Abstract
Cell preparations to be used in clinical practice must be free of infectious agents. Safety concerns are especially elevated upon the use of human fetal tissues, which are otherwise highly advantageous in cell therapy. We demonstrate that treating fetal samples with antibiotic, extensive washing, and homogenization prior to cryoconservation efficiently removes microbes in general. Screening a large collection by an automatic culture system showed that 89.2% fetal tissue samples were sterile, while contamination was detected in 10.8% samples. Liver and chorion were contaminated more than the brain, kidney, lung, and soft tissues. Broad-range PCR from the bacterial 16s rRNA gene was adopted as a confirmatory assay; however, the concordance between the culture-based and PCR assays was weak. Taxonomic identification was done for contaminated samples by bacteriological methods and sequencing 16s rRNA PCR products. The two approaches revealed different spectra of taxonomic groups sharing only Lactobacillus, the most frequently found genus. In addition, other representatives of vaginal microbiota were detected by culture-based identification, while PCR product sequencing has also revealed a subset of nosocomial microorganisms. Importantly, species known to cause sepsis were identified by both techniques, arguing for their indispensability and mutual complementarity. We suggest that most contaminations are taken up during collection of fetal material rather than originating from an in utero infection. In conclusion, a rigorous microbiological control by culture and PCR is a prerequisite for safe clinical use of fetal tissue suspensions.
Keywords: Fetal tissues, Broad-range PCR, 16s rRNA sequencing, BacT/ALERT, Microbial contamination
Introduction
The therapeutic potential of fetal cell preparations is widely acclaimed. An extensive success record exists for treatment of conditions that result from regeneration defects at the cellular level1,2. Particularly, fetal cell therapy was suggested to be efficient in autism3, Parkinson's disease4, heart failure5, diabetes6, lateral sclerosis7, etc. The advantage of fetal cells could be explained by their high proliferative potential, availability in high quantities, and low antigenicity. Fetal preparations contain a subpopulation of stem cells considered to be analogous to embryonic stem cells (ESCs) in terms of pluripotency and primitiveness, yet no tumorigenicity of fetal stem cells has been reported8.
Safety issues arise when considering fetal cell transplantation for therapy because fetal material originates from an environment highly populated by microorganisms. In most cases, fetal material is harvested upon voluntarily elected termination of pregnancy and does not insure complete sterility. We and others9,10 use stringent washing in antibiotic-containing medium upon processing crude fetal material. A study10 to specifically assess the efficacy of washing for microbial decontamination was carried out previously and showed a complete removal of cultivable microorganisms in 21 fetal samples.
Therefore, questions remain to be answered: How are fastidious uncultivable microorganisms detected? What are the species that occasionally make it through the processing procedure? How dangerous are they in terms of sepsis, abscess, and other posttransplantation complications? Additional methods are needed to approach these questions and to expand the safety control toolbox. Polymerase chain reaction (PCR) targeting conservative regions of the bacterial 16s rRNA (and fungal 18s rRNA) gene has recently gained widespread use in many areas where fast and sensitive detection of microbial contamination is required11–14. This broad-range PCR showed a good concordance with culture-based techniques in many12, but not all14, experimental settings depending on the constitution of the material tested. As a culture-independent method, PCR allows for detection of a much wider range of species. The extreme sensitivity of PCR is definitely a plus yet sometimes exceeding the necessary level of clinical significance. In addition, DNA from dead microorganisms could be detected leading to overdiagnosis and rejection of therapeutically valuable preparations. Proper quantification of PCR results would help to rightly place this method among others. The next dimension of microbiological analysis opens with the implementation of sequencing 16s rRNA PCR products, allowing for taxonomic identification15,16.
We assayed the efficiency of washing for microbial decontamination by fluorescence microscopy and broad-range PCR, thus expanding the culture-based approach used earlier for this purpose by Piroth et al.10. We summarized the data accumulated after 2 years of the routine use of the BacT/ALERT automatic system to screen for contaminated samples in our collection. We identified the taxonomic groups predominant in contaminated samples by means of bacteriology and sequencing 16s rRNA PCR products. As a result, we adopted more stringent criteria for the release of a sample for clinical use, thus strongly reducing the risk of sepsis after transplantation of fetal tissues.
Materials and Methods
Fetal Tissue Samples
Human fetal samples were collected after elective termination of pregnancy. Each woman donating fetal material gave informed consent and signed an appropriate form. The research and clinical practice at our center are performed according to the Law of Ukraine “on transplantation of organs and other anatomical materials to the person” and the Law of Ukraine “on licensing types of economic activity,” under the license of Health of Ukraine for medical practice and the license for the activities of the bank of umbilical cord blood, other human tissues, and cells issued by the State Service of Ukraine on the prevention of HIV infection/AIDS and other publicly dangerous diseases (No. 222-VIII, 02.03.2105). According to the licensing conditions, an ethical approval is required and was granted. The full list of regulatory documents and original patents covering the activity of our center can be found at www.emcell.com.
Processing fetal tissues was performed as described previously3. Two hundred fifteen aborted fetuses (aged 6–12 weeks) were harvested at the site of operation in accordance with aseptic surgical protocol. Fetal material was placed in a sterile transport medium made of Dulbecco's modified Eagle's medium (DMEM) without l-glutamine with gentamicin (100 mg/ml; Thermo Fisher Scientific, Waltham, MA, USA). Whole fetal organs were placed in Hank's balanced salt solution (HBSS) without calcium and magnesium (Sigma-Aldrich, St. Louis, MO, USA), and then were dissociated and homogenized mechanically. This produced a collection of 938 tissue samples used in this study. Cell suspensions were filtered using individual 100-μm filters (Becton-Dickinson, Franklin Lakes, NJ, USA), and isolated cells were cryopreserved using 5% dimethyl sulfoxide (DMSO; Sigma-Aldrich) in HBSS using a computer-controlled rate freezer ICE Cube 14 (Sy-LAB Geräte GmbH, Neupurkersdorf, Austria). Tissue samples were transferred to liquid nitrogen for long-term storage.
Assaying Bacterial Contamination by Fluorescence In Situ
Samples were assayed with Molecular Probes Cell Culture Contamination Detection Kit (C-7028; Thermo Fisher Scientific). This kit uses the following dyes: SYTO-9 (staining nucleic acids and emitting green fluorescence) and N-acetylglucosamine-specific Texas Red (claimed to bind stronger to the cell wall of Grampositive bacteria). Briefly, 20 μl of sample (or supernatant after centrifugation at 800 × g, 5 min for preclearing) was placed on the slide, dried for 10 min at room temperature (RT), flame fixed, and washed with bovine serum albumin (BSA)-saline solution (0.25% in 0.15 M NaCl) (Sigma-Aldrich). Then solutions of SYTO-9 (10 μl) or Texas Red (50 μl) were applied, and slides were incubated for 5 min. Texas Red slides were washed once again with BSA-saline. Visualization was done on an Axio Vert. A1 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). A Lactobacillus rhamnosus culture was used as a positive control (Probiotical SpA, Novara, Italy).
Semiautomatic Microbial Culture and Taxonomic Identification
A portion of cryoconserved cell suspension was transferred to BacT/ALERT® Pediatric FAN® assay bottles (containing peptone-enriched tryptic soy broth supplemented with brain heart infusion solids and activated charcoal; bio-Mérieux, Marcy-l'Etoile, France) and incubated on a BacT/ALERT® 3D instrument (bio-Mérieux). In this system, the automatic monitoring of growth is based on the colorimetric detection of CO2 emitted by metabolically active micro organisms. If no growth was detected during 7 days of incubation, the sample was deemed to be sterile, otherwise the incubation was stopped at the day of detection.
Taxonomic identification was done at the Laboratory for Microbiology, Kyiv Military Hospital (Kyiv, Ukraine). Culture from BacT/ALERT-positive bottles was transferred to rich solid media (HiMedia Laboratories, Mumbay, India), Gram stained (Filicit, Dnipro, Ukraine), preanalyzed on a PrimoVert light microsope (Carl Zeiss Microscopy GmbH), and identified using MIKROLATEST® ID kits (Erba-Lachema, Brno, Czech Republic). Pure cultures were than analyzed on a VITEK® 2 microbial identification system (bio-Mérieux). Cultures were normalized to the appropriate turbidity measured by a DensiChek Nephelometer (bio-Mérieux). Suspensions were transferred to test cards (bio-Mérieux) containing proprietary reagents for identification in 30 wells. The following cards were used: GNI+ (Gram-negative and nonfermenting bacteria), GPI (Gram-positive bacteria), YBC (fungi), BAC (bacilli), ANI (anaerobes and coryneform bacteria), and NHI (Haemophilus and Neisseria). Results were processed by VITEK® 2 software (bio-Mérieux), and the taxonomic group was determined along with the estimate of confidence.
DNA Extraction, PCR, and Sequencing
DNA was extracted using Molysis Complete 5 (Molysis, Bremen, Germany) specifically tailored for broad-range 16s rRNA PCR. The procedure consists of DNAse pretreatment to reduce human DNA content, enzymatic lysis of bacterial cell walls, and spin column purification. Indeed, we noted that the presence of human DNA interfered with 16s rRNA PCR. Moreover, another spin column kit (nondisclosed) added a detectable amount of 16s rRNA templates to mock samples, which was not observed with Molysis Complete 5. In addition, 16s rRNA PCR was cleaner from DNA prepared by Molysis Complete 5.
Broad-range real-time PCR was performed with the following primer/probe sets: BactQuant (forward primer, 5′-CCTACGGGDGGCWGCA-3′; reverse primer, 5′-G GACTACHVGGGTMTCTAATC-3′; probe, 6-FAM-5′-C AGCAGCCGCGGTAATACGGAGG-3′-BHQ1, which is an extended version of the probe in the original article13) and Jordan (forward primer, 5′-AACTGGAGGA AGGTGGGGAT-3′; reverse primer, 5′-AGGAGGTGA TCCAACCGCA-3′; probe, 5′-TACAAGGCCCGGGA ACGTATTCACCG-3′-BHQ2)11. For human DNA, PCR was done using primers to the b-Globin gene, 5′-GGCA GGTTGGTATCAAGGTTAC-3′, 5′-CCTAAGGGTGGG AAAATAGACC-3′ and probe, HEX-5′-ACTGGGCATG TGGAGACAGA-3′-BHQ2. The reaction mix for these reactions was 1× Tris-KCl Taq buffer, 0.2 mM deoxynucleotide (dNTP), 4 mM MgCl2 (Thermo Fisher Scientific), 2.5 U Taq polymerase (Gene and Cell Technologies, Vallejo, CA, USA), total volume 25 μl. Real-time PCR for Lactobacillus 16s rRNA was done with the for ward 5′-TGGAAACAGATGCTAATACCG-3′ and reverse 5′-GTCCATTGTGGAAGATTCCC-3′ primers published previously17. The reaction was done in the M-427 SYBR Green reaction mix (Syntol, Moscow, Russia) with the addition of MgCl2 to 4 mM. All the PCRs were run using the following touchdown program: predenaturation, 95°C for 5 min; touchdown cycling (10 cycles): 95°C for 10 s, 69–64°C (-2°C/cycle) for 15 s, 72°C for 30 s; cycling with constant annealing t°C (30 cycles): 95°C for 10 s, 63°C for 15 s, 72°C for 30 s. PCR products were sequenced at commercial facilities using the Jordan reverse primer. The readable part of the sequence [260–320 base pairs (bp) in length] was used for searching the bacterial database by BLASTN algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). For taxonomic identification, only BLAST hits with the Identity >80% were taken into account and ranged by the Identity. In cases when this parameter was not sufficient for the identification, hits leading to lower taxon were given preference.
Results
Microbial Decontamination of Fetal Tissues
Fetal cell suspensions that passed through the processing procedure consisting of incubation with anti biotic, washing, homogenizing, and freezing are expected to be essentially free of microorganisms10. To screen for microbes that could still remain in our preparations, we examined more than 40 nitrogen-frozen tissue samples by fluorescence microscopy using the Texas Red dye that targets bacterial cell wall components (see Materials and Methods). We found entities resembling bacilli (Fig. 1A and B) only in rare cases (two samples). We then prepared precleared samples of transportation medium in which the fetal material had been collected right after the surgery (“entry”). The precleared tissue samples and “entry” were stained with the DNA-binding dye SYTO-9 (Fig. 1C–F). Round-shaped entities resembling bacterial cells (Fig. 1E) were always found in “entry” (Fig. 1C) and, rarely, in tissue samples (Fig. 1D). Most samples did not have such entities (Fig. 1F), suggesting an efficient elimination of bacteria by the processing. However, it should be noted that nonspecific fluorescent dyes are of limited usefulness for fetal samples that contain high amounts of human cells and cell clumps. In addition, the precise quantitative analysis is hampered because of subjective image perception.
Figure 1.
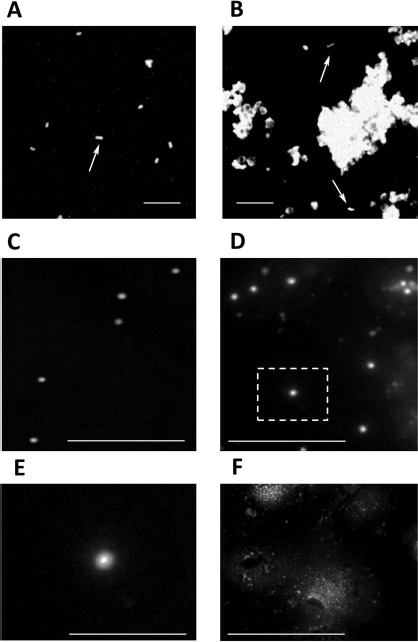
Microbial contamination in fetal tissue samples assayed by fluorescence in situ. Fluorescence staining with Texas Red (A, B) and SYTO-9 (C–E). (A) Lactobacillus rhamnosus culture (positive control). (B) Processed fetal tissue. Rod-shaped bacterial cells (arrows in A and B) occur among human cells and cell clumps in rare samples. (C) “Entry” sample (transportation medium used for harvesting the fetal material at the operation site). (D) A rare contaminated sample. Note round-shaped bacterial cells with the brighter area in the center likely representing the DNA. (E) The magnified portion of (D) outlined by a dashed square to show an individual bacterial cell. (F) A typical noncontaminated sample. Scale bars: 10 μm.
To better demonstrate the efficiency of microbe removal, we used real-time PCR, offering relative quantification of bacterial DNA load. We used two previously published primer/probe sets, BactQuant13 and Jordan11, that target different conservative regions of the bacterial 16s rRNA gene, thus covering a broad range of bacterial species. First, we analyzed the “entry” samples. A strong real-time PCR signal was registered (Ct = 23) (Fig. 2A and B), pointing to a high bacterial DNA content. At the same time, the “mock” sample showed a much weaker signal (Ct around 30). This background amplification was likely templated by traces of bacterial DNA (i) inevitably picked up during DNA isolation and/or (ii) always present in the reaction as an admixture to Taq polymerase. Fetal tissues (liver, brain, and soft tissues) prepared from “entry” produced results close to “mock” (Ct = 30).
Figure 2.
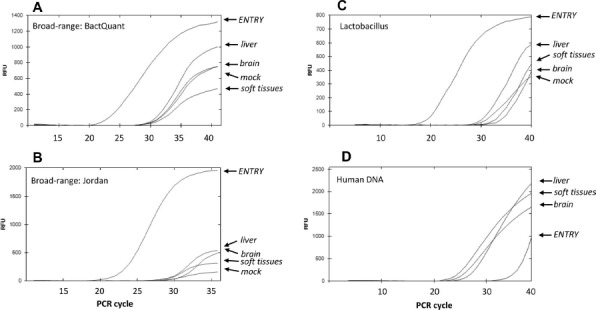
Polymerase chain reaction (PCR) showing an efficient removal of bacterial contamination by the processing procedure. Real-time PCR against 16s rRNA regions conservative for the entire bacterial kingdom [targeted by the BactQuant (A) and Jordan (B) primers], and regions specific for Lactobacillus (C) and human DNA (D). Abscissa: PCR cycle. Ordinate: relative fluorescence units (RFU). ENTRY: transportation medium used for harvesting the fetal material at the operation site. Liver, brain, soft tissues: samples prepared from the entry material. Mock: a water sample passed through the DNA isolation procedure to serve as a negative control. Note the much earlier rise of 16s rRNA PCR kinetics for “entry” in comparison to the prepared samples witnessing a dramatic reduction of bacterial DNA content after processing. The experiment has been repeated more than three times.
The major component of the normal vaginal microbiome is Lactobacillus. Thus, it could be used as an indicator genus for monitoring the efficiency of decontamination of fetal tissues. Now we used primers specific to Lactobacillus 16s rRNA17 and registered results mirroring those of the broad-range PCR: much less Lactobacillus DNA in processed samples than in “entry” (Fig. 2C). Noteworthy, the content of fetal material estimated by PCR for human DNA was actually higher in the processed tissues than in the “entry” (Fig. 2D). This rebukes the argument that the observed loss of bacterial DNA could have been simply due to diluting the “entry” sample. Together, the PCR results further argue that our processing procedure is efficient for microbe removal.
Routine Screening for Contaminated Fetal Samples
To screen for samples that might remain contaminated after processing, we utilized the BacT/ALERT automated microbial detection system. Of 932 samples tested, microbial growth was detected in 10.8% (Fig. 3A). A various number of tissues could be prepared from one fetus. That is why the n values of tissues are so different. In most cases, four tissues were available: liver, brain, soft tissues, and chorion. Liver and chorion samples were more contaminated, whereas the brain and lung appear to be the least contaminated. If a tissue sample turns out to be nonsterile, it does not mean that other tissues from the same fetus are so as well. Rather, one fetus might provide both clean and contaminated samples (Fig. 3B), arguing against a systemic fetal infection. In most of such fetuses, only one of the four harvested tissues has produced growth in BacT/ALERT falling in the category “25%–40%.” Two nonsterile samples of the four available would place the fetus in the category “50%–80%.” There were only 5 of the 47 nonsterile fetuses in which all tissues (100%) were contaminated.
Figure 3.
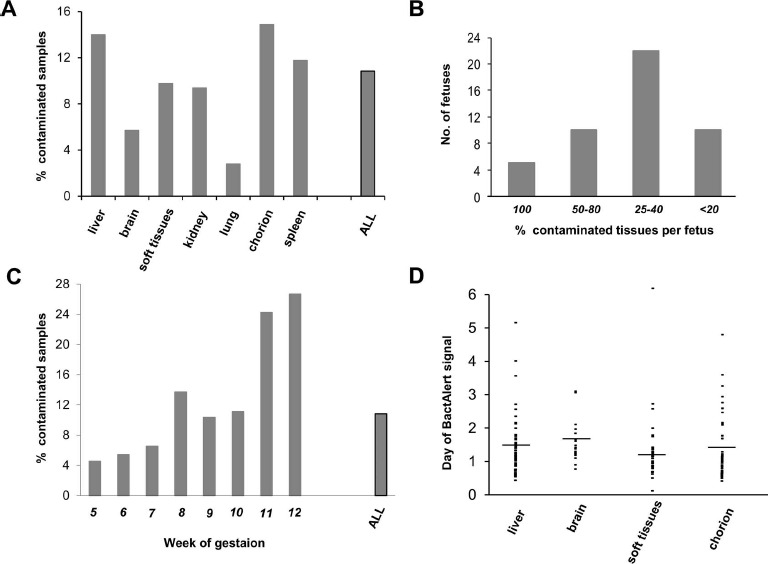
BacT/ALERT automatic culture from fetal samples. (A) Percentage of culture-positive samples of fetal liver (n = 200), brain (n = 193), soft tissues (n = 205), kidney (n = 32), lung (n = 36), chorion (n = 215), spleen (n = 51), and all samples (n = 932). (B) Tissue samples were first grouped by fetuses they were obtained from. Only fetuses providing four to eight tissues were considered. Then fetuses were ranged by the percentage of contaminated tissues as shown under the abscissa. Number (No.) of fetuses per each range is at the ordinate. (C) Percentage of culture-positive samples by the week of gestation: 5 (n =110), 6 (n = 111), 7 (n = 184), 8 (n = 241), 9 (n = 184), 10 (n = 45), 11 (n = 33), 12 (n = 30), and all samples with an identifiable age (n = 938). (D) The day at which the microbial growth was detected in BacT/ALERT culture bottles containing samples from the liver (mean = 1.34, n = 62), brain (mean = 1.57, n = 16), soft tissues (mean = 1.08, n = 25), and chorion (mean = 1.26, n = 59). The mean is indicated by the dash.
Classification by the age of gestation (932 samples from Fig. 3A plus 6 with mixed tissues) showed that tissues originating from older fetuses have a tendency to be more contaminated (Fig. 3C). Thus, among 11- and 12-week fetuses, the percentage of contaminated samples reached 20% and above, while the average in all fetuses was only 10.8%. In contrast, “younger” samples (5–7 weeks) produced BacT/ALERT growth only in less than 10% of the cases. The BacT/ALERT system allows for the rough estimation of the microbial load by the time at which growth becomes detectable. Most samples grew within 1–2 days of incubation (Fig. 3D), with few rare exceptions producing growth after up to 6 days. No significant difference in the microbial load was found between tissues.
To detect microorganisms, which for whatever reason were unable to grow in BacT/ALERT conditions, we recently adopted broad-range PCR with the BactQuant and Jordan primer/probes. The dynamic range of these sets is wide enough to ensure the proper detection of bacterial DNA in samples (Fig. 4A and B, gray PCR curves from 10-fold dilutions of purified Escherichia coli DNA). The sensitivity was as low as five genome equivalents per reaction. The PCR system appears to be oversensitive as it detects trace amounts of DNA even in the “mock” sample (Fig. 4, insets). This argues that only quantitative real-time PCR could be used to estimate the bacterial DNA content. We established an arbitrary cutoff criterion: to call a sample “contaminated with bacterial DNA,” Ct sample must be at least two cycles less than Ct mock in at least one of the two PCRs (BactQuant or Jordan). The plateau relative fluorescence units (RFU) value in contaminated samples may sometimes be lower than in non-contaminated ones. This could be due to (i) an incomplete match of our primers to the bacterial DNA predominant in the given “contaminated” sample and (ii) large amounts of human DNA introduced to reaction with the tissue sample (despite the DNAse treatment step during DNA purification; see Materials and Methods). Therefore, we do not take the plateau height in consideration when deeming a sample contaminated.
Figure 4.
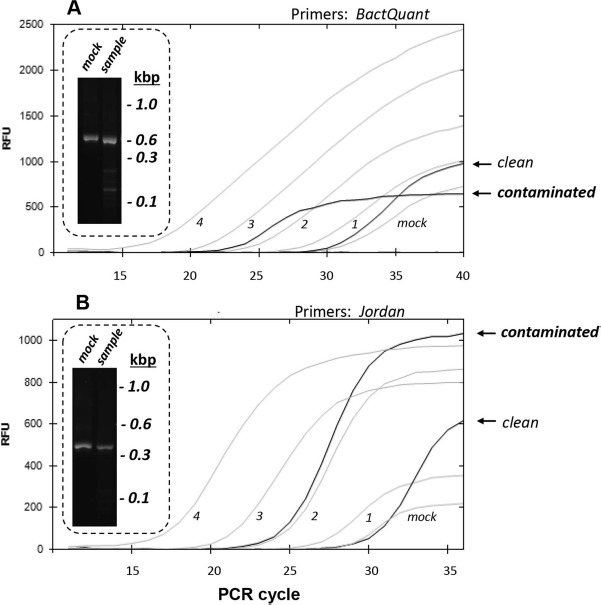
Detection of bacterial DNA in processed fetal preparations by PCR. PCR against 16s rRNA regions conservative for the entire bacterial kingdom [targeted by the BactQuant (A) and Jordan (B) primers]. Abscissa: PCR cycle. Ordinate: relative fluorescence units (RFU). Gray curves: dilutions of Escherichia coli (E. coli) DNA were used as standards. 1, 2, 3, 4: log genome equivalents per reaction. Dark curves: PCR from clinical samples labeled as “contaminated” and “clean” following the criteria described in the text. Mock: a water sample passed through the DNA isolation procedure to serve as a negative control. Insets: gel electrophoresis with BactQuant (447 bp) and Jordan PCR (371 bp) products from mock and clinical samples. Molecular weight is given in kilobases (kbp).
Another kind of broad-range PCR that we adopted for the safety control is aimed to detect the fungal 18s rRNA using the previously published FungiQuant system18. We optimized this primer/probes set to fetal material; however, fungi-positive samples occur extremely rarely, and hence insufficient data have been accumulated.
Of 85 BactT/ALERT culture-positive samples, PCR confirmed the presence of bacterial DNA in 54 samples. On the other hand, we detected a positive PCR signal in 2 of 24 BactT/ALERT culture-negative samples. Thus, for fetal tissue samples, the concordance between BacT/ALERT and PCR is not that remarkable as reported earlier for blood samples12. Interestingly, when the two assays were used for platelet concentrates (which are also rich in human cells), the analytic sensitivity of PCR was not high enough either14. Yet the advantages offered by BacT/ALERT and PCR encourage using both techniques, which increases the accuracy of contamination screening.
Microbial Identification in Contaminated Samples
This study could not bypass two questions: (i) where does microbial contamination originate from (in utero fetus infection, female reproductive tract, handling procedure, etc.), and (ii) how dangerous are the bacteria found in tissue samples in terms of the risk of sepsis? We set out to identify the prevalence of taxonomic groups and species of microorganisms that most frequently make it through the processing procedure. It is crucial to stress that we did not aim to reveal the composition of the entire microbiome but rather outline the most abundant groups and categorize them by the clinical importance.
First, we analyzed microbial cultures grown in BacT/ALERT using a series of differentiating and chromogenic media and stainings (see Materials and Methods). Eleven groups were identified among 56 samples at the level of genus and species (Table 1). The most prevalent geni were Staphylococcus (represented by three species), Streptococcus (two species), and Corynebacterium (no special identification), followed by Lactobacillus sp, Enterococcus sp, and Escherichia coli. Rare samples showed the presence of Micrococcus and the fungus Candida albicans. The revealed set of microorganisms suggests a strong connection between fetal tissue contamination and the female urogenital tract19.
Table 1.
Microbial Identification by Culture-Based Methods and 16s rRNA Sequencing
| Method | Order | Family | Genus | Species | No. Samples | Assessed Risk of Sepsis |
|---|---|---|---|---|---|---|
| Culture | Saccharomycetales | Saccharomycetaceae | Candida | albicans | 1 | High |
| Actinomycetales | Corynebacteriaceae | Corynebacterium | ND | 10 | Low | |
| Actinomycetales | Corynebacteriaceae | Micrococcus | ND | 2 | Low | |
| Bacillales | Staphylococcaceae | Staphylococcus | haemolyticus | 7 | High | |
| Bacillales | Staphylococcaceae | Staphylococcus | epidermidis | 2 | High | |
| Bacillales | Staphylococcaceae | Staphylococcus | saprophyticus | 1 | Low | |
| Lactobacillales | Enterococcaceae | Enterococcus | ND | 7 | High | |
| Lactobacillales | Lactobacillaceae | Lactobacillus | ND | 8 | Low | |
| Lactobacillales | Streptococcaceae | Streptococcus | mitis | 10 | Low | |
| Lactobacillales | Streptococcaceae | Streptococcus | intermedicus | 1 | High | |
| Enterobacteriales | Enterobacteriaceae | Escherichia | coli | 7 | High | |
| 16s rRNA sequencing | Bifidobacteriales | Bifidobacteriaceae | Gardnarella | vaginalis | 1 | Low |
| Clostridiales | Syntrophomonadaceae | Syntrophomonas | ND | 1 | Low | |
| Clostridiales | Lachnospiraceae | ND | ND | 4 | Low | |
| Lactobacillales | Lactobacillaceae | Lactobacillus | crispatus | 7 | Low | |
| Lactobacillales | Lactobacillaceae | Lactobacillus | iners or gasseri | 3 | Low | |
| Lactobacillales | Lactobacillaceae | Lactobacillus | ND | 9 | Low | |
| Sphingomonadales | Sphingomonadaceae | Sphingomonas | echinoides | 1 | Low | |
| Burkholderiales | Alcaligenaceae | Alcaligenes | ND | 1 | Low | |
| Burkholderiales | Burkholderiaceae | Burkholderia | cepacia | 1 | High | |
| Burkholderiales | Burkholderiaceae | Burkholderia | ND | 1 | High | |
| Burkholderiales | Comamonadaceae | Comamonas | ND | 1 | Low | |
| Burkholderiales | Comamonadaceae | Comamonas | testosteroni | 1 | Low | |
| Burkholderiales | Comamonadaceae | Delftia | ND | 2 | Low | |
| Pasteurellales | Pasteurellaceae | Haemophilus | haemolyticus | 3 | Low | |
| Pasteurellales | Pasteurellaceae | Haemophilus | influenzae | 3 | High | |
| Pseudomonadales | Moraxellaceae | Acinetobacter | bereziniae | 1 | High | |
| Pseudomonadales | Moraxellaceae | Acinetobacter | calcoaceticus | 1 | High | |
| Pseudomonadales | Moraxellaceae | Acinetobacter | ND | 3 | High | |
| Pseudomonadales | Pseudomonadaceae | Pseudomonas | ND | 5 | High |
Risk assessment was done on the basis of literature search. The risk was assigned as “high” if the higher taxon was known to have dangerous species. ND, the lower taxon identification was not possible.
Analysis of 16s rRNA sequence is another method of bacterial identification adoptable in clinical settings20. We sequenced Jordan PCR products (Fig. 4B, inset) by the Sanger method and confirmed that the PCR products indeed correspond to 16s rRNA. Despite the good quality of the analyzed DNA, there were quite a lot of ambiguous bases in the sequence reads suggesting that the target band on the agarose gel might represent amplicons from more than one species. Nevertheless, we were able to assign sequences to the species, genus, or family in 42 samples (Table 1). Interestingly, the reads from two “mock” samples were too mixed and did not produce any conclusive results in BLAST analysis. At the same time, 16s rRNA in “entry” samples (Fig. 2) belonged to the Lactobacillus sp in two cases (not included in Table 1), and one was unrecognizable. Most tissue samples contained Lactobacillus sp as well. Other species normally occurring in the female reproductive tract were detected too, such as Haemaphilus sp, Gardnerella vaginalis, and Lachnospiracea (similar to other studies of the vaginal microbiome15,21). Pseudomonadales and Burkholderiales were represented by soil/water inhabitants, some of which are still poorly investigated (Comamonas), while the others are known as ventilation-born/hospital infections (Pseudomonas and Acinetobacter22).
With the exception of Lactobacillus, there was no overlap between the culture-based and sequencing identification, even in samples analyzed by both techniques. Tissues originating from the same fetus might often show the prevalence of more than one group. More often, this would be Lactobacillus in one tissue and another species in the other tissue. Other interesting combinations found were Pseudomonas + Acinetobacter (by sequencing) and Streptococcus mitis + Staphylococcus haemolyticus + Escherichia coli + Candida albicans + Enterococcus sp (by culture). Fewer fetuses gave more than one tissue with the same taxonomic group (e.g., Streptococcus mitis in one case assayed by culture; Acinetobacter, Lachnospiraceae, and Haemophilus, one case each, assayed by sequencing; and, of course, Lactobacillus in several cases). Although we did not have a goal of studying the penetrance and tissue distribution of microorganisms, the obvious conclusion rises that, in most cases, the contamination is picked up upon the surgery rather than carried over with fetal material infected in utero.
Summarizing the results of microbial identification, we attempted to assess the risk of sepsis should a contaminated sample be, nevertheless, erroneously used for therapy (Table 1, the rightmost column, and Fig. 5). We assigned a risk to each group of microorganisms after a literature search for the reports on bacteremia and sepsis. One can hardly define strict criteria for this matter, yet we followed the major rule: if a species or a group was suggested to cause sepsis in some systematic multicenter survey with strong statistics, we assigned the risk as high. If, however, a microorganism appears only in case reports and/or negative clinical consequences of the bacteremia have not been convincingly shown, we ranked the risk as low. The use of these criteria and, of course, numerous discussions with specialists in the relevant medical fields prompted us to assign the high risk to Escherichia coli, Enteroccoccus sp, Candida albicans, Acinetobacter, Pseudomonas23, Haemophilus influenza24, Burkholderia cepacia25, Staphylococcus haemolyticus and epidermidis26, and Streptococcus intermedicus (known to belong to the so-called milleri group27). Taxonomic groups containing these microorganisms and their low-risk relatives found in fetal tissues (Table 1) are shown as bar graphs in Figure 5. We would like to reiterate that the discrimination of microorganisms by the risk has been undertaken here solely for the purpose of research and analysis, not to give low-risk samples a chance to pass the contamination control.
Figure 5.
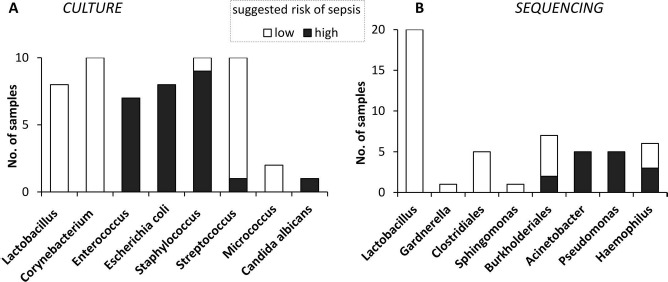
Microbial identification in fetal samples. (A) BacT/ALERT bottle culture was analyzed by bacteriological techniques. (B) Sequencing of PCR product from 16s rRNA gene (Fig. 4B, inset) followed by BLAST nucleotide database search. Dark portions of histograms correspond to the number (No.) of samples containing microorganisms known to pose a high risk of sepsis. White portions represent the samples bearing low-risk microorganisms only. Taxonomic grouping was done unevenly for a better representation of clinical importance.
Discussion
In this work, we analyzed the extent to which fetal suspensions are safe in terms of microbial sterility. While our processing procedure is efficient for the elimination of microorganisms (Figs. 1 and 2), traces of contamination remained in about 10.8% samples (Fig. 3A). Different, largely nonoverlapping subsets of taxonomic groups were found with the use of two methods of identification: automatic culture followed by bacteriological analysis and PCR followed by sequencing (Fig. 5). To explain this discordance, let us first remember that both methods have an amplification stage ensuring their high sensitivity. Indeed, microbial growth must occur in the BacT/ALERT bottle, and many cycles of target DNA synthesis must proceed in PCR before the respective signals become detectable. In both cases, the increase of the indicator parameter follows exponential kinetics. The fetal tissue sample is a highly heterogeneous system containing many species, and each of them may serve as a template for the signal amplification. Inevitably, the amplification processes from these templates compete to a certain extent so that only one of them will likely prevail by the moment of signal registration. The initial quantity of template is the major determinant of the success in this competition. Therefore, the final result will provide the information on the most abundant species only. Thus, in the majority of contaminated samples, we revealed Lactobacillus (Fig. 5) as the most abundant component of the normal vaginal microbiota. At the same time, both culture and PCR could have missed other species, some of which might be quite dangerous. Indeed, within the other groups, we found subsets featuring different potency for bacteremia upon intravenous administration (e.g., low- and high-risk species of Staphylococcus, Streptococcus, and Haemophilus). In view of this, we stay rather conservative in our routine screening, discarding all samples found to be contaminated by either method, regardless of the identification results.
Further, there is a plethora of fastidious noncultivated microorganisms, yet clinically important, which would not have a chance to be detected by BacT/ALERT. There are reports of both success and failure of automatic culture even for the same species28, some of which were also revealed here by sequencing, for example, Burholderia sp29, Pseudomonas sp30, and Haemophilus influenza31 (see also BacT/ALERT manual). Inconsistencies may arise due to differences at the preanalytical step: sometimes the presence of an antibiotic in the inoculum would affect growth32, and sometimes preincubation of samples at RT or 37°C would be beneficial30. This might be relevant to our practice because we inoculate the BacT/ALERT medium with an aliquot of cryoconserved sample.
Thus, we ultimately needed a supplementary technique to the automatic culture, and we hence added broad-range PCR as a relative quantitative assay for the bacterial (Fig. 4) and fungal (not shown) DNA load. We did not analyze the correlation between PCR and BacT/ALERT thoroughly enough, yet our data from the routine co-usage of the two techniques so far point to a weak concordance. This could be due to failure to grow in BacT/ALERT, PCR inhibition, primer/template mismatch, DNA loss, preanalytical contamination, etc. Yet the results of taxonomic identification by culture and PCR/sequencing (Fig. 5 and Table 1) strongly suggest that the two techniques complement each other and have a unique value for the contamination control.
As for the origin of contamination, we tend to think that most contaminants were taken up during the surgery and handling of fetal tissues. Indeed, the incomplete penetrance of contamination (Fig. 3B) together with the detection of low-risk bacteremic vaginal (Lactobaciaceae, Corynebacteriaceae, and others) (Table 1) and nosocomial ventilation-born microorganisms (Pseudomonas and Acinetobacter) supports this hypothesis. On the other hand, we observed a nonuniform distribution of the culture-positive samples by tissue (more in the chorion and liver, and less in the brain) (Fig. 3A) and the stage of gestation (Fig. 3C). These two pieces of data may suggest an infection in utero, which penetrates differently and is more likely to establish itself in older samples. Nevertheless, we believe that the share of operation-related contamination is much larger.
During 2 years after the introduction of the automatic culture system at our center, 790 patients received fetal tissue transplants following the safety control procedures approved by the license-issuing agencies (Materials and Methods). No case of bacteremia has been registered. However, we believe that a clinic must use as many approaches as possible to protect the patient from transplant-associated infections even if the reciprocal validation is incomplete. Having adopted broad-range PCR as a complementary test, we further strengthened the sample release criteria whereby the detection of contamination, either by culture or PCR, absolutely excludes the sample from any kind of clinical use.
Acknowledgments
The authors thank the personnel of the Lab for Microbiology, Kyiv Military Hospital for help with bacteriological analysis. All the authors are Emcell employees. The authors declare no conflicts of interest.
References
- 1.Ishii T, Eto K. Fetal stem cell transplantation: Past, present, and future. World J Stem Cells 2014; 6(4): 404–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya N. Fetal cell/tissue therapy in adult disease: A new horizon in regenerative medicine. Clin Exp Obstet Gynecol. 2004; 31(3): 167–73. [PubMed] [Google Scholar]
- 3.Bradstreet JJ, Sych N, Antonucci N, Klunnik M, Ivankova O, Matyashchuk I, Demchuk M, Siniscalco D. Efficacy of fetal stem cell transplantation in autism spectrum disorders: An open-labeled pilot study. Cell Transplant. 2014; 23(suppl 1): S105–12. [DOI] [PubMed] [Google Scholar]
- 4.Madrazo I, León V, Torres C, Aguilera MC, Varela G, Alvarez F, Fraga A, Drucker-Colín R, Ostrosky F, Skurovich M,. Transplantation of fetal substantia nigra and adrenal medulla to the caudate nucleus in two patients with Parkinson's disease. N Engl J Med. 1988; 318(1): 51. [DOI] [PubMed] [Google Scholar]
- 5.Benetti F, Peñherrera E, Maldonado T, Vera YD, Subramanian V, Geffner L,. Direct myocardial implantation of human fetal stem cells in heart failure patients: Long-term results. Heart Surg Forum 2010; 13(1): E31–5. [DOI] [PubMed] [Google Scholar]
- 6.Nasli-Esfahani E, Ghodsi M, Amini P, Keshtkar AA, Amiri S, Mojahed-Yazdi N, Tootee A, Larijani B,. Evaluation of fetal cell transplantation safety in treatment of diabetes: A three-year follow-up. J Diabetes Metab Disord. 2015; 14: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinelnyk AA, Klunnyk MO, Demchuk MP, Sych NS, Ivankova OV, Matiyashchuk IG, Skalozub MV, Novytska AV, Sorochynska KI. Combined therapy using fetal stem cells and a complex of physical exercises in treatment of patients with amyotrophic lateral sclerosis. Integr Mol Med. 2015; 2(6): 414–9. [Google Scholar]
- 8.Abdulrazzak H, Moschidou D, Jones G, Guillot PV. Biological characteristics of stem cells from foetal, cord blood and extraembryonic tissues. J R Soc Interface 2010; 7(suppl 6): S689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopyov OV, Jacques S, Lieberman A, Duma CM, Eagle KS. Safety of intrastriatal neurotransplantation for Huntington's disease patients. Exp Neurol. 1998; 149(1): 97–108. [DOI] [PubMed] [Google Scholar]
- 10.Piroth T, Pauly MC, Schneider C, Wittmer A, Mollers S, Dobrossy M, Winkler C, Nikkhah G. Transplantation of human fetal tissue for neurodegenerative diseases: Validation of a new protocol for microbiological analysis and bacterial decontamination. Cell Tranplant. 2013; 23(8): 995–1007. [DOI] [PubMed] [Google Scholar]
- 11.Jordan JA, Durso MB. Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis. J Mol Diagn. 2005; 7(5): 575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda K, Iwaki KK, Garcia-Gomez J, Hoffman J, Inderlied CB, Mason WH, Iwaki Y,. Bacterial identification by 16S rRNA gene PCR-hybridization as a supplement to negative culture results. J Clin Microbiol. 2011; 49(5): 2031–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu CM, Aziz M, Kachur S, Hsueh P-R, Huang Y-T, Keim P, Price LB,. BactQuant: An enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 2012; 12: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rood IGH, Pettersson A, Savelkoul PHM, de Korte D. Performance and suitability of polymerase chain reaction for early detection of bacteria in platelet concentrates. Transfusion 2011; 51(9): 2006–11. [DOI] [PubMed] [Google Scholar]
- 15.Muzny CA, Sunesara IR, Kumar R, Mena LA, Griswold ME, Martin DH, Lefkowitz EJ, Schwebke JR, Swiatlo E. Characterization of the vaginal microbiota among sexual risk behavior groups of women with bacterial vaginosis. PLoS One 2013; 8(11): e80254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motoshima M, Yanagihara K, Morinaga Y, Matsuda J, Hasegawa H, Kohno S, Kamihira S. Identification of bacteria directly from positive blood culture samples by DNA pyrosequencing of the 16S rRNA gene. J Med Microbiol. 2012; 61(Pt 11): 1556–62. [DOI] [PubMed] [Google Scholar]
- 17.Heilig HGHJ, Zoetendal EG, Vaughan EE, Marteau P, Akkermans ADL, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002; 68(1): 114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu CM, Kachur S, Dwan MG, Abraham AG, Aziz M, Hsueh P-R, Huang Y-T, Busch JD, Lamit LJ, Gehring CA, Keim P, Price LB,. FungiQuant: A broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 2012; 12: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokholm J, Schjørring S, Eskildsen CE, Pedersen L, Bischoff AL, Følsgaard N, Carson CG, Chawes BLK, Bønnelykke K, Mølgaard A, Jacobsson B, Krogfelt KA, Bisgaard H,. Antibiotic use during pregnancy alters the commensal vaginal microbiota. Clin Microbiol Infect. 2014; 20(7): 629–35. [DOI] [PubMed] [Google Scholar]
- 20.Schuurman T, de Boer RF, Kooistra-Smid AMD, van Zwet AA. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol. 2004; 42(2): 734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardines R, Daprai L, Giufrè M, Torresani E, Garlaschi ML, Cerquetti M,. Genital carriage of the genus Haemophilus in pregnancy: Species distribution and antibiotic susceptibility. J Med Microbiol. 2015; 64(7): 724–30. [DOI] [PubMed] [Google Scholar]
- 22.Drusano GL, Hope W, MacGowan A, Louie A. Suppression of emergence of resistance in pathogenic bacteria: Keeping our powder dry, Part 2. Antimicrob Agents Chemother. 2016; 60(3): 1194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004; 39(3): 309–17. [DOI] [PubMed] [Google Scholar]
- 24.Laupland KB, Schønheyder HC, Østergaard C, Knudsen JD, Valiquette L, Galbraith J, Kennedy KJ, Gradel KO, International Bacteremia Surveillance Collaborative. Epidemiology of Haemophilus influenzae bacteremia: A multinational population-based assessment. J Infect. 2011; 62(2): 142–8. [DOI] [PubMed] [Google Scholar]
- 25.Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J Appl Microbiol. 2008; 104(6): 1539–51. [DOI] [PubMed] [Google Scholar]
- 26.Czekaj T, Ciszewski M, Szewczyk EM. Staphylococcus haemolyticus—An emerging threat in the twilight of the antibiotics age. Microbiology 2015; 161(11): 2061–8. [DOI] [PubMed] [Google Scholar]
- 27.Weightman NC, Barnham MRD, Dove M. Streptococcus milleri group bacteraemia in North Yorkshire, England (1989–2000). Indian J Med Res. 2004; 119(suppl): 164–7. [PubMed] [Google Scholar]
- 28.Kocoglu ME, Bayram A, Balci I. Evaluation of negative results of BacT/Alert 3D automated blood culture system. J Microbiol. 2005; 43(3): 257–9. [PubMed] [Google Scholar]
- 29.Teerawattanasook N, Limmathurotsakul D, Day NPJ, Wuthiekanun V. Short report: Failure of Burkholderia pseudomallei to grow in an automated blood culture system. Am J Trop Med Hyg. 2014; 91(6): 1173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klaerner HG, Eschenbach U, Kamereck K, Lehn N, Wagner H, Miethke T. Failure of an automated blood culture system to detect nonfermentative gram-negative bacteria. J Clin Microbiol. 2000; 38(3): 1036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamal W, Tamaray G, Pazhoor A, Rotimi VO. Comparative evaluation of BacT/ALERT 3D and BACTEC systems for the recovery of pathogens causing bloodstream infections. Med Princ Pract. 2006; 15(3): 223–7. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan KV, Turner NN, Lancaster DP, Shah AR, Chandler LJ, Friedman DF, Blecker-Shelly DL. Superior sensitivity and decreased time to detection with the Bactec Peds Plus/F system compared to the BacT/Alert Pediatric FAN blood culture system. J Clin Microbiol. 2013; 51(12): 4083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


