Abstract
To investigate the role of Trim32 in traumatic brain injury (TBI), adult male Sprague Dawley (SD) rats and mice were randomly divided into sham (n = 6) and TBI groups (n = 24), respectively. Then, mice were assigned into Trim32 knockout mice (Trim32-KO [+/−]) and wild-type (WT) littermates. The TBI model used was the Feeney free-falling model, and neurological function was evaluated after TBI using a neurological severity score (NSS). Reverse transcription polymerase chain reaction (RT-PCR), Western blot, and immunohistochemistry were used to investigate the expression of Trim32 in the damaged cortex. Cell apoptosis in the cortex was detected by terminal-deoxynucleoitidyl transferase-mediated dUTP nick end labeling (TUNEL) staining. Moreover, Trim32-KO (+/−) mice were used to determine the effect of Trim in neurological repair after TBI. Results showed the NSS scores in TBI rats were significantly increased from day 1 to day 11 postoperation, compared with the sham group. Trim32 messenger RNA (mRNA) expression in the cortex was significantly increased at 7 d after TBI, while the level of Tnr and cytochrome c oxidase polypeptide 5A mRNA didn’t exhibit significant changes. In addition, Western blot was used to detect the level of Trim32 protein in the cortex. Trim32 expression was significantly increased at 7 d after TBI, and immunoreactive Trim32-positive cells were mainly neurons. Moreover, Trim32-KO (+/−) mice with TBI had lower NSS scores than those in the WT group from day 1 to day 11 postoperation. Meanwhile, Trim32-KO (+/−) mice had a decreased number of TUNEL-positive cells compared with the control group at 3 d postoperation. Protein 73 (p73) decreased at 7 d postoperation in Trim32-KO (+/−) mice with TBI, when compared with WT mice with TBI. Our study is the first to confirm that suppression of Trim32 promotes the recovery of neurological function after TBI and to demonstrate that the underlying mechanism is associated with antiapoptosis, which may be associated with p73.
Keywords: Trim32, motor function, traumatic brain injury, antiapoptosis
Introduction
With the development of transportation, the incidence of traumatic brain injury (TBI) is increasing with accidents.1 Although there has been continuous improvement in surgical treatments and innovative therapeutic drugs have been used to improve neural repair after TBI, the treatment results have been far from satisfactory for many patients. As a result, neurological impairments, such as motor dysfunction, learning, and memory loss, occur, which seriously affect the quality of life of TBI patients.1–3 Therefore, finding effective targets to reduce complications and further improve therapeutic outcomes are of vital importance for scientists and doctors.
Trim32, which belongs to the tripartite motif (Trim) protein family, is characterized with the RING finger, B-box, and coiled-coil domain structures that exist in this protein family, along with an additional NHL domain at the C terminus.4–6 It has been reported that Trim 32 acts as an E3 ligase for actin, a protein inhibitor of activated signal transducers and activators of transcription (STAT) y, dysbindin, and c-Myc. Mutations in Trim32 are responsible for several hereditary disorders including limb girdle muscular dystrophy type 2H, sarcotubular myopathy, and Bardet-Biedl syndrome,6–16 and it is also associated with diseases such as muscular dystrophy and epithelial carcinogenesis. In addition, a previous study showed that the expression of Trim32 is elevated in skin carcinogenesis, thus protecting keratinocytes from apoptosis, up to date by ultraviolet radiation b and tumor necrosis factor α.17 Recently, it has been reported that Trim32 has important and diverse functions in brain physiology and diseases, such as Alzheimer disease,18 neurogenesis,19 mood disorders.20 However, the role of Trim32 in TBI has not yet been reported.
Here, we screened the expression of Trim32 in TBI rats and TBI mice and then used Trim32 knockout mice (Trim32-KO [+/−]) to determine the role of Trim32 in neurological repair and possible mechanisms of action in an effort to ascertain whether Trim32 is a viable target for TBI therapy in the future.
Materials and Methods
Animals
Adult Sprague Dawley (SD) rats (weighing 180 to 220 g) and wild-type mice (weighing 20 to 40 g) were provided by the Animal Experimental Center of Kunming Medical University. Trim32-KO (+/−) mice (weighing 20 to 40 g) were provided by the molecular clinic institution (laboratory of Dr. Xiao). This research was approved by the Kunming Medical University. Animal Care and all procedures were performed in accordance with the guidelines of the Chinese Academy of Sciences. Animals were randomly divided into different groups as indicated in Tables 1 and 2.
Table 1.
Producer for Gene Expression between Sham and TBI in the Traumatic Penumbra Area of Rats or Mice.
| Groups | n | 1 hpo | 6 hpo | 24 hpo | 7 dpo |
|---|---|---|---|---|---|
| Sham | 6 | RT-PCR | |||
| WB | |||||
| TBI | 24 | RT-PCR | RT-PCR | RT-PCR | RT-PCR |
| WB | WB | WB | WB |
Abbreviations: TBI, traumatic brain injury; RT-PCR, reverse transcription polymerase chain reaction; WB, Western blot; hpo, hour(s) postoperation; dpo, day(s) postoperation.
Table 2.
Mice Grouping for Behavior, Morphology, and Molecule in Sham-, WT- and Trim32-KO(+/−)-TBI in the Traumatic Penumbra Area.
| Groups | n | 1 dpo | 3 dpo | 5 dpo | 7 dpo | 9 dpo | 11 dpo |
|---|---|---|---|---|---|---|---|
| Sham | 12 | NSS | NSS | NSS | NSS | NSS | NSS |
| TUNEL | PCR/WB | ||||||
| Trim32-KO (+/−)-TBI | 12 | NSS | NSS | NSS | NSS | NSS | NSS |
| TUNEL | PCR/WB | ||||||
| WT-TBI | 12 | NSS | NSS | NSS | NSS | NSS | NSS |
| TUNEL | PCR/WB |
Abbreviations: WT, wild type; TBI, traumatic brain injury; dpo, day(s) postoperation; WB, Western blot; Trim32-KO (+/−), Trim32 knockout mice; NSS, neurological severity score; PCR, polymerase chain reaction; TUNEL, terminal-deoxynucleoitidyl transferase-mediated dUTP nick end labeling.
TBI Model Preparation and Sample Collection
Experimental animals were anesthetized with 3.6% chloral hydrate (Sigma-Aldrich, St Louis, MO) by intraperitoneal (ip) injection (1 mL/100 g), and the hair from the top of the head was shaven using an electric shaving knife. The head skin was disinfected through routine use of medical iodine solution. The scalp was incised along the sagittal suture line and then bregma, after fontanelle and sagittal sutures, was clearly exposed. A bone window (5 mm in diameter) was opened with a dental drill via the midpoint of the bregma connection with the posterior fontanelle in the left margin 1 mm of the sagittal line. The animals were placed below free-fall hammer, and a cushion was placed into the bone window so as to touch the brain tissue. Then a 50-g hammer was allowed to fall down from a 30-cm high position to induce TBI in rats. After percussion, the crash lever was removed immediately, and the skin was sutured after hemostasis. Rats or mice in the sham group were only exposed to surgery for creating a bone window, without TBI. After surgery, mice were given ampicillin (dose 5 IU; Sigma-Aldrich) every day and rats were given daily ampicillin (dose 15 IU; Sigma-Aldrich) via ip injection for 3 d.
A total of 30 rats and 30 mice were euthanized at 1 h, 6 h, 24 h, and 7 d postoperation. The traumatic penumbra area from the ipsilateral injured brain and a similar area from the sham tissues were reserved for Western blot and reverse transcription polymerase chain reaction (RT-PCR; Table 1). A total of 18 mice were euthanized for standard gross at day 3 postoperation. All mice were transcardially perfused with 0.01 M phosphate-buffered saline (PBS), followed by 4% paraformaldehyde (PFA; Suolai Po Biological Technology Co., Ltd., Shanghai, China) in PBS for immunofluorescence. Then other mice were harvested freshly for PCR and Western blot (Table 2).
Behavioral Testing
The behavioral deficits after TBI were assessed according to neurological severity score (NSS). The traumatized rats were evaluated at 1, 3, 5, 7, and 11 d postoperation, and the traumatized mice were tested at 1, 3, 5, 7, 9, and 11 d postoperation. All behaviors were evaluated at 9:00 AM. Three observers were asked to perform NSS evaluation using a double-blind method, and the average score from 3 observers was calculated.
Reverse Transcription Polymerase Chain Reaction
Total RNA was isolated from the traumatic penumbra area from the ipsilateral injured brain of rats using TRIzol reagent (Invitrogen, Carlsbad, CA). Complementary DNA was synthesized by using Oligo (dT) 18 and moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI). Primers were designed by Primer Premier 5.0 software (Premier, Palo Alto, CA) and synthesized by Takara Biological Company (TaKaRa, Shiga, Japan), which are described in Table 3.
Table 3.
Primer Names, Sequences, and PCR Reaction Conditions.
| Gene | Primers | Temperature (°C) |
|---|---|---|
| Trim32 (rats) | Forward: 5′ CGGAGCATGGAAGTCACAG 3′ | 60 |
| Reverse: 5′ ACCACAGCCAGGAAACCC 3′ | ||
| Tnr (rats) | Forward: 5′ CCT GGT CGG GAA CAA AGT 3′ | 60 |
| Reverse: 5′ TTC ATC TGC CGC ACA ATT 3′ | ||
| COX5A (rats) | Forward: 5′ AGT TGG GAG GCT ATG TTG C 3′ | 60 |
| Reverse: 5′ TGC GGT TGG AAA TCT GTC 3′ | ||
| Trim32 (mouse) | Forward: 5′ GCATCCAGGAAGAGCTAG 3′ | 60 |
| Reverse: 5′ CTCTACCACTTGACTGTTG 3′ | ||
| TAp73 (mouse) | Forward: 5′ GCACCTACTTTGACCTCCCC 3′ | 60 |
| Reverse: 5′ GCACTGCTGAGCAAATTGAAC 3′ | ||
| GAPDH (rats) | Forward: 5′ GTC CTT GAT CAC CCG ATT C 3′ | 60 |
| Reverse: 5′ TCC TGT GTG CTT TCC ATT C 3′ | ||
| β-Actin (rats) | Forward: 5′ GAAGATCAAGATCATTGCTCCT 3′ | 52 |
| Reverse: 5′ TACTCCTGCTTGCTGATCCA 3′ | ||
| GAPDH (mouse) | Forward: 5′ TGTGTCCGTCGTGGATCTGA 3′ | 60 |
| Reverse: 5′ CCTGCTTCACCACCTTCTTGA 3′ | ||
| β-Actin (mouse) | Forward: 5′ TCACTATTGGCAACGAGCGGTTC 3′ | 52 |
| Reverse: 5′ GCACTGTGTTGGCATAGAGGTCTT 3′ |
Abbreviation: GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Western Blot
Standardized Western blot protocols were used to perform quantification of protein levels for each time point in the ipsilateral injured brain and a similar area from the sham tissues. After carefully rinsing in cooled PBS, these tissues were homogenized on ice in a lysis buffer (Biyuntian, Shanghai, China) containing 0.05 M Tris–HCl (pH7.4; Amresco, Solon, OH, USA), 0.5 M ethylenediaminetetraacetic acid (Amresco), 30% Triton X-100 (Amresco, Solon, OH, USA), NaCl (Amresco, Solon, OH, USA), 10% sodium dodecyl sulfate (Sigma-Aldrich), and 1 mM phenylmethanesulfonyl fluoride (Amresco, Solon, OH, USA) and centrifuged at 12,000 rpm (13362 g) for 15 min at 4 °C. The supernatant samples were frozen at −80 °C for later use. The protein concentrations were determined with all the samples on the same plate using the bicinchoninic acid protein assay kit and measuring absorbance at 562 nm with a SkanIt software 3.2 of ThemoType1510.
For quantification of the protein, 100 g of protein was loaded per well, then protein separation was performed using a 4% stacking gel followed by a 10% separation gel. Gels were run for 2 h at 100 V and 4 °C with constant stirring. Following gel separation, proteins were transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA), a wet transfer setup at 350 mA for 2.5 hours. Following protein transfer to PVDF membrane, blots were washed 3 times with Tris-buffered saline (TBS) and blocked with 5% milk in TBS for 2 h at room temperature. Then, PVDF membrane was washed 3 times with 0.1% Tween-20 + TBS (TBST) at room temperature with constant agitation. The primary antibody for Trim32 (1:300; Abcam, Cambridge, United Kingdom) and protein 73 (p73) (1:500; Abcam) was applied and incubated overnight at 4 °C. Following incubation with the primary antibody, the membrane was washed 3 times with TBST for 10 min each, again. Lastly, the membrane was incubated with a horseradish peroxidase–conjugated goat antirabbit immunoglobulin G (IgG) (1:5,000; Vector Laboratories, Lowellville, Ohio, USA) for 2 h at room temperature and washed as described above. Then membrane was developed in ECL kit, and pictures were captured by Bio-Gel Imagining system (Horcules, CA, USA) equipped with Image Lab software (NIH, Bethesda, MD, USA). Densitometry analysis for Trim32 and p73 protein was performed. β-Tubulin (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as internal control.
Immunohistochemistry
To detect the localization of Trim32 in the cortex, rats were deeply anesthetized and transcardially perfused with 4% PFA in 0.05 M PBS. Then brain was harvested and fixed in PFA at 4 °C. For immunofluorescent staining, frozen sections of 15-mm thickness were cut with a vibratome microtome (Leica CM1900, Watzlar, Hesse-Darmstadt, Germany), and sections were collected in wells containing 0.05 M PBS. Trim32 antibody (1:500, rabbit; ZSGB, Bei Jing, China) was used to label cells in the cortex, and NeuN antibody (1:500, mouse; ZSGB, Bei Jing, China) was used, also. After incubation with primary antibody, the sections were incubated with fluorescent secondary antibody working solution 488 (1:400, antirabbit; ZSGB, BeiJing, China; 1:400, antimouse; Invitrogen) for 30 min at 37 °C. Sections were rinsed as described above and coverslipped with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). Images were acquired with a Leica DMI 6000 B inverted microscope.
TUNEL Staining
Sections from each group at 3 d after TBI (Trim32-KO [+/−], n = 3; wild type, n = 3; sham-TBI, n = 3) were analyzed by terminal-deoxynucleoitidyl transferase-mediated dUTP nick end labeling (TUNEL) assay. The broken fragment was labeled using the In Situ Cell Death Detection Kit (fluorescence; Roche, Mannheim, Germany), according to the manufacturer’s instructions. TUNEL-positive nuclei were counted in the cortex in 3 coronal sections for each animal, with 3 animals per group.
Statistical Analysis
All data were expressed as the mean ± standard deviation . They were analyzed using 1-way analysis of variance and least significant difference (LSD) q test using the SPSS 17.0 software package (IBM, Armonk, NY). Statistical significance was defined as P < 0.05.
Results
Neurobehavioral Evaluation in TBI-Induced Rats
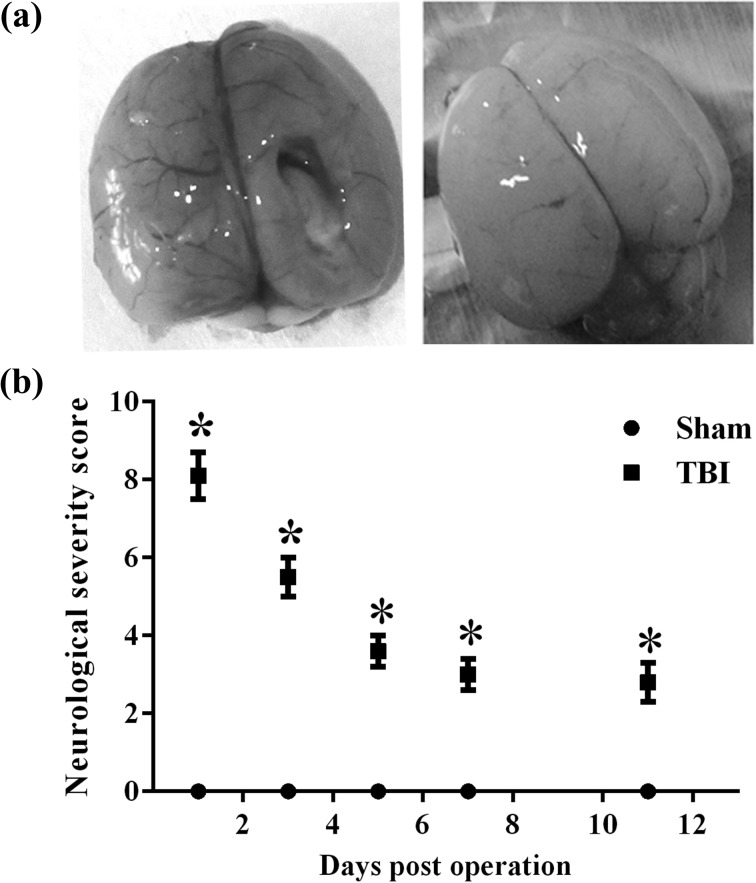
The neurological function deficits in both sham and TBI groups in rats were recorded. A leading increase in NSS scores was found in the TBI group, compared to the sham group, especially at 1, 3, and 7 d post-TBI, P < 0.05 (Fig. 1). However, there was a restorative tendency in NSS scores as time passed after injury.
Figure 1.
The neurological severity score (NSS) score of rats at different time points after traumatic brain injury (TBI). (a) The morphologic change of traumatic brain injury (TBI) in rats. (b) The NSS score of the TBI group was significantly increased, especially from 1 to 7 d after TBI. *P < 0.05.
Upregulation of Trim32 in Rats and Mice
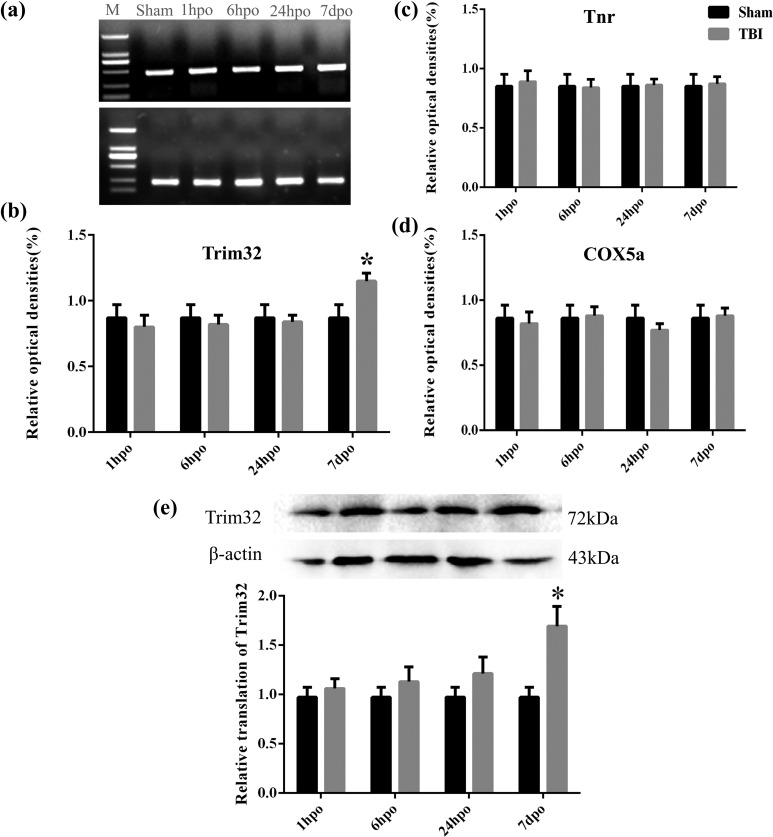
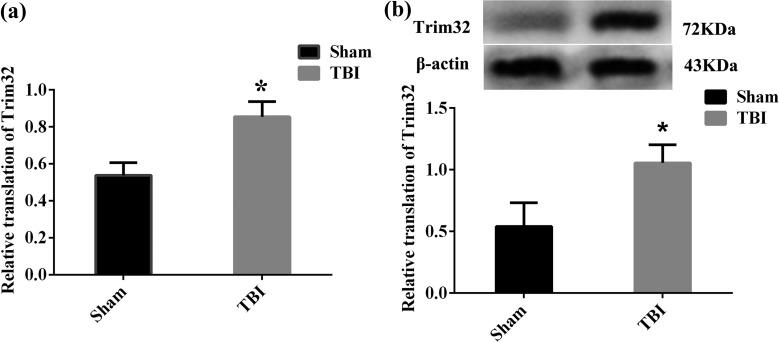
The messenger RNA (mRNA) expression of Trim32 in the cortex in the TBI group was detected by RT-PCR. Results indicated that it was enhanced at 7 d postoperation, compared with the sham group, and that it was significantly increased at 7 d postoperation, compared with 6 and 24 h postoperation, P < 0.05 (Fig. 2a, b). However, the mRNA expression of Tnr and cytochrome c oxidase polypeptide 5A (COX5A) was not significantly changed at 1 h, 6 h, 24 h, and 7 d postoperation (Fig. 2c, d). Moreover, the protein level of Trim32 in injured cortex detected by Western blot showed that it was significantly increased at 7 d postoperation, compared with other time points, P < 0.05 (Fig. 2e). Meanwhile, the mRNA and protein levels of Trim32 in TBI mice also increased at 7 d postoperation, compared to the sham group, P < 0.05 (Fig. 3a, b).
Figure 2.
The expression of Tnr, Trim32, and cytochrome c oxidase polypeptide 5A (COX5A) at different time points after traumatic brain injury (TBI) in rats. (a) Reverse transcription polymerase chain reaction (RT-PCR) results in injured cortex and sham-operated rats. β-Actin was used as control. Marker was in the left side and then from left to right is sham, 1 h, 6 h, 24 h, and 7 d postoperation, respectively. The bands (7 d postoperation) are obviously brighter than others. (b to d) The messenger RNA (mRNA) expression of Trim32, Tnr, and COX5A in the cortex. β-Actin was used as control. (e) This is to exhibit the protein level of Trim32. β-Actin was used as control. Lane 1 in the left side displays the protein band in the sham-operated group, and lanes 2 to 5 show the protein band at 1 h, 6 h, 24 h, and 7 d postoperation, respectively. The relative translation of Trim32 was higher in 7 d postoperation. *P < 0.05 compared with the sham group.
Figure 3.
The expression of Trim32 at 7 d after traumatic brain injury (TBI) in mice. (a) Reverse transcription polymerase chain reaction (RT-PCR) results in the injury cortex and sham-operated mice. β-Actin was used as control. (b) This is to exhibit the protein level of Trim32. β-Actin was used as control. Lane 1 in the left side displays the protein band in the sham group, and lane 2 shows the protein band at 7 d postoperation. The relative translation of Trim32 was higher at 7 d postoperation. *P < 0.05 compared with the sham group.
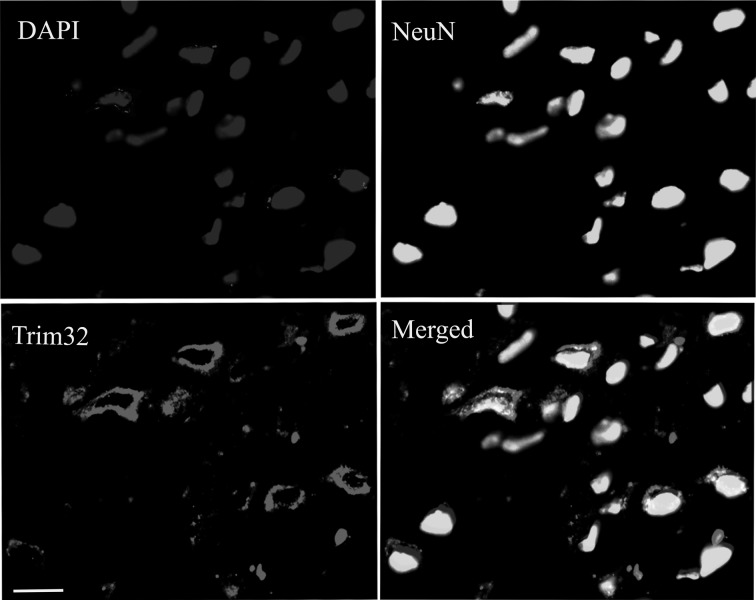
Immunofluorescent Staining for Trim32 and NeuN
Immunofluorescent staining showed that Trim32-positive reactants were mainly distributed in the cytoplasm of neurons in the cortex. Most of neurons in the cortex expressed Trim32, according to the results of the cytoplasm staining (Fig. 4).
Figure 4.
The results of immunohistochemical staining for 4′,6-diamidino-2-phenylindole (DAPI), NeuN, and Trim32. The distribution of NeuN and Trim32 in damage cortex at 11 d posttrauma of mice. Bars = 20 μm (shown in Trim32), also applied to other panels.
Neurobehavioral Evaluation of Mice
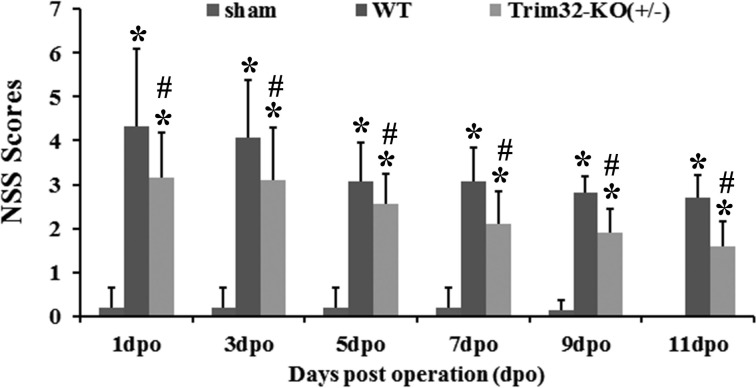
Deficits in neurobehavioral function in TBI-induced mice were present but gradually improved over time. Moreover, we can see that the NSS scores of Trim32-KO (+/−) mice exhibited a significant improvement at 1, 3, 7, 9, 11 d after injury compared to the scores of WT-TBI mice (P < 0.05 compared with the sham group, P < 0.05 compared with the WT-TBI group; Fig. 5).
Figure 5.
The neurological severity score (NSS) score at different time points after traumatic brain injury (TBI) from contusion mice and Trim32 knockout mice (Trim32-KO [+/−]) mice. The NSS in wild-type (WT)-TBI mice was significantly higher than in Trim32 knockout mice (Trim32-KO [+/−]) mice at 1, 3, 5, 7, 9, 11 d postoperation. *P < 0.05 compared with the sham group; #P < 0.01 compared with the WT-TBI group.
TUNEL Detection
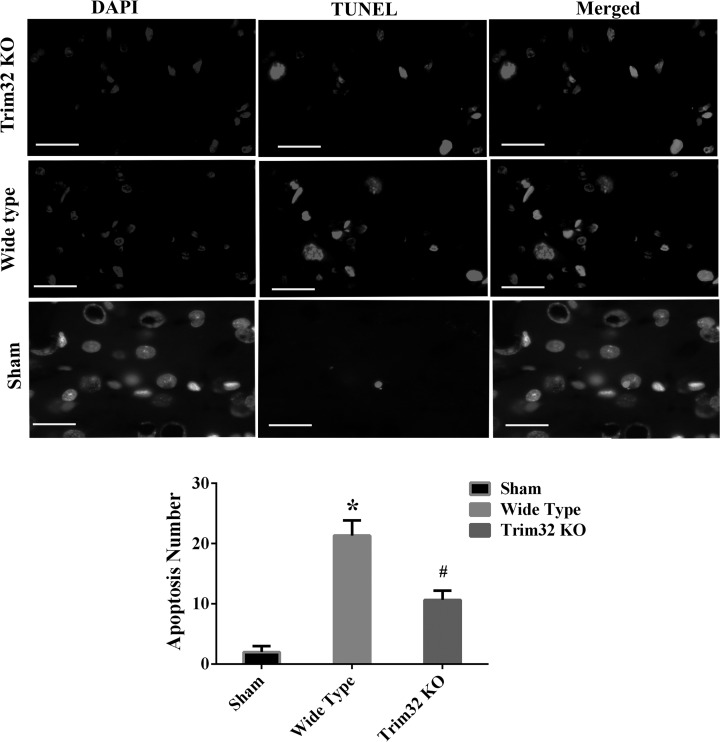
In the sham-operated cortex, there were hardly any apoptotic cells that could be seen. However, the TUNEL-stained positive cells from TBI mice presented a significant increase compared to sham-operated mice at 3 d, which showed a statistic significance postinjury (P < 0.05). The number of TUNEL-stained positive cells in Trim32-KO (+/−) mice was significantly suppressed compared to WT-TBI mice (P < 0.05; Fig. 6).
Figure 6.
TUNEL staining in the cortex at 3 d from traumatic brain injury (TBI) and Trim32 knockout mice (Trim32-KO [+/−]) TBI mice. This is to exhibit the results of TUNEL staining in sham-operated, wide-type TBI, and Trim32 KO (+/−) TBI mice. Bars = 50 μm in the pictures of Trim32 KO and wild-type (WT) group. Bars = 20 μm in the images of sham group, applied to all panels. *P < 0.05 compared with the sham group; #P < 0.05 compared with the WT-TBI group; TUNEL, terminal-deoxynucleoitidyl transferase-mediated dUTP nick end labeling.
The Expression of p73
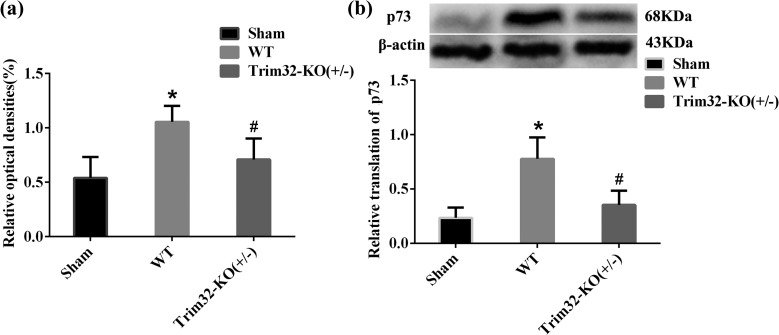
The mRNA expression of p73 in the cortices of WT mice was detected by RT-PCR, and results indicated that it was enhanced at 7 d postoperation, compared to the sham group. The mRNA expression of p73 in the cortex in Trim32-KO (+/−) mice decreased at 7 d postoperation compared to WT mice (P < 0.05; Fig. 7).
Figure 7.
The expression of p73 at 7 d after traumatic brain injury (TBI) from TBI mice and Trim32 knockout mice (Trim32-KO [+/−]) TBI mice. (A) Reverse transcription polymerase chain reaction (RT-PCR) results of p73 in TBI mice and Trim32 KO (+/−) TBI mice. β-Actin was used as control. (B) This is to exhibit the protein level of p73. β-Actin was used as control. Lane 1 in the left side displays the protein band in the sham group, lane 2 shows the protein band in wild-type (WT)-TBI mice, and lane 3 shows the protein band in Trim32 KO (+/−) TBI mice. The relative translation of p73 was lower in Trim32 KO (+/−) TBI mice. *P < 0.05 compared with the sham group; #P < 0.05 compared with the WT-TBI group.
Discussion
In this study, using free-fall hammer-controlled cortical impact device, we induced a TBI model, which can cause impairment in the animals’ motor function. Neurobehavioral assessment indicated that the motor function of rats and mice was substantially damaged after TBI, as evidenced by NSS scores. Moreover, Trim32, a crucial molecule that has not been previously reported in the context of TBI, has been found to be enhanced in the cortical regions of the brain. These findings show that trauma may deteriorate brain cells, which results in a substantial impairment of brain function. The underlying mechanism for this process may be related to the expression of multiple genes, and Trim32 may be an important gene to evaluate. In order to determine the role of Trim32 in TBI events, we utilized the Trim32 knockout mouse to explore the function of Trim32 in TBI. We found that knockout of Trim32 inhibited neuronal apoptosis and improved behavior in TBI mice. The findings in the present study therefore suggest that Trim32 may play a crucial role in TBI and that administration of Trim32 could be useful for the treatment of neurological disease.
Neurological Deficits Were Induced in TBI Condition
In this study, TBI caused motor deficits that were a significant behavioral problem. Enhanced NSS scores indicated a severe deterioration in neurological function. Previous studies have confirmed that TBI results in neurological deficits and deterioration of physiological function that contribute to decreased quality of life in mice, rats, monkeys, and human beings.2,3 Therefore, controlling the development of TBI-induced deficits in the cortex and protecting neurons from secondary injury to maintain neurological function are very important objectives for doctors and scientists. Before clinical trials commence, mechanistic studies of TBI-induced secondary injury are important, and key molecular events should be elucidated. In the present study, we screened the expressions of Trim32, Tnr, and COX5A and determined that Trim32 may be an important molecular target in TBI-induced cell injury.
Indication of Trim32 Expression in Cortex after TBI
The level of mRNA for Trim32 in the trauma penumbra of the cortex in rats and mice was distinctly elevated at 7 d postoperation, while TNR and COX5A mRNA expression level had no overt change at each time point after injury; Western blot also exhibited an increased Trim32 protein level in the cortex of both rats and mice at 7 d postoperation. This experiment showed that endogenous Trim32 was triggered after TBI. It has been reported that the expression of Trim32 is elevated during mouse skin carcinogenesis17 and that Trim32 is elevated both in the epidermal lesions of human psoriasis21 and in muscle undergoing remodeling due to changes in weight bearing.22 Here, we found that both mRNA and protein levels of Trim32 were upregulated at 7 d postoperation, which demonstrated that Trim32 might play an important role in the lesions or recovery process in the brain after TBI. Our findings are the first to show changes in Trim32 in the cortex after TBI, suggesting that Trim32 may be a critical molecule in TBI events.
Trim32 Knockout Improves Neurological Function in TBI Mice
The NSS scores of both WT and Trim32-KO (+/−) mice with TBI were gradually reduced at different time points after injury, but the NSS scores of Trim32-KO (+/−) transgenic mice with TBI exhibited significant lower scores than that of WT-TBI mice at 1, 3, 7, 9, and 11 d posttrauma. This demonstrated Trim32 had a neuroprotective effect on TBI-induced mice. As no literature could be found on the relationship between Trim32 and TBI, we hypothesized the effect of Trim32 in TBI may be related to cell proliferation and apoptosis, as Trim32 is involved in cell differentiation, oncogenesis, and apoptosis. Here, we first discovered that knockout of the Trim32 gene can significantly improve neurological function in TBI mice, and the underlying mechanism is regulation of cell apoptosis.
TUNEL Staining Deduced the Role of Trim32 in TBI
We showed that TUNEL-stained positive cells in trauma penumbra of WT-TBI mice were significantly increased compared to those of sham TBI mice at 3 d after injury. In Trim32-KO (+/−) mice, TUNEL-stained positive cells were significantly decreased compared to WT-TBI mice at the same time postinjury. This suggests that Trim32 downregulation can effectively reduce cellular apoptosis in the cortex after TBI. Previous studies have shown that members of the Trim protein family are involved in various cellular processes, such as cell proliferation, differentiation, development, oncogenesis, and apoptosis.23 Trim32 overexpression enhances X-linked inhibitor of apoptosis ubiquitination and subsequent proteasome-mediated degradation, whereas Trim32 knockdown has the opposite effect24; TAp73 binds to the Trim32 promoter and activates its expression,25 and Trp73, a member of the protein 53 (p53) gene family, plays a crucial role in neural development.26 Both WT p73a and p73b polypeptide expression can induce cellular apoptosis in SAOS2 of osteosarcoma cells and BHK cell lines of baby hamster kidney cells.27,28 Therefore, we believe that TAp73 can activate p53 in the same way as the p53 gene p21,26 inhibiting cell proliferation and inducing apoptosis. Trim32 downregulation reduced apoptosis, possibly through feedback inhibition of TAp73, which is consistent with our result pertaining to p73.
Trim32-Positive Reactants Were Mainly Localized in Neurons
Our experimental results show that Trim32-positive reactants were mainly located in neurons. Various reports indicated that Trim32 was present in the mitochondria and endoplasmic reticulum,29 which are plentiful in neurons. Therefore, there exists an important link between neurons and Trim32. Trim32 binds to the protein kinase C and is retained in the cytoplasm of neural cells.30 In addition, it was found that the Trim32 protein was localized in the skeletal muscle cytosol.31,32 Our finding is consistent with the literature, as we showed that Trim32 is located in neurons in the central nervous system.
A recent study found that overexpression of Trim32 in HL60 cells suppressed cellular proliferation,33 while overexpression of Trim32 in mouse neuroblastoma cells resulted in enhancement of neural differentiation.34 Moreover, Trim32 deficiency induced more neural progenitor cell proliferation and less cell death,19 and Trim32 facilitated cell growth and migration via degradation of Abl-interactor 2.35 Therefore, the role of Trim32 varies under different conditions.
Conclusion
In summary, TBI causes severe motor deficits and downregulation of Trim32 facilitated recovery of neurological function. The mechanism underlying recovery most likely involves suppression of neuronal apoptosis in the injured cortex.
Footnotes
Author Contributions: Zi-Bin Zhang and Liu-Lin Xiong contributed equally to this work.
Ethical Approval: The protocols in this study were approved by the relevant ethics committee (see Materials and Methods).
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: This research was supported by the Program for IRTSTYN, together with the program Innovative Research Team in Science and Technology in Yunnan province and a grant from the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (CN; No. 2014BAI01B10).
References
- 1. Chang TP, Nager AL. Pediatric traumatic brain injury: the utility of beta-natriuretic peptide. J Trauma. 2010;68(6):1401–1405. [DOI] [PubMed] [Google Scholar]
- 2. Namjoshi DR, Good C, Cheng WH, Panenka W, Richards D, Cripton PA, Wellington CL. Towards clinical management of traumatic brain injury: a review of models and mechanisms from a biomechanical perspective. Dis Model Mech. 2013;6(6):1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dewall J. The ABCs of TBI. Evidence-based guidelines for adult traumatic brain injury care. JEMS. 2010;35(4):54–61; quiz 63. [DOI] [PubMed] [Google Scholar]
- 4. Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci. 1998;23(12):474–475. [DOI] [PubMed] [Google Scholar]
- 5. El-Husseini AE, Vincent SR. Cloning and characterization of a novel RING finger protein that interacts with class V myosins. J Biol Chem. 1999;274(28):19771–19777. [DOI] [PubMed] [Google Scholar]
- 6. Frosk P, Weiler T, Nylen E, Sudha T, Greenberg CR, Morgan K, Fujiwara TM, Wrogemann K. Limb-girdle muscular dystrophy type 2 H associated with mutation in Trim32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet. 2002;70(3):663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kudryashova E, Struyk A, Mokhonova E, Cannon SC, Spencer MJ. The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum Mol Genet. 2011;20(20):3925–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liewluck T, Tracy JA, Sorenson EJ, Engel AG. Scapuloperoneal muscular dystrophy phenotype due to TRIM32-sarcotubular myopathy in South Dakota Hutterite. Neuromuscul Disord. 2013;23(2):133–138. [DOI] [PubMed] [Google Scholar]
- 9. Shieh PB, Kudryashova E, Spencer MJ. Limb-girdle muscular dystrophy 2 H and the role of TRIM32. Handb Clin Neurol. 2011;101:125–233. [DOI] [PubMed] [Google Scholar]
- 10. Muller J, Stoetzel C, Vincent MC, Leitch CC, Laurier V, Danse JM, Helle S, Marion V, Bennouna-Greene V, Vicaire S, et al. Identification of 28 novel mutations in the Bardet-Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Hum Genet. 2010;127(5):583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borg K, Stucka R, Locke M, Melin E, Ahlberg G, Klutzny U, Hagen M, Huebner A, Lochmuller H, Wrogemann K, et al. Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum Mutat. 2009;30(9):E831–E844. [DOI] [PubMed] [Google Scholar]
- 12. Cossee M, Lagier-Tourenne C, Seguela C, Mohr M, Leturcq F, Gundesli H, Chelly J, Tranchant C, Koenig M, Mandel JL. Use of SNP array analysis to identify a novel TRIM32 mutation in limb-girdle muscular dystrophy type 2 H. Neuromuscul Disord. 2009;19(4):255–260. [DOI] [PubMed] [Google Scholar]
- 13. Saccone V, Palmieri M, Passamano L, Piluso G, Meroni G, Politano L, Nigro V. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2 H. Hum Mutat. 2008;29(2):240–247. [DOI] [PubMed] [Google Scholar]
- 14. Frosk P, Del Bigio MR, Wrogemann K, Greenberg CR. Hutterite brothers both affected with two forms of limb girdle muscular dystrophy: LGMD2H and LGMD2I. Eur J Hum Genet. 2005;13(8):978–982. [DOI] [PubMed] [Google Scholar]
- 15. Schoser BG, Frosk P, Engel AG, Klutzny U, Lochmuller H, Wrogemann K. Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2 H. Ann Neurol. 2005;57(4):591–595. [DOI] [PubMed] [Google Scholar]
- 16. Nigro V. Molecular bases of autosomal recessive limb-girdle muscular dystrophies. Acta Myol. 2003;22(2):35–42. [PubMed] [Google Scholar]
- 17. Albor A, El-Hizawi S, Horn EJ, Laederich M, Frosk P, Wrogemann K, Kulesz-Martin M. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2 H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J Biol Chem. 2006;281(35):25850–25866. [DOI] [PubMed] [Google Scholar]
- 18. Yokota T, Mishra M, Akatsu H, Tani Y, Miyauchi T, Yamamoto T, Kosaka K, Nagai Y, Sawada T, Heese K. Brain site-specific gene expression analysis in Alzheimer’s disease patients. Eur J Clin Invest. 2006;36(11):820–830. [DOI] [PubMed] [Google Scholar]
- 19. Hillje AL, Pavlou MA, Beckmann E, Worlitzer MM, Bahnassawy L, Lewejohann L, Palm T, Schwamborn JC. TRIM32-dependent transcription in adult neural progenitor cells regulates neuronal differentiation. Cell Death Dis. 2013;19(4):e976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hillje AL, Beckmann E, Pavlou MA, Jaeger C, Pacheco MP, Sauter T, Schwamborn JC, Lewejohann L. The neural stem cell fate determinant TRIM32 regulates complex behavioral traits. Front Cell Neurosci. 2015;18(9):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y, Lagowski JP, Gao S, Raymond JH, White CR, Kulesz-Martin MF. Regulation of the psoriatic chemokine CCL20 by E3 ligases Trim32 and Piasy in keratinocytes. J Invest Dermatol. 2010;130(5):1384–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2 H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354(2):413–424. [DOI] [PubMed] [Google Scholar]
- 23. Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3(10):799–808. [DOI] [PubMed] [Google Scholar]
- 24. Ryu YS, Lee Y, Lee KW, Hwang CY, Maeng JS, Kim JH, Seo YS, You KH, Song B, Kwon KS. TRIM32 protein sensitizes cells to tumor necrosis factor (TNFalpha)-induced apoptosis via its RING domain-dependent E3 ligase activity against X-linked inhibitor of apoptosis (XIAP). J Biol Chem. 2011;286(29):25729–25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez-Cano L, Hillje AL, Fuertes-Alvarez S, Marques MM, Blanch A, Ian RW, Irwin MS, Schwamborn JC, Marin MC. Regulatory feedback loop between TP73 and TRIM32. Cell Death Dis. 2013;4:e704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. El Husseini N, Schlisser AE, Hales BF. Editor’s highlight: hydroxyurea exposure activates the p53 signaling pathway in murine organogenesis-stage embryos. Toxicol Sci. 2016;152(2):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90(4):809–819. [DOI] [PubMed] [Google Scholar]
- 28. Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389(6647):191–194. [DOI] [PubMed] [Google Scholar]
- 29. Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287(34):28646–28655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hillje AL, Worlitzer MM, Palm T, Schwamborn JC. Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells. 2011;29(9):1437–1447. [DOI] [PubMed] [Google Scholar]
- 31. Guglieri M, Magri F, Comi GP. Molecular etiopathogenesis of limb girdle muscular and congenital muscular dystrophies: boundaries and contiguities. Clin Chim Acta. 2005;361(1-2):54–79. [DOI] [PubMed] [Google Scholar]
- 32. Vainzof M, Zatz M. Protein defects in neuromuscular diseases. Braz J Med Biol Res. 2003;36(5):543–555. [DOI] [PubMed] [Google Scholar]
- 33. Sato T, Okumura F, Iguchi A, Ariga T, Hatakeyama S. TRIM32 promotes retinoic acid receptor alpha-mediated differentiation in human promyelogenous leukemic cell line HL60. Biochem Biophys Res Commun. 2012;417(1):594–600. [DOI] [PubMed] [Google Scholar]
- 34. Sato T, Okumura F, Kano S, Kondo T, Ariga T, Hatakeyama S. TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J Cell Sci. 2011;124(Pt 20):3492–3502. [DOI] [PubMed] [Google Scholar]
- 35. Kano S, Miyajima N, Fukuda S, Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68(14):5572–5580. [DOI] [PubMed] [Google Scholar]