Abstract
MicroRNAs (miRNAs) are small regulatory RNAs processed from primary miRNA transcripts, and plant miRNAs play important roles in plant growth, development, and response to infection by microbes. Microbial infections broadly alter miRNA biogenesis, but the underlying mechanisms remain poorly understood. In this study, we report that the Rice stripe virus (RSV)-encoded nonstructural protein 3 (NS3) interacts with OsDRB1, an indispensable component of the rice (Oryza sativa) miRNA-processing complex. Moreover, the NS3-OsDRB1 interaction occurs at the sites required for OsDRB1 self-interaction, which is essential for miRNA biogenesis. Further analysis revealed that NS3 acts as a scaffold between OsDRB1 and pri-miRNAs to regulate their association and aids in vivo processing of pri-miRNAs. Genetic evidence in Arabidopsis showed that NS3 can partially substitute for the function of double-stranded RNA binding domain (dsRBD) of AtDRB1/AtHYL1 during miRNA biogenesis. As a result, NS3 induces the accumulation of several miRNAs, most of which target pivotal genes associated with development or pathogen resistance. In contrast, a mutant version of NS3 (mNS3), which still associated with OsDRB1 but has defects in pri-miRNA binding, reduces accumulation of these miRNAs. Transgenic rice lines expressing NS3 exhibited significantly higher susceptibility to RSV infection compared with non-transgenic wild-type plants, whereas the transgenic lines expressing mNS3 showed a less-sensitive response. Our findings revealed a previously unknown mechanism in which a viral protein hijacks OsDRB1, a key component of the processing complex, for miRNA biogenesis and enhances viral infection and pathogenesis in rice.
Author summary
MicroRNAs (miRNAs) regulate gene expression at the transcriptional or post-transcriptional level and have emerged as key players in regulating plant growth, development and response to biotic and abiotic stresses. Accumulating evidences suggest that miRNAs are pivotal modulators of host–virus interactions, but how virus regulates miRNA accumulation remains poorly understood. Here, we report that NS3 protein encoded by Rice stripe virus (RSV) regulates the processing of several primary miRNA transcripts (pri-miRNAs) by acting as an intermediary to modulate the association of pri-miRNAs and OsDRB1, a key factor of the pri-miRNA processing complex. NS3 increases recruitment of pri-miRNA to the processing complex by its association with OsDRB1 at the sites required for OsDRB1 dimer formation and induces several miRNAs accumulations as well as target genes repression, promoting the sensitivity of rice to RSV infection. Together these findings reveal a novel mechanism by which RSV regulates pri-miRNA processing, leading to enhanced viral infection.
Introduction
MicroRNAs (miRNAs), a class of endogenous small RNAs processed from their primary transcripts (pri-miRNAs), are crucial for plant development and responses to abiotic and biotic stresses [1–3]. Invading pathogens can manipulate the biogenesis and stability of many miRNAs to promote infection, or affect plant defense. For example, the accumulation of miR168 is elevated by infections with Cymbidum ringspot virus (CymRSV), crucifer-infecting Tobacco mosaic virus (crTMV), Potato virus X (PVX), and Tobacco etch virus (TEV) in Nicotiana benthamiana and by Rice stripe virus (RSV) and Rice dwarf virus (RDV) in rice (Oryza sativa). Indeed, this induced accumulation was found to promote the infection process of these viruses [4–9]. Rice ragged stunt virus (RRSV) and Rice black streaked dwarf virus (RBSDV) infections in rice increase the level of miR319, which suppresses jasmonic acid mediated antiviral defense in rice [10]. Overexpression of miR528 in transgenic rice plants reduces the accumulation level of reactive oxygen species (ROS) compared with that of wild type (WT), and these plants are more sensitive to RSV infection [11]. Arabidopsis miR393 suppresses auxin signaling in response to bacterial infection [12], while miR398b is involved in defense against fungal pathogens [13]. In addition, some miRNAs that function in basal metabolism are regulated by pathogen invasion. For instance, miR395 and miR399, which are involved in the regulation of sulfur assimilation [14,15] and response to phosphorus starvation [16–18], respectively, are up-regulated by RSV infection in rice [19].
In plants, the miRNA biogenesis pathway is regulated by the cooperation of several host factors [20]. Work in Arabidopsis showed that miRNAs are processed from primary transcripts that contain partially complementary fold-back regions of variable lengths (pri-miRNAs) by a processing complex consisting of the RNAse III enzyme DICER-LIKE 1 (AtDCL), the double-stranded RNA (dsRNA) binding protein HYPONASTIC LEAVES1 (AtDRB1/AtHYL1), and the zinc finger protein SERRATE (AtSE) [21–23]. AtHYL1 and AtSE are essential for the accurate and efficient cleavage of pri-miRNAs by AtDCL1 [21]. Homodimerization/self-interaction of AtHYL1 (orthologous to the OsDRB1) ensures the correct selection of cleavage sites on pri-miRNAs [24]. After processing, the miRNA/miRNA* duplex is methylated by HUA ENHANCER 1 (AtHEN1) at the 3’-terminus to ensure the stability of mature miRNA [25,26]. The miRNA strand is recruited by diverse ARGONAUTE (AtAGO) proteins to form miRNA-induced silencing complexes to mediate post-transcription gene silencing or translation repression [27–29]. Although the miRNA biogenesis pathway in plants has been well documented and many studies have indicated that miRNAs are involved in host–virus interactions, little is known about how pathogens regulate miRNA processing and accumulation.
Rice stripe virus (RSV), the type member of the genus Tenuivirus, causes severe disease and yield losses in many Asian rice cultivars. The RSV genome comprises four negative-sense, single-stranded RNA segments, RNA1, 2, 3, and 4. RNA1 uses a negative sense coding strategy while RNA2, 3, and 4 use ambisense coding strategy [30–33]. RNA1 encodes RNA-dependent RNA polymerase (RdRp, 337 kDa) [34]. RNA2 encodes NS2 (22.8 kDa), a weak suppressor of RNA silencing [35] and NSvc2 (94.2 kDa), a glycoprotein that targets the Golgi body and the endoplasmic reticulum (ER) [36]. RNA3 encodes nonstructural protein 3 (NS3, 23.9 kDa) and the coat protein (CP, 35.1 kDa) [37,38]. NS3 was identified as a viral-encoded RNA silencing suppressor (VSR) which suppresses post-transcriptional gene silencing (PTGS) in N. benthamiana and binds single- or double-stranded RNA without sequence preference [37,38]. RNA4 encodes disease specific protein (SP, 20.5 kDa) which interferes with photosynthesis by interaction with an oxygen-evolving complex protein [30] and NSvc4 (32.4 kDa), a cell-to-cell movement protein [39]. Several studies have demonstrated that RSV infection perturbs miRNA accumulation, for example, 38 miRNAs including miR167, miR168, miR395, miR399 etc., were induced upon RSV infection between 7 to 15 dpi [9,19], but the underlying mechanism for this is unclear. In this study, we found that RSV-encoded NS3 is responsible for the over-accumulation of several miRNAs, many of them known to regulate biotic or abiotic stress response genes, in a dsRNA-binding activity-dependent manner both in rice and Arbidopsis. In vivo experiments demonstrated that NS3 enhanced pri-miRNA processing through its dsRNA binding domain. Additionally, NS3 interacts specifically with the second dsRNA-binding domain (dsRDB2) of OsDRB1. Importantly, the NS3-interacting sites in OsDRB1 are also required for the homodimerization of the OsDRB1 and NS3 acts as a scaffold to regulate the association of OsDRB1 and pri-miRNA. Genetic analysis showed that NS3-ΔdsRBD-AtHYL1 fusion protein could partially rescue the phenotype of hyl1-2, but NS3 or ΔD1-AtHYL1 could not do so. NS3-overexpressing transgenic rice lines enhanced RSV pathogenicity compared with the control transgenic lines expressing a mutant form of NS3 (mNS3) and the WT plants. Our data revealed that the RSV NS3 protein regulates the association between OsDRB1 and pri-miRNAs, induces accumulation of a number of miRNAs, and enhances viral pathogenicity in rice.
Results
NS3 is responsible for the RSV-induced over-accumulation of a set of miRNAs
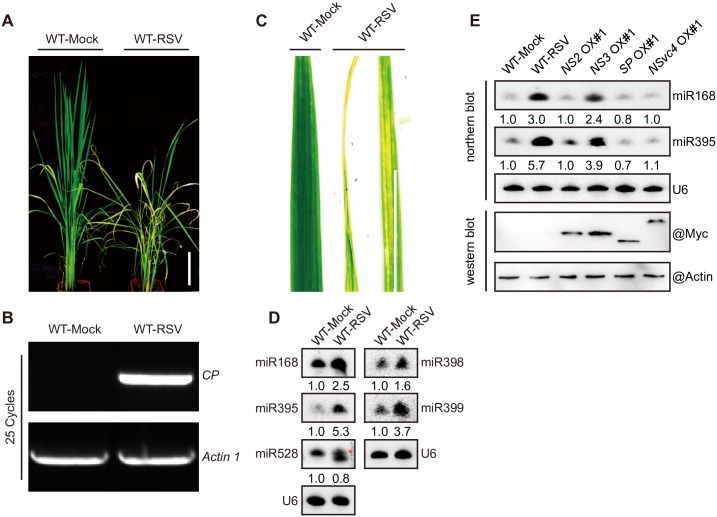
RSV-infected rice plants show stunting, rolled-leaf and chlorotic mottling in leaves symptom compared to mock-infected rice plants at early stage (Fig 1A to 1C). In a previous study, we found that RSV infection resulted in an increased accumulation of several rice miRNAs, including miR168, miR395, miR398, miR399 and miR528 (20-nt and 21-nt forms) [5,9,11,19]. To confirm the RSV-induced increase in the accumulation of these miRNAs in a different set of plants, we analyzed the differential expression of these rice miRNAs in mock-inoculated and RSV-infected rice plants by northern blotting. All tested miRNAs, with the exception of miR528 (21-nt), were up-regulated by RSV infection (Fig 1D), which was consistent with previous reports [5,11,40,41].
Fig 1. RSV symptoms and miRNAs induced by NS3 in rice.
(A) Images of whole plants exhibiting stunted phenotypes. Scale bar = 15 cm. (B) Detection of the RSV CP gene by RT-PCR. (C) Images of RSV-infected leaves exhibiting chlorotic mottling and rolled-leaf phenotypes. Scale bar = 5 cm. (D) Detection of miRNA (miR168, 395, 398, 399, and 528) accumulations in healthy (mock-infected) and RSV-infected rice plants by northern blotting. Red star, 21-nt miR528. (E) Northern and western blot assay detection of miRNA (miR168 and miR395) accumulations and protein expression in mock-infected, RSV-infected (RSV), NS2 OX#1, NS3 OX#1, SP OX#1, and NSvc4 OX#1 rice plants. In (D and E), U6 served as a loading control, the expression levels in the WT-Mock plants are set to a value of 1.0 and the expression levels in the other plants are relative to this reference value.
To determine which RSV-encoded protein triggers the up-regulation of miRNAs in the RSV-infected rice plants, we measured the expression of these miRNAs in rice plants overexpressing various RSV-encoded proteins, driven by the ACTIN 1 promoter, with a 4×Myc epitope tag at the N-terminus. Western blot assays confirmed the expression of myc-NS2, myc-NS3, myc-SP, and myc-NSvc4 in the corresponding transgenic rice lines (Fig 1E) (bottom two panels). As shown in Fig 1E, (top three panels), the levels of miR168 and miR395, measured by northern blot hybridizations, were strongly elevated by RSV infection and transgenic expression of NS3 (Fig 1E, lane 2 and 4), but not other RSV proteins. Also, RSV infection and NS3 transgene expression is produced similar elevations of miRNA levels (Fig 1E, lane 2 and 4). Therefore, expression of NS3 alone fully recapitulates the perturbation of miRNA levels caused by RSV infections.
NS3 triggers the accumulation of several miRNAs through its dsRNA binding domain
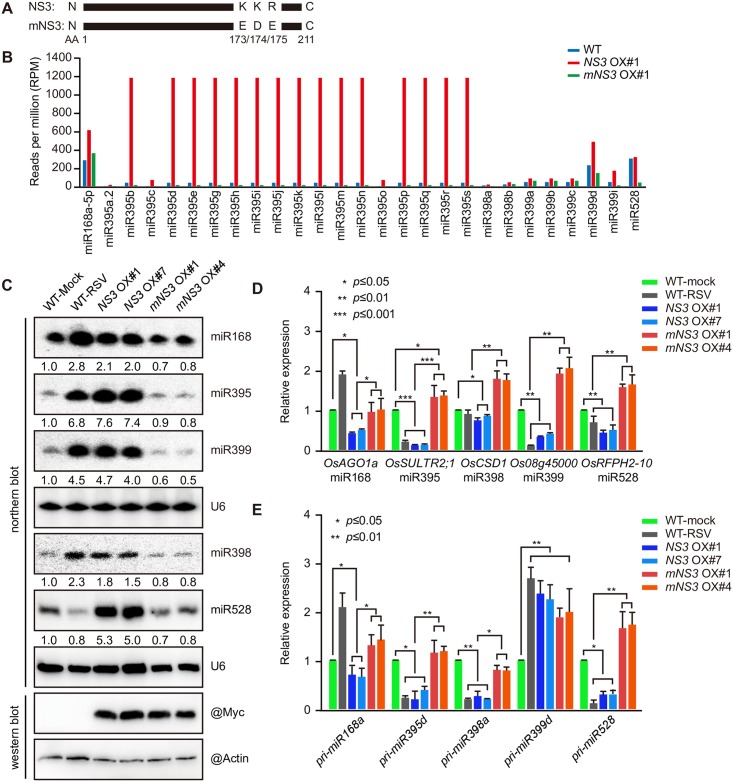
The dsRNA binding domain of NS3 is important for its activity [37]. To determine whether NS3-promoted miRNA accumulation depends on its dsRNA binding activity, we constructed a transgenic rice line overexpressing Myc-tagged mNS3, in which the dsRNA binding domain was disrupted by the replacement of a 173K174K175R motif with173E174D175E (Fig 2A). We then conducted de novo sequencing of small RNAs in the WT, and the NS3 overexpression (OX) #1 and mNS3 OX#1 rice lines. The results showed that miR168, miR395, miR398, miR399, and miR528 were all up-regulated in the NS3-overexpressing rice line, but down-regulated or unchanged in the mNS3-overexpressing rice line compared with the WT (See S1 Table for more details) (Fig 2B). To verify the de novo sequencing results, we carried out northern blot and western blot assays in two independent overexpression lines each for NS3 and mNS3 (NS3 OX#1 and #7 and mNS3 OX#1 and #4). As shown in Fig 2C, most miRNAs accumulated at higher levels in the NS3 overexpression lines compared with the mock-inoculated WT, whereas lower levels were detected in the mNS3 overexpression lines. These results indicated that NS3 plays a critical role in the positive regulation of miRNA accumulations in rice and this regulation is dependent on its dsRNA binding activity.
Fig 2. Expression of NS3 promotes the accumulation of several miRNAs and reduces the expression of their targets.
(A) Mutation site of the mutant NS3 (mNS3). (B) Measurement of miRNA (miR168, 395, 398, 399, and 528) accumulation in the WT, NS3 OX#1, and mNS3 OX#1 rice lines by small RNA sequencing. (C) Detection of miRNA (miR168, 395, 398, 399, and 528) accumulations in mock-infected, RSV-infected (RSV), NS3 OX#1, NS3 OX#7, mNS3 OX#1, and mNS3 OX#4 rice plants by northern blotting. U6 served as a loading control, the expression levels in the WT-Mock plants are set to a value of 1.0 and the expression levels of in the other plants are relative to this reference value. (D) Relative expression levels of the target genes of the miRNAs (miR168, 395, 398, 399, and 528), including OsAGO1a, OsSULTR2;1, OsCSD1, Os08g45000, and OsRFPH2-10 in the mock-infected, RSV-infected (RSV), NS3 OX#1, NS3 OX#7, mNS3 OX#1, and mNS3 OX#4 rice plants. (E) Relative expression levels of the miRNA (miR168, 395, 398, 399, and 528) precursors, including pri-miR168a, pri-miR395d, pri-miR398a, pri-miR399b, and pri-miR528 in the mock-infected, RSV-infected (RSV), NS3 OX#1, NS3 OX#7, mNS3OX#1, and mNS3 OX#4 rice plants. Average (± SD) values based on RT-qPCR analysis of three biological replicates are shown. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05.
To verify that the increased accumulation of these miRNAs further reduced the expression levels of their target genes, we examined the expression levels of their target mRNAs by quantitative RT-PCR (RT-qPCR). As shown in Fig 2D, mRNA levels of OsAGO1a (miR168), OsSULTR2;1 (miR395), OsCDS1 (miR398), Os08g45000 (miR399) and OsRFPH2-10 (miR528) were all reduced in the NS3 overexpression lines compared with the mock-inoculated WT plants, whereas expression levels in the mNS3 overexpression lines were relatively unchanged compared to the WT.
To further test whether NS3 triggers accumulation of miRNAs through its dsRNA binding domain in dicots, we constructed NS3 and mNS3 overexpression transgenic Arabidopsis plants. Northern blot showed that the levels of miR168 and miR395 were up-regulated by RSV infection and NS3 overexpression in these Arabidopsis plants (S1A and S1B Fig). Although overexpression of NS3 in rice did not result in disease-like symptoms, NS3-overexpressing Arabidopsis plants exhibited a severely stunted phenotype that phenocopied the disease symptoms of the RSV infected Arabidopsis (S1C and S1D Fig). In contrast, overexpression of mNS3 had no influence on the growth or the levels of miR168 and miR395 in the Arabidopsis plants (S1B and S1D Fig). These results indicate that induction of miRNA accumulations by NS3 overexpression is conserved in dicots and monocots.
To determine whether RSV infection and NS3 overexpression increases the accumulation of these miRNAs through promoting primary miRNA (pri-miRNA) processing, we measured the primary transcript levels of miR168, miR395, miR398, miR399, and miR528 by RT-qPCR. The results demonstrated that NS3, but not mNS3, down-regulated all of the pri-miRNAs, except for pri-miR399d, which suggests that NS3 is involved in pri-miRNA processing in a dsRNA-binding activity-dependent manner (Fig 2E).
NS3 aids in vivo processing of pri-miRNA
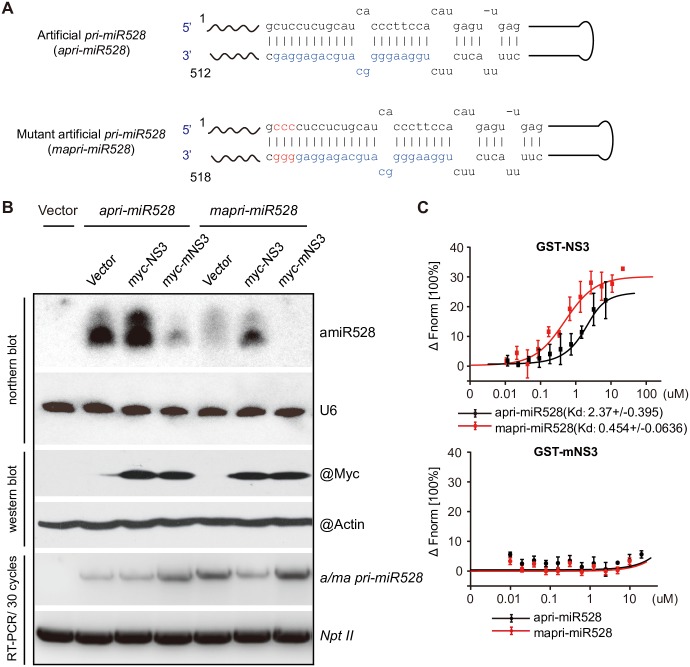
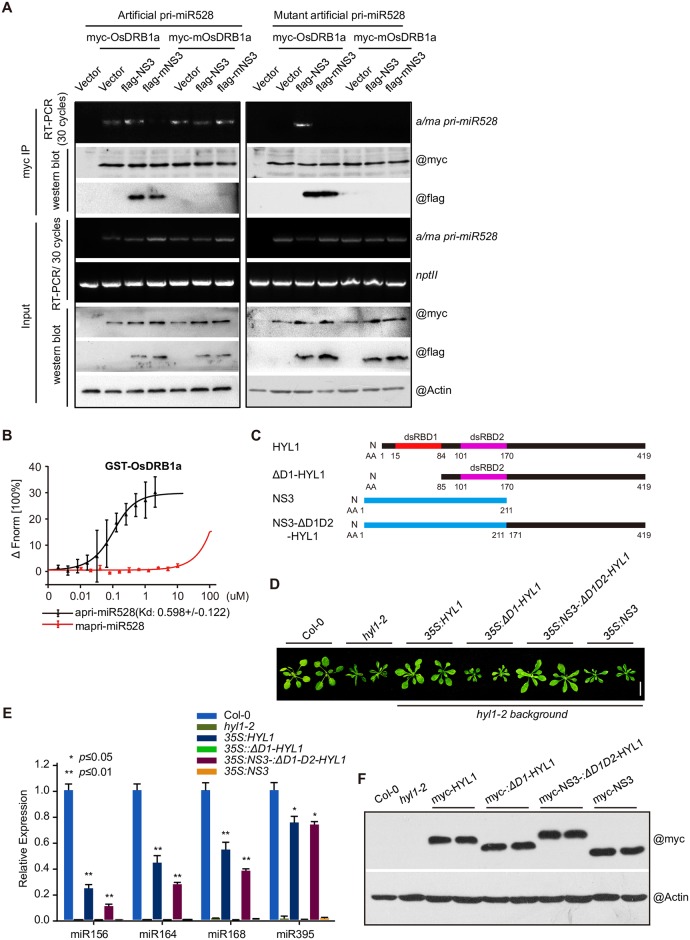
Given that NS3 reduces the accumulation of a set of pri-miRNAs but induces the accumulation of corresponding mature miRNAs, we suggested that NS3 may promote the recruitment of pri-miRNAs by affecting the miRNA-processing complex. Since the thermo stability of the end of the miRNA/miRNA* duplex is important for mature miRNA accumulation [42], we designed an experiment in which a pri-miRNA can be recognized by NS3, but not by the processing complex, which would indicate that NS3 directly assists in the association of the processing complex with pri-miRNAs when NS3 is co-expressed with the pri-miRNA. We constructed a 35S promoter-driven artificial primary miR528 (apri-miR528) and mutant artificial primary miR528 (mapri-miR528) with three additional C/G pairs at the end of the miRNA/miRNA* duplex using an Arabidopsis primary miR159a backbone (Fig 3A). miR528 is only expressed in monocot plants; therefore, we carried out an in vivo transient expression assay to co-express apri-miR528 and vector/NS3/mNS3 and also mapri-miR528 and vector/NS3/mNS3 in the leaves of the dicots N. benthamiana and measured mature artificial miR528 (amiR528) levels in each group by northern blotting at 3 days post-infiltration (dpi). We found that expression of NS3 had little influence on the accumulation of amiR528 in the apri-miR528 group, but expression ofmNS3 reduced the levels of miR528 relative to the vector control. As expected, no miR528 accumulation was observed in the negative control. Additionally, no mature miR528 accumulation was detected in the mapri-miR528 group in the absence of NS3 expression. The accumulation of mature miRNA was only detected in the leaves co-expressing mapri-miR528 with NS3 (Fig 3B). To test if NS3 interacts with apri-miR528 and mapri-miR528, an in vitro microscale thermophoresis assay with GST-mNS3 serving as the negative control was used to reveal that both apri-miR528 and mapri-miR528 were recognized by GST-NS3 (Fig 3C). These results provide further evidence for the role of NS3 in mature miRNA biogenesis.
Fig 3. NS3 promotes pri-miRNA processing.
(A) Structures of artificial pri-miR528 (apri-miR528) and mutated artificial pri-miR528 (mapri-miR528). (B) Northern blot, western blot, and RT-PCR detection of the products of transiently co-expressed pri-miRNAs (apri-miR528 or mapri-miR528) and proteins (empty vector, NS3, or mNS3) in N. benthamiana. (C) Results of a microscale thermophoresis assay shows the interactions between pri-miRNA (apri-miR528 or mapri-miR528) and protein (GST-NS3 or GST-mNS3).
Association of NS3 with the dsRBD2 domain of OsDRB1
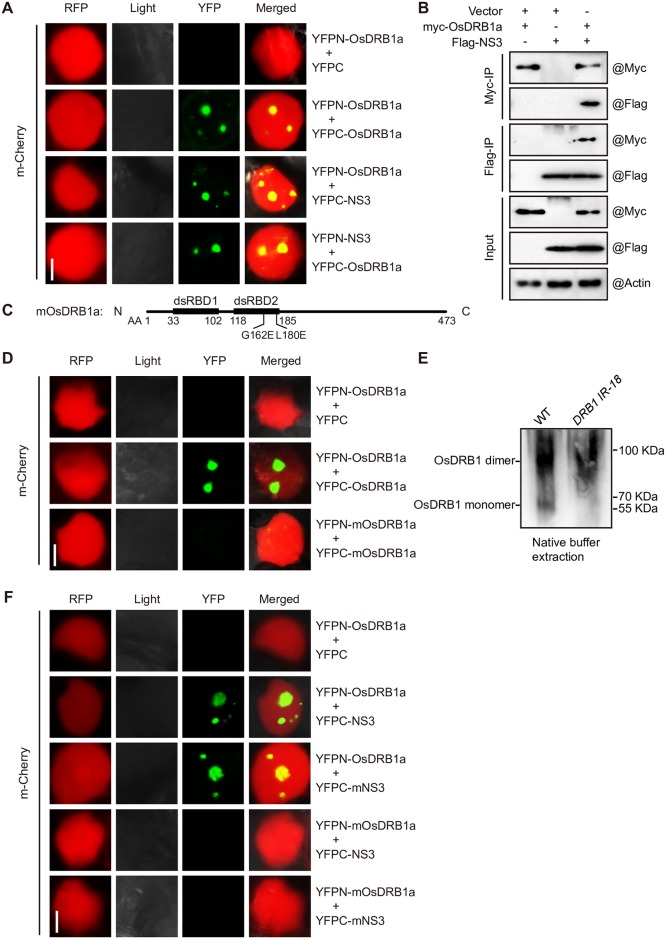
NS3 does not contain an RNase III domain; therefore, it cannot promote miRNA accumulation by itself. We hypothesized that NS3 may function in miRNA processing through its association with components of the Dicing body (D-body) to promote the recruitment of the pri-miRNA by the processing complex. To test this hypothesis, we performed bimolecular fluorescence complementation (BiFC) assays to co-express Arabidopsis DCL1 (AtDCL1), SE (AtSE), HYL1 (AtHYL1), or CBP20 (AtCBP20) with NS3 in N. benthamiana leaves. NS3 associated with AtHYL1 and AtSE but not with AtDCL1 or AtCPB20. In addition, we found that AtHYL1, but not AtSE, specifically interacted with NS3 in the D-body, and the interaction between NS3 and AtHYL1 may involve in miRNA maturation (S2A Fig). Using the basic local alignment search tool (BLAST) and the UniProt protein database (uniprot.org), we found six AtHYL1 homologs in rice, OsDRB1a, OsDRB1b, OsDRB1c, OsDRB2, OsDRB3 and OsDRB4 (S2B Fig).To confirm that OsDRB1 has the same function as AtHYL1, we measured the levels of miR164, miR166, and miR168 by small-RNA RT-qPCR in an OsDRB1-knockdown rice line and found that these miRNAs were down-regulated (S2C Fig). We also analyzed OsDRB1 protein levels, and found that the level of OsDRB1 was reduced in the OsDRB1-knockdown line (S2D Fig). OsDRB1a contains all the domains of the other two OsDRB1s, as well as a unique C-terminus, so we chose OsDRB1a to test the interaction between NS3 and OsDRB1, BiFC and co-immunoprecipitation (CoIP) assays demonstrated that OsDRB1a does interact with NS3 (Fig 4A and 4B). We also tested interactions between NS3 and the other OsDRBs by BiFC assay, and found that only OsDRB2 has a weak interaction with NS3 in cytoplasm (S2E Fig). Previous studies have shown that homodimerization of AtHYL1 is required for AtDCLs to locate the correct cleavage sites in pri-miRNAs, while the G147 and L165 residues of AtHYL1 are critical for homodimer formation [24]. The G162 and L180 residues of OsDRB1a correspond to the G147 and L165 residues of AtHYL1 and may be essential for homodimer formation according to amino acid alignment (S2F Fig). To test our hypothesis, we constructed an OsDRB1a mutant (mOsDRB1a) by replacing the G162 and L180 residues with E162 and E180, respectively (Fig 4C). In a subsequent BiFC assay, we found that mOsDRB1a could not interact with itself (Fig 4D), as expected. We further confirmed that wild-type OsDRB1a, like AtHYL1, formed a homodimer (Fig 4E) [24,43]. Also, mOsDRB1a could not interact with NS3 or mNS3 (Fig 4F), but NS3 interacts with itself, mNS3 has weak interaction with NS3, and OsDRB1a has weak interaction with mOsDRB1a too (S2G Fig). We summarize the interaction of each pair between NS3, mNS3, OsDRB1a and mOsDRB1a in Table 1. These results demonstrated that the association between NS3 and OsDRB1a depends on the dsRBD2 domain of OsDRB1a and is essential for miRNA processing.
Fig 4. NS3 interacts with OsDRB1, a pri-miRNA processing factor.
The BiFC assay was conducted in N. benthamiana epidermal cells, mCherry is a nuclear localization marker fused with red florescent protein (RFP). (A) Results of a BiFC assay showing the interaction between NS3 and OsDRB1a. Scale bar = 0.1 μm. (B) Results of a co-immunoprecipitation analysis showing the interaction between NS3 and OsDRB1a. (C) The accuracy of mutation sites of mOsDRB1a. (D) Results of a BiFC assay showing the accuracy of interaction sites between OsDRB1a. Scale bar = 0.1 μm (E) Formation of OsDRB1a dimers in vivo. Total rice protein extracts from the WT and OsDRB1-knockdown lines were treated with ‘‘native” buffer and detected using anti-HYL1antibodies. (F) Results of a BiFC assay showing the accuracy of interaction sites between NS3 and DRB1a. Scale bar = 0.1 μm.
Table 1. Interactions of each two proteins as shown below.
√√: strong interaction;
√: weak interaction;
×: no interaction.
NS3 mimics the dsRBD domain of DRB1 in miRNA processing
Given that the dsRNA-binding activity of NS3 is important for the processing of pri-miRNAs and the accumulation of miRNA, and NS3 interacts with DRB1, we deduced that NS3, rather than DRB1, recognizes pri-miRNAs during NS3–DRB1 interactions. To test this hypothesis, we transiently co-expressed OsDRB1a/mOsDRB1a, apri-miR528/mapri-miR528, or empty vector/NS3/mNS3 in N. benthamiana and detected the CoIP products of OsDRB1a/mOsDRB1a by RT-PCR. We found that both OsDRB1a and mOsDRB1a associated with apri-miR528, but neither of them recognized mapri-miR528, and NS3 and mNS3 associated with OsDRB1a instead of mOsDRB1a. With the expression of NS3, both OsDRB1a and mOsDRB1a associated with apri-miR528. However, with the expression of mNS3, only mOsDRB1a associated with apri-miR528. Additionally, only co-expression with NS3 resulted in an OsDRB1a interaction with mapri-miR528 (Fig 5A), see Table 2 for more details. Using a microscale thermophoresis assay, we also found that OsDRB1a associated with apri-miR528 but not mapri-miR528 (Fig 5B). We also found that NS3, but not mNS3, could bind with the endogenous miRNA precursors (pre-miR168a, pre-miR395d, pre-miR398a, pre-miR399d, and pre-miR528) in an electrophoretic mobility shift assay (EMSA) (S3A Fig). These results indicated that by interacting with DRB1, NS3 replaced the dsRNA-binding activity of DRB1. To test this hypothesis, we overexpressed AtHYL1, ΔD1-HYL1, a double-stranded RNA binding domain 1 deletion form of AtHYL, NS3-ΔD1D2-HYL1, a fusion protein of NS3 and ΔD1D2-HYL1 and NS3 (Fig 5C) in Arabidopsis hyl1-2 mutant background, the transgenes were all driven by a 35S promoter with the proteins tagged with a myc-epitope tag at the N-terminus, we found that the fusion protein NS3-ΔD1D2-AtHYL1 could ameliorated the phenotype (Fig 5D) and miRNA (miR156, miR164, miR168 and miR395) levels (Fig 5E) of hyl1-2 mutant as HYL1 did but NS3 or ΔD1-HYL1 could not (Fig 5D and 5E), we test these transgenic Arabidopsis plants by western blotting (Fig 5F), and this results indicated that NS3 could substitute for the dsRBD domain of AtHYL1 in miRNA processing.
Fig 5. NS3 acts as a scaffold between DRB1 and pri-miRNA.
(A) RT-PCR and western blot detection of co-immunoprecipitated and input products of transiently co-expressed protein (DRB1a or mDRB1a), pri-miRNA (apri-miR528 or mapri-miR528), and protein (empty vector, NS3, or mNS3) in N. benthamiana. (B) Results of a microscale thermophoresis assay shows the interactions between pri-miRNA (apri-miR528 or mapri-miR528) and GST-OsDRB1a. (C) Gene structure of HYL1, ΔD1-HYL1, NS3 and NS3-ΔD1D2-HYL1. (D) Phenotype of Col-0, hyl1-2 and transgenic Arabidopsis plants overexpressed AtHYL1, ΔD1-HYL1, NS3-ΔD1D2-HYL1 and NS3 with the 35S promoter in the hyl1-2 mutant background. (E) miRNA (miR156, miR164, miR168 and miR395) levels in Col-0, hyl1-2 and transgenic Arabidopsis plants overexpressing AtHYL1, ΔD1-HYL1, NS3-ΔD1D2-HYL1 and NS3 with the 35S promoter in the hyl1-2 mutant background. (F) Western blot of AtHYL1, ΔD1-HYL1, NS3-ΔD1D2-HYL1 and NS3 transgenic Arabidopsis plants. Fnorm, normalized fluorescence.
Table 2. Interaction of apri-miR528 or mapri-miR528 with single protein or protein complex as shown below.
| NS3 | mNS3 | OsDRB1a | OsDRB1a+NS3 | OsDRB1a+NS3 | mOsDRB1a | mOsDRB1a+NS3 | mOsDRB1a+NS3 | |
|---|---|---|---|---|---|---|---|---|
| apri-miR528 | √ | × | √ | √ | × | √ | √ | √ |
| mapri-miR528 | √ | × | × | × | × | × | √ | × |
√: Yes;
×: No.
NS3 enhances virus pathogenicity in rice
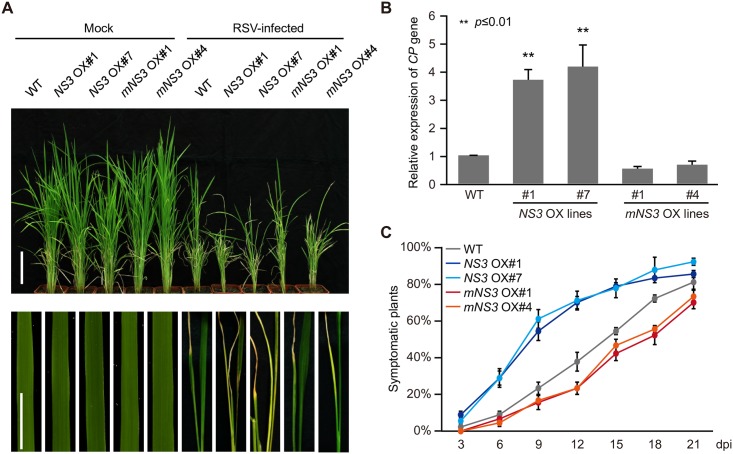
Because NS3 induced miRNA accumulation along with a decreased antiviral defense response in rice, we speculated that NS3 may play a role in regulating viral pathogenicity. We used virus-free (mock) and viruliferous (RSV) planthoppers (Laodelphax striatellus) to inoculate the WT, and NS3- and mNS3-overexpression rice lines (NS3 OX#1 and OX#7 and mNS3 OX#1 and OX#4) and found that the NS3 OX#1 and OX#7 lines, but not the mNS3 OX#1 and OX#4 lines, were hypersensitive to RSV infection compared with WT plants, with most serious stunted and chlorisis phenotypes (Fig 6A). To examine whether the increased susceptibility of the NS3 OX lines was due to the increased accumulation of RSV, we used RT-qPCR to measure the transcript levels of the RSV CP gene. We found that the expression of CP mRNA in the NS3 OX#1 and #7 lines was much higher than that in the WT plants, with no obvious changes detected in the mNS3 OX#1 and #4 lines when compared with the WT (Fig 6B). We also monitored differences in RSV infection rates among the WT and the NS3 OX#1, NS3 OX#7, mNS3 OX#1, and mNS3 OX#4 lines every 3 days until 21 dpi. These observations indicated that NS3 increased RSV pathogenicity in rice (Fig 6C and S3 Table). To test if NS3 OX plants are also more sensitive to infection with other viruses, we used Rice ragged stunt virus (RRSV), a member of the genus Oryzavirus, to infect the WT, and the NS3 OX#1, NS3 OX#7, mNS3 OX#1, and mNS3 OX#4 lines. The results showed that NS3 OX plant lines displayed a stunted phenotype (S4A Fig) and accumulated more RRSV CP genes (S4B Fig) compared with other rice plant lines.
Fig 6. Disease symptoms induced by RSV infection in WT, NS3 OX, and mNS3 OX rice lines.
(A) Images of whole plants and details showing stunted or folded-leaf phenotypes of the wild-type, NS3 OX#1, NS3 OX#7, mNS3 OX#1 and mNS3 OX#4 rice plants. Scale bars = 15 cm (upper panel) and 5 cm (lower panel). (B) Detection of the RSV CP gene by RT-PCR. (C) Time course of RSV symptom development in the WT and NS3 OX or mNS3 OX transgenic plants. Values represent the percentage of RSV-infected plants at various days post inoculation (dpi). Thirty plants were used for each treatment. **, P ≤ 0.01. Average (± SD) values from three biological replicates are shown.
Discussion
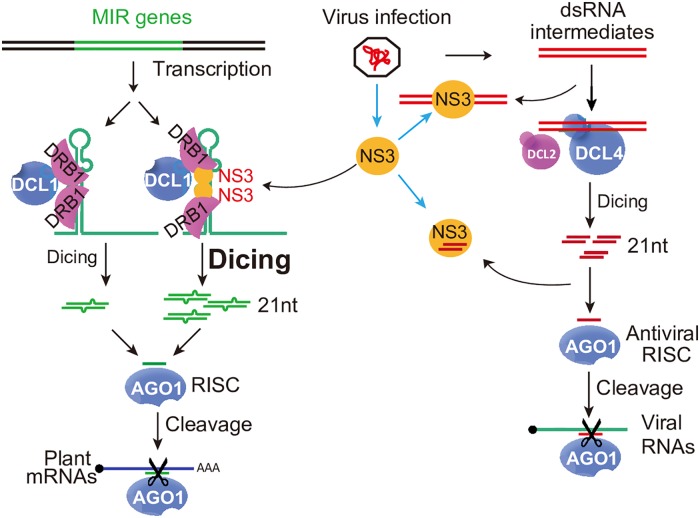
RNA interference (RNAi) is a conserved and effective antiviral mechanism in plants and insects [44]. To counter the host’s antiviral defense mechanisms, viruses encode VSR (s) to interfere with the host’s RNAi. Most reported VSRs act to suppress the host’s RNA silencing system, such as by inhibiting viral RNA recognition, blocking dicing, suppressing assembly of the RNA-induced silencing complex (RISC), and preventing siRNA amplification [44–50]. RSV NS3, a reported RNA silencing suppressor, suppresses post-transcriptional gene silencing (PTGS) in N. benthamiana through its dsRNA binding ability [38]. Our previously obtained small RNA sequencing data of changes associated with RSV infection revealed that many miRNAs are induced by RSV infection [9,19,41]. Up to now, only a few studies have focused on how a virus hijacks the RNA silencing pathway to regulate miRNA accumulation and advance its own pathogenicity, with very limited reports focusing on how virus regulates miRNA processing. In the present study, we revealed that NS3 exploits OsDRB1, a key component of the D-body, to promote the processing of pri-miRNA along with the regulation of miRNA target gene expression (Fig 7). NS3 showed a weak interaction with OsDRB2 in the cytoplasm (S2G Fig). Since miRNA processing is occurred in the nucleus, we deduced that the association of NS3 and OsDRB2 would not affect NS3-mediated miRNA processing.
Fig 7. Proposed model.
NS3 represses the PTGS pathway. Viral-derived long double-stranded RNAs (dsRNA) are processed into 21-nt siRNAs, the 21-nt siRNAs are loaded into AGO protein complex for cleavage of viral RNA genome RNAs. However, NS3 binds viral long dsRNA or siRNAs and inhibits siRNA production or the loading of siRNAs into AGO complex. Therefore, the PTGS pathway is blocked by NS3. NS3 enhances the miRNA biogenesis pathway. In mock-infected rice plants, a given pri-miRNA has a low probability of being recognized by dimeric DRB1 and then being processed into a mature miRNA by the processor complex. Consequently, the mature miRNA is insufficient to repress its target gene, which plays a crucial role in antiviral activity or development. Under RSV infection, pri-miRNA-bound NS3 interacts with DRB1 in sites required for DRB1 self-interaction and acts as a scaffold to regulate the association between DRB1 and the pri-miRNA. This increases the chance that this pri-miRNA will be processed into a mature miRNA. In addition, this compromises the expression of the target gene and enhances RSV pathogenicity.
Previous studies showed that NS3 suppression of the PTGS pathway may depend on its siRNA and long dsRNA binding activity [37,38], which is different from the results of this study showing that NS3 enhanced the miRNA pathway through its function of bridging pri-miRNA and OsDRB1, a key player in miRNA processing complex. These two different functions may act in parallel. These results widen our knowledge of the molecular mechanisms underlying viral-host interactions.
Maturation of miRNAs is a complex process. miRNA-coding genes (MIRs) are transcribed by DNA-dependent RNA polymerase II (Pol II) and subjected to splicing and addition of a 5’ 7-methyguanosine cap and a 3’ polyadenylated tail [51]. Besides Pol II, many other Pol II-associated factors such as RNA-binding proteins PLEIOTROPIC REGULATORY LOCUS 1 and DAWDLE, CAP BINDING PROTEIN 20 and 80, SE, CAM33/XAP CIRCADIAN TIMEKEEPER and TOUGH have been reported to regulate miRNA transcription [52–59]. The structures of pri-miRNAs also affect pri-miRNA processing [60,61]. RSV infection results in the differential expression of pri-miRNAs in rice. For example, pri-miR168a and pri-miR399d were up-regulated, while other pri-miRNAs were down-regulated (Fig 2E). We revealed that NS3 mainly decreases the accumulation of pri-miRNAs, but the mechanism by which NS3 influences pri-miRNA expression levels remains unclear.
De novo smallRNA sequencing showed that NS3 and mNS3 have a strong influence on the accumulation of miRNAs, but not on other types of smallRNAs. The total number of miRNA reads in the NS3 OX lines was higher than that in the WT, and the total number of miRNA reads in the mNS3 OX lines was lower than that in the WT (S3B Fig), suggesting that NS3 may directly promote pri-miRNA recruitment by the processing complex. This hypothesis was supported by the finding that NS3 aids in in vivo pri-miRNA processing (Fig 3B). Further investigations showed that NS3 associated with OsDRB1a at sites required for OsDRB1a self-interaction (Fig 4F) and promoted the association of OsDRB1a with pri-miRNAs (Fig 5A). SmallRNA sequencing showed that the lengths and sequences of miRNAs were not changed by NS3 or mNS3 overexpression in rice (S1 Table). These results suggested that NS3 promotes recruitment of pri-miRNAs by the processor complex through an interaction with DRB1. The observation that NS3 overexpressing rice plants showed no notable developmental abnormities at vegetative stages under normal growth conditions may relate to the presence of target genes of miRNAs, regulated by NS3, in a complex genome, and a similar phenomenon was previously described [3,56,62]. A study in Arabidopsis showed that a set of miRNAs is regulated by DRB1 [63]. NS3 up-regulates several miRNAs in a DRB1 dependent manner (Fig 5D). These results may explain why NS3 could not enhance some miRNA which are DRB1-independent. But it’s still unknown how the preferential regulation of DRB1-dependent miRNA by NS3, we did not find similarities of sequences or secondary structure between precursors of DRB1-dependent miRNA.
Understanding the effects of NS3 of miRNA biogenesis will widen our understanding of the functions of virus-encoded proteins in regulating miRNA metabolism. Our findings of the association of NS3 with miRNA processing and accumulation provides insights into RSV pathogenicity and helps identify important targets for RSV infection, which can offer new strategies for the breeding or genetic engineering of RSV-resistant rice.
Materials and methods
Plant growth and virus inoculation
Rice (Oryza sativa spp. japonica) seedlings were grown in a greenhouse at 28–30°C and 60 ± 5% relative humidity under natural sunlight for 4 weeks. Arabidopsis thaliana plants were grown at 22°C under long-day conditions (16-h light/8-h dark). When the seedlings were about 10 days old, they were inoculated using viruliferous (RSV) or virus-free (mock) planthoppers (L. striatellus) at a ratio of four insects per plant for 72 h. After removal of the insects, the inoculated plants were returned to the greenhouse and monitored daily for the appearance of viral symptoms. The number of symptomatic rice plants of each line was recorded (Fig 6C).
Vector construction and plant transformation
The Gateway system (Invitrogen) and the enzyme digestion connection method were used to make binary constructs. Binary gateway vectors pSAT4A-DEST-N(1–174)EYFP-N1, pSAT5A-DEST-C(175-END)EYFP-N1, pEarleyGate202, and pEarleyGate203 [64] were used for transient expression in N. benthamiana and stable transformation of Arabidopsis, while binary vectors pGEX-4T-1 and pCam2300:Actin1::OCS were used for Escherichia coli and stable rice transformations. Most cDNA and miRNA genes were cloned into pENTR/D and pEASY Blunt Zero vectors, and pENTR/D-mNS3 and mOsDRB1a clones were prepared using a Quik Change site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). After confirmation by sequencing, all clones were transferred to the appropriate destination vector by recombination using the Gateway LR Clonase II Enzyme mix (Invitrogen) or T4 DNA ligase (TransGen Biotech, Beijing, China). Agrobacterium tumefaciens-mediated rice transformation was carried out at Weiming Kaituo Co., Ltd. (Beijing, China). Transgenic Arabidopsis plants were selected by their resistance to Basta on soil.
RNA extraction, quantitative RT-PCR, RT-PCR, and northern blot analysis
Total RNAs were extracted from plants using Trizol (Invitrogen). The extracted RNAs were treated with RNase-free DNase I (Promega, Madison, WI, USA) to remove DNA contamination and then reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen) using oligo (dT) primers and a Mir-X miRNA first-strand synthesis kit (Clontech Laboratories). The resulting cDNAs were then used as templates for RT-qPCR and RT-PCR. RT-qPCR was performed using SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan). The rice OsEF-1a gene was detected in parallel and used as an internal control. Northern blot analyses were performed as previously described [65]. Briefly, RNA samples were separated by 15% denaturing gel electrophoresis and transferred onto Hybond-N+ membranes (Amersham, Fairfield, CT, USA). Membranes were UV cross-linked and hybridized to 32P end-labeled oligonucleotide probes. Sequences of primers used in RT–qPCR and northern blot assays are listed in S2 Table.
Microscale thermophoresis assay
The Microscale Thermophoresis assays were performed as previously described [66,67]. GST-NS3, GST-OsDRB1a, and GST-mNS3 proteins were individually labeled with NHS red fluorescent dye according to the instructions of the RED-NHS Monolith NT Protein Labeling kit (NanoTemper Technologies GmbH, München, Germany). In protein and RNA interaction assays, the concentration of NHS-labeled protein was maintained at 100 nM, whereas RNA concentrations were gradient-diluted (20,000 nM, 10,000 nM, 5,000 nM, and then 2-fold dilutions until 10 nM). The RNA was denatured and annealed to form dsRNA, followed by addition of RNase inhibitor (1 U per group). After a short incubation, the samples were loaded into MST standard-treated glass capillaries. Measurements were performed at 25°C and LED and MST powers of 20% in buffer containing 20 mM Tris (pH 8.0) and 150 mM NaCl. The assays were repeated two times for each affinity measurement. Data analyses were performed using Nanotemper Analysis and OriginPro 8.0 software provided by the manufacturer.
Electrophoretic mobility shift assays
The pGEX-4T-1-NS3, pGEX-4T-1-mNS3, and pGEX-4T-1-OsDRB1a constructs, as well as empty pGEX-4T-1 vectors, were individually transformed into E. coli Transetta (DE3) (Transgene, Beijing, China) with protein expression induced by IPTG. The soluble GST fusion proteins were extracted and immobilized onto glutathione sepharose beads (Amersham). RNAs were labeled with 32P–UTP using a T7 RNA Production system (Promega) and purified with a MEGAclear kit (Ambion, Waltham, MA, USA). Approximately 2,000 cpm of radioactive RNA was denatured and then annealed to form dsRNA. The dsRNA was incubated with individual GST fusion proteins in the presence of RNase inhibitor (1 U per group) for 30 min on ice. The mixtures were then run on an RNase-free native PAGE gel for 1 h at 4°C.
Protein preparation and western blot analysis
The procedures used for protein extraction and western blotting have been described previously [5]. The following antibodies were purchased from commercial sources: anti-flag-peroxidase (Sigma, St. Louis, MO, USA), anti-Myc-peroxidase (Sigma), anti-Myc (Sigma), anti-FLAG (Sigma), anti-Actin (Easybio, Beijing, China), and anti-HYL1 (Agrisera AB Box 57, SE-911 21 Vännäs, Sweden).
Small RNA sequencing
Small RNA cloning for Illumina sequencing was carried out at Bainuodacheng Co., Ltd (Beijing, China). Rice miRNA annotations were obtained from miRBase (http://microrna.sanger.ac.uk/sequences, Release 14). Statistical analysis of the small RNA data sets was performed using in-house Perl scripts [5].
Bimolecular fluorescence complementation
BiFC assays were performed as previously described [22]. Briefly, we transformed the recombinant constructs into A. tumefaciens strain EHA105 and injected the Agrobacterium cultures into N.benthamiana leaves. After 3 days, we observed the tobacco epidermal cells with a confocal laser scanning microscope (LSM 710 NLO & DuoScan System, Zeiss).
RNA immunoprecipitation
RNA immunoprecipitation was carried out essentially as previously described [68] using 1% formaldehyde-treated leaves, transiently expressed protein, and RNA for 72 h. RIP was carried out with the following modifications: nuclei were isolated, resuspended in high salt nuclear lysis buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 4 mM MgCl2, 0.2% NP-40). The chromatin supernatant was diluted five times with dilution buffer (20 mM Tris-HCl, pH 7.5, 4 mM MgCl2, 0.2% NP-40) and immunoprecipitated using antibody. Pri-miRNAs were detected in the immunoprecipitates by RT-PCR using the artificial primers miR528-5p and -3p.
Accession numbers
Sequence data generated in this study can be found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), the Rice Genome Database (http://rice.plantbiology.msu.edu/) and the Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: NS3 (AY284945.1), AGO1a (LOC_Os02g45070), SULTR2;1 (LOC_Os03g09930), CDS1 (LOC_Os07g46990), LOC_Os08g45000, OsRFPH2-10 (LOC_Os06g06050), OsDRB1a (LOC_Os11g01869), OsDRB1b (LOC_Os12g01916), OsDRB1c(LOC_Os05g24160), OsDRB2(LOC_Os10g33970), OsDRB3 (LOC_Os09g33460), OsDRB4 (LOC_Os01g56520), AtHYL1 (At1g09700),AtDCL1 (At1g01040), AtSE (At2g27100), and AtCBP20 (At5g44200), AtDRB2 (At2g28380), AtDRB3 (At3g26932), AtDRB4 (At3g62800), AtDRB5 (At5g41070).
Supporting information
(A) Detection of miR168 and miR395 in mock-infected and RSV-infected Arabidopsis plants by northern blotting. (B) Detection of miR168 and miR395 in NS3 OX and mNS3 OX transgenic Arabidopsis plants by northern blotting. (C) Phenotypes of RSV-infected Arabidopsis plants and detection of RSV genomic RNA1 and RNA3 accumulation by northern blotting. Scale bar = 1 cm. (D) Phenotypes of NS3 OX and mNS3 OX transgenic Arabidopsis plants. Scale bar = 1 cm. In (A) and (B), U6 served as a loading control, the expression levels in Col-0 mock-inoculated plants were set to a value of 1.0 and the expression in other plants are relative to this reference value.
(TIF)
(A) Results of a BiFC assay showing the specific interaction of NS3 and Arabidopsis HYL1 in the D-body. Scale bar = 0.1 μm. (B) Phylogenetic tree of all Arabidopsis and rice DRBs. (C) Detection of miRNA levels in an OsDRB1-knockdown line by small-RNA RT-qPCR. (D) Detection of OsDRB1 levels in the WT and an OsDRB1-knockdown line by western blotting. (E) BiFC assays test interaction of NS3 between OsDRB2, OsDRB3 or OsDRB4. Scale bar = 1 μm. (F) Amino acid alignment of OsDRB1a, AtHYL1 and dsRBD2 of AtHYL1. (G) BiFC assays of NS3-NS3, mNS3-mNS3, NS3-mNS3 and OsDRB1a-OsDRB1a interactions. Scale bar = 5 μm. The BiFC assay was conducted in N. benthamiana epidermal cell, mCherry is a nuclear marker fused with red florescence protein (RFP).
(TIF)
(A) Results of an EMSA showing that NS3 protein interacts with precursor miRNAs (pre-miR168a, pre-miR395d, pre-miR398a, pre-miR399d, and pre-miR528) whereas mNS3 does not. (B) Global effect of NS3 and mNS3 on small RNA accumulations.
(TIF)
(A) Images of whole plants showing stunted phenotypes of the wild-type, NS3 OX#1, NS3 OX#7, mNS3 OX#1 and mNS3 OX#4 rice plants. Scale bars = 15 cm. (B) Detection of the RRSV CP gene by RT-qPCR.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Dr. Yijun Qi (Center for Plant Biology, School of Life Sciences, Tsinghua University) for providing DCL1-, SE-, and HYL1-related constructs, OsDRB1 knockdown rice materials and the constructive discussion and assistance from other laboratory members from Qi’s laboratory and Dr. Xiaofeng Cao (State Key Laboratory of Plant Genomics and National Center for Plant Gene Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for generously providing the CBP20 construct.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Natural Science Foundation of China (31530062, 31420103904, http://www.nsfc.gov.cn/), National Basic Research Program 973 (2014CB138400, http://program.most.gov.cn/) and Transgenic Research Program (2016ZX08010-001, http://program.most.gov.cn) to YL. Natural Science Foundation of Fujian Province of China and Outstanding Young Scientific Research Plan in Higher Education of Fujian Province (JA3091 and 2014J06011, http://fjkjt.gov.cn/) to JW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363. doi: 10.1038/nature02874 [DOI] [PubMed] [Google Scholar]
- 2.Iwakawa H-o, Tomari Y (2015) The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biology 25: 651–665. [DOI] [PubMed] [Google Scholar]
- 3.Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44. doi: 10.1146/annurev.cellbio.042308.113417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Várallyay É, Válóczi A, Ágyi Á, Burgyán J, Havelda Z (2010) Plant virus—mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J 29: 3507–3519. doi: 10.1038/emboj.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Yang Z, Wang Y, Zheng L, Ye R, et al. (2015) Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4: e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Wu Z, Li Y, Wu J (2015) Biogenesis, Function, and Applications of Virus-Derived Small RNAs in Plants. Front Microbiol 6: 1237 doi: 10.3389/fmicb.2015.01237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426. doi: 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pumplin N, Voinnet O (2013) RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11: 745–760. doi: 10.1038/nrmicro3120 [DOI] [PubMed] [Google Scholar]
- 9.Du P, Wu J, Zhang J, Zhao S, Zheng H, et al. (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog 7: e1002176 doi: 10.1371/journal.ppat.1002176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C, Ding Z, Wu K, Yang L, Li Y, et al. (2016) Suppression of Jasmonic Acid-Mediated Defense by Viral-Inducible MicroRNA319 Facilitates Virus Infection in Rice. Mol Plant 9: 1302–1314. doi: 10.1016/j.molp.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Yang R, Yang Z, Yao S, Zhao S, et al. (2017) ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants 3: 16203 doi: 10.1038/nplants.2016.203 [DOI] [PubMed] [Google Scholar]
- 12.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. doi: 10.1126/science.aae0382 [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Lu YG, Shi Y, Wu L, Xu YJ, et al. (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164: 1077–1092. doi: 10.1104/pp.113.230052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605. doi: 10.1104/pp.102.019802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, et al. (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant Journal 23: 171–182. [DOI] [PubMed] [Google Scholar]
- 16.Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, et al. (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18: 412–421. doi: 10.1105/tpc.105.038943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043. doi: 10.1016/j.cub.2005.10.016 [DOI] [PubMed] [Google Scholar]
- 18.Hu B, Wang W, Deng K, Li H, Zhang Z, et al. (2015) MicroRNA399 is involved in multiple nutrient starvation responses in rice. Front Plant Sci 6: 188 doi: 10.3389/fpls.2015.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian S, Cho WK, Kim SM, Choi H, Kim KH (2016) Time-Course Small RNA Profiling Reveals Rice miRNAs and Their Target Genes in Response to Rice Stripe Virus Infection. PLoS One 11: e0162319 doi: 10.1371/journal.pone.0162319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.F-M AG, K P, Kim M-H, Eo HJ, Oh SA, et al. (2015) THO2, core member of the THO/TREX complex, is required for micro RNA production in Arabidopsis. Plant Journal 82: 1018–1029. doi: 10.1111/tpj.12874 [DOI] [PubMed] [Google Scholar]
- 21.Dong Z, Han MH, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A 105: 9970–9975. doi: 10.1073/pnas.0803356105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823. doi: 10.1016/j.cub.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y (2007) Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol 48: 1243–1253. doi: 10.1093/pcp/pcm099 [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Ren W, Zhao Q, Zhang P, Wu F, et al. (2014) Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res 42: 12224–12236. doi: 10.1093/nar/gku907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507. doi: 10.1016/j.cub.2005.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu B, Yang Z, Li J, Minakhina S, Yang M, et al. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935. doi: 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui Y, Fang X, Qi Y (2016) TRANSPORTIN1 Promotes the Association of MicroRNA with ARGONAUTE1 in Arabidopsis. Plant cell 28: 2576–2585. doi: 10.1105/tpc.16.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djuranovic S, Nahvi A, Green R (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240. doi: 10.1126/science.1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang X, Qi Y (2016) RNAi in Plants: An Argonaute-Centered View. Plant Cell 28: 272–285. doi: 10.1105/tpc.15.00920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong Lingfang, Wu Jianxiang, Lu, et al. (2014) Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant 7: 691–708. doi: 10.1093/mp/sst158 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi M, Toriyama S, Hamamatsu C, Ishihama A (1993) Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J Gen Virol 74 (Pt 4): 769–773. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Hayakawa T, Toriyama S, Takahashi M (1991) Complete nucleotide sequence of RNA 3 of rice stripe virus: an ambisense coding strategy. J Gen Virol 72 (Pt 4): 763–767. [DOI] [PubMed] [Google Scholar]
- 33.Jiang L, Qian D, Zheng H, Meng LY, Chen J, et al. (2012) RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res 163: 512–519. doi: 10.1016/j.virusres.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 34.Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A (1994) Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol 75 (Pt 12): 3569–3579. [DOI] [PubMed] [Google Scholar]
- 35.Du Z, Xiao D, Wu J, Jia D, Yuan Z, et al. (2011) p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol Plant Pathol 12: 808–814. doi: 10.1111/j.1364-3703.2011.00716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao M, Liu X, Li S, Xu Y, Zhou Y, et al. (2014) Rice Stripe Tenuivirus NSvc2 Glycoproteins Targeted to the Golgi Body by the N-Terminal Transmembrane Domain and Adjacent Cytosolic 24 Amino Acids via the COP I-and COP II-Dependent Secretion Pathway. J Virol 88: 3223–3234. doi: 10.1128/JVI.03006-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen M, Xu Y, Jia R, Zhou X, Ye K (2010) Size-independent and noncooperative recognition of dsRNA by the Rice stripe virus RNA silencing suppressor NS3. J Mol Biol 404: 665–679. doi: 10.1016/j.jmb.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 38.Xiong R, Wu J, Zhou Y, Zhou X (2009) Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 387: 29–40. doi: 10.1016/j.virol.2009.01.045 [DOI] [PubMed] [Google Scholar]
- 39.Xiong R, Wu J, Zhou Y, Zhou X (2008) Identification of a movement protein of the tenuivirus rice stripe virus. J Virol 82: 12304 doi: 10.1128/JVI.01696-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Z, Xiao D, Wu J, Jia D, Yuan Z, et al. (2011) p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol Plant Pathol 12: 808–814. doi: 10.1111/j.1364-3703.2011.00716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Zhang F, Li J, Chen JP, Zhang HM (2016) Integrative Analysis of the microRNAome and Transcriptome Illuminates the Response of Susceptible Rice Plants to Rice Stripe Virus. PLoS One 11: e0146946 doi: 10.1371/journal.pone.0146946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM (2009) The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235. doi: 10.1261/rna.1646909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang SW, Chen HY, Yang J, Machida S, Chua NH, et al. (2010) Structure of Arabidopsis HYPONASTIC LEAVES1 and Its Molecular Implications for miRNA Processing. Structure 18: 594–605. doi: 10.1016/j.str.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding SW (2010) RNA-based antiviral immunity. Nat Rev Immunol 10: 632–644. doi: 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- 45.Burgyan J, Havelda Z (2011) Viral suppressors of RNA silencing. Trends Plant Sci 16: 265–272. doi: 10.1016/j.tplants.2011.02.010 [DOI] [PubMed] [Google Scholar]
- 46.Cao X, Zhou P, Zhang X, Zhu S, Zhong X, et al. (2005) Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J Virol 79: 13018–13027. doi: 10.1128/JVI.79.20.13018-13027.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18: 1179–1186. doi: 10.1101/gad.1201204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duan CG, Fang YY, Zhou BJ, Zhao JH, Hou WN, et al. (2012) Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24: 259–274. doi: 10.1105/tpc.111.092718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li F, Ding SW (2006) Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60: 503–531. doi: 10.1146/annurev.micro.60.080805.142205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren B, Guo Y, Gao F, Zhou P, Wu F, et al. (2010) Multiple functions of Rice dwarf phytoreovirus Pns10 in suppressing systemic RNA silencing. J Virol 84: 12914–12923. doi: 10.1128/JVI.00864-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, et al. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154. doi: 10.1104/pp.105.062943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baek D, Kim MC, Chun HJ, Kang S, Park HC, et al. (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161: 362–373. doi: 10.1104/pp.112.205922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang X, Cui Y, Li Y, Qi Y (2015) Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat Plants 1: 15075 doi: 10.1038/nplants.2015.75 [DOI] [PubMed] [Google Scholar]
- 54.Kim YJ, Zheng B, Yu Y, Won SY, Mo B, et al. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822. doi: 10.1038/emboj.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. doi: 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- 56.Wang L, Song X, Gu L, Li X, Cao S, et al. (2013) NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25: 715–727. doi: 10.1105/tpc.112.105882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu G, Park MY, Conway SR, Wang JW, Weigel D, et al. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. doi: 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 21: 347–361. doi: 10.1105/tpc.108.060137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S, Xie M, Ren G, Yu B (2013) CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc Natl Acad Sci U S A 110: 17588–17593. doi: 10.1073/pnas.1310644110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, et al. (2010) Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci U S A 107: 466–471. doi: 10.1073/pnas.0913203107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim W, Kim HE, Jun AR, Jung MG, Jin S, et al. (2016) Structural determinants of miR156a precursor processing in temperature-responsive flowering in Arabidopsis. J Exp Bot 67: 4659–4670. doi: 10.1093/jxb/erw248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, et al. (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725. doi: 10.1038/ng1804 [DOI] [PubMed] [Google Scholar]
- 63.Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible W-R, et al. (2009) Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res 37: 3083–3093. doi: 10.1093/nar/gkp189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154. doi: 10.1104/pp.107.106989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428. doi: 10.1016/j.molcel.2005.06.014 [DOI] [PubMed] [Google Scholar]
- 66.Jin L, Qin Q, Wang Y, Pu Y, Liu L, et al. (2016) Rice Dwarf Virus P2 Protein Hijacks Auxin Signaling by Directly Targeting the Rice OsIAA10 Protein, Enhancing Viral Infection and Disease Development. PLoS Pathog 12: e1005847 doi: 10.1371/journal.ppat.1005847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1: 100 doi: 10.1038/ncomms1093 [DOI] [PubMed] [Google Scholar]
- 68.Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. doi: 10.1016/j.cell.2008.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Detection of miR168 and miR395 in mock-infected and RSV-infected Arabidopsis plants by northern blotting. (B) Detection of miR168 and miR395 in NS3 OX and mNS3 OX transgenic Arabidopsis plants by northern blotting. (C) Phenotypes of RSV-infected Arabidopsis plants and detection of RSV genomic RNA1 and RNA3 accumulation by northern blotting. Scale bar = 1 cm. (D) Phenotypes of NS3 OX and mNS3 OX transgenic Arabidopsis plants. Scale bar = 1 cm. In (A) and (B), U6 served as a loading control, the expression levels in Col-0 mock-inoculated plants were set to a value of 1.0 and the expression in other plants are relative to this reference value.
(TIF)
(A) Results of a BiFC assay showing the specific interaction of NS3 and Arabidopsis HYL1 in the D-body. Scale bar = 0.1 μm. (B) Phylogenetic tree of all Arabidopsis and rice DRBs. (C) Detection of miRNA levels in an OsDRB1-knockdown line by small-RNA RT-qPCR. (D) Detection of OsDRB1 levels in the WT and an OsDRB1-knockdown line by western blotting. (E) BiFC assays test interaction of NS3 between OsDRB2, OsDRB3 or OsDRB4. Scale bar = 1 μm. (F) Amino acid alignment of OsDRB1a, AtHYL1 and dsRBD2 of AtHYL1. (G) BiFC assays of NS3-NS3, mNS3-mNS3, NS3-mNS3 and OsDRB1a-OsDRB1a interactions. Scale bar = 5 μm. The BiFC assay was conducted in N. benthamiana epidermal cell, mCherry is a nuclear marker fused with red florescence protein (RFP).
(TIF)
(A) Results of an EMSA showing that NS3 protein interacts with precursor miRNAs (pre-miR168a, pre-miR395d, pre-miR398a, pre-miR399d, and pre-miR528) whereas mNS3 does not. (B) Global effect of NS3 and mNS3 on small RNA accumulations.
(TIF)
(A) Images of whole plants showing stunted phenotypes of the wild-type, NS3 OX#1, NS3 OX#7, mNS3 OX#1 and mNS3 OX#4 rice plants. Scale bars = 15 cm. (B) Detection of the RRSV CP gene by RT-qPCR.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.