Abstract
The mechanisms by which hypoxic tumors evade immunological pressure and anti-tumor immunity remain elusive. Here, we report that two hypoxia-responsive microRNAs, miR25 and miR93, are important for establishing an immunosuppressive tumor microenvironment by down-regulating expression of the DNA-sensor cGAS. Mechanistically, miR25/93 targets NCOA3, an epigenetic factor that maintains basal levels of cGAS expression, leading to repression of cGAS upon hypoxia. This allows hypoxic tumor cells to escape immunological responses induced by damage-associated molecular pattern molecules (DAMPs), specifically the release of mtDNA. Moreover, restoring cGAS expression results in an anti-tumor immune response. Clinically, decreased levels of cGAS are associated with poor prognosis for patients with breast cancer harboring high levels of miR25/93. Together, these data suggest that inactivation of the cGAS pathway plays a critical role in tumor progression, and reveals a direct link between hypoxia-responsive miRNAs and adaptive immune responses to the hypoxic tumor microenvironment, thus unveiling potential new therapeutic strategies.
Introduction
The ability of cancer cells to evade immune responses is critical for the emergence of malignant phenotypes1. Cancer cells can escape both innate and adaptive immune responses by deregulating immune effector cells and immunosuppressive cells, as well as molecules responsible for cancer cell recognition and elimination2. Hypoxia contributes to immunosuppression within tumors, helping shield cancer cells from immune attack and inhibiting immune killing functions3–5. For example, immunosuppressive cells (e.g., regulatory T cells) accumulate within hypoxic regions of tumors where they promote tumor progression6–9 and activate immune tolerance mechanisms that enable cancer cells to evade host immunosurveillance. Despite recent advances, the molecular mechanisms underlying hypoxia-induced immune escape are not yet fully understood.
MicroRNAs (miRNAs) regulate a variety of cellular processes, including immune cell differentiation, immune responses to infection, and the development of immune disorders10–11. In the context of tumorigenesis, dysregulated miRNAs promote tumor progression by regulating cancer pathways associated with tumor malignancy12. A recent screen for hypoxia-regulated miRNAs in breast cancer cells revealed that hypoxia modulates miRNA function during breast cancer progression13. Hypoxia-responsive miRNAs regulate a complex spectrum of candidate target genes, including those involved in proliferation, apoptosis, metabolism, and migration14. Here we identify hypoxia-responsive miR25 and miR93 as critical factors in suppressing the expression of cGAS through a pathway that requires TET1 and NCOA3. In this way, these miRNAs promote immune escape of hypoxic tumors from DAMP-induced immunological stress, ultimately leading to the formation of an immunosuppressive tumor microenvironment.
Results
Hypoxia signaling induces immunosuppressive phenotypes
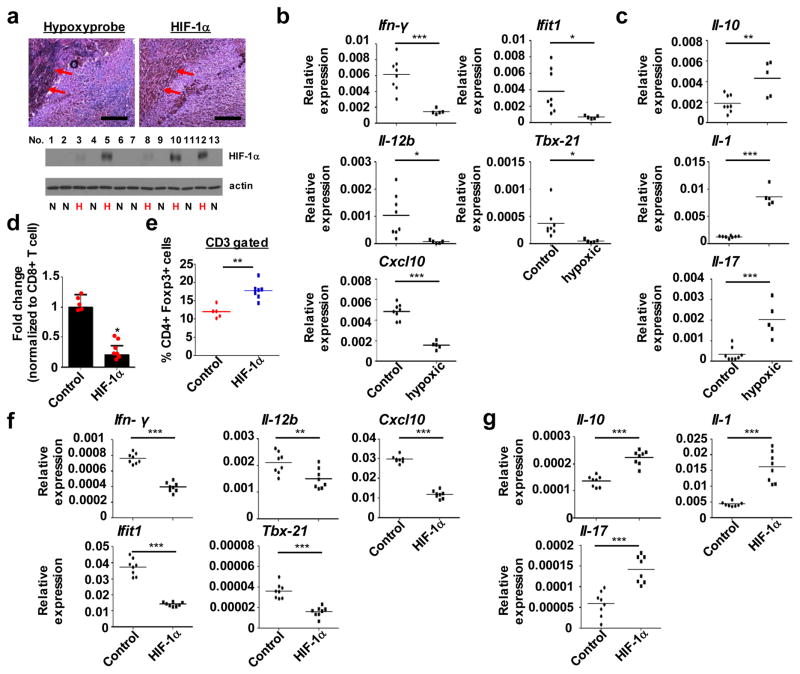
Recent findings suggest that hypoxia/HIF-1 signaling suppresses host defense against cancer9,15. To prove this concept, we examined associations between hypoxic tumors and immunosuppression. We identified hypoxic breast tumor samples based on levels of HIF-1α and assessed the expression of an array of inflammatory and immune mediators. This analysis revealed decreased levels of Ifn-γ, an anti-tumor factor, as well as decreased levels of several factors known to drive anti-tumor immunity, such as Il-12, Cxcl10, Ifit1, and Tbx-2116. In contrast, Il-10, a tumor-promoting factor, as well as Il-1β and Il-17, were increased (Fig. 1a–c). As a second means of studying hypoxic cancer cells, we subcutaneously inoculated wild type mice with murine breast cancer cells (E0771) that overexpress a constitutively active form of HIF-1α17. Because hypoxia results in the recruitment of immunosuppressive cells4, we characterized tumor infiltrating immune cells (TICs). FACS analysis of tumor biopsies indicated an increase in CD11b+ Ly6G− Ly6Chigh monocytic myeloid-derived suppressor cells (MDSCs) in HIF-1α overexpressing tumors (compared to control tumors), whereas HIF-1α did not affect levels of CD11b+ F4/80+ tumor associated macrophages (TAMs) or CD11b+ Ly6G+ Ly6Clow granulocytic MDSCs (Fig. S1a–c). The population of effector CD8+ T cells was decreased in HIF-1α overexpressing tumors, accompanied by a change in the percentage of tumor infiltrating CD8+ T cells, whereas the percentage of CD4+ T cells remained the same (Fig. 1d; Fig. S1d and e). Furthermore, we found greater numbers of CD4+FoxP3+ Tregs in HIF-1α overexpressing tumors (Fig. 1e), consistent with previous reports showing enhanced recruitment of Tregs by hypoxia9,18. We also analyzed these tumors and consistently observed immunosuppressive gene expression profiles, as shown by decreased levels of anti-tumor immunity factors and increased levels of tumor-promoting factors (Fig. 1f and g). Collectively, these results indicate that hypoxia/HIF-1αpromotes immunosuppression during breast cancer formation in vivo.
Figure 1. miR25/93 induced by hypoxia drives immunosuppression.
(a) Upper: Correlation between regions of hypoxia (defined by hypoxyprobe) and regions of high HIF-1α in murine tumor samples. Lower: Screening for hypoxic murine tumor samples. Endogenous HIF-1α was used as an indicator for hypoxia. E0771 cells were used for tumor formation. The scale bars represent 50 μm. (b and c) Gene expression analysis for selected genes responsible for either anti-tumor immunity or tumor-promoting effects in hypoxic tumor samples, compared with non-hypoxic tumor samples. Anti-tumor immunity factors include Ifn-γ, Il-12, Cxcl10, lfit1, and Tbx21. Tumor-promoting factors include Il-10, Il-1, and Il-17. (d) The percentage of effector CD8+ cytotoxic T cells was decreased in HIF-1α overexpressing tumors. Changes in effector CD8+ cytotoxic T cells normalized to total CD8+ T cell proportions. Effector CD8+ cytotoxic T cells were defined by the production of TNF-α and IFN-γ. E0771 cells expressing HIF-1α or empty vector were used for tumor formation. (e) HIF-1α-overexpressing tumors showed significantly higher recruitment of CD4+FoxP3+Tregs. (f) Gene expression analysis for anti-tumor immunity factors in whole-tumor homogenates derived from HIF-1α-overexpressing tumors or control tumors. (g) The level of tumor promoting factors was increased in HIF-1α-overexpressing tumors vs. control tumors. For panels b and c, control: n=8 tumors from 8 different animals, hypoxic: n=5 tumors from 5 different animals. For panels d and e, control: n=5 tumors from 5 different animals, HIF-1α: n=7 tumors from 7 different animals. For panels f and g, control: n=8 tumors from 8 different animals, HIF-1α: n=8 tumors from 8 different animals. For panel a, N: non-hypoxic sample. H: hypoxic sample. Samples were compared using two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. The image shown in panel a is representative of three independent experiments. Unprocessed scans of western blot analysis are available in Supplementary Figure 9. Statistics source data are available in Supplementary Table 4. Data shown in panels b, c, e, f, and g represent the mean. For panel d, data is represented as mean ± SD.
Hypoxia-responsive miR25/93 functions as an immunosuppressive factor
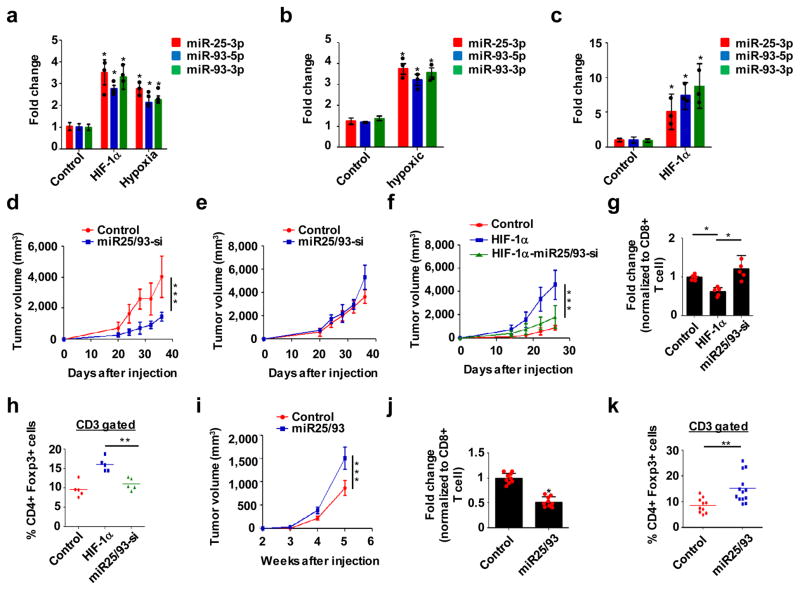
To identify bona fide hypoxia-regulated miRNAs, we performed small RNA sequencing using different cellular models, including hypoxic cells and HIF-1αoverexpressing cells, as described19 (Fig. S2a). Ingenuity pathway analysis (IPA) for selected miRNA candidates revealed correlations between these miRNAs and breast cancer formation, as well as oncogenic signaling pathways. (Fig. S2b and c). These data led us to focus on themiR-106b-25/miR-17-92 clusters, which are deregulated in human cancer20. It is well established that themiR-17-92 cluster plays roles in the development of the immune system and cancer; however, the role of themiR-106b-25 cluster remains largely unknown. We identified miR25 and miR93 as potential targets of hypoxia, as expression of these miRNAs was markedly increased in hypoxic conditions (Fig. 2a–c; Fig. S2d). However, hypoxia did not affect levels of miR106b, another member of the miR106b-25 cluster. shRNA-mediated knockdown of HIF-1αabolished the ability of hypoxia to induce miR25/93 expression (Fig. S2e), thereby indicating that miR25/93 are hypoxia-responsive miRNAs.
Figure 2. Hypoxia-responsive miR25/93 functions as an immunosuppressive factor.
(a) Up-regulation of miR25/93 in hypoxic and HIF-1α-overexpressing E0771 cells. (b) Elevated levels of miR25/93 in hypoxic murine breast tumors. (c) miR25/93 levels in HIF-1α-overexpressing tumors vs. control tumors. (d) Inhibiting miR25/93 reduced tumor growth in wild type mice. (e) Reduced tumor growth by the inhibition of miR25/93 was diminished in immune-deficient mice (NOD-Rag1nullIL2rgnull). (f) Suppression of miR25/93 reduced HIF-1α-enhanced tumor growth. (g) Restoration of effector CD8+ cytotoxic T cells in HIF-1α-overexpressing tumors by suppressing miR25/93. (h) Levels of CD4+FoxP3+ Tregs in control tumors, HIF-1α-overexpressing tumors, and HIF-1α-miR25/93-si tumors. (i) Overexpression of miR25/93 led to increased tumor proliferation. (j) The percentage of effector CD8+ cytotoxic T cells was decreased in miR25/93-overexpressing tumors. Changes in effector CD8+ cytotoxic T cells normalized to total CD8+ T cell proportions. Effector CD8+ cytotoxic T cells were defined by the production of TNF-α and IFN-γ. (k) miR25/93-overexpressing tumors attracted more CD4+FoxP3+ Tregs than control tumors. For panel a, n=3 independent experiments. For panels b and c, n=3 tumors from 3 different animals. For panels g and h, n=5 tumors from 5 different animals. For panels j and k, control: n=10 tumors from 10 different animals, miR25/93: n=13 tumors from 13 different animals. For panel i, n = 8 tumors from 8 different mice for each group. Data shown in panels a–g, i and j represent the mean ± SD of indicated sample numbers. Data in panels h and k represent the mean. Samples were compared using two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. Statistics source data are available in Supplementary Table 4.
Inhibiting miR25/93 slowed tumor growth in wild type mice. Importantly, this effect was compromised in immune-deficient mice, suggesting that miR25/93inhibitshost immune responses against the tumor(Fig. 2d and e). To explore the role of miR25/93 in hypoxia-relevant tumor immune responses, we inhibited miR25/93 in tumors overexpressing HIF-1α. This compromised the ability of HIF-1α to: 1) enhance tumor growth (Fig. 2f), 2) decrease levels of effector CD8+ T cells, and 3) increase recruitment of Tregs (Fig. 2g and h). Furthermore, we analyzed the profile of TICs in tumors constitutively overexpressingmiR25/93. Monocytic MDSCs consistently accumulated in miR25/93 overexpressing tumors; this was accompanied by an increased rate of tumor growth (Fig. 2i; Fig. S3a and b). In contrast, miR25/93 overexpression did not affect TAMs or granulocytic MDSCs (Fig. S3c and d). Overexpression of miR25/93 decreased the population of effector CD8+ T cells, alongside changes in the percentage of tumor infiltrating CD4+ T cells and CD8+ T cells (Fig. 2j; Fig. S3 e and f). Similar to hypoxic tumors, miR25/93 overexpression resulted in a greater number of Tregs (Fig. 2k). Gene expression analysis ofmiR25/93-overexpressing tumors consistently indicated an immune regulatory gene signature associated with hypoxia (Fig. S3g and h). Together, these results indicate that elevated levels of miR25/93 in tumor cells drive the establishment of an immunosuppressive environment.
TET1 is important for the up-regulation of miR25/93 in response to hypoxia
To investigate the molecular mechanism underlying hypoxia-induced miR25/93 expression, we first examined the role of HIF-1α.HIF-1α expression was required for hypoxia to induce the expression ofmiR25/93 (Fig. S2e). However, we did not observe significant HIF-1α binding to the miR25/93 genomic region, as compared to the VEGF promoter, which HIF-1α bound tightly upon hypoxia (Fig. S4a). This suggests that hypoxia regulates miR25/93 in a HIF-1α-independent manner. It is known that hypoxia can regulate miRNA biogenesis via epigenetic mechanisms21. Recent findings indicate that TET-mediated local 5-hydroxymethylcytosine (5-hmC) changes are important for the regulation of hypoxia-responsive genes19,22. Thus, we examined the role of 5hmC and TET proteins in this context. hMeDIP-seq analysis as well as conventional hMeDIP-qPCR analysis showed an increase in 5hmC levels within the miR25/93 genomic loci in hypoxic cells (Fig. S4b and c). ChIP-qPCR analysis for TET1 and TET3 indicated increased binding of TET1, but not TET3, to these regions (Fig. S4d). In support of these findings, silencing TET1, but not TET3, prevented hypoxia-induced upregulation of miR25/93 expression (Fig. S4e–h). Furthermore, the expression of wild type TET1, but not an inactive form of TET1, restored hypoxic epigenetic phenotype (Fig. S4i). Together, these results indicate an epigenetic layer of regulation in determining miR25/93 levels in hypoxic conditions.
A hypoxia-TET1-miR25/93 signaling axis represses cGAS-dependent immunity
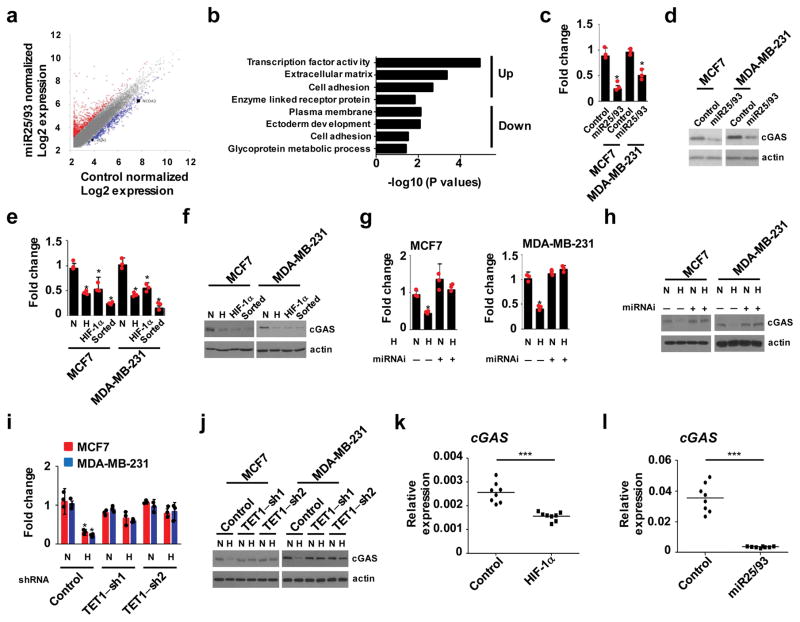
Next, we sought to identify the downstream signaling pathways that contribute to miR25/93-driven immunosuppression. RNA-seq analysis in breast cancer cells overexpressing miR25/93 was conducted to identify miRNA-responsive genes (Fig. 3a). Gene ontology analysis of these downstream genes highlighted biological processes related to transcription factor activity, cell adhesion, etc. (Fig. 3b). Validation of these potential target genes by qPCR indicated that cyclic-GMP-AMP synthase (cGAS), which is critical for the innate immune system to detect cytosolic DNA23–24, was a promising downstream target for miR25/93. Decreased levels of cGAS were detected in breast cancer cells overexpressing miR25/93orin hypoxic conditions (Fig. 3c–f). In line with this, shRNA-mediated inhibition ofmiR25/93restored cGAS expression in hypoxic cancer cells (Fig. 3g and h). Because TET1 regulates miR25/93 expression in hypoxia, we examined the expression of cGAS in TET1-deficient breast cancer cells. As expected, loss of TET1 inhibited hypoxia-induced down-regulation of cGAS expression (Fig. 3i and j). Furthermore, in tumors overexpressingHIF-1α or miR25/93, cGAS levels were consistently down-regulated (Fig. 3k and l). Collectively, these results indicate a role for the hypoxia-TET1-miR25/93 signaling axis in regulating cGAS.
Figure 3. Identification of cGAS as a target of miR25/93.
(a) Comparison of gene expression profiles in miR25/93-overexpressing cells vs. control cells. MCF7 cells expressing miR25/93 or empty vector were used in RNA-seq analysis. Data is presented by normalized Log2 fold change. Up-regulated genes (2,277) are highlighted red; down-regulated genes (1330) are highlighted blue. (b) Gene ontology results for up- and down-regulated genes show the top ranked functional gene cluster correlated with miR25/93. (c) cGAS mRNA levels were down-regulated in miR25/93-overexpressing cells vs. control cells. (d) Western blot analysis for cGAS protein levels in MCF7 or MDA-MB-231 cells stably expressing miR25/93. (e and f) Decrease level of cGAS in hypoxic, HIF-1α overexpressing, and BTIC cells, compared to normoxic cells. (g and h) shRNA-mediated miR25/93 inhibition reduced the ability of hypoxia to repress cGAS. (i and j) Gene expression analysis for cGAS in TET1 deficient breast cancer cells upon hypoxia. Control cells express shRNA targeting EGFP. (k) Decreased level of cGAS in mouse breast tumors derived from E0771 stable transfectants expressing HIF-1α. (l) Overexpression of miR25/93 correlated with decreased levels of cGAS. N: normoxia H: hypoxia. Sorted cells are BTICs isolated from MCF7 or MDA-MB-231 cells. All gene expression analysis data (panels c, e, g, i) are presented as means ± SD (n=3 independent experiments). Data shown in panels k and l represent the mean of n=8 tumors from 8 different animals. Samples were compared using two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. The images shown in panels f, h, and j are representative of three independent experiments. Unprocessed scans of western blot analysis are available in Supplementary Figure 9. Statistics source data are available in Supplementary Table 4.
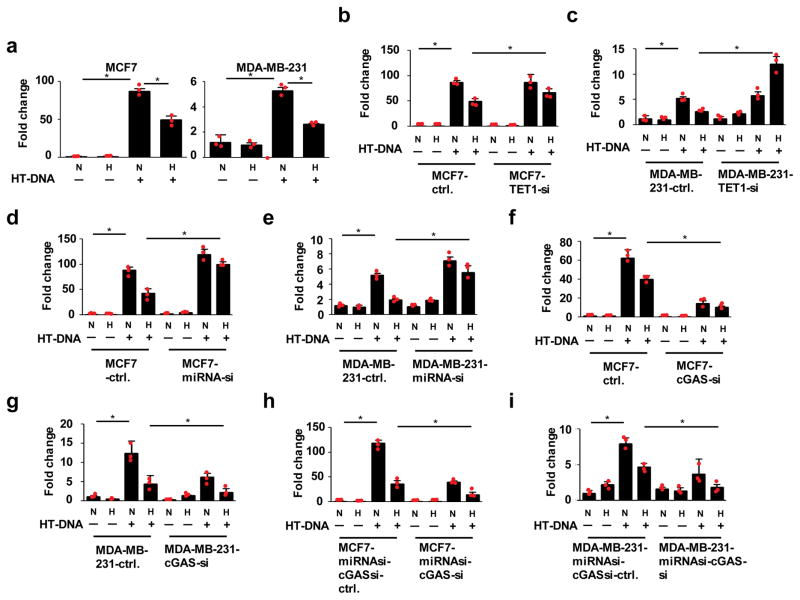
cGAS serves as a sensor for cytosolic DNA, inducing the production of type I interferons, which lead to innate immunity24. Hence, we investigated whether hypoxia attenuates the innate immune response against cytosolic DNA. We used qPCR analysis to assess levels of IFN-β expression, an indicator of cGAS-mediated immune response24. Hypoxia treatment inhibited the induction of IFN-β in breast cancer cells (Fig. 4a). Given that the TET1-miR25/93 signaling pathway is required to repress cGAS expression in response to hypoxia, this pathway may also be important for hypoxia-induced repression of innate immunity responses to cytosolic DNA. Indeed, we observed the restoration of hypoxia-induced immune responses in TET1-deficient cells (Fig. 4b and c). Likewise, inhibition of miR25/93 in hypoxic cells restored the induction of IFN-β in the presence of cytosolic DNA (Fig. 4d and e). In support of these observations, we demonstrated that down-regulation of cGAS both decreased the innate immune response to cytosolic DNA and enhanced hypoxia-induced immunosuppression (Fig. 4f and g). Furthermore, the cytosolic DNA-induced immune response was significantly reduced by knocking down cGAS expression in either MCF7 or MDA-MB-231 cells undergoing miR25/93 inactivation (Fig. 4h and i). Together, these results suggest that repression of cGAS by the TET1-miR25/93 pathway is crucial for hypoxia to suppress the immune response to cytosolic DNA.
Figure 4. Hypoxia-TET1-miR25/93 signaling axis reduces cGAS-mediated immune response.
(a) MCF7 or MDA-MB-231 cells were transfected with HT-DNA, followed by hypoxia treatment for 18 h. IFN-β expression was measured by RT-qPCR as an indicator of cGAS-mediated immune response. (b and c) Down-regulation of TET1 restored IFN-β expression in the presence of HT-DNA upon hypoxia. (d and e) The inhibitory effect of hypoxia on immune response was reduced by inactivation of miR25/93. (f and g) cGAS deficiency significantly impaired HT-DNA-induced immune response. (h and i) The immune phenotype of MCF7 or MDA-MB-231 cells stably expressing shRNAs against miR25/93 was reversed by knocking down cGAS expression. Control cells express shRNA targeting EGFP. N: normoxia H: hypoxia. All data presented here are mean ± SD (n=3 independent experiments). Samples were compared using two-tailed Student’s t test. *p< 0.05. Statistics source data are available in Supplementary Table 4.
Levels of endogenous DNA, considered damage-associated molecular pattern molecules (DAMPs), are increased by hypoxia during cancer progression and can trigger the rejection of tumors by the immune system25–26. Recent studies have demonstrated that the cGAS-dependent DNA sensing pathway can also recognize mtDNA released by apoptosis27–28. In line with this, hypoxia induces mitophagy and the extensive fragmentation of mitochondria26,29–30, which may lead to the release of mtDNA into the cytoplasm. We therefore hypothesized that to relieve immune pressure driven by the accumulation of cytosolic mtDNA, which occurs in the hypoxic tumor microenvironment during cancer progression, cancer cells must suppress the cGAS-dependent DNA sensing pathway via TET1 and miR25/93. Thus, we examined whether hypoxia leads to the accumulation of cytosolic mtDNA in cancer cells. qPCR analysis demonstrated increased levels of cytosolic mtDNA in hypoxic breast cancer cells compared to normoxic controls (Fig. S5a). Consistent with previous findings29, hypoxia treatment induced LC3-II and decreased levels of p62, markers of autophagy/mitophagy (Fig. S5b). To ensure that mtDNA activates the cGAS-dependent immune response, we transfected mtDNA into breast cancer cells and analyzed IFN-β levels. The induction of IFN-β in this context indicated that cytosolic mtDNA serves as a trigger for innate immunity (Fig. S5c). Notably, repression of miRNA25/93 restored the induction of IFN-β by cytosolic mtDNA in hypoxic cells (Fig. S5d and e). Furthermore, down-regulation of cGAS attenuated mtDNA-induced immune response (Fig. S5f and g), confirming that cGAS acts as an important senor for cytosolic mtDNA. We reasoned that manipulating miR25/93 or cGAS levels may alter levels of cytosolic mtDNA, leading to compromised immunosuppression. Thus, we analyzed cytoplasmic levels of mtDNA and found similar levels of hypoxia-induced cytoplasmic mtDNA in either miRNA25/93-inactivated or cGAS-deficient cells, compared to control cells (Fig. S6a and b). Together, these results reveal that the TET1-miR25/93 signaling pathway represses a cGAS-dependent immune response that results from hypoxia-mediated release of mtDNA.
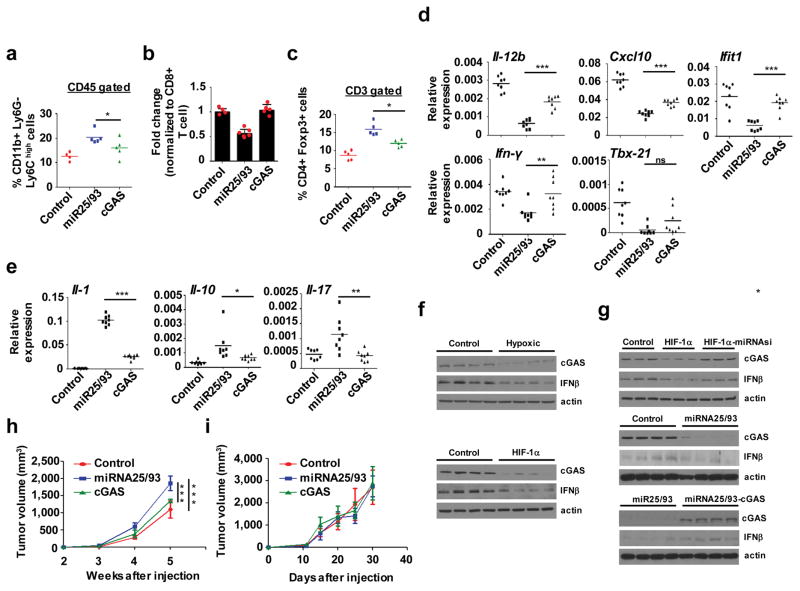
To ensure that cGAS functions to dampen the immunosuppressive microenvironment, we constitutively expressed cGAS in miR25/93 overexpressing tumors. FACS analysis demonstrated that re-expression of cGAS reversed immunosuppressive phenotypes, as indicated by significantly decreased populations of monocytic MDSCs, along with an increase in effector CD8+ T cells (Fig. 5a and b). Likewise, Tregs were decreased in cGAS re-expressing tumors compared with miR25/93 overexpressing tumors (Fig. 5c). Furthermore, gene expression analysis indicated increased levels of antitumor immunity factors in cGAS re-expressing tumors, whereas levels of tumor promoting factors were significantly decreased (Fig. 5d and e). As a key effector of cGAS signaling, type I interferons (IFNs) mediate anti-tumor immunity via their immunostimulatory functions31. As the immunosuppressive function of miR25/93 may rely on restrained IFN-stimulated immunosurveillance, we examined levels of INF-β in vivo. We observed decreased levels of INF-β and cGAS in hypoxic tumors andHIF-1α overexpressing tumors (Fig. 5f). INF-β levels were restored in HIF-1α-overexpressing tumors in which miR25/93 were inhibited, as well as in miR25/93-overexpressing tumors in which cGAS was re-expressed (Fig. 5g). To ascertain the role of cGAS-IFN signaling in tumor development, we assessed tumor growth in mice lacking the interferon (alpha and beta) receptor 1 (IFNAR1). Forced expression of miR25/93 increased tumor cell proliferation in wild type mice; this effect was reversed by re-expressing cGAS (Fig. 5h). Loss of IFNAR1 compromised the ability of miR25/93 to promote tumor progression, supporting the notion that cGAS-IFN signaling inhibits tumor progression (Fig. 5i).
Figure 5. miR25/93 signaling axis dampens anti-tumor immunity by down-regulating the cGAS/IFNs pathway.
(a) FACS analysis of CD11b+ Ly6G− Ly6Chigh monocytic MDSC in miR25/93-overexpressing, cGAS-re-expressing, and control tumors. E0771 cells were used for these gene manipulations.(b) Constitutive expression of cGAS restored the percentage of effector CD8+ cytotoxic T cells in miR25/93-overexpressing tumors. Changes in effector CD8+ cytotoxic T cells normalized to total CD8+ T cell proportions. Effector CD8+ cytotoxic T cells were defined by the production of TNF-α and IFN-γ. (c) Decreased levels of CD4+FoxP3+ Tregs in cGAS re-expressing tumors, as compared to miR25/93-overexpressing tumors. (d) Reverted gene expression profile for anti-tumor immunity factors in cGAS re-expressing tumors vs. miR25/93-overexpressing tumors. (e) The expression of tumor promoting factors was down-regulated by cGAS re-expression. (f) Western blot analysis for IFNβ and cGAS levels in either hypoxic tumors (upper) or HIF-1α-overexpressing tumors (lower). (g) Levels of IFNβ were regulated by theHIF-1α-miR25/93-cGAS signaling axis. (h) Tumor growth rate in E0771 cells overexpressing miRNA25/93 was higher than either cGAS re-expression cells or control cells in wild type mice. (i) Tumor growth analysis in IFN-αβR−/− mice injected with control cells or cells overexpressing miR25/93 with or without cGAS re-expression. N: normoxia H: hypoxia. For panels a–c, control: n=4 tumors from 4 different animals, miR25/93: n=5 tumors from 5 different animals, cGAS: n=5 tumors from 5 different animals. For panel b, data represent the mean ± SD. For panels a, c, d, and e, data represent the mean. Data shown in panel h and i are mean ± SD of n=10 tumors from 10 different animals for each group. Samples were compared using two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. Images shown in panels f and g are representative of three independent experiments. Unprocessed scans of western blots are available in Supplementary Figure 9.
In addition, we found that tumors lacking cGAS were more proliferative than control tumors. (Fig. S7a). As expected, the analysis of TIC profiles in cGAS-deficient tumors revealed a pattern similar to miR25/93- or HIF-1-overexpressing tumors (Fig. S7b–d). Silencing cGAS decreased the percentage of effector CD8+ T cells; this is accompanied by changes in the levels of tumor infiltrating CD4+ T cells and CD8+ T cells (Fig. S7e–g). Likewise, FACS analysis indicated a significant increase in Tregs in cGAS-deficient tumors (Fig. S7h). Gene expression analysis further showed that down-regulation of cGAS recapitulated the immune gene signature that characterizes hypoxic tumors (Fig. S7i–k).
miR25/93 indirectly regulates cGAS expression by targeting NCOA3 upon hypoxia
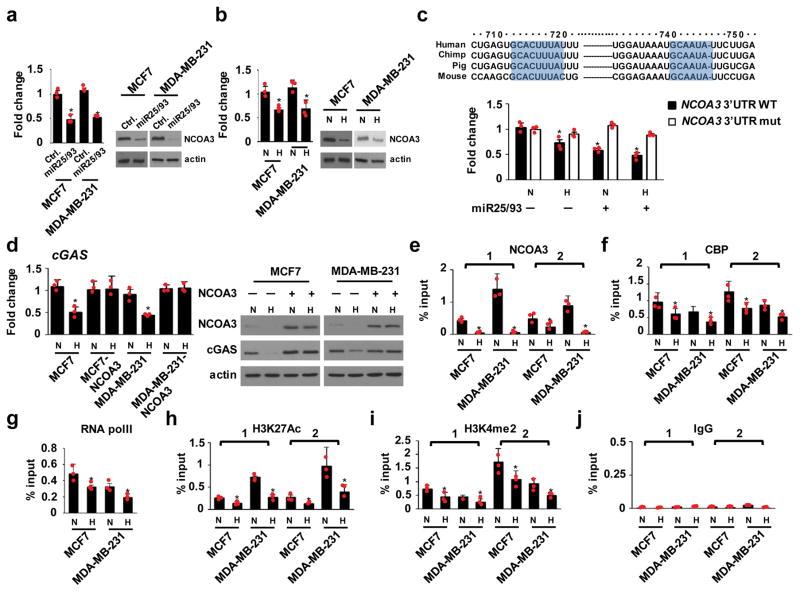
Next, we sought to characterize the molecular mechanisms by which miR25/93 represses cGAS. Analysis of the 3′-UTR of the cGAS gene did not identify a putative miR25/93 target sequence, which led us to conclude that miR25/93 suppresses cGAS expression by targeting other regulators. In our search for additional factors that regulate cGAS, we noticed that the cGAS promoter region contains an AP-1 biding site. It is unlikely that AP-1 is a key factor in the hypoxia-mediated repression of cGAS, but our RNA-seq data indicated that NCOA3, a member of the nuclear receptor co-activator (Ncoa) family that coordinates with AP-132, was down-regulated in miR25/93 overexpressing cells (Fig. 6a). Likewise, hypoxia treatment resulted in decreased levels of NCOA3 (Fig. 6b). Analysis of the 3′-UTR associated with the NCOA3 transcript identified conserved target sequences for miR25/93. A 3′-UTR reporter assay showed that miR25/93 suppressed NCOA3 reporter activity. Mutating the miR25/93 target sequences within the NCOA3 3′-UTR abrogated miR25/93-mediated repression (Fig. 6c). Together, these results indicate that NCOA3 is a downstream target of miR25/93.
Figure 6. Hypoxia-miR25/93 regulates cGAS signaling via targeting NCOA3.
(a) NCOA3mRNA and protein levels in miR25/93-overexpressing cells vs. control cells. Left: RT-qPCR analysis. Right: western blot analysis. MCF7 and MDA-MB-231 cells were used, as indicated. (b) Hypoxia down-regulated the expression of NCOA3 in MCF7 and MDA-MB-231 breast cancer cells. (c) Upper: Multiple species sequence alignment of the NCOA3 3′-UTR for putative miR25 (right) and miR93 (left) target sites, as indicated. Lower: relative reporter activity of the wild-type and mutant NCOA3 reporter in HEK 293T cells transfected with miR25/93 expression vector or empty vector upon hypoxia treatment. (d) Re-expressing NCOA3 restored cGAS level in response to hypoxia. Left: real-time PCR analysis. Right: western blot analysis. MCF7 and MDA-MB-231 cells were used, as indicated. (e) ChIP analysis for NCOA3 binding to the proximal promoter region (~−1 kbp) and transcription start site (TSS) of the cGAS gene upon hypoxia. (f) Decreased binding of CBP to cGAS genomic regions in hypoxic cells. (g) RNA pol II binding at the TSS of cGAS was decreased upon hypoxia. (h) Levels of H3K27Ac in the cGAS genomic region were reduced by hypoxia. (i) Hypoxia down-regulated H3K4me2 levels in cGAS genomic regions. (j) IgG control for ChIP assay. N: normoxia H: hypoxia. For ChIP assay, two primer sets were used to target the proximal promoter region and TSS of the cGAS gene. All data presented here are mean ± SD (n=3 independent experiments). Samples were compared using two-tailed Student’s t test. *p< 0.05, compared with controls. Images shown in panels a, b, and d are representative of three independent experiments. Unprocessed scans of western blots are available in Supplementary Figure 9. Statistics source data are available in Supplementary Table 4.
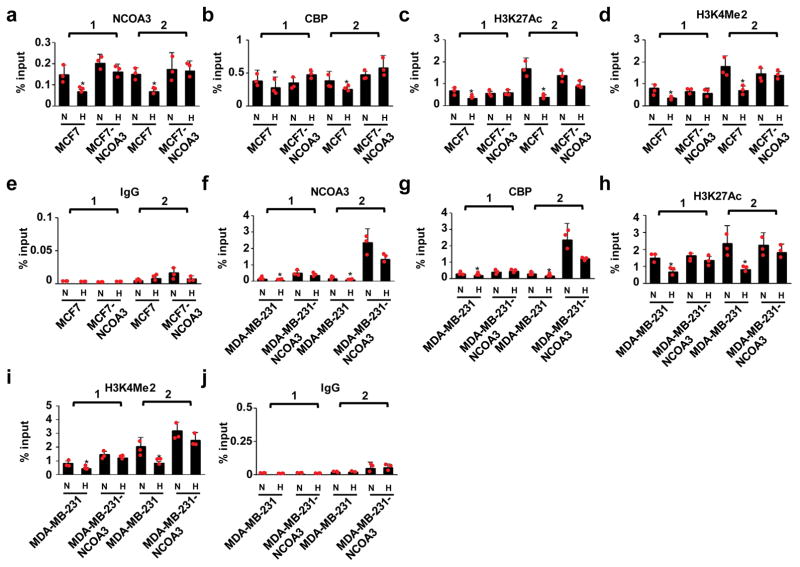
Because NCOA3 functions as a lysine acetyltransferase involved in transcriptional activation33, we asked whether NCOA3 regulates cGAS expression in response to hypoxia. We found that ectopic expression of NCOA3 restored the expression of cGAS upon hypoxia (Fig. 6d). ChIP-qPCR analysis revealed decreased levels of NCOA3 binding to the cGAS promoter in hypoxic cancer cells, compared with normoxic controls. This was associated with decreased levels of CBP binding, a protein that interacts with NCOA3, as well as decreased binding of RNA polII to the cGAS transcription start site (Fig. 6e–g). Moreover, two epigenetic modifications indicative of active transcription, H3K27Ac and H3K4me2, are associated with NCOA3 function34 and were consistently downregulated in hypoxic cells (Fig. 6h–j). Re-expression of NCOA3 restored levels of CBP binding to cGAS genomic regions in hypoxic cancer cells (Fig. 7a,b, f, g). Likewise, levels of H3K27Ac and H3K4Me2 were restored by re-expression of NCOA3 (Fig. 7c–e, h–j). Collectively, these results indicate that repression of NCOA3 by miR25/93 is required for the suppression of cGAS in response to hypoxia.
Figure 7. Re-expression of NCOA3 restores the epigenetic context required for cGAS expression.
(a and b) ChIP assay of MCF7-NCOA3 cells for NCOA3 and CBP protein bindings on genome regions of cGAS gene compared with control cells. (c–e) H3K27Ac and H3K4Me2 level within genome regions of the cGAS gene in MCF7-NCOA3 cells vs. control cells. IgG was used as a control for ChIP assay. (f and g) NCOA3 and CBP binding were restored in MDA-MB-231-NCOA3 cells, compared to control cells. (h–j) ChIP analysis for H3K27Ac, H3K4Me2, and IgG on genome regions of cGAS gene in MDA-MB-231-NCOA3 cells vs. control cells. Two primer sets were used, as indicated in Figure 6. N: normoxia H: hypoxia. All data presented here are mean ± SD (n=3 independent experiments). Samples were compared using two-tailed Student’s t test. *p< 0.05, compared with controls. Statistics source data are available in Supplementary Table 4.
The biological significance for miR25/93 and cGAS in clinical tissues
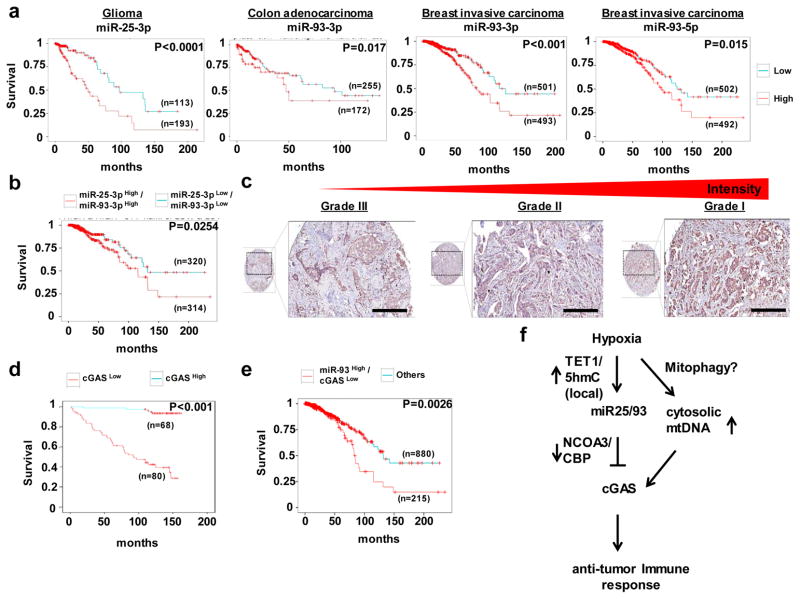
To test the relevance of these findings to human cancer, we first examined the prognostic impact of miR25/93. Levels of these miRNAs were correlated with poor prognosis for patients with different types of hypoxia-relevant tumors, such as brain, colon, and breast (Fig. 8a). Notably, higher levels of miR25 and miR93 were associated with reduced overall survival in patients with invasive breast carcinoma (Fig. 8b). We next examined the prognostic impact of cGAS levels for 148 patients with invasive ductal carcinoma (IDC). Levels of cGAS were inversely associated with poor prognosis for these patients (Fig. 8c and d). Consistent with this result, lower levels of cGAS were correlated with poor survival for cancer patients harboring elevated levels of miR93 (Fig. 8e), whereas other identified miRNAs targets (e.g., Bim and PTEN) did not exhibit clinical significance (Fig. S8a and b), suggesting that the down-regulation of cGAS is a biologically relevant factor in miR25/93-driven tumor progression. Collectively, these results support our hypothesis that repression of cGAS by hypoxia-responsive miR25/93 is critical for establishing an immunosuppressive environment that is associated with cancer progression.
Figure 8. The correlation of miR25/93-cGAS with prognosis for patients with cancer.
(a) TCGA analysis for miR25 or miR93 indicated that levels of these miRNAs correlated with poor survival of patients with brain, colon, or breast tumors. (b) The clinical relevance of levels of miR25/93 in cancer patients with breast invasive carcinoma. (c) IHC staining for cGAS levels for different grades of IDC revealed an inverse correlation between cGAS levels and tumor grades. The scale bars represent 50 μm. (d) Overall survival analysis for 148 patients with IDC showed an inverse correlation between cGAS levels and poor survival of cancer patients. The intensity of cGAS staining from low to high was divided into two group (0 and 1+, 2+ and 3+). n= 12 for 0, negative; n=56 for 1+, low expression; n=74 for 2+, intermediate expression; n=6 for 3+, high expression. n represents the number of patient samples for each group. (e) TCGA analysis indicated reduced overall survival of breast cancer patients harboring lower levels of cGAS and higher levels of miR93. (f) A model of miR25/93-mediated immune escape of hypoxic tumors from DAMP-induced immune stress. Sample numbers and p values are indicated in the panel for TCGA analysis and IHC study. The prognostic significance was assessed by Kaplan–Meier analysis.
Discussion
The mechanisms by which tumor cells maintain immunological self-tolerance and hinder effective tumor immunity are critical for tumor progression. Here, we uncover a mechanism for hypoxia-induced escape from immunological stress caused by DAMPs (Fig. 8f). Hypoxia induced the expression of miR25/93 by increasing 5hmC levels (i.e., DNA demethylation) near the miR25/93 gene via the TET1 protein. Differential regulation of the miR-106b-25 cluster in response to hypoxia, as shown by our data and previous reports35–36, suggests coordination between epigenetic factors and HIF-mediated post-transcriptional machinery37 to differentially regulate miR25/93 levels. These data also reveal the complexity of the role played by 5hmC and TET in regulating cellular responses to hypoxia. This is also supported by our observations (Fig. S8c), as well as recent reports38–40, indicating an un-coupling of expression of the MCM7 gene (host gene for themiR-106b-25 cluster) with the differential expression of members of themiR-106b-25 cluster. Recently, miR25/93 has emerged as an important onco-miRNA during tumorigenesis41, and here we further reveal its function in inducing immunosuppressive phenotypes. To promote immune avoidance, miR25/93 repressed cGAS by disrupting the epigenetic machinery that maintains basal levels of cGAS (this machinery includes NCOA3). Activation of the cGAS-dependent cytosolic DNA sensing pathway within tumor-resident immune cells results in antitumor immunity, which contributes to tumor regression42. We observed consistent repression of cGAS by hypoxia in different cell types, although miR25/93 is likely not essential for this effect in every case. Thus, we conclude that the repression of cGAS-IFN signaling in tumor cells is a critical event during cancer progression, indicating a critical role for hypoxia-induced cell-intrinsic immune tolerance in the development of tumors. Interestingly, difference in tumor-infiltrating immune cells (e.g. CD4+ T cells and MDSCs) between different experimental settings (e.g. Fig. S1 and S3) suggest that hypoxia-responsive molecules regulate a diversity of immunosuppressive pathways. Thus, further analyses are required to understand the interplay between the tumor microenvironment and tumor immune responses.
Excessive DAMPs influence immunity and are associated with inflammatory responses, which contribute to many chronic diseases, including cancer 43. mtDNA represents one type of DAMP. During cancer progression, hypoxia induces the intracellular translocation and release of mtDNA26, which may contribute to immunological stress. We observed increased levels of cytoplasmic mtDNA in hypoxic cells, leading to the induction of an immune response. This immune response was inhibited by the hypoxia-TET1-miR25/93 signaling axis, suggesting a direct link between hypoxia-responsive miRNAs and immune tolerance to DAMP-induced immunological stress. In summary, our results highlight the link between cGAS down-regulation and tumor immunosuppression, and implicate miR25/93 as a central regulator that acts in concert with hypoxia to regulate immune escape from DAMPs-induced immunological pressure. These findings have potential implications for anti-cancer immunotherapies.
Methods
Cells, plasmids, stable transfection, and oxygen deprivation
The E0771 murine breast cancer cell line was maintained in RPMI medium supplemented with 10% FBS and 1%penicillin/streptomycin. MCF7 and MDA-MB-231 cell lines were cultured in DMEM medium supplemented with10% FBS at 37°C in the presence of 5% CO2. A pcDNA3 mHIF-1α-MYC (P402A/P577A/N813A) (addgene) plasmid was used for generating HIF-1α overexpression clones in E0771 cells. A pBabe-NCOA3 expression vector was used for NCOA3 re-expression in human breast cancer cells. The plasmids for gene knockdown experiments were generated by inserting an oligonucleotide targeting a specific gene sequence into the pSUPER vector (oligoengine). These vectors were then used to establish stable tranfectants. For miRNA inhibition, MISSION LentimiRNA inhibitors (Sigma) targeting miR25 and miR93 were used to generate stable transfectants in MCF7 or MDA-MB-231 cells. For miRNA overexpression, the MDH1-PGK-GFP vector (addgene) was used to express human or mouse miR25/93 in human breast cancer cells or murine breast cancer cell, respectively. Oxygen deprivation was conducted in the hypoxic incubator with 1% O2, 5% CO2and 94% N2for 18 or 24 h.
Western blot analysis, RNA extraction, quantitative real-time PCR
Western blot was performed following standard protocols, as described19. Briefly, cells were harvested and lysed using RIPA buffer (50mMTris, 150mMNaCl, 0.1% SDS, 0.5% Deoxycholate, 1% NP-40). Protein extracts were subjected to SDS-PAGE analysis. The membranes were blocked with 5% nonfat milk followed by antibody hybridization and the signals were visualized on X-ray film. For RNA extraction, total RNAs from cultured cells were extracted using the Trizol reagent (Invitrogen Life Technologies), and 1 μg of RNA was used for cDNA synthesis. Quantitative real-time PCR was carried out to quantify gene expression levels by using the CFX384 Touch Real-Time PCR Detection System (BIO-RAD Laboratories, Inc.).
For measuring mtDNA in cytosol, as described44, the Mitochondria Isolation Kit (Thermo Fisher Scientific) was used to isolate cytosolic, mitochondrial, and nuclear fractions from cancer cells. Primers for human cytochrome B (and), human cytochrome C oxidase subunit III (and), human NADH dehydrogenase (and) were used in qPCR analysis. Cytoplamic mtDNA levels were normalized to nuclear DNA encoding 18S ribosomal RNA. The antibodies and primers used in the experiments are listed in supplementary table 3.
Luciferase reporter assays
Cells were seeded onto 6-well plates and transfected with the following: psiCheck2 luciferase reporter plasmid containing wild type or mutated Ncoa3 3′-UTR and expression vector encoding human miR25/93. Transfected cells were exposed to 20% or 1% O2 for 24 h. Luciferase activity in cell lysates was measured using Dual-Luciferase Reporter Assay System (Promega).
qPCR-ChIP and hMe-DIP
qPCR-ChIP assay was performed as described45. Briefly, crosslinked cell lysate was sonicated and subjected to an immunoprecipitation reaction with specific antibody conjugated beads. The immunoprecipitated DNA was purified through a phenol-chloroform DNA extraction protocol and then subjected to qPCR analysis. For the hMe-DIP assay, genomic DNA was prepared with the genomic DNA extraction kit (Promega) and sonicated into fragments of ~500bp. Fragmented DNA was denatured for 10 min at 95°C and immunoprecipitated overnight at 4°C with anti-5hmC antibody (Active Motif) in 500 μl IP buffer (10 mM sodium phosphate, 140 mM NaCl, 0.05% Triton X-100). DNA was eluted from the beads followed by purification with the chromatin IP DNA purification kit (Active Motif).
Flow cytometry and cytokines secretion assays
Cell surface marker staining and flow cytometric analysis for CD3, CD4, CD8, CD45, F4/80, and CD11b (Biolegend) expression was performed as described46. For MDSC analysis, mouse MDSC flow cocktail kit (Biolegend) was used. For intracellular staining for FOXP3 (Biolegend), True-Nuclear Transcription Factor Buffer Set (Biolegend) was used. To measure T cell cytokine production for effector CD8+ cytotoxic T cells, TICs were treated with Cell Activation Cocktail (Biolegend) for at least 4 hr at 37° before staining for TNF-α and IFN-γ (Biolegend).
In vivo models
All animal studies were performed under protocols approved by the Institutional Review Board of the Salk Institute for Biological Studies, La Jolla, U.S.A. This study is compliant with all relevant ethical regulations regarding animal research. Cells were trypsinized and resuspended in PBS/Matrigel (1:1; BD Biosciences). Suspended cells were then subjected to mammary fat pad injection or subcutaneous injection of C57BL/6 mice or IFNαβR−/− mice. Tumor size was measured by caliper. Tumor biospecimens were collected at a similar size for immune cell profiling analysis or gene expression analysis. After six weeks, all of the tumors were collected and the animals were sacrificed.
High-throughput whole transcriptome (mRNA-Seq) and small RNA-Seq
Total RNA was isolated by the Trizol reagent (Invitrogen Life Technologies) and treated with DNase. Invitrogen Qubit and Agilent Tape Station were used to determine RNA concentration and RNA integrity (RIN) numbers respectively, prior to library preparation. Stranded mRNA-Seq libraries were prepared using the Illumina TruSeq Stranded mRNA Library Prep Kit according to the manufacturer’s instructions. Briefly, RNA with poly-A tail was isolated using magnetic beads conjugated to poly-T oligos. mRNA was then fragmented and reverse-transcribed into cDNA. dUTPs were incorporated, followed by second strand cDNA synthesis. dUTP-incorporated second strand was not amplified. cDNA was then end-repaired, index adapter-ligated and PCR amplified. AMPure XP beads (Beckman Coulter) were used to purify nucleic acid after each steps of the library prep. Small RNA libraries were generated using NEB Next Multiplex Small RNA Library Prep Set for Illumina according to vendor’s instructions (New England Biolabs). Briefly, 3′ SR Adaptor was ligated to short RNAs at 25C for 1 hr. SR RT Primer was added to hybridize any un-ligated 3′SR adaptor to prevent adaptor-dimer formation. 5′ SR Adaptor was then ligated, followed by reverse transcription and PCR amplification to add indexes to each library. Amplified libraries were TBE-PAGE gel purified (130–150bp). All sequencing libraries were then quantified, pooled and sequenced at single-end 50 base-pair (bp) on Illumina HiSeq 2500 at the Salk NGS Core. Raw sequencing data was de-multiplexed and converted into FASTQ files using CASAVA (v1.8.2).
RNA-Seq Analysis
Sequenced reads were quality-tested using FASTQC and aligned to the hg19 human genome using the STAR aligner. Mapping was carried out using default parameters (up to 10 mismatches per read, and up to 9 multi-mapping locations per read). The genome index was constructed using the gene annotation supplied with the hg19 Illumina iGenomes collection and sjdbOverhang value of 100. FPKM normalized expression was quantified across all gene exons (RNA-Seq), using the top-expressed isoform as proxy for gene expression, and differential genes were defined as having a log2 fold change > 0.5 after the addition of a pseudocount of 5 to control for low-expressing noisy genes. The normalized expression table for all genes, and lists of differential genes is provided as supplementary data.
Clinical sample collection and evaluation of immunohistochemistry
Adequate surgical specimens from 148 breast invasive duct carcinoma (IDC) patients with relevant pathological information were collected from 1992 to 2005. This study was approved by The Institutional Review Board, Salk Institute for Biological Studies. The study is compliant with all relevant ethical regulations and informed consent was obtained by all participants. Tissue microarray slides were comprised of 45 of grade I cases, 53 of grade II cases and 50 of grade III cases. The histopathological differentiation of breast invasive duct carcinoma was determined on the basis of the WHO classification criteria for tumors. The pathological diagnosis for each cases was under blind examination by at least two senior pathologists. Immunohistochemistry (IHC) was performed using an EnVision Dual Link System-HRP (DAB+) kit. To evaluate the immunoreactivity of cGAS, cases were scored from 0 to 3+ (0, negative; 1+, low expression; 2+, intermediate expression; 3+, high expression) on the basis of the intensity of cGAS staining.
Statistics and reproducibility
For animal experiments no statistical method was used to predetermine sample size and the experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. All data were reported as mean ± SD. Statistical analysis was performed by using Student’s t test and p<0.05 was considered significant. The prognostic significance of the clinicopathological parameters related to survival rate was assessed by Kaplan–Meier analysis. All statistical analyses were performed using SPSS version 20 (SPSS, Inc., Chicago, IL, USC). The results relative to miRNAs expression in human breast cancer are based on data generated by The Cancer Genome Atlas pilot project (TCGA) established by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov.
Data availability
Sequencing data, including small RNA sequencing and RNA sequencing, have been deposited at the Gene Expression Omnibus (GEO) with accession number GSE79789. Source data for Figure 1, 2, 3, 4, 6, 7 and Supplementary Figure 1, 4, 5 have been provided as Supplementary Table 4. Previously published hMe-DIP-seq data is available under accession GSE60434. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgments
We are grateful to Drs. Manching Ku, Christopher Benner and Maxim Shokhirev of the Next Generation Sequencing Core and Integrative Genomics and Bioinformatics Core, respectively, at Salk Institute for Biological Studies for technical assistance. We would also like to thank David O’Keefe for his help in preparing the manuscript. This work was supported by The Razavi Newman Integrative Genomics and Bioinformatics Core Facility and the NGS Core Facility of the Salk institute with funding from NIH-NCI CCSG: P30014195, the Chapman Foundation and the Helmsley Charitable Trust. Work in the laboratory of J.C.I.B. was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002), The Moxie Foundation and UCAM.
References
- 1.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. 331/6024/1565 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6:715–727. doi: 10.1038/nri1936. nri1936 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. nrc2618 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palazon A, Aragones J, Morales-Kastresana A, de Landazuri MO, Melero I. Molecular pathways: hypoxia response in immune cells fighting or promoting cancer. Clin Cancer Res. 2012;18:1207–1213. doi: 10.1158/1078-0432.CCR-11-1591. 1078-0432.CCR-11-1591 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Noman MZ, et al. Microenvironmental hypoxia orchestrating the cell stroma cross talk, tumor progression and antitumor response. Crit Rev Immunol. 2011;31:357–377. doi: 10.1615/critrevimmunol.v31.i5.10. 47d4cd1f0e2c889b,64d7de0664c089e1 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Lewis C, Murdoch C. Macrophage responses to hypoxia: implications for tumor progression and anti-cancer therapies. Am J Pathol. 2005;167:627–635. doi: 10.1016/S0002-9440(10)62038-X. S0002-9440(10)62038-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corzo CA, et al. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207:2439–2453. doi: 10.1084/jem.20100587. jem.20100587 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noman MZ, et al. Tumor-Promoting Effects of Myeloid-Derived Suppressor Cells Are Potentiated by Hypoxia-Induced Expression of miR-210. Cancer Res. 2015;75:3771–3787. doi: 10.1158/0008-5472.CAN-15-0405. 0008-5472.CAN-15-0405 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Facciabene A, et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature. 2011;475:226–230. doi: 10.1038/nature10169. nature10169 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. S0092-8674(09)00008-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. nri2708 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. nrc1840 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Shen J, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. nature12080 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–671. doi: 10.1038/sj.cdd.4402310. 4402310 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Noman MZ, et al. The cooperative induction of hypoxia-inducible factor-1 alpha and STAT3 during hypoxia induced an impairment of tumor susceptibility to CTL-mediated cell lysis. J Immunol. 2009;182:3510–3521. doi: 10.4049/jimmunol.0800854. 182/6/3510 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 17.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. ncb1691 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Clambey ET, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A. 2012;109:E2784–2793. doi: 10.1073/pnas.1202366109. 1202366109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu MZ, et al. Hypoxia Drives Breast Tumor Malignancy through a TET-TNFalpha-p38-MAPK Signaling Axis. Cancer Res. 2015;75:3912–3924. doi: 10.1158/0008-5472.CAN-14-3208. 0008-5472.CAN-14-3208 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. 68/20/8191 [pii] [DOI] [PubMed] [Google Scholar]
- 21.van den Beucken T, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. ncomms6203 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariani CJ, et al. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014;7:1343–1352. doi: 10.1016/j.celrep.2014.04.040. S2211-1247(14)00342-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. science.1240933 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. science.1232458 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059296/5566/301. [pii] [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. J Hepatol. 2015;63:114–121. doi: 10.1016/j.jhep.2015.02.009. S0168-8278(15)00083-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rongvaux A, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. S0092-8674(14)01514-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. S0092-8674(14)01513-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. ncb2422 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Kim H, et al. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44:532–544. doi: 10.1016/j.molcel.2011.08.045. S1097-2765(11)00849-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng L, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. S1074-7613(14)00395-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J, et al. Steroid receptor coactivator-3 and activator protein-1 coordinately regulate the transcription of components of the insulin-like growth factor/AKT signaling pathway. Cancer Res. 2006;66:11039–11046. doi: 10.1158/0008-5472.CAN-06-2442. 66/22/11039 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Chen H, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. S0092-8674(00)80516-4 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Percharde M, et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–2298. doi: 10.1101/gad.195545.112. gad.195545.112 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. MCB.01395-06 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hazarika S, et al. MicroRNA-93 controls perfusion recovery after hindlimb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation. 2013;127:1818–1828. doi: 10.1161/CIRCULATIONAHA.112.000860. CIRCULATIONAHA.112.000860 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J, et al. MiR-215 Is Induced Post-transcriptionally via HIF-Drosha Complex and Mediates Glioma-Initiating Cell Adaptation to Hypoxia by Targeting KDM1B. Cancer Cell. 2016;29:49–60. doi: 10.1016/j.ccell.2015.12.005. S1535-6108(15)00469-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldar S, Roy A, Banerjee S. Differential regulation of MCM7 and its intronic miRNA cluster miR-106b-25 during megakaryopoiesis induced polyploidy. RNA Biol. 2014;11:1137–1147. doi: 10.4161/rna.36136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramalingam P, et al. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2014;20:76–87. doi: 10.1261/rna.041814.113. rna.041814.113 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camps C, et al. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol Cancer. 2014;13:28. doi: 10.1186/1476-4598-13-28. 1476-4598-13-28 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrocca F, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. S1535-6108(08)00049-4 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Corrales L, Gajewski TF. Molecular Pathways: Targeting the Stimulator of Interferon Genes (STING) in the Immunotherapy of Cancer. Clin Cancer Res. 2015;21:4774–4779. doi: 10.1158/1078-0432.CCR-15-1362. 1078-0432.CCR-15-1362 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. nature08780 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu MZ, et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43:811–822. doi: 10.1016/j.molcel.2011.07.012. S1097-2765(11)00535-1 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Lu LF, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. S0092-8674(10)00919-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data, including small RNA sequencing and RNA sequencing, have been deposited at the Gene Expression Omnibus (GEO) with accession number GSE79789. Source data for Figure 1, 2, 3, 4, 6, 7 and Supplementary Figure 1, 4, 5 have been provided as Supplementary Table 4. Previously published hMe-DIP-seq data is available under accession GSE60434. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.