was sharpened using B of −50 Å2 and Fourier low-pass filtered to 3.4 Å; the recommended contour level is 3.2σ. B factors for all atoms were set to 0.0 A2. Some parts of the structure, for example the C terminus of molecule A, do not appear to have been properly fitted to the density, since residues 232–234 are missing despite the presence of quite significant uninterpreted density in this region. The density covering the substrate analog, covalently bound to the N-terminal threonine residues in six protomers, is fragmented and unclear (Fig. 3F). As expected due to the much lower resolution, the appearance of the map is much less clear than in the other structures discussed above.
While we did not attempt to compare directly the cryo-EM maps with the reference electron density maps, it is possible to look at the appearance of some most distinctive residues to obtain visual clues of whether the maps represent well the resolution claim. In Fig. 4, a single tyrosine residue (Tyr2) in ferredoxin (Fig. 4A) is compared with a well-ordered tyrosine (TyrA382) in glutamate dehydrogenase, in the original (Fig. 4B) and sharpened (Fig. 4C) maps. The nominal resolution of all maps is the same 1.8 Å, but the residue shown in the electron density map seems to be slightly better outlined than in the cryo-EM maps. As expected, there is clear difference between the original and sharpened cryo-EM maps, with the latter appearing to be at a higher resolution than the former.
Figure 4.
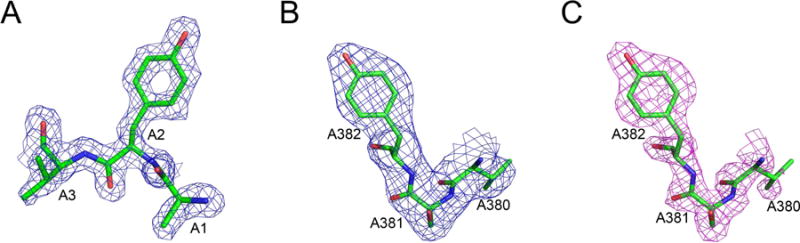
A comparison of the maps showing well-defined tyrosine residues in representative crystal and cryo-EM structures at 1.8 Å resolution. A) The N terminal residues Ala1-Tyr2-Val3 in ferredoxin, with a map contour level at 1σ. B–C) Residues ValA380-SerA381-TyrA382 in glutamate dehydrogenase. The original cryo-EM map (B) and the sharpened map (C) were both contoured at 3σ.
Whereas the appropriate contour level for displaying X-ray derived maps is about 1σ (where σ is the root-mean-square deviation from the average map level in the whole unit cell), such a level is about 3σ for nine of the inspected cryo-EM maps, and only for the structure 5t5h the appropriate level is 1.3σ. This difference seems to be related to the size of the “map box” (i.e. the P1 unit cell) surrounding the atomic model in the cryo-EM maps. If this box is much larger than the molecule and encompasses large regions with low, flat density, the average overall map variation (expressed as σ) is small and for the proper display of the model features the map level has to be raised to about 3σ. In the crystal structures the flat regions of bulk solvent are usually smaller, and the molecular features are prominent at the level close to 1σ. Indeed, the ribosomal subunit in the model 5t5h is placed in much tighter map box than in the other structures, resulting in the lower display level. The question of what is the appropriate map “margin” around the atomic model seems not to be generally agreed upon.
Discussion
It is well known that the definition of map resolution is not completely unambiguous even in the case of crystallographically determined electron density maps, and is even less clear in the case of cryo-EM maps. The appearance of the maps based on crystallographic data depends on both the extent of diffraction data and on the quality of the indirectly determined phases. Thus it is not surprising that maps calculated by us at the same 1.8 Å resolution appear to be better for insulin and ferredoxin, the two structures phased with data extending to truly atomic resolution (Wlodawer and Dauter, 2017), than for sedolisin and thioesterase, where the limit of the observable data has already been reached. Also, whereas a single descriptor of the resolution of crystallographic maps is routinely utilized, the quality of the maps is not uniform in space and the electron density is often less clear in some areas, especially on the surface of proteins, in places not involved in crystal contacts. The concept of variable resolution is better defined for the cryo-EM maps where the less clear areas are often referred to as corresponding to lower resolution, although a single descriptor relating to the apparent resolution limit in the best-defined regions of the maps is usually reported in the publications.
A comparison of the original and sharpened cryo-EM maps clearly shows that the former appear closer to lower-resolution electron density maps, whereas the appearance of the latter maps is more in line with crystallographic maps claiming the same resolution. However, the sharpened cryo-EM maps appear to have some less regular features and more noise than the crystallographic counterparts. The differences are most apparent for the bound solvent – whereas many solvent molecules can be seen in high-resolution electron density maps contoured at standard levels, contour levels need to be lowered in many cryo-EM maps to visualize the ordered solvent.
Some of the observations based on our analysis of the highest-resolution cryo-EM structures go beyond the question of their resolution, but reflect on the general quality of the final models deposited in the PDB. We were initially quite confused by not being able to reproduce the published features of some cryo-EM maps, for example for glutamate dehydrogenase, until we realized that the original map was available from the PDB, whereas both the original and the sharpened maps (the latter used by (Merk et al., 2016) to generate the figures) could be found in EMDB. We would like to postulate that, as long as the PDB links the coordinates to only one map, it should be the one that was interpreted in the publication. We are in agreement that deposition of four maps to EMDB (two maps computed with half of the data each, the full map, and the sharpened map) might allow fully independent validation of the structures, but might not be absolutely necessary, whereas even though map sharpening could be performed by the user since the value of the negative B factor can be usually found in the publications, such technical manipulations are clearly better performed by the original authors. Map sharpening may also become more difficult to reproduce if the use of different B factors for different parts of the map becomes an established practice.
The treatment of the atomic displacement factors (B factors) differs among these high-resolution cryo-EM structures, but most B factors found in the deposited models do not seem to represent any physical reality. It is well established that the meaning of B factors is quite clear in crystallography – they are correlated to the accuracy of the determination of the positions of individual atoms, and their values are influenced by the individual atomic motions, by global motions that may lead to local disorder, and by the presence of multiple conformations. The values of B factors are computed during the usual reciprocal-space refinement and they are normally restrained, such that they do not differ very significantly between covalently bonded atoms. However, if only real-space refinement is applied during creation of cryo-EM models, the values of B factors may not be explicitly computed (unless through correlation with map density) and may represent remainders of the initially placed models. It is quite clear that not much attention was paid to estimation of B factors in a majority of the structures analyzed by us, although methods for calculation of meaningful B factors from cryo-EM data have been described (DiMaio et al., 2015). In some models the B factors were set at zero or a fixed number, clearly indicating that they were not being considered, which might actually have been a reasonable approach. A completely different scale of B factors for the molecule of grapevine fanleaf virus and for the nanobody complexed with it makes no physical sense, and neither do the huge differences between the B factors of bonded atoms, found in the highest-resolution model of glutamate dehydrogenase. We would like to postulate that it might be better to leave B factors undefined than to deposit their values that cannot represent any physical reality. The treatment of B factors may improve in the future if reciprocal-space refinement of the cryo-EM models becomes a standard procedure (Hryc et al., 2017).
Another observation common to almost all the deposited models based on high-resolution maps is that they seem to lack the final quality control. The presence of very doubtful multiple conformations of the side chains, poor geometry of the model in comparatively clear regions of the maps, location of the side chains outside of the clear density, or the occurrence of interatomic clashes may indicate the difficulty of manual inspection of these very large structures, sometimes exceeding 100,000 independent atomic positions. Nevertheless, more attention needs to be paid to such problems that are not easily solved by purely automated means.
The analysis of the high-resolution cryo-EM structures presented here indicated that whereas some authors are trying to push the resolution to its limits, this is accomplished at the cost of paying less attention to the quality of the resulting models, and not only the maps. We would like to postulate that the standards of deposition of cryo-EM data should be strengthened – for example, by requiring that the map that was actually used for structure determination be deposited in the PDB, and both the original and sharpened map (and maybe also half-maps) be deposited in the EMDB. The final models should be evaluated by humans (and not only by computer programs) to assure the best fit of the model to the map, and the departures from standard geometry, as well as interatomic clashes, should be avoided. Finally, although the use of cryo-EM for the purpose of identification of ligands bound to their targets is one of the important stated aims of current research, the claims that the technique is now ready to make crystallography obsolete need to be taken with some skepticism. It is clear that cryo-EM has emerged during the last four years as a major breakthrough technology, but the expectations of what it could accomplish should be made more realistic.
STAR+METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information or reagents, including unpublished structure factors corresponding the crystallographic models, may be directed to the Lead Contact, Alexander Wlodawer (wlodawer@nih.gov)
METHODS DETAILS
Analyzed cryo-EM Datasets
The coordinates and electron density maps for selected highest-resolution cryo-EM structures were downloaded from the PDB in May 2017, and in several cases additional maps were downloaded from the Electron Microscopy Data Bank at the PDBe (EMDB) site. The 20S proteasome core (5a0q) was selected for comparison despite its lower resolution. With a single exception, the cryo-EM structures have been published and the methods for data collection and processing can be found in the relevant publications (see Table 1 for a list of references). The models and maps were displayed with the program Coot (Emsley and Cowtan, 2004), whereas the figures were prepared with PyMol (DeLano, 2002). Contour levels of the maps were usually set as recommended in EMDB, but on occasion were adjusted to clarify the images. The maps corresponding to only the internal parts of the macromolecules (expected to best represent the claimed resolution limits) were considered for the analysis.
Crystallographic Procedures and Data
The experimental electron density maps were calculated for the following four crystal X-ray structures, previously solved and published with the participation of the present authors:
Ferredoxin at 0.94 Å resolution (Dauter et al., 1997).
Insulin at 1.0 Å resolution (Dauter et al., 2002).
Sedolisin at 1.8 Å resolution (Dauter et al., 2001).
Thioesterase at 1.8 Å resolution (Devedjiev et al., 2000).
Maps for all four proteins were obtained after SAD phasing based on identification of appropriate anomalous scatterers by SHELXD (Sheldrick, 2008). The phase estimation and density modification was performed by SHELXE (Sheldrick, 2008) for ferredoxin, insulin, and sedolisin, and by PHASER (McCoy et al., 2007) for thioesterase.
Eight atoms of Fe (located within two 4Fe-4S clusters) were found in each molecule of ferredoxin, crystallized in space group P43212. Density modification was performed with data extending to 0.94 Å resolution, with solvent fraction set at 0.38, obtaining the final figure-of-merit (FOM) of 0.82.
Two Zn atoms of rhombohedral insulin (space group R3) were identified on the crystallographic 3-fold axis. Density modification was performed at 1.0 Å resolution with solvent content of 0.45, resulting in the final FOM of 0.81.
Unlike the above-mentioned two structures, which were phased based on anomalous scattering of intrinsically present atoms, sedolisin and thioesterase were derivatized by bromide soaking (Dauter et al., 2000). A hexagonal crystal of sedolisin (space group P62) was soaked for 30 s in a solution containing 1 M NaBr. Thirteen Br sites were identified and the density modification was performed at 1.8 Å resolution, with solvent content of 0.55, resulting in the final FOM of 0.73.
Crystals of thioesterase were monoclinic, space group P21. They were soaked for 20 s in a solution containing 1 M of NaBr. The number of Br sites found was 28, and phasing by PHASER at 1.8 Å resolution resulted in the final FOM of 0.43.
All electron density maps were calculated from the appropriate sets of amplitudes and phases for each of these structures, after truncating the data to the desired resolution limit.
Highlights.
We compare high-resolution cryo-EM and crystallographic electron density maps
Sharpened cryo-EM maps resemble their crystallographic counterparts
Most published high-resolution cryo-EM models have not to been refined to completion
We provide data-driven argument for defining & enforcing cryo-EM deposition standards
Acknowledgments
We would like to thank Professor Edward Egelman (University of Virginia) for his critical comments on this manuscript. This work was supported in part with Federal funds from the National Cancer Institute, NIH, under contract HHSN261200800001E and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Author contribution
AW and ZD conceived and performed this study. ML analyzed the data and created the figures.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, et al. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JL, Subramaniam S. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Vos MR, Kalisman N, Sherman NE, Rachel R, Wirth R, Schroder GF, Egelman EH. Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc Natl Acad Sci U S A. 2016;113:10352–10357. doi: 10.1073/pnas.1607756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Morris EP. Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nat Commun. 2015;6:7573. doi: 10.1038/ncomms8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauter Z, Dauter M, Dodson E. Jolly SAD. Acta Crystallogr. 2002;D58:494–506. doi: 10.1107/s090744490200118x. [DOI] [PubMed] [Google Scholar]
- Dauter Z, Dauter M, Rajashankar KR. Novel approach to phasing proteins: derivatization by short cryo soaking with halides. Acta Crystallogr. 2000;D56:232–237. doi: 10.1107/s0907444999016352. [DOI] [PubMed] [Google Scholar]
- Dauter Z, Li M, Wlodawer A. Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics. Acta Crystallogr. 2001;D57:239–249. doi: 10.1107/s0907444900015249. [DOI] [PubMed] [Google Scholar]
- Dauter Z, Wilson KS, Sieker LC, Meyer J, Moulis JM. Atomic resolution (0.94 A) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein. Biochemistry. 1997;36:16065–16073. doi: 10.1021/bi972155y. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Devedjiev Y, Dauter Z, Kuznetsov SR, Jones TL, Derewenda ZS. Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure Fold Des. 2000;8:1137–1146. doi: 10.1016/s0969-2126(00)00529-3. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y, Baker D. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryc CF, Chen DH, Afonine PV, Jakana J, Wang Z, Haase-Pettingell C, Jiang W, Adams PD, King JA, Schmid MF, et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc Natl Acad Sci U S A. 2017;114:3103–3108. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Pentelute BL, Collier RJ, Zhou ZH. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature. 2015;521:545–549. doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, Shore VC. Structure of myoglobin. A three-dimensional fourier synthesis at 2 Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, Wang Y. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168:121–134 e112. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hill MG, Klose T, Chen Z, Watters K, Bochkov YA, Jiang W, Palmenberg AC, Rossmann MG. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc Natl Acad Sci U S A. 2016a;113:8997–9002. doi: 10.1073/pnas.1606595113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford JL, Jr, et al. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc Natl Acad Sci U S A. 2016b;113:12174–12179. doi: 10.1073/pnas.1614594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallograhic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JL, et al. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Schneider TR, Cramer P. Core mediator structure at 3.4 A extends model of transcription initiation complex. Nature. 2017;545:248–251. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Schulte KW, Green E, Wilz A, Platten M, Daumke O. Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure. 2017;25:1025–1033 e1023. doi: 10.1016/j.str.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Shalev-Benami M, Zhang Y, Matzov D, Halfon Y, Zackay A, Rozenberg H, Zimmerman E, Bashan A, Jaffe CL, Yonath A, et al. 2.8-A Cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite leishmania. Cell Rep. 2016;16:288–294. doi: 10.1016/j.celrep.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Earl LA, Falconieri V, Milne JL, Egelman EH. Resolution advances in cryo-EM enable application to drug discovery. Curr Opin Struct Biol. 2016;41:194–202. doi: 10.1016/j.sbi.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wlodawer A, Dauter Z. ‘Atomic resolution’: a badly abused term in structural biology. Acta Crystallogr D Struct Biol. 2017;73:379–380. doi: 10.1107/S205979831700225X. [DOI] [PMC free article] [PubMed] [Google Scholar]


