Abstract
Immune cells of the myeloid lineage are key players in skin wound healing, since they secrete various cytokines and growth factors that orchestrate the repair process. In addition, they are crucial for the defense against invading pathogens through their capacity to produce high levels of reactive oxygen species (ROS). To limit the toxicity of ROS, cells have developed antioxidant defense strategies, including expression of the cytoprotective NRF2 transcription factor. Here we show that murine neutrophils and to a lesser extent macrophages strongly express Nrf2 already when present in the circulation and in particular at the wound site. To determine the role of Nrf2 in neutrophils and macrophages for wound repair, we generated mice with a gain- or loss-of-function of this transcription factor in the myeloid cell lineage. Expression of a constitutively active Nrf2 mutant in myeloid cells did not further enhance the overall Nrf2 activity in these cells due to the already high steady-state activity of endogenous Nrf2. Surprisingly, deletion of Nrf2 in myeloid cells only mildly affected the levels of ROS and the expression of pro-inflammatory cytokines by these cells. In particular, various parameters of wound healing, including wound closure, reepithelialization, wound contraction and the presence of myeloid cells at the wound site were not affected. These results reveal that Nrf2 in myeloid cells is dispensable for wound healing and suggest the presence of additional antioxidant defense strategies of these cells that compensate for the loss of Nrf2, even in the harsh environment of skin wounds.
Introduction
Wounding of the skin initiates a complex repair process that encompasses several temporally overlapping and highly coordinated events, including blood clotting, inflammation, new tissue formation and finally extensive tissue remodeling. These processes involve interaction among different cell types, including skin resident cells like keratinocytes, fibroblasts and endothelial cells, as well as infiltrating immune cells [1, 2]. Unfortunately, repair of skin wounds in adult mammals is imperfect and results in scar formation and consequent functional impairments. In addition, aged individuals as well as patients treated with anti-inflammatory steroids, chemo- or radiotherapy, and those suffering form certain major diseases, e.g. diabetes and cancer, frequently suffer from impaired healing [2]. This may result in the formation of chronic, non-healing ulcers, which strongly impair the quality of life and cause an enormous financial burden to the health care system [3]. Therefore, there is a strong need to improve the healing process and this requires a thorough understanding of the underlying cellular and molecular mechanims.
Myeloid cells, which include neutrophils, monocytes and macrophages, play important roles during different phases of wound healing, in particular during the early inflammatory phase. These cells produce growth factors and enzmyes that directly contribute to the healing response, together with large amounts of ROS, which are required for the defense against invading pathogens [4]. While this effect of ROS is beneficial, it has been suggested that excessive levels of ROS produced in skin wounds cause tissue damage, delay the healing response, and favour the development of non-healing ulcers and of hypertrophic scars [5]. Therefore, it is important to tightly control the levels of ROS at the wound site. A key player in the control of the cellular ROS balance is the nuclear factor erythroid derived 2-related factor 2 (NRF2; NFE2L2) transcription factor. Under normal conditions the NRF2 inhibitor KEAP1 retains NRF2 in the cytoplasm and also mediates its ubiquitination and subsequent proteasomal degradation. Activation of NRF2 occurs through conformational changes of KEAP1, which are induced by covalent coupling of electrophilic compounds to this protein. These conformational changes result in the weakening of the NRF2-KEAP1 interaction and stabilization of NRF2. Furthermore, it has been suggested that ROS activate certain kinases that phosphorylate NRF2, thereby affecting its binding to KEAP1. As a consequence of these events, the NRF2 turnover is modified and newly synthetized NRF2 is stabilized and accumulates in the nucleus. Upon dimerization with small Maf proteins, NRF2 binds to antioxidant response elements (AREs) in the promoters or enhancers of NRF2 target genes to activate their expression [6, 7]. Among the major NRF2 targets are genes encoding ROS-detoxifying enzymes and other antioxidant proteins, as well as different transporters and proteins involved in the production of the low molecular weight antioxidant glutathione [6].
We and others recently identified important roles of Nrf2 in wound repair of mice. While wound closure in normal mice was not affected in the absence of Nrf2, Nrf2 knockout mice showed a prolonged inflammatory response after wounding, with macrophages still being abundant during the tissue remodelling phase [8]. However, upon induction of diabetes in mice with a global Nrf2 knockout, the wound healing process was significantly delayed compared to diabetic wild-type controls [9]. Conversely, pharmacological activation of Nrf2 or treatment of wounds from diabetic mice with Keap1 siRNA promoted wound healing in diabetic mice [9–11]. It is unclear, however, which cell types require normal Nrf2 levels for efficient wound healing and which cell types would benefit from an increase in Nrf2 activity. In this study we addressed this question and determined the effect of Nrf2 activation and deletion specifically in myeloid cells for wound repair in mice.
Materials and methods
Genetically modified mice
CMVcaNrf2 mice have been described previously [12, 13]. By crossing CMVcaNrf2 mice (FVB/N1 genetic background) with LysM-Cre transgenic mice (C57BL/6 genetic background), constitutive expression of the caNrf2 mutant was induced in cells of the myeloid lineage. Mice lacking Nrf2 in myeloid cells were obtained by mating of LysM-Cre transgenic mice with mice carrying floxed alleles for Nrf2 (C57BL/6 genetic background) [14]. Mice were housed under optimal hygiene conditions and received food and water ad libitum. Mouse maintenance and all animal experiments had been approved by the local veterinary authorities of Zurich, Switzerland (Kantonales Veterinäramt Zürich).
Wounding and preparation of wound tissue
Female mice (8–10 weeks old) were anaesthetized by inhalation of isoflurane prior to and during wound surgery. Four full-thickness excisional wounds of 5 mm diameter were generated on the back of mice using disposable 5mm biopsy punches (Stiefel Laboratorium, Winterthur, Switzerland). Wounds were allowed to heal without dressing. After surgery and during the healing phase, mice were closely observed for any signs of pain or discomfort. In very rare cases where symptoms were observed the mice were sacrificed by CO2 inhalation.
For the analysis of the wound healing process, mice were sacrificed by CO2 inhalation at different time points after injury, and wounds were excised with a 5mm biopsy punch. Two of the four wounds were placed in RPMI 1640/HEPES medium for analytical flow cytometric analysis. The remaining two wounds were bisected and one half of each wound was snap frozen for RNA isolation and the other half was prepared for histological/morphometric analysis as described below. For fluorescence-activated cell sorting (FACS) experiments we used all the four wounds to obtain enough isolated cells.
We used 3–4 mice per time point for FACS sorting, 3–6 mice (5–12 wounds) per time point and genotype for the analytical flow cytometry analysis and 4–5 mice (6–8 wounds) per genotype for the morphometric analysis of the wounds.
Histology
For histological analysis of the excised wounds and normal skin, the tissue was fixed overnight in 4% paraformaldehyde (PFA) in PBS or in 95% ethanol / 1% acetic acid, followed by paraffin embedding. Paraffin sections (7μm) from the middle of the wounds were stained using the Herovici procedure [15]. Sections from normal skin were stained with hematoxylin/eosin (H/E). Stained sections were photographed using a Zeiss Axioskop 2 microscope with an Axiocam HRc camera or with the Panoramic 250 Slide Scanner (3D Histech, Budapest, Hungary), and histomorphometrical measurements were quantified using FIJI software [16].
Isolation of RNA from mouse wounds and from sorted cells
Total RNA was isolated from skin wounds using the RNeasy fibrous tissue mini kit, including a proteinase K digestion step and on-column DNase treatment (Qiagen, Hilden, Germany).
For RNA isolation from sorted cells, they were collected in Eppendorf tubes containing flow buffer (0.2%BSA, 5 mM EDTA in 1 x PBS) supplemented with RNAsin Plus RNase inhibitor (Promega, Madison, WI). The sorted cells were immediately centrifuged, and RNA was isolated using the RNease micro kit with on-column DNase treatment.
RT-qPCR
cDNA was synthesized using the iScript kit (Bio-Rad Laboratories, Hercules, CA). Quantitative real-time RT-PCR (RT-qPCR) was performed according to the manual of the Light Cycler 480 II (Roche, Rotkreuz, Switzerland). After an initial denaturation step at 95°C for 10min the reaction was performed in 50 cycles (95°C for 10s, 60°C for 20s and 72°C for 20s for each cycle). Samples were analyzed in duplicates. Rps29 was used as a reference gene. Target gene expression levels were quantified by second derivative method. To determine relative expression levels in the FACS-sorted cells the value from a lymphocyte RNA sample was arbitrarily set to 1 and all the values of the other samples are given relative to this lymphocyte sample. When lymphocytes were not included, a sample from the macrophage population was set to 1. For the RNA data from wound samples, the value from one control mouse was set to 1.
Primers used for RT-qPCR are listed below in Table 1.
Table 1. RT-qPCR primer list.
| Target gene | Forward sequence 5'->3' | Reverse sequence 5'->3' |
|---|---|---|
| Nrf2 | 5’-CCAGCTACTCCCAGGTTGC-3’ | 5’-CCAAACTTGCTCCATGTCCT-3’ |
| caNrf2 | 5’-TGTTCCTTGTTCCCAAAAGC-3’ | 5’-CCGGAATATTAATAGGGACGGTA-3’ |
| Nrf2 Exon5 | 5’- TCCATTCCCGAATTACAGTGTCTTA-3’ | 5’-CGCCAAAATCTGTGTTTAAGGTG-3’ |
| Nqo1 | 5’- TTTGAGAGAGTGCTCGTAGC-3’ | 5’- GGTCTTCTTATTCTGGAAAGG-3’ |
| Gclc | 5’-AACAAGAAACATCCGGCATC-3’ | 5’-CGTAGCCTCGGTAAAATGGA-3’ |
| Srxn1 | 5’-CGGTGCACAACGTACCAAT-3’ | 5’-TTGATCCAGAGGACGTCGAT-3’ |
| Slpi | 5’-GGGCAAATACAAGTGCTGTG-3’ | 5’-CCTGGGAGCAGGGAAGTAGT-3’ |
| Il1b | 5’-GGACAGAATATCAACCAACAAGTG-3’ | 5’-TGCTGATGTACCAGTTGGGG-3’ |
| Il6 | 5’-CCGGAGAGGAGACTTCACAG-3’ | 5’-TTCTGCAAGTGCATCATCGT-3’ |
| Tnfa | 5’-GACCCTCACACTCAGATCATCTTCT-3’ | 5’-CCACTTGGTGGTTTGCTACGA-3’ |
| Vegfa | 5’-GTACCTCCACCATGCCAAGT-3’ | 5’-CTGCATGGTGATGTTGCTCT-3’ |
| Rps29 | 5'-GGTCACCAGCAGCTCTACTG-3' | 5'-GTCCAACTTAATGAAGCCTATGTCC-3' |
Isolation of cells from normal and wounded skin for flow cytometry
Mice were sacrificed, shaved, and the non-wounded back skin or wound biopsies were harvested. In case of normal back skin, the subcutaneous fat was scraped off, and epidermis and dermis were separated using 0.25% dispase (Gibco, Zug, Switzerland) for 50min at 37°C under continuous shaking. The epidermis was treated with 100 KUnits/ml DNase (Sigma) in RPMI supplemented with penicillin/streptomycin (P/S), 10% fetal bovine serum and 20mM HEPES for 45 min at 37°C under continuous shaking. The dermis as well as the wound biopsies were cut into small pieces and treated with 0.25mg/ml TL Liberase (Roche) in RPMI supplemented with P/S and 20 mM HEPES for 1h at 37°C under continuous shaking. Finally, single cell suspensions were prepared by straining the mixture through 70 μm cell strainers, and cells were washed with 1x PBS.
Flow cytometry
Unspecific binding sites were blocked with anti-CD16/CD32 antibody. Dead cells were labeled with Zombie Aqua™ dye (BioLegend, San Diego, CA) and gated out. Expression of different antigens was analyzed using BD LSRII Fortessa equipped with BD FACSDivaTM 6.0 software (both from BD Biosciences, San Jose, CA). Data analysis was performed using FlowJo v8.7 or FlowJo v10 software (FlowJo, Ashland, OR). Compensation was performed using single color controls prepared from compensation beads (BD Biosciences) and skin samples. Samples were acquired using Fortessa’s High Throughput Sampler plate reader option with an event rate below 20,000 events/s. Compensation matrices calculated by BD FACSDivaTM 6.0 software were applied and cell populations were defined by manual gating. Doublets and dead cells were excluded from the analysis. The dyes and antibodies used for flow cytometry are listed below in Table 2:
Table 2. Antibodies and reagents used for flow cytometry.
| Antigen | Clone | Fluorophore | Dilution | Source |
|---|---|---|---|---|
| CD16/32 | 2.4G2 | 1:200 | BD Biosciences, Franklin Lakes, NJ | |
| CD45 | 30-F11 | BV 605 | 1:500 | BD Biosciences |
| 30-F11 | PB | 1:400 | BD Biosciences | |
| CD11b | M1/70 | PE | 1:200 | BD Biosciences |
| M1/70 | BV 711 | 1:500 | BioLegend, San Diego, CA | |
| CD3 | 17A2 | BV 785 | 1:300 | BioLegend |
| 145-2C11 | PE | 1:400 | BD Biosciences | |
| F4/80 | Cl:A3-1 | AF 647 | 1:200 | AbD Serotec, Kidlington, UK |
| Ly6C | AL-21 | PE | 1:200 | BD Biosciences |
| HK1.4 | PE-Cy7 | 1:500 | BioLegend | |
| HK1.4 | PerCP-Cy5.5 | 1:500 | BioLegend | |
| Ly6G | 1A8-Ly6g | AF 700 | 1:100 | BioLegend |
| 1A8-Ly6g | FITC | 1:200 | BD Biosciences | |
| 1A8-Ly6g | PerCP-Cy5.5 | 1:200 | BioLegend | |
| Live/Dead | Zombie Aqua | 1:400 | BioLegend |
Determination of intracellular ROS levels
Mice were euthanized by CO2 inhalation and femurs and tibias were removed. The marrow cavities were flushed with sterile Dulbecco’s Modified eagle Medium (DMEM; Sigma-Aldrich, Saint Louis, MO) and single cells were obtained by filtering through 70 μm cell strainers. Cells were plated at equal numbers and then pre-incubated at 37°C for 15 min with 5 μM dichloro-dihydro-fluorescein diacetate (DCFH-DA) and subsequently incubated with the H2O2-producing enzyme glucose oxidase (85 mU) along with the flow cytometry antibodies for another 15 min and immediately analyzed by flow cytometry using the BD LSRII Fortessa.
Statistical analysis
Statistical analyses were performed using PRISM software v7 (Graph pad Software Inc., La Jolla, CA. For normal data sets, Student’s t-test or one-way ANOVA tests were performed, depending on whether two or more than two groups were compared. Tukey’s-corrected multiple comparisons with adjusted P value reporting was applied in the case of one-way ANOVA. For non-normal data sets, pairwise comparisons were performed using the Mann-Whitney U rank test. *P ≤0.05; **P ≤0.01; ***P ≤0.001, **** P ≤0.0001.
Results
Nrf2 is strongly expressed in neutrophils in the circulation and in the wound tissue
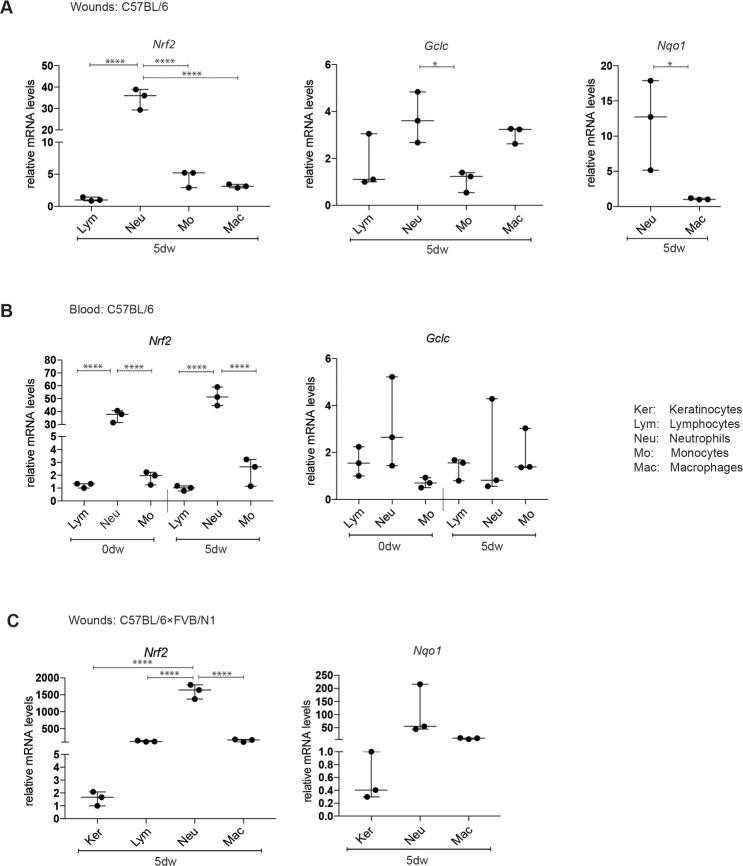
To investigate the expression of Nrf2 in myeloid cells during wound healing, we generated full-thickness excisional wounds on the back of wild-type mice of C57BL/6 genetic background and isolated different types of immune cells from the wound tissue at day 5 after injury when neutrophils and macrophages are abundant. For this purpose we established an optimized procedure based on gentle dissociation of the skin samples followed by fluorescence activated cell sorting (FACS) with a pre-established gating strategy (S1 Fig). In the same experiment, we isolated different populations of immune cells from the blood prior to wounding and during the healing phase. The isolated cells included neutrophils (CD45+CD11b+Ly6G+), a macrophage-enriched population (CD45+CD11b+F4/80+ that also includes some Langerhans cells and monocytes [17]), monocytes/inflammatory macrophages (CD45+CD11b+Ly6C+) and lymphocytes (CD45+CD11b-). Interestingly, Nrf2 mRNA levels were extremely high in neutrophils of the wound tissue and of the blood and more than 10-fold higher compared to monocytes, macrophages or non-myeloid cells (lymphocytes) (Fig 1A and 1B). We also analyzed at least one of the classic Nrf2 target genes NAD(P)H dehydrogenase quinone 1 (Nqo1) and glutamate-cysteine ligase catalytic subunit (Gclc) [7]. Indeed, the different Nrf2 expression levels between wound neutrophils and macrophages were reflected by similar differences in the expression of Nqo1, while the difference between neutrophils and macrophages was less pronounced for Gclc (Fig 1A and 1B). Since we subsequently used a C57BL/6 × FVB/N1 background for the generation of Nrf2 mutant mice, we repeated this experiment in the mixed background and confirmed the strong expression of Nrf2 and Nqo1 in wound neutrophils. Remarkably, Nrf2 mRNA levels were more than 500-fold higher in neutrophils compared to keratinocytes that had been freshly isolated by FACS (CD49f+CD140a-CD45-CD31-) (Fig 1C). This is remarkable, since Nrf2 plays an important role in keratinocytes under different conditions [18]. This experiment also revealed that wound lymphocytes express higher levels of Nrf2 compared to keratinocytes, but still much less compared to neutrophils (Fig 1C).
Fig 1. Expression of endogenous Nrf2 and its target genes in keratinocytes and immune cells.
Immune cells from the blood or the wound tissue were isolated by FACS based on the following markers: CD45+CD11b- (lymphocytes), CD45+CD11b+Ly6G+ (neutrophils), CD45+CD11b+F4/80+ (mainly macrophages, but also some Langerhans cells and monocytes) and CD45+CD11b+Ly6C+ (monocytes/inflammatory macrophages). (A) RT-qPCR analysis using RNA from sorted cells of 5-day wounds (dw) from C57BL/6 mice for Nrf2, Gclc and Nqo1 relative to Rps29. (B) Blood was collected prior to wounding (0dw) and at day 5 after wounding (5dw) of C57BL/6 mice, and RNA from FACS-sorted immune cells was analyzed by RT-qPCR analysis for Nrf2 and Gclc. (C) RT-qPCR analysis using RNA from sorted immune cells and keratinocytes (CD49f+CD140a-CD45-CD31-) of 5-day wounds (dw) from C57BL/6 x FVB/N1 mice for Nrf2, Gclc and Nqo1 relative to Rps29. N = 3–6 mice. Bars indicate median with 95% confidence interval (CI). Each data point represents the result from one mouse. *P ≤0.05; **P ≤0.01; ***P ≤0.001, **** P ≤0.0001 (one-way ANOVA for multiple group comparisons and Student’s t-test for two-group comparison).
Genetic activation of Nrf2 in myeloid cells does not affect cutaneous wound healing
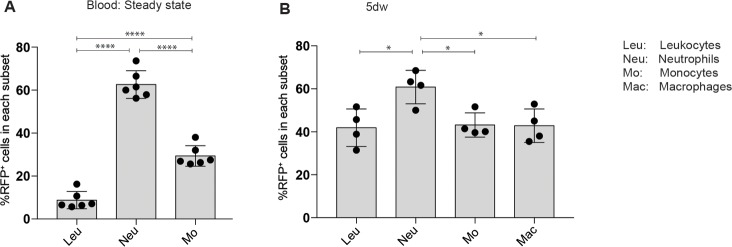
We next determined if modification of Nrf2 expression in myeloid cells affects the wound healing process. To target these cells we made use of LysM-Cre transgenic mice [19]. To verify the specificity of this promoter for myeloid cells in wound tissue, we used LysM-Cre/ROSA-RFP mice, which express red fluorescence protein (RFP) in cells in which the Lysozyme M promoter is active. Flow cytometry revealed that approximately 60% of the neutrophils were RFP positive in the blood or in 5-day full-thickness excisional wounds, but only 30–40% of the monocytes and macrophages (Fig 2A and 2B and S2 Fig).
Fig 2. Characterization of deletion efficiency of LysM-Cre mice in blood and wound leukocytes.
Flow cytometry analysis to quantify the percentage of red fluorescent protein positive (RFP+) cells among all leukocytes or among neutrophils, monocytes or macrophages (A) in the blood of non-injured mice (N = 6 mice) or (B) in 5-day wounds (n = 4 wounds obtained from two different mice (N = 2)). Bars indicate mean ±SD. *P ≤0.05; **P ≤0.01; ***P ≤0.001, **** P ≤0.0001 (one-way ANOVA).
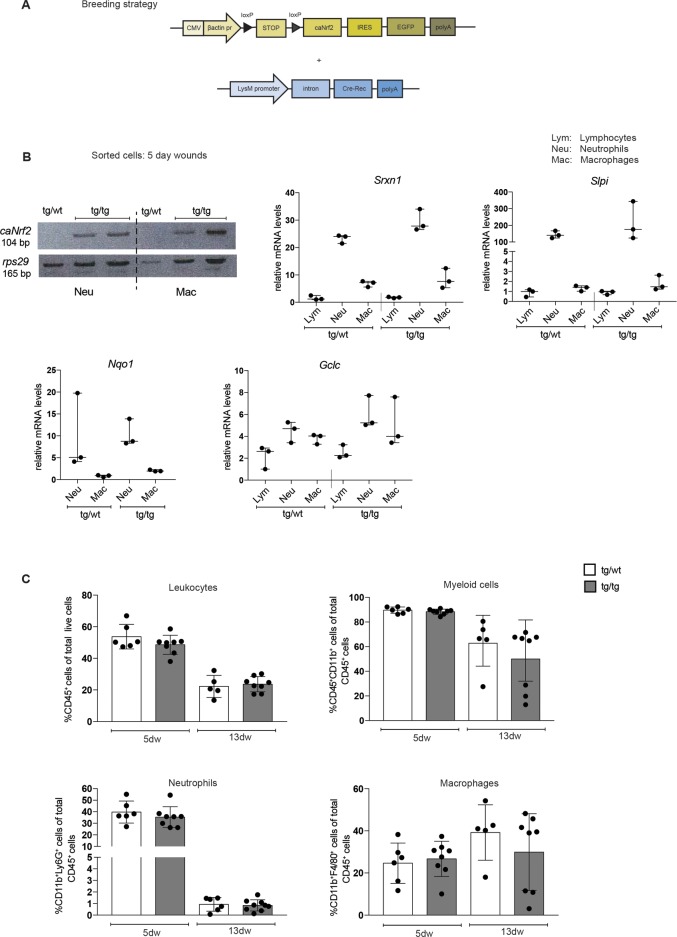
Being aware of the only partial targeting of the wound myeloid cell population by the LysM-Cre transgenic mice, we used them to express a constitutively active Nrf2 mutant (caNrf2) in myeloid cells by mating them with mice expressing caNrf2 under control of a cytomegalovirus enhancer and a β-actin promoter. To avoid constitutive expression of the transgene, it is preceded by a transcription/translation STOP cassette flanked by loxP sites (Fig 3A) [12]. Upon mating with LysM-Cre mice, myeloid cells are expected to express the caNrf2 transgene.
Fig 3. Genetic activation of Nrf2 in myeloid cells.
(A) Schematic representation of the transgenes used for the generation of LysM-Cre CMVcaNrf2 mice. (B) Left upper panel: RNA from sorted neutrophils or macrophages from 5dw of control (tg/wt) and LysM-cre CMVcaNrf2 mice (tg/tg) were analyzed for caNrf2 transgene expression and for Rps29 by RT-qPCR. PCR products were analyzed by agarose gel electrophoresis. Other panels: RT-qPCR using RNA from lymphocytes, neutrophils and macrophages isolated from 5d wounds for the Nrf2 target genes Gclc, Srxn1, Slpi, and Nqo1 relative to Rps29. N = 3 mice. Bars indicate median with 95% CI. (C) Flow cytometry analysis of 5dw and 13dw of tg/wt and tg/tg mice for total leukocytes, myeloid cells, neutrophils and macrophages. N = 3–5 mice, n = 5–10 wounds. Bars indicate mean ±SD. *P ≤0.05; **P ≤0.01; ***P ≤0.001, **** P ≤0.0001 (Mann-Whitney U test).
The double transgenic mice were viable and fertile and did not reveal obvious macroscopic abnormalities. Histological analysis of their non-injured skin did not uncover obvious abnormalities. Furthermore, flow cytometry analysis of skin immune cells and blood cells did not reveal a significant difference between caNrf2-transgenic mice and their littermate controls (S3 Fig).
To verify the specific expression of the transgene in myeloid cells, we isolated lymphocytes, neutrophils, and macrophages from 5-day wounds of double transgenic LysM-Cre CMV-caNrf2 mice (designated tg/tg) and single transgenic LysM-Cre control mice (designated tg/wt) using FACS. RT-PCR analysis revealed that caNrf2 was indeed only expressed in neutrophils and macrophages of the double mutant mice (Fig 3B). Surprisingly, however, expression of Nrf2 target genes, including Gclc, sulfiredoxin-1 (Srxn1), secretory leukocyte protease inhibitor (Slpi), and Nqo1, was only slightly, but non-significantly higher in tg/tg mice (Fig 3B). This experiment was performed with mice in a mixed genetic background (C57BL/6 × FVB/N1) and with mice in a pure C57BL/6 background (S4 Fig). Furthermore, flow cytometry revealed no differences in the number of total leukocytes, total myeloid cells, neutrophils and macrophages, in the wounds at day 5 and day 13 after injury (Fig 3C). These findings suggest that the caNrf2 mutant cannot further increase the high activity of Nrf2 in these cells, and we therefore did not further characterize the wound healing process in these animals.
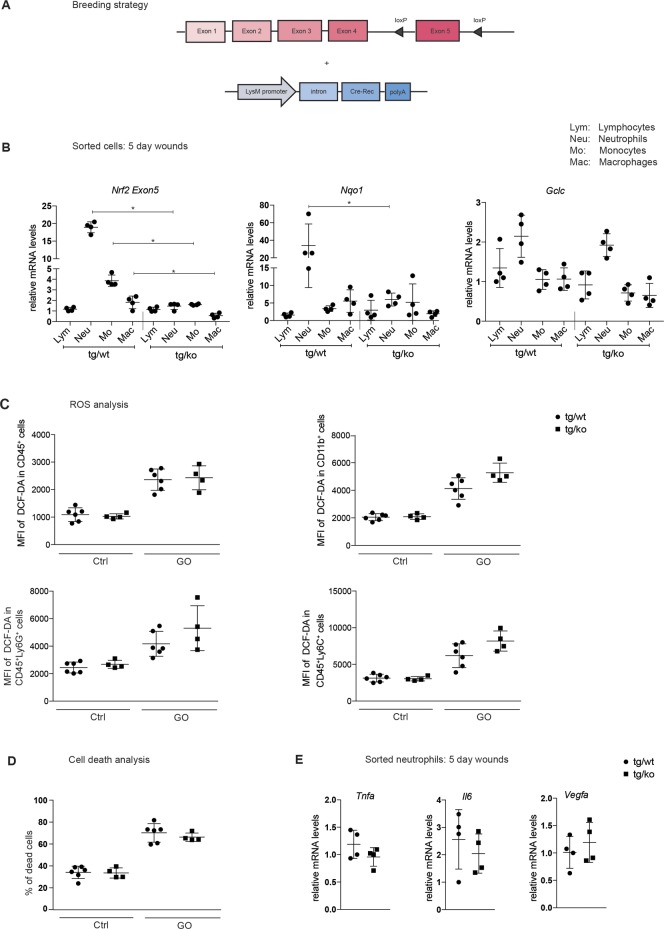
Generation of mice lacking Nrf2 in myeloid cells
Due to the strong expression of Nrf2 in myeloid cells, it was particularly interesting to determine the consequences of a loss of Nrf2 in these cells for wound repair. For this purpose, we mated mice with floxed Nrf2 alleles [14] with LysM-Cre mice (Fig 4A). The double transgenic LysM-Cre-Nrf2-ko mice (designated tg/ko) were viable and fertile, and did not exhibit obvious abnormalities, including no alterations in the histological appearance of the skin. Flow cytometry analysis demonstrated that loss of Nrf2 in myeloid cells did not affect major immune cell populations in the non-injured skin (S5 Fig).
Fig 4. Characterization of LysM-Cre Nrf2-ko mice.
(A) Schematic representation of the breeding strategy for LysM-Cre Nrf2-ko mice. (B) RNA from different immune cells sorted from 5dw of control (tg/wt) and LysM-Cre Nrf2-ko (tg/ko) mice were analyzed by RT-qPCR for Nrf2 (exon5), Nqo1 and Gclc relative to Rps29. N = 4 mice per genotype. *P ≤0.05 (Mann-Whitney U test). (C) Bone marrow derived cells (BMDCs) from tg/wt and tg/ko mice were pre-incubated for 15 min with 5 μM DCFH-DA and subsequently stimulated with glucose oxidase (GO; 85 mU) or left untreated (control) for another 15 min. Cells were analyzed by flow cytometry based on DCF fluorescence (reflecting ROS levels). Results shown are representatives of at least three independent experiments. N = 2–3 mice per experiment, two data points represent each mouse. (D) Survival of BMDCs after GO treatment. The percentage of dead cells as determined by flow cytometry is shown. (E) RNA from sorted neutrophils of 5dw was analyzed by RT-qPCR for Tnfa, Il6 and Vegfa relative to Rps29. N = 4 mice. Bars indicate mean ±SD. *P ≤0.05; **P ≤0.01; ***P ≤0.001, **** P ≤0.0001 (Mann-Whitney U test).
We next isolated different types of immune cells from 5-day wounds by FACS and analyzed the expression of Nrf2 and selected target genes. As expected, there was a significant reduction in the levels of the full-length Nrf2 mRNA in neutrophils and to a lesser extent in monocytes and macrophages of tg/ko mice as determined by RT-qPCR with primers for exon 5, which is targeted in the mutant mice. This correlated with significantly reduced expression of Nqo1, and a milder reduction of Gclc expression was also detectable (Fig 4B). By contrast, and as expected from the LysM promoter activity, expression of Nrf2 and Nrf2 target genes was not affected in lymphocytes (Fig 4B).
Since the knockout of Nrf2 was particularly efficient in neutrophils, we further analyzed the consequences of the Nrf2 knockout for these cells. Due to the important role of Nrf2 in the control of the cellular redox balance, we treated bone marrow derived cells (BMDC) from tg/ko and tg/wt mice with glucose oxidase, which results in continuous production of hydrogen peroxide. The levels of ROS in these cells were then measured using a 2`,7`- DCFH-DA-based assay. However, the loss of Nrf2 only caused a very mild, but non-significant increase in ROS levels in the different myeloid cell populations (Fig 4C). There was also no difference in the percentage of dead cells before or after GO treatment of the BMDCs (Fig 4D).
Finally, loss of Nrf2 in neutrophils did not affect the expression of the pro-inflammatory cytokines tumor necrosis factor alpha (Tnfa) and interleukin 6 (Il6) or of the pro-angiogenic factor vascular endothelial growth factor A (Vegfa) (Fig 4E).
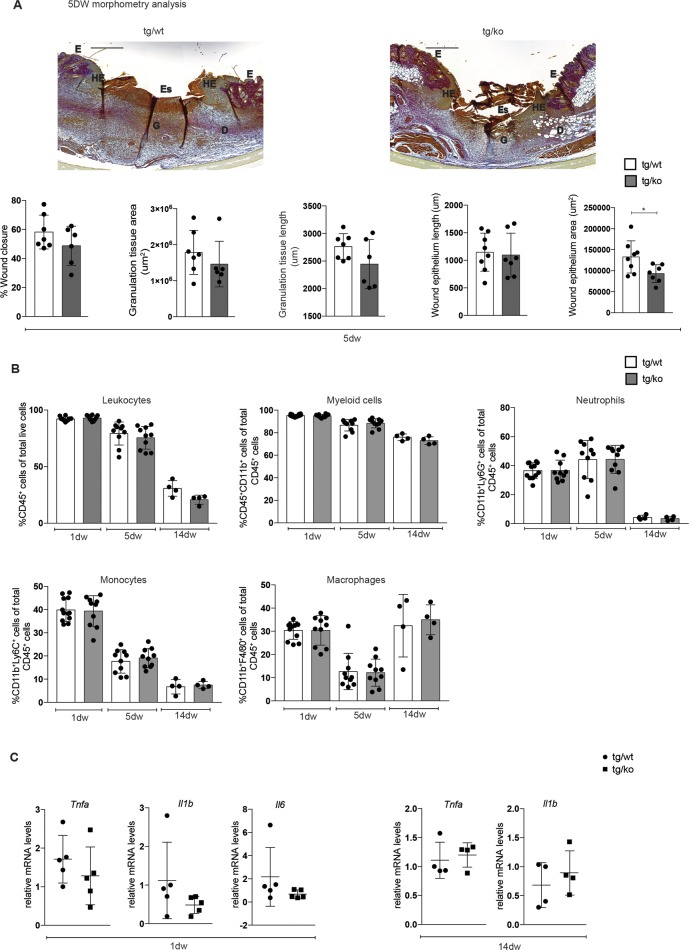
Nrf2 in myeloid cells is dispensable for wound healing in mice
Since loss of Nrf2 in all cells affected the inflammatory response in healing wounds [8], we investigated if this phenotype results directly from the loss of Nrf2 in myeloid cells. Histomorphometric analysis of sections from the middle of the wounds did not reveal significant alterations in wound closure, granulation tissue area and length, or length of wound epithelium in 5-day wounds of tg/ko mice compared to their littermate controls (Fig 5A). The area of wound epithelium was significantly smaller in the tg/ko compared to control mice, indicating that Nrf2-deficient myeloid cells that are present in the wound bed affect keratinocytes in a paracrine manner. However, the rate of wound closure was not significantly reduced. Furthermore, the immune cell composition at day 1, day 5 and day 14 after wounding was not obviously affected, suggesting that the lack of Nrf2 in myeloid cells neither affected the infiltration of immune cells nor their resolution (Fig 5B). Consistent with this finding, expression of the pro-inflammatory cytokines Tnfa, Il1b and Il6, which are highly expressed in neutrophils early after injury [20], was not affected in 1-day wounds of tg/wt mice, and Tnfa and Il1b mRNA levels were also similar in 14-day wounds of mice of both genotypes (Fig 5C).
Fig 5. Wound healing is not affected in LysM-Cre Nrf2-ko mice.
(A) Representative pictures of Herovici-stained sections from 5dw of tg/wt and tg/ko mice. Scale bar: 500 μm. D: Dermis; E: Epidermis, Es: Eschar; G: Granulation tissue; HE: Hyperproliferative wound epidermis. Quantitative data from a morphometric analysis of 5dw of tg/wt and tg/ko mice for wound closure, area and length of the granulation tissue, and area and length of wound epithelium are shown below (N = 3–4 mice, n = 6–8 wounds). The raw data of the morphometric analysis of the wounds are shown in S1 Table. (B) Flow cytometry analysis of 1dw, 5dw and 14dw of tg/wt and tg/ko mice for total leukocytes, myeloid cells, neutrophils, monocytes and macrophages. N = 4–6 mice, n = 8–12 wounds. (C) RNA from 1dw and 14dw from control and conditional knockout mice was analyzed for Tnfa, Il1b and Il6 relative to Rps29. Bars indicate mean ±SD. *P ≤0.05; **P ≤0.01; ***P ≤0.001, **** P ≤0.0001 (Mann-Whitney U test).
Together, these results show that loss of Nrf2 in myeloid cells does not affect the wound healing capability in normal mice. Therefore, the altered inflammatory response that occurred during wound healing in mice with global Nrf2 knockout is most likely an indirect effect that is mediated via skin resident cells.
Discussion
Hematopoietic cells of the myeloid lineage play an irreplaceable role in host defense mechanisms. They are also key regulators of wound healing, a process that involves an intricate and highly orchestrated interplay between various innate and adaptive immune cells along with the skin resident epithelial and mesenchymal cells [1, 4, 21]. A major task of myeloid cells in wound healing is the production of high levels of ROS as a mechanism to defend invading pathogens. However, these reactive compounds can severely damage cells in the wound bed, including the myeloid cells themselves. Therefore, these cells had to develop various antioxidant defense strategies, including expression of the cytoprotective transcription factor Nrf2 [22–24]. Indeed, abnormal expression/activation of Nrf2 in myeloid cells had severe effects in different mouse models for major human diseases, including, but not limited to cancer, atherosclerosis, and emphysema [25]. Therefore, we speculated that Nrf2 in myeloid cells has an important role in wound healing.
Consistent with this assumption we show here that myeloid cells, in particular neutrophils, display very high Nrf2 expression not only during wound healing, but also while circulating in the blood under physiological conditions. This finding suggests that Nrf2 protects these cells from oxidative damage in injured/inflamed tissues and allows them to rapidly adapt to inflammatory conditions. This beneficial activity may be further promoted by activation of endogenous Nrf2, since it has previously been shown in in vitro and ex vivo settings that chemical activation of Nrf2 in macrophages and neutrophils can have anti-inflammatory or anti-oxidative effects, respectively [26, 27]. On the other hand, loss of Nrf2 in myeloid cells was detrimental in different settings. For example, knockout of Nrf2 in myeloid cells aggravated the symptoms in a mouse model of systemic sepsis and resulted in dramatically reduced survival of the mice, whereas deletion of the Nrf2 antagonist Keap1 in myeloid cells had a protective function in this model. This finding was explained by enhanced ROS detoxification by myeloid cells in the absence of Keap1 [14]. However, Keap1 has also Nrf2-independent functions [28, 29], which may contribute to this phenotype. This limitation can be overcome by the use of a constitutively active Nrf2 mutant. Using this approach, it was shown that genetic activation of Nrf2 in myeloid cells leads to less inflammation and ultimately less tissue injury in the liver after hepatic ischemia/reperfusion injury [30]. Furthermore, genetic activation of Nrf2 in myeloid cells aggravated acute colitis in mice, while it had a beneficial effect in a chronic colitis model [31]. These findings suggest that the effect of Nrf2 activation may depend on the affected organ, the injury model and on the duration of the inflammation.
To our surprise, however, neither Nrf2 activation nor loss of this protein in myeloid cells affected the wound healing process in mice. The lack of an effect of Nrf2 activation may be explained by the already high expression and activity of this transcription factor in myeloid cells of the wound tissue, which could not be further enhanced by the caNrf2 mutant. This may be different in myeloid cells of the liver or intestine. Alternatively, caNrf2 may be able to upregulate other Nrf2 target genes that we have not analyzed and which may be important in repair of the liver and intestine, but not in murine wound repair. Finally, it may well be that the targeting by the LysM promoter is more efficient in myeloid cells of the liver and intestine compared to wound myeloid cells. The rather inefficient targeting of myeloid cells, in particular of macrophages, may explain at least in part why we did not observe an effect on wound repair in mice lacking Nrf2 in myeloid cells. Thus, myeloid cells with wild-type Nrf2 may still be present in the wound at significant levels and they could be sufficient to allow normal healing. However, the very low levels of Nrf2 that we detected in isolated wound neutrophils of the knockout mice suggest that at least Nrf2-deficient neutrophils are not out-competed by cells that have escaped recombination.
We had previously shown a prolonged presence of macrophages in wounds of mice with a global Nrf2 knockout [8]. Surprisingly, this was not the case in mice lacking Nrf2 only in myeloid cells, suggesting that expression of Nrf2 in other skin cell types indirectly affects the duration of the inflammatory phase during wound healing. Consistent with this hypothesis, Nrf2 from non-myeloid cells contributed to high fat diet-induced tissue inflammation and insulin resistance, whereas loss of myeloid-derived Nrf2 had no effect [32].
While loss of Nrf2 in myeloid cells and even in all cells is obviously dispensable for wound healing in normal mice, Nrf2 seems to be crucial for wound healing in diabetic mice, which are characterized by impaired healing and enhanced oxidative stress [33]. Thus, treatment of wounds in diabetic mice with siRNA against the Nrf2 antagonist Keap1 reduced the oxidative stress and hence, resulted in accelerated wound closure [10, 11]. Vice versa, diabetic mice with global Nrf2 knockout showed delayed wound healing compared to their wild-type littermates as a consequence of more severe oxidative DNA damage, reduced cell proliferation and migration and increased apoptosis [9]. Therefore, it may well be that loss of Nrf2 in myeloid cells also aggravates the wound healing defect in diabetic mice and this should be addressed in future studies. The normal healing in healthy mice lacking Nrf2 in myeloid cells suggests, however, that these cells have developed additional and potent antioxidant defense mechanisms that are not dependent on Nrf2. Such a redundancy may be required for their survival under various harsh conditions, such as in a wound environment. The identification of these additional mechanisms will be an important goal for the future.
Supporting information
Representative flow cytometry gating plots for sorting different populations of immune cells from 5dw. The live single cells were selected for myeloid cells (CD45+CD11b+) and these cells were further gated for different myeloid cell subtypes, including neutrophils, macrophages and monocytes. All gates are based on fluorescence minus one control.
(PDF)
Representative pseudocolor plots for the gating strategy to quantify the percentage of RFP-positive cells among different immune cell populations of control and transgenic mice in (A) blood in the steady state and in (B) 5dw.
(PDF)
(A) Representative pictures of H/E-stained sections from the back skin of 10-week-old female mice. Scale bar: 200 μm. (B) Flow cytometry analysis of dissociated single cells from the back skin epidermis or dermis or from the blood of tg/wt and tg/tg mice for the quantification of different immune cell populations as indicated. Bars indicate mean ±SD.
(PDF)
(A) RT-qPCR using RNA from lymphocytes, neutrophils and macrophages isolated from 5d wounds for the classical Nrf2 target genes Gclc and Nqo1 relative to Rps29. N = 3 mice. Bars indicate median with 95% CI.
(PDF)
(A) Representative pictures of H/E- stained sections from the back skin of 10-week-old female mice. Scale bar: 200 μm. (B) Flow cytometry analysis of dissociated single cells from the back skin epidermis or dermis of tg/wt and tg/ko mice for the quantification of different immune cell populations as indicated. Bars indicate mean ±SD.
(PDF)
(XLSX)
Acknowledgments
We thank Dr. Manfred Kopf, ETH Zurich, for helpful suggestions and for providing LysM-Cre-ROSA/RFP reporter mice, Dr. Shyam Biswal, Johns Hopkins University, Baltimore, MD, for the conditional Nrf2 knockout mice, and Drs. Eric Haertel, Michael Meyer and Paul Hiebert, ETH Zurich, for experimental help and helpful suggestions. N.J. is a member of the Molecular and Translational Biomedicine PhD program of the Life Science Zurich Graduate School, Zurich.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by Swiss National Science Foundation Grant PDFMP3_137197/1 to SW; http://www.snf.ch.
References
- 1.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453(7193):314–21. doi: 10.1038/nature07039 [DOI] [PubMed] [Google Scholar]
- 2.Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med. 2014;6(265):265sr6 doi: 10.1126/scitranslmed.3009337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17(6):763–71. doi: 10.1111/j.1524-475X.2009.00543.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127(3):514–25. doi: 10.1038/sj.jid.5700701 [DOI] [PubMed] [Google Scholar]
- 5.Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9(8):628–38. doi: 10.1038/nrm2455 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T, Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic Biol Med. 2015;88(Pt B):93–100. doi: 10.1016/j.freeradbiomed.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 7.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3(112):re3 doi: 10.1126/scisignal.3112re3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun S, Hanselmann C, Gassmann MG, auf dem Keller U, Born-Berclaz C, Chan K, et al. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol Cell Biol. 2002;22(15):5492–505. doi: 10.1128/MCB.22.15.5492-5505.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long M, Rojo de la Vega M, Wen Q, Bharara M, Jiang T, Zhang R, et al. An Essential Role of NRF2 in Diabetic Wound Healing. Diabetes. 2016;65(3):780–93. doi: 10.2337/db15-0564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soares MA, Cohen OD, Low YC, Sartor RA, Ellison T, Anil U, et al. Restoration of Nrf2 Signaling Normalizes the Regenerative Niche. Diabetes. 2016;65(3):633–46. doi: 10.2337/db15-0453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabbani PS, Zhou A, Borab ZM, Frezzo JA, Srivastava N, More HT, et al. Novel lipoproteoplex delivers Keap1 siRNA based gene therapy to accelerate diabetic wound healing. Biomaterials. 2017;132:1–15. doi: 10.1016/j.biomaterials.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 12.Schafer M, Farwanah H, Willrodt AH, Huebner AJ, Sandhoff K, Roop D, et al. Nrf2 links epidermal barrier function with antioxidant defense. EMBO Mol Med. 2012;4(5):364–79. doi: 10.1002/emmm.201200219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafer M, Willrodt AH, Kurinna S, Link AS, Farwanah H, Geusau A, et al. Activation of Nrf2 in keratinocytes causes chloracne (MADISH)-like skin disease in mice. EMBO Mol Med. 2014;6(4):442–57. doi: 10.1002/emmm.201303281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong X, Thimmulappa R, Craciun F, Harvey C, Singh A, Kombairaju P, et al. Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am J Respir Crit Care Med. 2011;184(8):928–38. doi: 10.1164/rccm.201102-0271OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herovici C. [Picropolychrome: histological staining technic intended for the study of normal and pathological connective tissue]. Rev Fr Etud Clin Biol. 1963;8:88–9. [PubMed] [Google Scholar]
- 16.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose S, Misharin A, Perlman H. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A. 2012;81(4):343–50. doi: 10.1002/cyto.a.22012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafer M, Werner S. Nrf2—A regulator of keratinocyte redox signaling. Free Radic Biol Med. 2015;88(Pt B):243–52. doi: 10.1016/j.freeradbiomed.2015.04.018 [DOI] [PubMed] [Google Scholar]
- 19.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–77. [DOI] [PubMed] [Google Scholar]
- 20.Hubner G, Brauchle M, Smola H, Madlener M, Fassler R, Werner S. Differential regulation of pro-inflammatory cytokines during wound healing in normal and glucocorticoid-treated mice. Cytokine. 1996;8(7):548–56. doi: 10.1006/cyto.1996.0074 [DOI] [PubMed] [Google Scholar]
- 21.Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 2005;15(11):599–607. doi: 10.1016/j.tcb.2005.09.002 [DOI] [PubMed] [Google Scholar]
- 22.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275(21):16023–9. [DOI] [PubMed] [Google Scholar]
- 23.Kong X, Thimmulappa R, Kombairaju P, Biswal S. NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J Immunol. 2010;185(1):569–77. doi: 10.4049/jimmunol.0902315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoetzenecker W, Echtenacher B, Guenova E, Hoetzenecker K, Woelbing F, Bruck J, et al. ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-associated immunosuppression. Nat Med. 2011;18(1):128–34. doi: 10.1038/nm.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi E, Suzuki T, Yamamoto M. Roles nrf2 plays in myeloid cells and related disorders. Oxid Med Cell Longev. 2013;2013:529219 doi: 10.1155/2013/529219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dias IH, Chapple IL, Milward M, Grant MM, Hill E, Brown J, et al. Sulforaphane restores cellular glutathione levels and reduces chronic periodontitis neutrophil hyperactivity in vitro. PLoS One. 2013;8(6):e66407 doi: 10.1371/journal.pone.0066407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Abdalrahman A, Lai Y, Janicki JS, Ward KW, Meyer CJ, et al. Dihydro-CDDO-trifluoroethyl amide suppresses inflammatory responses in macrophages via activation of Nrf2. Biochem Biophys Res Commun. 2014;444(4):555–61. doi: 10.1016/j.bbrc.2014.01.101 [DOI] [PubMed] [Google Scholar]
- 28.Dinkova-Kostova AT, Kostov RV, Canning P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch Biochem Biophys. 2017;617:84–93. doi: 10.1016/j.abb.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal. 2010;22(11):1645–54. doi: 10.1016/j.cellsig.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 30.Lee LY, Harberg C, Matkowskyj KA, Cook S, Roenneburg D, Werner S, et al. Cell-specific overactivation of nuclear erythroid 2 p45-related factor 2-mediated gene expression in myeloid cells decreases hepatic ischemia/reperfusion injury. Liver Transpl. 2016;22(8):1115–28. doi: 10.1002/lt.24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerstgrasser A, Melhem H, Leonardi I, Atrott K, Schafer M, Werner S, et al. Cell-specific Activation of the Nrf2 Antioxidant Pathway Increases Mucosal Inflammation in Acute but Not in Chronic Colitis. J Crohns Colitis. 2017;11(4):485–99. doi: 10.1093/ecco-jcc/jjw172 [DOI] [PubMed] [Google Scholar]
- 32.Meher AK, Sharma PR, Lira VA, Yamamoto M, Kensler TW, Yan Z, et al. Nrf2 deficiency in myeloid cells is not sufficient to protect mice from high-fat diet-induced adipose tissue inflammation and insulin resistance. Free Radic Biol Med. 2012;52(9):1708–15. doi: 10.1016/j.freeradbiomed.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhall S, Do DC, Garcia M, Kim J, Mirebrahim SH, Lyubovitsky J, et al. Generating and reversing chronic wounds in diabetic mice by manipulating wound redox parameters. J Diabetes Res. 2014;2014:562625 doi: 10.1155/2014/562625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative flow cytometry gating plots for sorting different populations of immune cells from 5dw. The live single cells were selected for myeloid cells (CD45+CD11b+) and these cells were further gated for different myeloid cell subtypes, including neutrophils, macrophages and monocytes. All gates are based on fluorescence minus one control.
(PDF)
Representative pseudocolor plots for the gating strategy to quantify the percentage of RFP-positive cells among different immune cell populations of control and transgenic mice in (A) blood in the steady state and in (B) 5dw.
(PDF)
(A) Representative pictures of H/E-stained sections from the back skin of 10-week-old female mice. Scale bar: 200 μm. (B) Flow cytometry analysis of dissociated single cells from the back skin epidermis or dermis or from the blood of tg/wt and tg/tg mice for the quantification of different immune cell populations as indicated. Bars indicate mean ±SD.
(PDF)
(A) RT-qPCR using RNA from lymphocytes, neutrophils and macrophages isolated from 5d wounds for the classical Nrf2 target genes Gclc and Nqo1 relative to Rps29. N = 3 mice. Bars indicate median with 95% CI.
(PDF)
(A) Representative pictures of H/E- stained sections from the back skin of 10-week-old female mice. Scale bar: 200 μm. (B) Flow cytometry analysis of dissociated single cells from the back skin epidermis or dermis of tg/wt and tg/ko mice for the quantification of different immune cell populations as indicated. Bars indicate mean ±SD.
(PDF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.