Abstract
Chemosensory proteins (CSPs) play various roles in insect physiology including olfaction and development. The brown planthopper, Nilaparvata lugens Stål , is one of the most notorious rice pests worldwide. The wing-from variation and annually long distance migration imply that olfaction would play a key role in N. lugens behavior. In this study, full-length cDNAs of nine CSPs were cloned by the rapid amplification of cDNA ends procedure, and their expression profiles were determined by the quantitative real-time Polymerase Chain Reaction (qPCR), with regard to developmental stage, wing-form, gender, and tissues of short-wing adult. These NlugCSP genes showed distinct expression patterns, indicating different roles they play. In particular, NlugCSP5 was long wing form biased and highly expressed in female wings among tissues; NlugCSP1 was mainly expressed in male adults and abdomen; NlugCSP7 was widely expressed in chemosensory tissues but little in the nonchemosensory abdomen. The function of NlugCSP7 in olfaction was further explored by the competitive fluorescence binding assay using the recombinant protein. However, the recombinant NlugCSP7 showed no obvious binding with all tested volatile compounds, suggesting that it may participate in physiological processes other than olfaction. Our results provide bases and some important clues for the function of NlugCSPs .
Keywords: Nilaparvata lugens, chemosensory protein, wing-form, expression profile
Chemosensation is crucial for insect behaviors, such as locating food sources, recognizing mates, and finding oviposition sites. It is widely accepted that the volatile and lipophilic semiochemicals are transported to the olfactory receptors (ORs) by ligand carrier proteins ( Vogt et al. 2002 , Pelosi et al. 2006 ). These proteins included at least two main families: odorant-binding proteins (OBPs) and chemosensory proteins (CSPs; Zhou 2010 ). CSPs belong to OS-D superfamily and are thought to be involved in chemosensation in insects ( Ozaki et al. 2005 ). CSPs have four conserved cysteines that form two disulphide bonds ( Campanacci et al. 2003 ).
The first CSP (OS-D or A10) was identified in Drosophila melanogaster using subtractive hybridization ( McKenna et al. 1994,Pikielny et al. 1994 ). To date, CSPs have been reported in many insect species of different orders including Lepidoptera ( Maleszka and Stange 1997 , Qiao et al. 2013 ), Hymenoptera ( Briand et al. 2002 ), Blattoidea ( Kitabayashi et al. 1998 ), Orthoptera ( Picimbon et al. 2000 , Zhou et al. 2012 ), and Hemiptera ( Gu et al. 2012 ). Some CSPs are strictly expressed in the antennae, such as those in Adelphocoris lineolatus ( AlinCSP1-3 ; Gu et al. 2012 ), Apolygus lucorum ( AlucCSP1 ; Hua et al. 2012 ), and Linepithema humile ( LhumCSP ; Yuko et al. 2002 ), suggesting a function in the chemosensation. However, most CSPs such as those in Bombyx mori ( Qiao et al. 2013 ), Plutella xylostella ( Liu et al. 2010 ), and Sesamia inferens ( Zhang et al. 2013 ) are widely expressed in both chemosensory and nonchemosensory tissues. These nonchemosensory organs include female sex pheromone gland ( Jacquin-Joly et al. 2001 ), female reproductive organ ( Zhou et al. 2012 ), and male ejaculatory bulb ( Dyanov and Dzitoeva 1995 ), suggesting functions other than chemosensation.
The brown planthopper, Nilaparvata lugens Stål (Hemiptera: Delphacidae), is one of the most notorious rice pests, posing heavy damage to rice by sucking phloem sap and transmitting stunt virus and ragged stunt virus ( Cheng et al. 2013 ). N. lugens is a monophagous species restricted to cultivate and wild rice plants, suggesting that N. lugens applies chemical clues of rice to forage suitable parts and find oviposition sites. N. lugens adults have dual wing-form called the brachypterous (short wing) and the macropterous (long wing). The brachypterous mainly occurs when the environment is suitable for reproduction. In contrast, when the N. lugens population is too crowded and the nutrition of rice plants is not suitable, the macropterous will be produced and start to migrate. The annual long-distance migrations of the N. lugens from tropical into subtropical and temperate area have long been observed and studied ( Bottrella and Schoenlyb 2012 , Cheng et al. 2013 ). These biological characteristics imply a critical role of chemosensation in the behavior of this insect. However, the genes involved in the chemoreception are rarely addressed.
Previously, we identified three NlugOBP and nine putative NlugCSP cDNA fragments by combined use of bioinformatics and molecular biology methods ( Xu et al. 2009 ). Further study revealed that three NlugOBP s, particularly the NlugOBP3, might be involved in the olfaction of N. lugens ( He et al. 2011 ). In this study, to explore the functions of the CSPs, we cloned the full-length cDNAs and investigated the expression patterns of the nine NlugCSP s. On the basis of the expression results, we further expressed NlugCSP7 in vitro and determined its ligand binding characteristics. Our results suggested that some of these NlugCSP s may have chemosensory function, while others have nonchemosensory functions.
Materials and Methods
Insects and Sample Collection
N. lugens wild population was collected from a rice field of Jiangsu Academy of Agricultural Science, China. N. lugens were then raised in laboratory on rice seedlings in plastic box (40 by 30 by 40 cm) under 26 ± 1°C, 16:8 (L:D) h cycle, and 70–90% RH.
Nymphs of different instars and virginal long- and short-wing adults of 1–3-d-old were collected to determine the temporal expression pattern of the CSP genes, and various tissues from male and female adults (1–3-d-old short wing) were dissected and collected to determine the tissue expression pattern. Before sample collection, the age of the nymph or adult was checked based on the insect morphology (mainly the wing-bug and antenna) under a stereoscopic microscope according to literature ( Ding et al. 2012 ). After the collection, samples were immediately stored at −70°C until use. For the temporal expression pattern investigation, 20 individuals were collected per replication; for the tissue expression pattern study, ∼ 600 antennae, 20 heads, 10 abdomen, 300 legs, and 200 wings were collected per replication. All samples were collected in three biological replications.
RNA Isolation and cDNA Synthesis
Total RNA was extracted by SV 96 Total RNA Isolation System (Promega, Madison, USA. www.promega.com ) following the manufacturer’s protocol, including a step of DNase I treatment of the RNA extracts. RNA quality was checked with a spectrophotometer NanoDrop 1000 (Thermo Fisher Scientific, Pittsburgh, USA. www.thermofisher.com ). The single-stranded cDNA template was synthesized using 1 μg total RNA with oligo (dT) 18 primer as the anchor primer, by M-MLV reverse transcriptase (TaKaRa, Dalian, China. www.takara-bio.com ) at 42°C for 1 h, according to the protocol provided. The synthesized cDNA template was stored at −20°C for future use.
RACE Amplification and Sequences Alignment
A rapid amplification of cDNA ends (RACE) procedure was employed to amplify the 3′ and 5′ ends of the NlugCSPs using GeneRacer kit (Invitrogen, Carlsbad, USA. www.invitrogen.com ). The full-length sequences were assembled with RACE results and confirmed by end-to-end PCR using the high fidelity polymerase PrimeSTAR HS DNA Polymerase (TaKaRa, www.takara-bio.com ). The PCR reaction was 30 cycles of 98°C for 5 s, 55°C for 15 s, and 68°C 1 min. Three positive clones for each gene PCR product were sequenced. All primer sequences were designed by Primer Premier 5.0 (PREMIER Biosoft International, www.premierbiosoft.com ) and listed in Table 1 . Alignments of NlugCSP sequences were performed using ClustalW ( Clamp et al. 2004 ). The signal peptides were predicted by SignalP 4.1 ( www.cbs.dtu.dk/services/SignalP/ ; Petersen et al. 2011 ). The phylogenetic tree was constructed by the neighbor-joining method ( Tamura et al. 2011 ) after removing the highly divergent signal peptide, with a bootstrap analysis of 1,000 replications.
Table 1.
Primers used for RACE, qPCR, and vector construction
| Purpose | Gene name | Primer (5′→3′) |
|---|---|---|
| RACE | NlugCSP1- 5′ | tgcactagaccttggctgggaagc |
| NlugCSP1- 3′ | tccaactacatacgacgatg | |
| NlugCSP2- 5′ | cggaaaatgccctgaggatcg | |
| NlugCSP2- 3′ | gactgtctactcgataagggt | |
| NlugCSP3- 5′ | agccctcgcccatgagacacttgatg | |
| NlugCSP3- 3′ | gcctgatgcaatccagtccaactgctc | |
| NlugCSP4- 5′ | atcagctggggcggaccaaacctg | |
| NlugCSP4- 3′ | accaaatgcagtccccaacagaagaagg | |
| NlugCSP5- 5′ | gaagtagctgtcgaacagacgctggttgc | |
| NlugCSP5- 3′ | cagcgagaagcagaaagaaggcactgag | |
| NlugCSP6- 5′ | cgttggctgaaggcgccataagc | |
| NlugCSP6- 3′ | gctgccaaaattagccaagacgatgtgc | |
| NlugCSP7- 5′ | gaacagcctctggttgttgaggatctc | |
| NlugCSP7- 3′ | acatcgacctcgacgagatcctcaaca | |
| NlugCSP8- 5′ | tgttcagtcaagcttgatgcctttcatctc | |
| NlugCSP8- 3′ | gccaagtgcagcgaagctcagaag | |
| NlugCSP9- 5′ | cctcccgttgtgaacatttgggacatc | |
| NlugCSP9- 3′ | tgggaaataagccctgcgaccatg | |
| qPCR | NlugCSP1- F | tgcactagaccttggctgggaagc |
| NlugCSP1- R | tccaactacatacgacgatg | |
| NlugCSP2- F | cggaaaatgccctgaggatcg | |
| NlugCSP2- R | gactgtctactcgataagggt | |
| NlugCSP3- F | agccctcgcccatgagacacttgatg | |
| NlugCSP3- R | gcctgatgcaatccagtccaactgctc | |
| NlugCSP4- F | atcagctggggcggaccaaacctg | |
| NlugCSP4- R | accaaatgcagtccccaacagaagaagg | |
| NlugCSP5- F | gaagtagctgtcgaacagacgctggttgc | |
| NlugCSP5- R | cagcgagaagcagaaagaaggcactgag | |
| NlugCSP6- F | cgttggctgaaggcgccataagc | |
| NlugCSP6- R | gctgccaaaattagccaagacgatgtgc | |
| NlugCSP7- F | gaacagcctctggttgttgaggatctc | |
| NlugCSP7- R | acatcgacctcgacgagatcctcaaca | |
| NlugCSP8- F | tgttcagtcaagcttgatgcctttcatctc | |
| NlugCSP8- R | gccaagtgcagcgaagctcagaag | |
| NlugCSP9- F | cctcccgttgtgaacatttgggacatc | |
| NlugCSP9- R | tgggaaataagccctgcgaccatg | |
| Vector construction | NlugCSP7- V-F | cg ggatcc aagcccaagccagctgagaag |
| NlugCSP7- V-R | cc ctcgag ttagaccttgatgcctcgcttggcg |
F , forward; R , reverse. Restriction sites are underlined.
Quantitative Real-Time PCR
The quantitative real-time PCR (qPCR) was performed on an ABI 7500 (Applied Biosystems, New York, USA. www.lifetechnologies.com ) with SYBR Premix Ex Taq (TaKaRa, www.takara-bio.com ). The gene-specific primers were designed by Beacon Designer 7.9 (PREMIER Biosoft International, www.premierbiosoft.com ), and were listed in Table 1 . The reaction condition for the ABI 7500 was 10 s at 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. This was followed by the measurements of fluorescence, which resulted in a melting curve used to determine the primer specificity and primer dimers. The cDNA templates in fivefold dilution series were used to construct a relative standard curve to determine the PCR efficiency (>99%). Each template was run in three technical replicates. mRNA level was quantified in relation to the expression of the reference gene 16 S ribosomal RNA (FJ810191; Bao et al. 2010 ), using the 2 −ΔΔCt method ( Livak and Schmittgen 2001 ).
In vitro Expression and Purification of the Recombinant Protein
The NlugCSP7 sequence encoding mature protein was amplified by primers including BamH I and Xho I restriction sites. The sequences of the 2 primers are listed in Table 1 . The plasmids containing the inserts were digested by BamH I and Xho I FastDigest restriction enzymes (Thermo Scientific, www.thermofisher.com ). The expected bands were purified from agarose gel and ligated into the expression vector pET-30 a (+) (Novagen, Darmstadt, Germany. www.emdmillipore.com ), which was previously digested by the same enzymes. After ligation, expression of recombinant protein was carried out in Luria–Bertani (LB; 100 μg/mL, kanamycin) with Escherichia coli BL21 (DE3) cells at 37°C following recommended protocols. The positive clones were validated by PCR test and sequencing. The expression of recombinant protein was induced by addition of IPTG to a final concentration of 1 mM when the LB medium culture reached a OD600 value of 0.6. The protein was purified by XK-16 Column with Ni Sepharose High performance (GE Healthcare Life Sciences, www.gelifesciences.com ). The His-tag was cleavaged by enterokinase (Genscript Biology Company, Nanjing, China. www.genscript.com.cn ). The cleavaged protein was further purified by column mentioned above to remove the His-tag. This purified protein was then desalinated by dialysis against five gradient buffers, in which NaCl concentration ranged from 0.4 M to zero. The buffer formula was listed in Supp Table S1 (online only). For each buffer, the protein was dialyzed for 5 h at 4°C according to the previous study ( Liu et al. 2012a ). The resulted protein was kept at −70°C after freeze-dry.
Competitive Fluorescence Binding Assay
To measure the affinity of the fluorescent probe N-phenyl-1-naphthylamine (1-NPN) to NlugCSP7, a 2 μM solution of the protein in 50 mM Tris–HCl buffer, pH 7.4, was titrated with aliquots of 1 mM 1-NPN dissolved in methanol to a final concentrations of 2–36 μM, assuming that the protein was 100% active, with a stoichiometry of 1:1 protein:ligand at saturation ( Ban et al. 2002 ). The solutions were excited at 337 nm and emission spectra were recorded between 380 to 480 nm. The affinities of other ligands were measured by competitive binding assays, using 1-NPN as the fluorescent reporter. In the assay, the final concentrations of the ligand ranged from 2–48 μM. For each test, it takes 2 min for the reaction and about 10 s to measure the fluorescence, presuming that all the protein could be bound ( Ban et al. 2003 ). Three technical replicates were carried out. The binding constants (K 1-NPN ) of 1-NPN to NlugCSP7 was calculated by the reported method ( Liu et al. 2012b ). The dissociation constants (Ki) of each competitor were calculated from the corresponding IC 50 values (the concentration of competitor halving the initial fluorescence intensity), using the equation: Ki = [IC 50 ]/(1 + [1-NPN]/K [1-NPN] ), where [1-NPN] is being the free concentration of 1-NPN and K 1-NPN being the dissociation constant of the complex protein/1-NPN.
All chemicals including 1-NPN and 21 ligand compounds were purchased from Sigma-Aldrich (purity ≥ 95%), and were diluted by high-performance liquid chromatography methanol to 10 μM as stock solution, and to 1 μM as working solution. All solutions were stored in −20°C and were kept on ice during experiments.
Data Analysis
The relative mRNA expression levels were presented as mean ± SE. The differences in expression level were analyzed by ANOVA followed by least significant difference (LSD) test using SPASS 18.0.
Results
Full-Length cDNAs of CSPs
The nine NlugCSP cDNA full-lengths were obtained by using RACE strategy in N. lugens . These deduced amino acid sequences possessed all hallmarks of classic CSP, including a signal peptide, four conserved cysteine residues, and considerable identities (39–65%) to reported CSPs in amino acid sequence ( Supp Table S2 [online only]). The identities among the nine NlugCSPs ranged from 10% between NlugCSP6 and NlugCSP9 to 77% NlugCSP5 and NlugCSP7 ( Supp Table S3 [online only]). Besides the four conserved cysteine residues, 9 NlugCSPs shared three more amino acids: one Asparagine and one Arginine before the first “C”, and one Proline between the second and the third “C”. It was noted that NlugCSP9 had an additional six amino acid before the first “C” and one before the third “C”; NlugCSP2, NlugCSP6, and NlugCSP7 contained an exceptionally long C-terminus ( Fig. 1 ).
Fig. 1.
Alignments of amino acid sequences of NlugCSPs. The predicted signal peptides were boxed. Conserved cysteines were highlighted by black and marked with “☆” below the alignment, and other conserved residues were highlighted by gray.
Phylogenetic Analysis
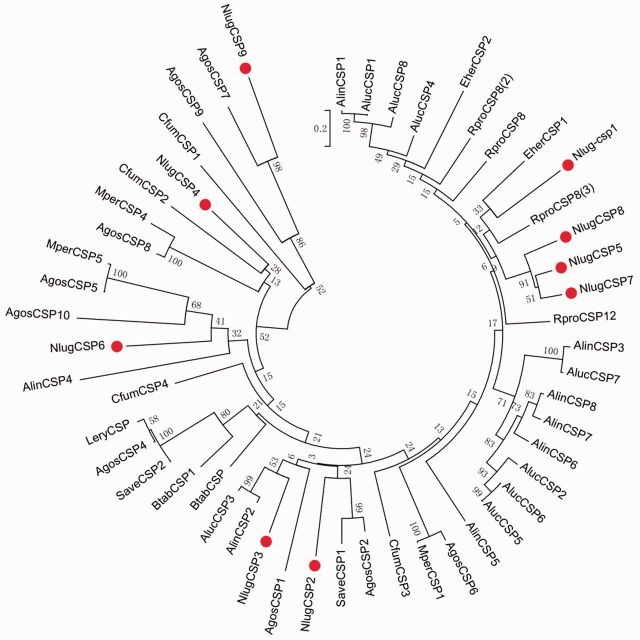
The amino acid sequences of these NlugCSP s and other hemipteran CSPs on GenBank were used to construct a phylogenetic tree ( Fig. 2 ). The tree showed that 9 NlugCSP s were scattered in six different groups, with NlugCSP5 , NlugCSP7, and NlugCSP8 in a same group.
Fig. 2.
Phylogenetic analysis of the amino acid sequences of NlugCSPs (indicated by “•”) in the context of various hemipteran CSPs. The tree was constructed by the neighbor-joining method after removing the highly divergent signal peptide, with a bootstrap of 1,000 replications. The bar indicated phylogenetic distance value. Abbreviations: Agos, Aphis gossypii ; Alin, Adelphocoris lineolatus ; Aluc, Apolygus lucorum ; Apis, Acyrthosiphon pisum ; Btab, Bemisia tabaci ; Eher, Euschistus heros ; Lery, Lipaphis erysimi ; Mper, Myzus persicae ; Rpro, Rhodnius prolixus ; Save, Sitobion avenae .
Expression Profiles of CSPs
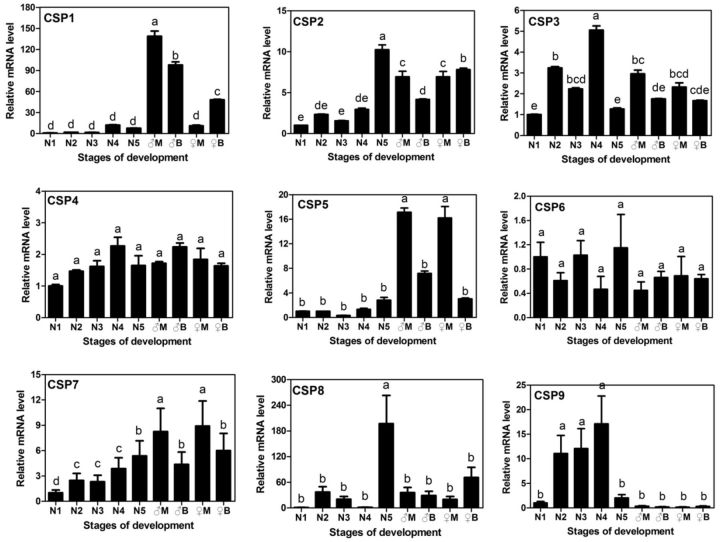
To understand roles of CSPs, mRNA expression levels of these genes in various stages of development ( Fig. 3 ), long- and short-wing forms ( Figs. 3 and 4 ), and various tissues from male and female adults of short wings ( Fig. 5 ) were determined by qPCR. The major results were summarized as follows.
Fig. 3.
Relative mRNA expression levels of NlugCSPs in nymph of different developmental stages and in adult of different wing-forms and genders, determined by qPCR. The relative expression level was indicated as mean ± SE (N = 3), with the first instar nymph as the calibrator. N1–N5, nymph of first to fifth instar; ♂M and ♂B, male macropterous and brachypterous adult, respectively; ♀M and ♀B, female macropterous and brachypterous adult, respectively. Different letter on the top of error bar means significant difference in expression among insects of developmental stages ( P < 0.05, LSD test).
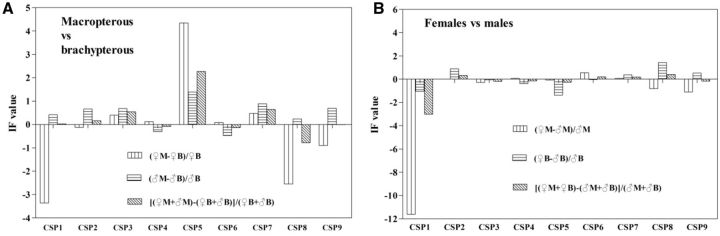
Fig. 4.
The relative expression level of CSP genes in adults, compared between long and short wing form ( A ), and between female and male ( B ). The relative expression level was indicated as “IF of expression level (M/B or ♀/♂)”.
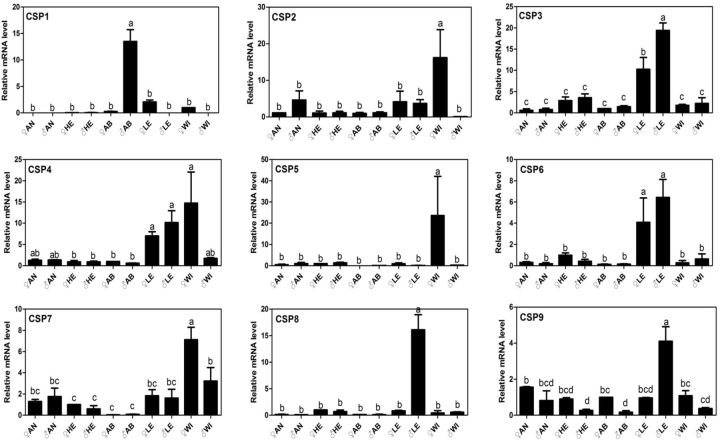
Fig. 5.
Relative expression levels of NlugCSPs in different tissues, determined by qPCR. ♀AN, female antennae; ♂AN, male antennae; ♀HE, female head; ♂HE, male head; ♀AB, female abdomen; ♂AB, male abdomen; ♀LE, female legs; ♂LE, male legs; ♀WI, female wings; ♂WI, male wings. Different letter on the top of error bar means significant difference in expression among tissues ( P < 0.05, LSD test).
Nymphs Versus Adults
In general, NlugCSP1, NlugCSP2, and NlugCSP5 showed significantly higher expression in adults than nymphs, while CSP9 was higher in nymphs than adults. Other five CSPs displayed similar expression levels between nymphs and adults. It was noticed that three CSPs (NlugCSP2, NlugCSP8, and NlugCSP9) showed much different levels in the fifth instar nymph (N5) with those lower instar nymphs. NlugCSP8 expressed in significantly higher level in fifth instar nymph than nymphs of other instars and adults, representing a distinct case ( Fig. 3 ).
Long Wing Versus Short Wing
Increased folds (IFs) of long-wing form than short-wing form in expression levels were calculated ( Fig. 4 A). Expression levels of NlugCSP2, NlugCSP3, NlugCSP4, NlugCSP6, and NlugCSP7 showed no great differences between long- and short-wing adults, with the absolute IF < 1. However, other genes presented obvious wing-form bias. The expression levels of NlugCSP5 were 4.35- and 1.39-folds higher in long-wing female and male adults than those in the short-wing adults, respectively, while three genes (NlugCSP1, NlugCSP8, and NlugCSP9) in females showed higher expressions in the short-wing than those in the long-wing N. lugens .
Females Versus Males
The IF of female/male were also calculated ( Fig. 4 B). Among the nine genes, NlugCSP1 showed a significantly highly male-biased expression, with IF value of −11.61 in long-wing adults and −1.04 in short-wing adults. NlugCSP5 in short-wing adults also showed certain male-biased expression (IF = −1.37). In contrast, NlugCSP8 in short-wing adults presented a female bias in expression (IF = 1.44), but in long-wing adults the IF value was not pronounced, indicating difference between short-wing and long-wing adults. Other genes were generally the same in expression level between females and males, with the absolute IF value < 1.
Tissue Expression Pattern
All nine NlugCSPs were expressed in a wide range of tissues but with different levels ( Fig. 5 ). NlugCSP2, 3, 4, 6, and 9 were expressed in all tested tissues with considerable levels. In contrast, NlugCSP1 was mainly expressed in male abdomen, while NlugCSP7 was expressed in all tissues except for male and female abdomen. In addition, NlugCSP5 was expressed in higher level in female wing than in other tested tissues, while NlugCSP8 was expressed in significantly higher level in male legs.
Ligand-Binding Experiments of NlugCSP7
The recombinant NlugCSP7 was highly expressed (19 mg/liter) as inclusion bodies in the E. coli expression system, by induction with IPTG. After purification and His-tag removal, the recombinant NlugCSP7 migrated as a single band of about 13 kDa ( Fig. 6 ).
Fig. 6.
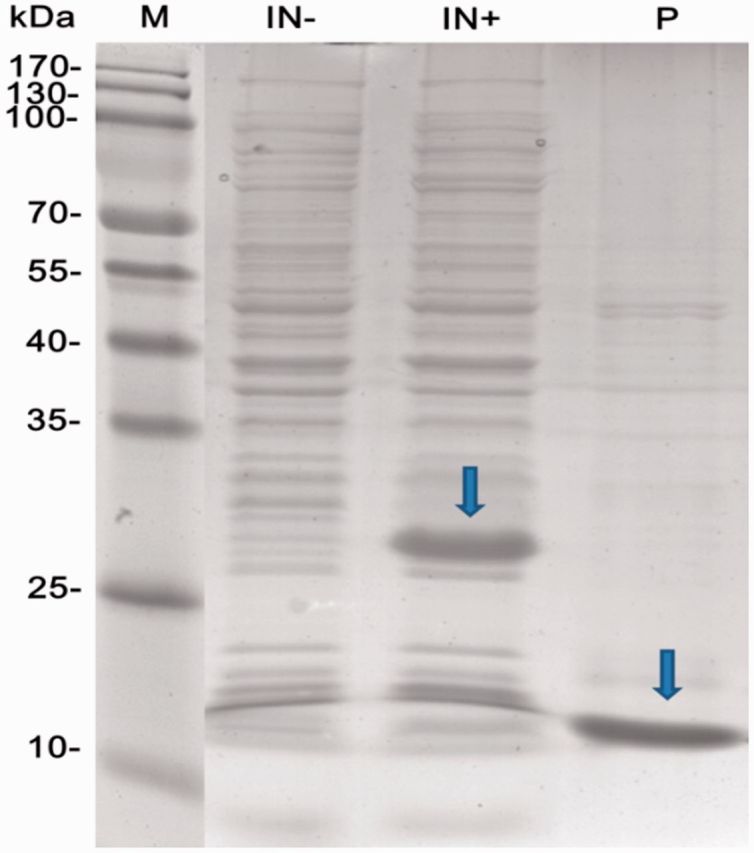
Expression and purification of NlugCSP7. A SDS-PAGE was used to detect the crude extracts from the bacterial pellets before (IN−) and after (IN+) induction with IPTG, and purified samples after His-tag cleavage by enterokinase (P).
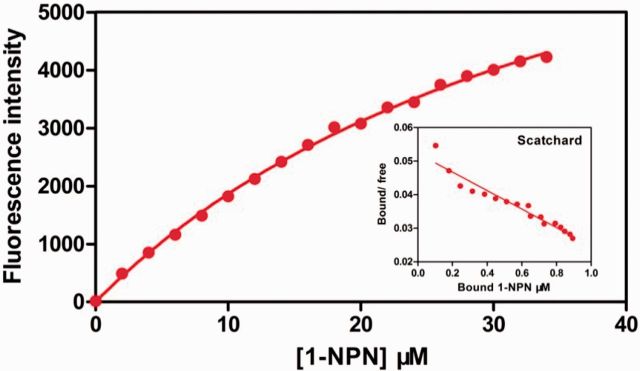
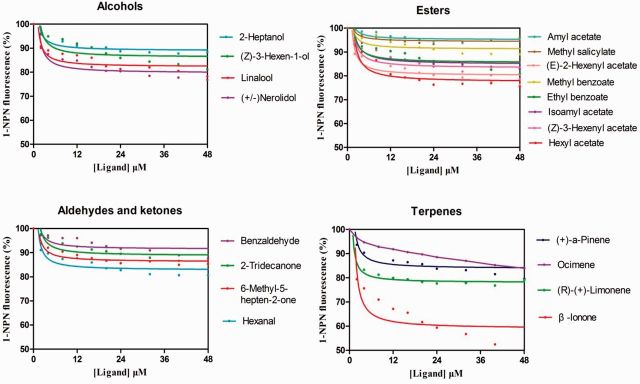
Twenty-nine compounds including 17 reported rice plant volatiles ( Visser and Yan 1995 , He et al. 2011 ) were selected to define the binding characteristics of the NlugCSP7. By titrating NlugCSP7 with increasing concentrations of 1-NPN allowed measurement of a dissociation constant of 40.7 μM ( Fig. 7 ). Typical displacement curves of 1-NPN by ligands with different function groups were observed ( Fig. 8 ). Among the tested plant volatiles, all ligands tested showed low binding affinity, with Ki > 50 μM ( Supp Table S4 [online only]).
Fig. 7.
Binding curve of 1-NPN to NlugCSP7 and relative Scatchard plot (inset).
Fig. 8.
Competitive binding curves of NlugCSP7 with 29 volatile compounds.
Discussion
The Amino Acid Sequences and Phylogenetic Analysis of NlugCSPs
In this study, nine NlugCSP full-length cDNAs were cloned and characterized based on the fragments obtained by our previous study ( Xu et al. 2009 ), providing an important step toward understanding their functions. Analysis of the amino acid sequences showed a great variation in identities (10–77%) among nine NlugCSPs. Similar diversifications in amino acid sequence of CSPs were also found in other insects, such as B. mori (10–50%; Qiao et al. 2013 ), P. xylostella (30–55%; Liu et al. 2010 ), and Papilio xuthus (20–70%; Ozaki et al. 2008 ). In agreement, nine NlugCSPs were distributed in six different groups in the phylogenetic analysis. The diversification in amino acid sequence suggests functional differentiation among these NlugCSP s.
Expression Patterns and Their Functional Implications of NlugCSPs
To explore the function, expression patterns were investigated, regarding to developmental stages, sexes, wing forms, and tissues. Some NlugCSP exhibited distinct expression patterns regarding to developmental stages, which may imply stage-related functions. For example, NlugCSP5 was mainly expressed in the adult, and was long-wing biased, whereas the NlugCSP9 was highly nymph (before fifth instar) biased, with a sharp reduction at fifth instar. In the natural conditions, N. lugens nymphs normally feeding on one or few rice plants in a closed range, but the adults often disperse in a relatively large area to find rice plants of high nutritional quality. Therefore, NlugCSP5 may play roles in perception of rice plant volatiles after the N. lugens dispersion. On the other hand, sharp reduction of expression at fifth instar suggests that NlugCSP9 is important for nymph development and possibly related to the nymph–adult metamorphosis, because after the fifth instar, the N. lugens will molt to the adult. In our previous study ( He et al. 2011 ), an OBP (NlugOBP3) with the similar expression pattern as NlugCSP9 is hypothesized to have nonolfactory functions, such as transporting the juvenile hormone. In contrast, NlugCSP8 was highly expressed only in fifth instar nymphs and is also possible to play roles in the nymph–adult metamorphosis.
For the tissue expression pattern, antenna-specific or highly expression often suggests a chemosensory function, as antennae is the most important chemosensory organ to the insect ( Vogt and Riddiford 1981 ). In addition, leg and wing also bear some olfactory and or gustory sensilla ( Zhou et al. 2008 , 2012 ). In this study, no NlugCSP was antenna specific or biased, similar as studies in many other insects, such as Locusta migratoria manilensis ( Zhou et al. 2012 ) and B. mori ( Gong et al. 2007 ). However, 8 NlugCSPs were specifically or highly expressed in legs, wings, or both. Notably, NlugCSP3 and NlugCSP6 were specifically expressed in female and male legs. Whether these CSPs play chemosensory roles need further studies.
In addition to the possible chemosensory role, CSPs has been reported to play function in leg regeneration in Periplaneta americana ( Nomura et al. 1992 , Kitabayashi et al. 1998 ). Among the tested tissues, abdomen is a nonchemosensory tissue. Some studies have showed that OBBs and CSPs in abdomen were often associated to reproductive organs. In Helicoverpa armigera, OBP10 was highly abundant in seminal fluid of males and was transferred to females during mating and eventually went on the surface of fertilized eggs, suggesting that it acts as a carrier for oviposition deterrents ( Sun et al. 2012 ), In L. migratoria , 17 OBPs were abundantly expressed in the female reproductive organs, and CSP91 was distinctly expressed in male organs ( Zhou et al. 2012 ). In our study, NlugCSP1 was pronouncedly expressed in abdomen in males and very weakly expressed in female abdomen and nymphs, which strongly suggests that this CSP is associated with reproduction events in N. lugens males. It would be interesting to clarify the function of NlugCSP1 .
Binding Ability of NlugCSP7 With Odors
NlugCSP7 was chosen to perform the binding assay, as it showed highly biased expression in antenna and other chemosensory organs (heads, legs, and wings). Although 17 rice plant volatiles and 12 other ligands were tested, none of them showed a high binding with NlugCSP7. It is possible that volatiles of high binding affinity would be found if more volatiles were tested. On the other hand, NlugCSP7 may possess physiological roles other than the chemosensation, as it was also expressed in nonchemosensory tissues.
Acknowledgments
This work was partly supported by the National 973 Project (2010CB126200) and the National Natural Science Foundation (31372264) of China.
References Cited
- Ban L. P., Zhang L., Yan Y. H., Pelosi P. . 2002. . Binding properties of a locust's chemosensory protein . Biochem. Biophys. Res. Commun. 293 : 50 – 54 . [DOI] [PubMed] [Google Scholar]
- Ban L., Scaloni A., D'ambrosio C., Zhang L., Yahn Y., Pelosi P. . 2003. . Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria . Cell. Mol. Life Sci. 60 : 390 – 400 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. Y., Li B. L., Liu Z. B., Xue J., Zhu Z. R., Cheng J. A., Zhang C. X. . 2010. . Triazophos up-regulated gene expression in the female brown planthopper, Nilaparvata lugens . J. Insect Physiol. 56 : 1087 – 1094 . [DOI] [PubMed] [Google Scholar]
- Bottrella D. G., Schoenlyb K. G. . 2012. . Resurrecting the ghost of green revolutions past: the brown planthopper as a recurring threat to high-yielding rice production in tropical Asia . J. Asia Pac. Entomol. 15 : 122 – 140 . [Google Scholar]
- Briand L., Swasdipan N., Nespoulous C., Bezirard V., Blon F., Huet J. C., Ebert P., Penollet J. C. . 2002. . Characterization of a chemosensory protein (ASP3c) from honeybee ( Apis mellifera L.) as a brood pheromone carrier . Eur. J. Biochem. 269 : 4586 – 4596 . [DOI] [PubMed] [Google Scholar]
- Campanacci V., Lartigue A., Hallberg B. M., Jones T. A., Giudici-Orticoni M. T., Tegoni M., Cambillau C. . 2003. . Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding . Proc. Nat. Acad. Sci. U S A 100 : 5069 – 5074 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Zhu L., He G. . 2013. . Towards understanding of molecular interactions between rice and the brown planthopper . Mol. Plant 6 : 621 – 634 . [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S. M., Barton G. J. . 2004. . The Jalview Java alignment editor . Bioinformatics 20 : 426 – 427 . [DOI] [PubMed] [Google Scholar]
- Ding J. H., Hu C. L., Fu Q., He J. C., Xie M. C. . 2012. . A colour atlas of commonly encountered delphacids in China rice regions . Zhejiang Sci. Technol. Press 2 : 22 – 26 . [Google Scholar]
- Dyanov H. M., Dzitoeva S. G. . 1995. . Method for attachment of microscopic preparations on glass for in situ hybridization, PRINS and in situ PCR studies . Biotechniques 18 : 822 – 824, 826 . [PubMed] [Google Scholar]
- Gong D. P., Zhang H. J., Zhao P., Lin Y., Xia Q. Y., Xiang Z. H. . 2007. . Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori . Insect Biochem. Mol. Biol. 37 : 266 – 277 . [DOI] [PubMed] [Google Scholar]
- Gu S. H., Wang S. Y., Zhang X. Y., Ji P., Liu J. T., Wang G. R., Wu K. M., Guo Y. Y., Zhou J. J., Zhang Y. J. . 2012. . Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition . PLoS One 7 : e42871 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Zhang J., Liu N. Y., Zhang Y. N., Yang K., Dong S. L. . 2011. . Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens Stal . PLoS One 6 : e28921 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J. F., Zhang S., Cui J. J., Wang D. J., Wang C. Y., Luo J. Y., Lv L. M. . 2012. . Identification and binding characterization of three odorant binding proteins and one chemosensory protein from Apolygus lucorum (Meyer-Dur) . J. Chem. Ecol. 38 : 1163 – 1170 . [DOI] [PubMed] [Google Scholar]
- Jacquin-Joly E., Vogt R. G., Francois M. C., Nagnan-Le Meillour P. . 2001. . Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae . Chem. Senses 26 : 833 – 844 . [DOI] [PubMed] [Google Scholar]
- Kitabayashi A. N., Arai T., Kubo T., Natori S. . 1998. . Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach) . Insect Biochem. Mol. Biol. 28 : 785 – 790 . [DOI] [PubMed] [Google Scholar]
- Liu X., Luo Q., Zhong G., Rizwan-Ul-Haq M., Hu M. . 2010. . Molecular characterization and expression pattern of four chemosensory proteins from diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) . J. Biochem. 148 : 189 – 200 . [DOI] [PubMed] [Google Scholar]
- Liu N. Y., He P., Dong S. L. . 2012a. . Binding properties of pheromone-binding protein 1 from the common cutworm Spodoptera litura . Comp. Biochem. Physiol. B Biochem. Mol. Biol. 161 : 295 – 302 . [DOI] [PubMed] [Google Scholar]
- Liu S. J., Liu N. Y., He P., Li Z. Q., Dong S. L., Mu L. F. . 2012b. . Molecular characterization, expression patterns, and ligand-binding properties of two odorant-binding protein genes from Orthaga achatina (butler) (Lepidoptera:Pyralidae) . Arch. Insect Biochem. Physiol. 80 : 123 – 139 . [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. . 2001. . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method . Methods 25 : 402 – 408 . [DOI] [PubMed] [Google Scholar]
- Maleszka R., Stange G. . 1997. . Molecular cloning, by a novel approach, of a cDNA encoding a putative olfactory protein in the labial palps of the moth Cactoblastis cactorum . Gene 202 : 39 – 43 . [DOI] [PubMed] [Google Scholar]
- Mckenna M. P., Hekmatscafe D. S., Gaines P., Carlson J. R. . 1994. . Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system . J. Biol. Chem. 269 : 16340 – 16347 . [PubMed] [Google Scholar]
- Nomura A., Kawasaki K., Kubo T., Natori S. . 1992. . Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach) . Int. J. Dev. Biol. 36 : 391 – 398 . [PubMed] [Google Scholar]
- Ozaki M., Wada-Katsumata A., Fujikawa K., Iwasaki M., Yokohari F., Satoji Y., Nisimura T., Yamaoka R. . 2005. . Ant nestmate and non-nestmate discrimination by a chemosensory sensillum . Science 309 : 311 – 314 . [DOI] [PubMed] [Google Scholar]
- Ozaki K., Utoguchi A., Yamada A., Yoshikawa H. . 2008. . Identification and genomic structure of chemosensory proteins (CSP) and odorant binding proteins (OBP) genes expressed in foreleg tarsi of the swallowtail butterfly Papilio xuthus . Insect Biochem. Mol. Biol. 38 : 969 – 976 . [DOI] [PubMed] [Google Scholar]
- Pelosi P., Zhou J. J., Ban L. P., Calvello M. . 2006. . Soluble proteins in insect chemical communication . Cell. Mol. Life Sci. 63 : 1658 – 1676 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., Von Heijne G., Nielsen H. . 2011. . SignalP 4.0: discriminating signal peptides from transmembrane regions . Nat. Methods 8 : 785 – 786 . [DOI] [PubMed] [Google Scholar]
- Picimbon J. F., Dietrich K., Angeli S., Scaloni A., Krieger J., Breer H., Pelosi P. . 2000. . Purification and molecular cloning of chemosensory proteins from Bombyx mori . Arch. Insect Biochem. Physiol. 44 : 120 – 129 . [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Hasan G., Rouyer F., Rosbash M. . 1994. . Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs . Neuron 12 : 35 – 49 . [DOI] [PubMed] [Google Scholar]
- Qiao H. L., Deng P. Y., Li D. D., Chen M., Jiao Z. J., Liu Z. C., Zhang Y. Z., Kan Y. C. . 2013. . Expression analysis and binding experiments of chemosensory proteins indicate multiple roles in Bombyx mori . J. Insect Physiol. 59 : 667 – 675 . [DOI] [PubMed] [Google Scholar]
- Sun Y. L., Huang L. Q., Pelosi P., Wang C. Z. . 2012. . Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species . PLoS One 7 : e30040 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. . 2011. . MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods . Mol. Biol. Evol. 28 : 2731 – 2739 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser J. H., Yan F. S. . 1995. . Electroantennogram responses of the grain aphids Sitobion avenae (F) and Metopolophium dirhodum (Walk) (Hom, Aphididae) to plant odor components . J. Appl. Entomol. Zeitschrift Fur Angewandte Entomologie 119 : 539 – 542 . [Google Scholar]
- Vogt R. G., Riddiford L. M. . 1981. . Pheromone binding and inactivation by moth antennae . Nature 293 : 161 – 163 . [DOI] [PubMed] [Google Scholar]
- Vogt R. G., Rogers M. E., Franco M. D., Sun M. . 2002. . A comparative study of odorant binding protein genes: differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera) . J. Exp. Biol. 205 : 719 – 744 . [DOI] [PubMed] [Google Scholar]
- Xu Y. L., He P., Zhang L., Fang S. Q., Dong S. L., Zhang Y. J., Li F. . 2009. . Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects . BMC Genom. 10 : 632 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuko I., Vicky C., Leal W. S. . 2002. . Protein that makes sense in the Argentine ant . Naturwissenschaften 89 : 505 – 507 . [DOI] [PubMed] [Google Scholar]
- Zhang Y. N., Jin J. Y., Jin R., Xia Y. H., Zhou J. J., Deng J. Y., Dong S. L. . 2013. . Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker) . PLoS One 8 : e69715 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. J. 2010. . Odorant-binding proteins in insects . Vitam Horm 83 : 241 – 272 . [DOI] [PubMed] [Google Scholar]
- Zhou S. H., Zhang J., Zhang S. G., Zhang L. . 2008. . Expression of chemosensory proteins in hairs on wings of Locusta migratoria (Orthoptera: Acrididae) . J. Appl. Entomol. 132 : 439 – 450 . [Google Scholar]
- Zhou X. H., Ban L. P., Iovinella I., Zhao L. J., Gao Q., Felicioli A., Sagona S., Pieraccini G., Pelosi P. P., Zhang L., et al. . 2012. . Diversity, abundance and sex-specific expression of chemosensory proteins in the reproductive organs of the locust Locusta migratoria manilensis . Biol. Chem. 1 : 43 – 54 . [DOI] [PubMed] [Google Scholar]