Abstract
Molecular testing represents a promising adjunct for the diagnosis of antibody-mediated rejection (AMR). Here we apply a novel gene expression platform in sequential formalin-fixed paraffin-embedded (FFPE) samples from nonhuman primate (NHP) renal transplants. We analyzed 34 previously-described gene transcripts related to AMR in humans in 197 archival NHP samples, including 102 from recipients that developed chronic AMR, 80 from recipients without AMR, and 15 normal native nephrectomies. Three endothelial genes (VWF, DARC, CAV1), derived from 10-fold cross-validation ROC curve analysis, demonstrated excellent discrimination between AMR and non-AMR samples (AUC=0.92). This 3-gene set correlated with classic features of AMR, including glomerulitis, capillaritis, glomerulopathy, C4d, and DSA (r=0.39–0.63, p<0.001). Principal component analysis confirmed the association between 3-gene set expression and AMR and highlighted the ambiguity of v-lesions and ptc-lesions between AMR and T-cell mediated rejection (TCMR). Elevated 3-gene set expression corresponded with the development of immunopathologic evidence of rejection and often preceded it. Many recipients demonstrated mixed AMR and TCMR suggesting that this represents the natural pattern of rejection. These data provide NHP animal model validation of recent updates to the Banff classification including the assessment of molecular markers for diagnosing AMR.
INTRODUCTION
RNA transcript analysis has the potential for precise, objective, and mechanism-based evaluation of allograft biopsies that can supplant routine pathology (1). Over the last decade, numerous microarray studies have begun to define the molecular phenotypes of allograft rejection (2–8). Despite several gene sets being described and validated, none have successfully been translated into routine clinical practice. A major barrier to clinical implementation has been the requirement for dedicated fresh tissue samples, which must be specially processed and procured in addition to the standard-of-care formalin-fixed paraffin-embedded (FFPE) biopsies used for histology. The consequences of this limitation are increased cost, complexity, and patient risk, as well as reduced tissue for routine histology (9).
NanoString® nCounter® (NanoString Technologies, Seattle, WA) is a novel gene expression platform that works reliably with FFPE tissue. It is able to analyze up to 800 customizable transcript targets per sample and is similar in sensitivity to real-time PCR and more sensitive than microarrays (10, 11). The utilization of FFPE material allows for molecular-histological correlation on the same tissue. Cellular populations submitted for molecular testing and potential histological features of significance can therefore be better integrated than with techniques requiring separately processed tissue. Furthermore, this technology represents the first opportunity to perform reliable retrospective gene expression testing on well-annotated archival FFPE tissue, facilitating immediate correlation with long-term clinical follow-up (1).
We have previously demonstrated the feasibility of the NanoString® system with routine FFPE tissue and have used it to assess a 34-gene set as a potential molecular diagnostic tool for antibody-mediated rejection (AMR) in human renal and cardiac allograft biopsies (12, 13). Here we apply this AMR 34-gene set to nonhuman primate (NHP) renal transplants developing chronic AMR off immunosuppression with analogous features to human renal allografts, including donor specific antibodies (DSA), C4d deposition, transplant glomerulopathy, and arteriopathy (14–19). Previous studies of protocol biopsies from these recipients have identified four stages of chronic AMR (16, 17), which have subsequently been confirmed and refined in humans (20). This study aims to use the NanoString® system to validate AMR-related transcripts from humans in a well-studied animal model representing the natural course of rejection and determine if molecular diagnostics can enhance our understanding of the pathogenesis and evolution of AMR.
MATERIALS AND METHODS
Subjects
The recipients were NHPs, Macaca fascicularis, given allogeneic bone marrow transplants (BMT) and kidney allografts from the same donor in conjunction with transient immunosuppression. Detailed treatment protocols and outcomes have previously been published (16, 17). In brief, the standard protocol included low-dose total body irradiation on days -6 and -5, thymic irradiation on day -1, pre-transplant anti-thymocyte globulin and BMT on day 0, followed by a one-month course of calcineurin inhibitor. Recipients after 2002 were also treated with co-stimulatory blockade with anti-CD154 monoclonal antibody or belatacept. In the delayed protocol, recipients initially underwent kidney transplantation alone with conventional immunosuppression, including tacrolimus, mycophenolate mofetil, and methylprednisolone, plus anti-CD8 monoclonal antibody or anti-thymocyte globulin. All immunosuppression was permanently discontinued one month after BMT in both protocols. The recipients were not treated for AMR or T-cell mediated rejection (TCMR). All surgical procedures and postoperative care were performed in accordance with National Institutes of Health guidelines for the care and use of primates and were approved by the Massachusetts General Hospital Subcommittee on Animal Research.
Samples
A total of 197 FFPE kidney samples from 81 recipients obtained between 1993 and 2013 were retrieved from the transplant pathology research tissue bank at Massachusetts General Hospital. These included 102 sequential allograft samples, consisting of protocol biopsies (n=78) and post-mortem samples obtained by euthanasia (n=24), from 29 recipients in which the treatment protocol failed and chronic active AMR developed. The only selection criteria were that 1) there were two or more samples per recipient and 2) there was enough tissue for histology and gene expression analysis. Comparison samples included 15 normal native nephrectomies from 15 recipients and a representative collection of 80 allograft biopsy (n=53) and euthanasia (n=27) samples from 37 recipients with pathological diagnoses of TCMR (n=33), borderline changes/suspicious for TCMR (n=13), suspicious for AMR (n=7), no pathological evidence of rejection (n=19), polyomavirus nephropathy (n=3), pyelonephritis (n=2), post-transplant lymphoproliferative disorder (n=2), and obstruction (n=1). None of the comparison samples came from recipients that developed AMR.
Clinical Data and Pathology Review
Treatment protocols, serum creatinine, and DSA results were retrieved. Histology slides (H&E, PAS, and C4d immunohistochemistry) were reviewed by at least one transplant pathologist (RNS, IAR) and diagnoses were assigned according to the 2015 Banff classification (21). The histological techniques have previously been described (16, 17).
RNA Isolation
A previously reported NanoString® workflow was utilized (12). In brief, three consecutive 20-µm sections were obtained from each FFPE block and sent to the University of Alberta. Xylene deparaffinization and RNA extraction were performed with the Ambion™ RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific, Waltham, MA). RNA concentration and purity were measured with a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
NanoString® Gene Expression Analysis
Oligonucleotide probes specific to Macaca fascicularis were manufactured for the mRNA sequences of 38 genes (Integrated DNA Technologies, Coralville, IA). These included a previously-described AMR 34-gene-set comprised of 18 endothelial, 6 NK cell, and 10 inflammation-related genes, as well as 4 housekeeping genes (12). Probe sequences are provided in Table S1. Gene expression was then quantified with the NanoString® nCounter® Gene Expression assay (NanoString Technologies, Seattle, WA) as per manufacturer instructions. To assess reproducibility, eight samples were randomly selected for duplicate analysis in separate runs. Quality control assessment and normalization of raw NanoString® gene expression results were performed with nSolver™ Analysis Software Version 3.0 (NanoString Technologies, Seattle, WA) using the manufacturer-recommended default parameters.
Retrospective Analysis of Human Microarray Data Set
A publicly-available human cDNA microarray data set (GSE36059) was retrieved from the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) to compare gene set performance in human samples. The data set included gene expression results (Affymetrix® Human Genome U133 Plus 2.0 Array) for 403 renal allograft indication biopsies with diagnostic labels of AMR (n=65), TCMR (n=35), mixed rejection (n=22), and non-rejection (n=281), as per Banff 2009 criteria (22). The raw data files were imported and normalized with BRB-ArrayTools Version 4.5.0 (23). Mean values were used for genes with repeat data points.
Data Analysis
Post-normalization statistical analysis and visualization were performed with R version 3.3.2 (R Foundation for Statistical Computing, Vienna, Austria). Normalized transcript counts (NanoString® data) and log intensity values (microarray data) were converted to z-scores (number of standard deviations away from the population mean for each gene) for individual gene analysis. Mean z-scores were used for aggregate gene set analysis. Gene expression and correlation heat map analyses (heatmap.2 function in gplots package) were performed with unsupervised hierarchical clustering by Euclidean distance. Spearman’s rank correlation coefficients (cor function in stats package) and unsupervised principal component analysis (PCA; prcomp function in stats package) were used to characterize inter-variable relationships. Mann-Whitney U-tests (wilcox.test function in stats package) were utilized for class comparison analyses. Receiver operating characteristic (ROC) curve analysis (roc function in pROC package) was used for assessment of diagnostic performance. Youden’s J-statistic (point on ROC curve farthest from diagonal index line) was utilized for defining diagnostic thresholds (24). Individual gene ranking, gene set construction, and gene set ranking were achieved with repeated 10-fold cross-validation analysis (train function in caret package) using three repeats and Naive Bayes model. P-values less than 0.05 were considered statistically significant.
RESULTS
RNA and Quality Control
The mean RNA yield obtained from three 20-µm sections per FFPE block was 6138 ng (range: 212–66508 ng) with a mean concentration of 153.5 ng/µL (5.3–1662.7 ng/µL) and a mean A260/A280 RNA purity ratio of 1.85 (1.54–2.20). No quality control or normalization flags were encountered during nSolver™ analysis for any of the 197 NHP samples included. The eight samples analyzed in duplicate demonstrated excellent reproducibility with a mean correlation coefficient of 0.990 (range: 0.953–0.999).
Individual Gene Expression vs. Diagnosis
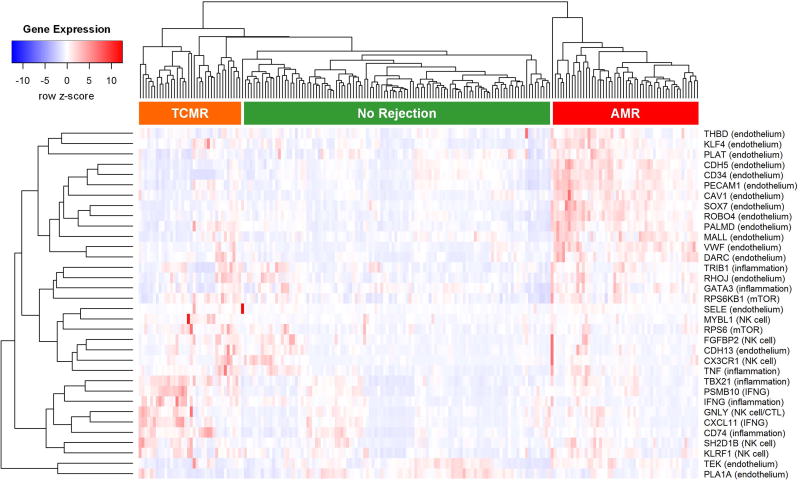
Following histological evaluation, the samples were assigned one of the following diagnostic labels according to Banff 2015 criteria: AMR (n=38), suspicious for AMR (n=15), mixed rejection (n=27), TCMR (n=41), borderline (n=21), no rejection (n=32), other (n=8), and normal native nephrectomy (n=15). Heat map analysis revealed general grouping of diagnostic categories based on individual gene expression patterns (Figure 1). Sixty-seven (74%) of the 91 samples diagnosed as normal, no rejection, other, borderline, or suspicious for AMR clustered within the larger ‘No Rejection’ group indicated in Figure 1. Thirty-five (54%) of the 65 samples diagnosed as pure AMR or mixed rejection clustered within the ‘AMR’ group. Five (33%) of the 15 samples diagnosed as suspicious for AMR also clustered within the ‘AMR’ group. Sixteen (39%) of the 41 samples diagnosed as TCMR clustered within the ‘TCMR’ group. Endothelium-associated transcripts generally exhibited higher expression in the ‘AMR’ group. Inflammation-related transcripts showed a tendency for higher expression in the ‘TCMR’ group as well as the ‘No Rejection’ samples with inflammatory diseases.
Figure 1. Gene expression heat map for 34 antibody-mediated rejection (AMR) related genes in 197 nonhuman primate samples.
There is general clustering of gene expression patterns with diagnostic categories, i.e. higher expression of endothelial genes in AMR cases, higher expression of inflammation-related genes in T-cell mediated rejection (TCMR) cases, and lower expression of all genes in cases lacking immunopathologic evidence of rejection.
Individual Gene Expression vs. Immunopathology
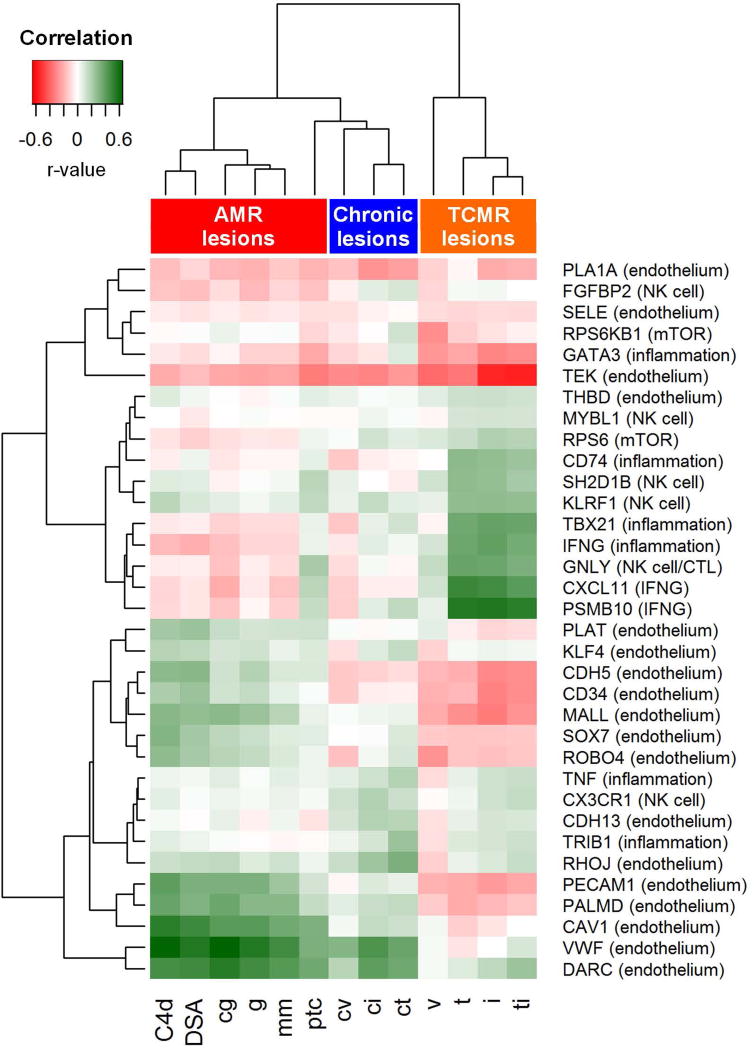
Figure 2 represents a correlation heat map demonstrating the correlation coefficients between gene expression and Banff histology scores/DSA status. Endothelium-associated transcripts exhibited strong correlation with DSA and traditional AMR lesions, including C4d deposition, transplant glomerulopathy (cg), glomerulitis (g), and peritubular capillaritis (ptc). Mesangial matrix expansion (mm) also correlated with endothelial gene expression. Histological lesions of chronic allograft injury, including arterial fibrous intimal thickening (cv), interstitial fibrosis (ci), and tubular atrophy (ct), clustered together and also demonstrated greater correlation with endothelial transcripts. Traditional TCMR-related lesions, including intimal arteritis (v), tubulitis (t), interstitial inflammation (i), and total inflammation (ti) exhibited the strongest correlation with inflammatory, NK cell, and interferon gamma-induced transcripts.
Figure 2. Correlation heat map demonstrating Spearman’s rank correlation coefficients comparing expression of 34 antibody-mediated rejection (AMR) related genes with Banff histology scores and DSA status.
Endothelial genes demonstrate the strongest correlation with lesions associated with antibody-mediated rejection (C4d, DSA, cg, g, ptc). Inflammation-related genes generally correlate with T-cell mediated rejection lesions (t, i, ti). Histologic lesions associated with chronic injury (cv, ci, ct) also correlate with endothelial genes. Abbreviations: AMR, antibody-mediated rejection; cg, transplant glomerulopathy; ci, interstitial fibrosis; ct, tubular atrophy; cv, arterial fibrous intimal thickening; DSA, donor specific antibody; g, glomerulitis; i, interstitial inflammation; mm, mesangial matrix increase; ptc, peritubular capillary margination; t, tubulitis; TCMR, T-cell mediated rejection; ti, total interstitial inflammation; v, intimal arteritis.
Diagnostic Performance of Gene Expression Analysis
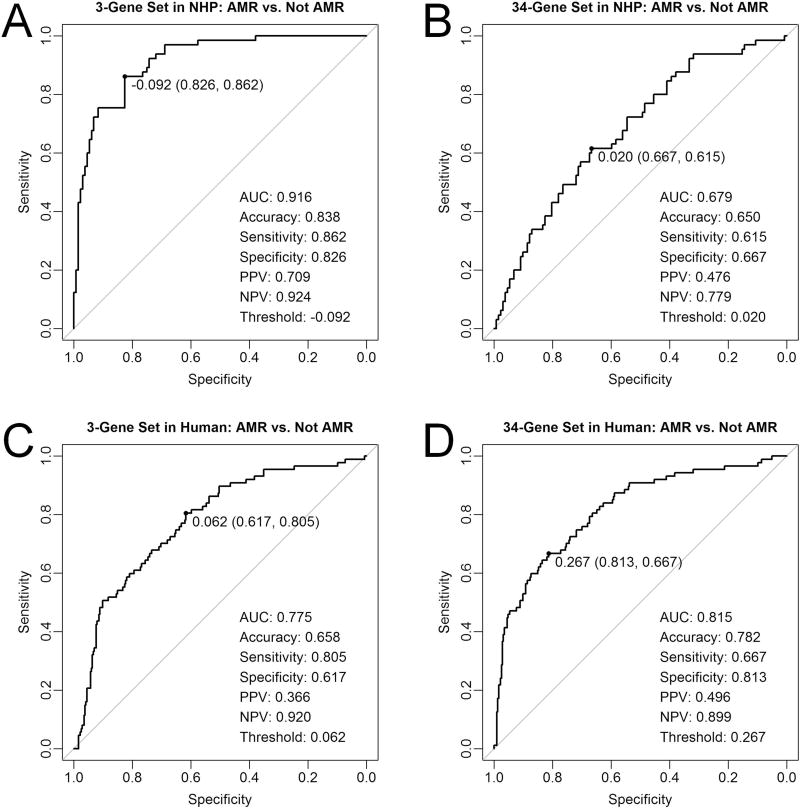
The ability of gene expression testing to discriminate AMR from non-AMR cases, as per Banff 2015 criteria, was assessed with repeated 10-fold cross-validation ROC curve analysis. Table S2 lists the mean area under the curve (AUC), mean accuracy, and mean Cohen’s kappa statistic between resampling repeats for each individual gene. Ranking the genes by mean AUC value, VWF was the strongest individual performer (AUC=0.917) while CX3CR1 was the weakest (AUC=0.435). There was general correlation between mean AUC, accuracy, and kappa, particularly for the top performing genes, with VWF also demonstrating the highest mean accuracy (0.881) and kappa (0.728) values. Detailed diagnostic performance parameters calculated from the study set as a single cohort are also provided in Table S2 and show generally good agreement with the cross-validation results, particularly for the top performing genes.
Based on the cross-validation AUC rank order, the genes were combined into successively larger gene sets as follows: 1-gene set includes VWF, 2-gene set includes VWF and DARC, 3-gene set includes VWF, DARC, and CAV1, …, 34-gene set includes all genes. Cross-validation and study set ROC curve analyses were then repeated for each of the 34 gene sets (Table S3). The 3-gene set demonstrated the highest AUC on both analyses (cross-validation AUC = 0.920, study set AUC = 0.916) and was thus used for subsequent analysis. Figure 3A shows the study set ROC curve for the 3-gene set, with the following diagnostic performance parameters: AUC = 0.916, accuracy = 0.838, sensitivity = 0.862, specificity = 0.826, positive predictive value = 0.709, and negative predictive value = 0.924. The 34-gene set demonstrated the weakest diagnostic performance with cross-validation and study set AUCs of 0.679 (Figure 3B). The diagnostic cut-off for the 3-gene set, as defined by Youden’s J-statistic, was a mean z-score of −0.092. The range of 3-gene set values in this NHP cohort was −0.948 to 3.831 (mean = 0.000, median = −0.278), with the diagnostic threshold of −0.092 representing the 60th percentile.
Figure 3. Receiver operating characteristic curves demonstrating diagnostic performance of gene expression testing in discriminating antibody-mediated rejection (AMR) cases from non-AMR cases, as defined by Banff 2015 criteria.
(A) Refined 3-gene set and (B) full 34-gene set in 197 nonhuman primate samples (NHP) analyzed with NanoString®. (C) Refined 3-gene set and (D) full 34-gene set in 403 human samples analyzed with microarray. Abbreviations: AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value.
A publicly-available human renal allograft microarray data set (n=403) was retrospectively analyzed to compare gene set performance in human versus NHP samples. The NHP-refined 3-gene set demonstrated inferior diagnostic performance in human samples with an AUC of 0.775 (Figure 3C). In contrast, the complete 34-gene set exhibited superior performance in human samples with an AUC of 0.815 (Figure 3D).
AMR 3-Gene Set vs. Diagnosis
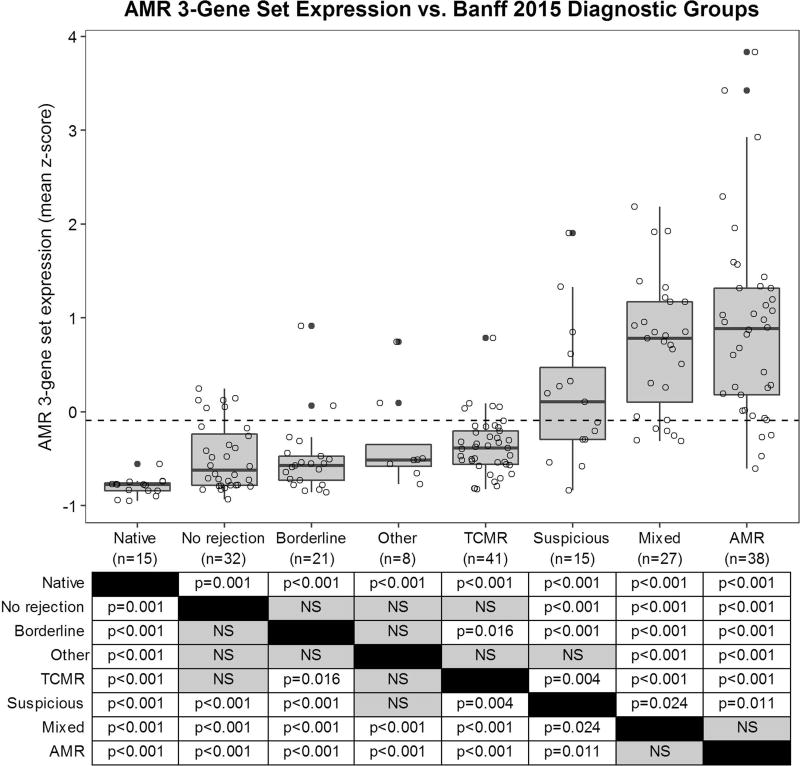
Figure 4 demonstrates AMR 3-gene set (VWF, DARC, CAV1) expression versus Banff 2015 diagnoses for all 197 NHP samples. The normal native nephrectomy group showed significantly lower gene set expression than all of the other groups (p≤0.001). The AMR and mixed rejection groups exhibited significantly higher gene set expression than all of the other groups (p≤0.024). The suspicious for AMR group demonstrated significantly higher expression than the remainder of the non-AMR groups (p≤0.004) with the exception of ‘other’ (p=0.076). The TCMR group showed significantly higher expression than the borderline group (p=0.016) but not the ‘other’ or no rejection groups. There was no significant difference in gene set expression between the no rejection, borderline, and ‘other’ groups.
Figure 4. Gene expression box plot demonstrating expression of antibody-mediated rejection (AMR) 3-gene set in 197 nonhuman primate samples according to Banff 2015 diagnostic groups.
P-values corresponding to Mann-Whitney U-tests between diagnostic groups are provided in the table. The ROC curve-derived diagnostic threshold for AMR (−0.092) is indicated by the dashed line. Abbreviations: AMR, antibody-mediated rejection; Mixed, mixed AMR and TCMR; Native, normal native nephrectomy; Suspicious, suspicious for AMR; TCMR, T-cell mediated rejection.
Thirty-four (89%) of the 38 pure AMR samples and 22 (81%) of the 27 mixed rejection samples demonstrated AMR 3-gene set expression above the ROC-derived diagnostic cut-off (dashed line in Figure 4). Only 23/132 (17%) non-AMR samples (eight suspicious for AMR, six no rejection, five TCMR, two borderline, one polyomavirus nephropathy, and one pyelonephritis) exhibited gene set expression above the diagnostic threshold. Twelve (52%) of these 23 samples (five suspicious for AMR, three no rejection, two TCMR, and two borderline), associated with a total of eight recipients, had subsequent biopsies diagnostic for AMR an average of 356 days later (range: 23–749 days). According to Banff 2015 criteria, two of these cases (i.e. those showing isolated g1/ptc1 lesions with DSA but no C4d) would be reclassified as AMR with the inclusion of the 3-gene set in the diagnostic algorithm.
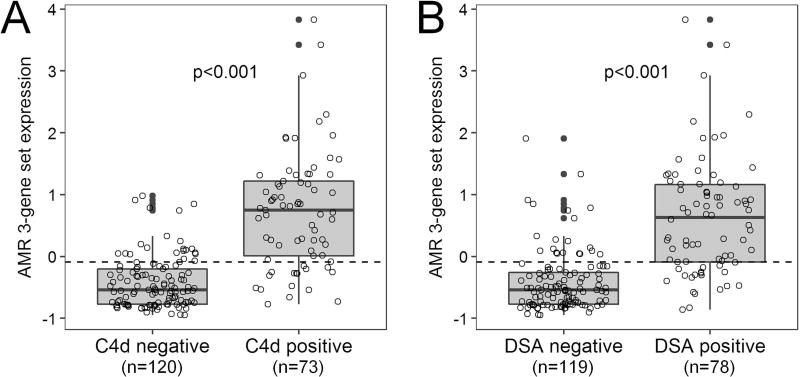
AMR 3-gene set expression was significantly higher in C4d-positive versus C4d-negative cases (p<0.001) as well as DSA-positive versus DSA-negative cases (p<0.001) (Figure 5). Gene set expression showed significant correlation with serum creatinine at the time of sample procurement (r=0.152, p=0.036). There was no significant difference in gene set expression between biopsy and euthanasia samples and no significant correlation with the number of days post-transplantation. There was also no significant difference in AMR incidence or gene set expression between standard and delayed protocol samples.
Figure 5. Gene expression box plots showing increased antibody-mediated rejection (AMR) 3-gene set expression in (A) C4d positive vs. negative cases and (B) donor specific antibody (DSA) positive vs. negative cases.
The ROC curve-derived diagnostic threshold for AMR (−0.092) is indicated by the dashed line.
AMR 3-Gene Set vs. Immunopathology
AMR 3-gene set expression exhibited significant correlation with AMR-related immunopathologic features, including C4d deposition (r=0.634, p<0.001), cg (r=0.620, p<0.001), DSA positivity (r=0.593, p<0.001), g (r=0.560, p<0.001), and ptc (r=0.392, p<0.001) (Table 1). Correlation with ptc was seen in both mononuclear (r=0.649, p<0.001) and polymorphonuclear (r=0.340, p<0.001) subtypes. Gene set expression also correlated with chronic injury lesions, including mm (r=0.487, p<0.001), ci (r=0.388, p<0.001), ct (r=0.336, p<0.001), and cv (r=0.199, p=0.006). It did not correlate with traditional TCMR lesions including i, ti, t, and v.
Table 1.
Correlation between antibody-mediated rejection (AMR) 3-gene set expression and histological/serological features.
| Histological/serological feature | Correlation coefficient | p-value |
|---|---|---|
| Antibody-mediated rejection-related lesions | ||
| C4d-positive | 0.634 | <0.001 |
| Transplant glomerulopathy (cg) | 0.620 | <0.001 |
| Donor specific antibodies | 0.593 | <0.001 |
| Glomerulitis (g) | 0.560 | <0.001 |
| Peritubular capillary margination (ptc) | 0.392 | <0.001 |
| T-cell mediated rejection-related lesions | ||
| Total interstitial inflammation (ti) | 0.135 | 0.061 |
| Interstitial inflammation (i) | 0.038 | 0.600 |
| Intimal arteritis (v) | 0.032 | 0.660 |
| Tubulitis (t) | −0.041 | 0.575 |
| Chronic injury-related lesions | ||
| Mesangial matrix increase (mm) | 0.487 | <0.001 |
| Interstitial fibrosis (ci) | 0.388 | <0.001 |
| Tubular atrophy (ct) | 0.336 | <0.001 |
| Arterial fibrous intimal thickening (cv) | 0.199 | 0.006 |
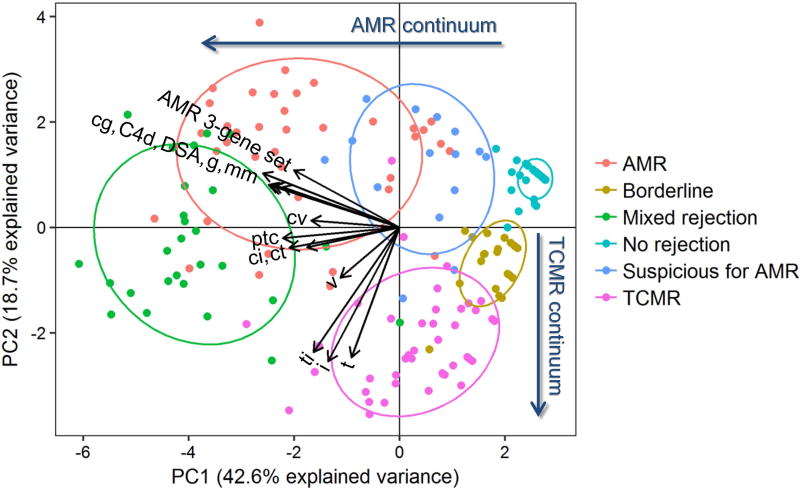
PCA was used to further characterize the relationship between Banff 2015 diagnoses and gene set expression, DSA, and histology. Figure 6 demonstrates distinct separation of the diagnostic categories with principal component (PC) 1 and PC2, together accounting for 61.3% of the explained variance. PC1 primarily shows an AMR continuum from ‘no rejection’ to ‘suspicious for AMR’ to ‘AMR/mixed rejection’. PC2 primarily demonstrates a TCMR continuum from ‘no rejection/AMR’ to ‘borderline’ to ‘TCMR’. Overlap between mixed rejection and pure AMR is partially discriminated with both PC1 and PC2. Mixed rejection partially overlaps with pure TCMR on PC2 but not PC1. The AMR 3-gene set vector is most closely oriented in the direction of the pure AMR group, indicating that it has the strongest association with this diagnostic category. A tight cluster of traditional AMR-related variables, including cg, C4d, DSA, g, and mm (in order of counterclockwise orientation), are also associated with the pure AMR group. A cluster of traditional TCMR-related lesions, including t, i, and ti, are most closely associated with the pure TCMR group. In between these two sets of more specific AMR and TCMR-related variables lies a group of more ambiguous histologic lesions, including those of chronicity (ci, ct, and cv) as well as ptc (closer to AMR) and v (closer to TCMR).
Figure 6. Principal component analysis demonstrating relationships between immunopathologic features, antibody-mediated rejection (AMR) 3-gene set expression, and Banff 2015 diagnostic categories.
Principal components (PC) 1 and 2 primarily separate the diagnostic categories along AMR and T-cell mediated rejection (TCMR) continuums, respectively. There is a general association between AMR-associated features (AMR 3-gene set, cg, C4d, DSA, g, mm) and AMR cases as well as between TCMR-associated features (t, i, ti) and TCMR cases. Intermediate between these two clusters are lesions of chronicity (cv, ci, ct) as well as ptc and v-lesions. Abbreviations: cg, transplant glomerulopathy; ci, interstitial fibrosis; ct, tubular atrophy; cv, arterial fibrous intimal thickening; DSA, donor specific antibody; g, glomerulitis; i, interstitial inflammation; mm, mesangial matrix increase; ptc, peritubular capillary margination; t, tubulitis; ti, total interstitial inflammation; v, intimal arteritis.
AMR 3-Gene Set in Sequential Samples
The temporal pattern of AMR 3-gene set expression for the 20 recipients that had at least four sequential samples is demonstrated in Figure S1. These included 13 recipients that developed AMR, five recipients that developed TCMR only, and two recipients that did not exhibit diagnostic evidence of rejection. All of the recipients with AMR demonstrated at least one episode of increased gene set expression above the ROC-derived diagnostic threshold (indicated by the dashed line). Some of these recipients had persistently rising expression levels over time (AMR #1) while others demonstrated a transient rise followed by a decline (AMR #6). Some recipients showed relatively stable expression (AMR #13), while others were more erratic (AMR #5). Elevated gene set expression generally corresponded with the development of immunopathologic evidence of AMR but it also occasionally preceded it (AMR #12). The single ABO-incompatible recipient included in the study (AMR #8) showed rapid development of AMR within the first two months of transplantation. Nine (69%) of the 13 AMR recipients demonstrated mixed rejection (AMR #2). Some of the euthanasia samples exhibited a sharp decline in expression (AMR #2) while others did not (AMR #7). The no rejection and TCMR only recipients generally showed consistently low levels of expression, although one from each group demonstrated transient elevation (No Rejection #2 and TCMR #2).
DISCUSSION
There is an urgent need for improved methods of detecting and grading clinical and subclinical AMR (25). Growing recognition of the potential diagnostic utility of gene expression testing is evidenced by its inclusion in the 2013 Banff classification (26) and its prioritization as a major focus of discussion at the 2015 Banff meeting (21). However, obstacles to the successful translation of molecular testing into routine clinical practice include: 1) lack of a true diagnostic ‘gold standard’ for validating potential molecular diagnostics, 2) heterogeneity of existing data associated with different Banff classification iterations, 3) absence of robust validation studies, and 4) lack of consensus on platforms to be used, transcripts to be assessed, and criteria for positivity (21).
We aimed to address these challenges by analyzing the expression of AMR-related transcripts in an animal model free from the confounding variables of inconsistent immunosuppression, rejection treatment, and patient compliance. The recipients in this study were also otherwise healthy and thus not complicated by the heterogeneity of medical comorbidities affecting human patients. These factors allowed for the molecular characterization of a relatively pure model for allograft rejection. Our analysis validated the molecular phenotype of AMR previously identified in humans by confirming the correlation between endothelium-associated transcripts (27) and immunopathologic criteria currently used in the Banff classification for diagnosing AMR (21). The inclusion of only AMR-related transcripts allowed for a focused evaluation of molecular testing in a specific diagnostic condition.
This study further demonstrated the technical robustness of the NanoString® platform through successful analysis of very old archival FFPE samples, some of which had been in storage for more than 20 years. This confirms the potential utility of this already clinically-approved platform for analyzing both retrospective and prospective samples in the validation of novel molecular diagnostics. Furthermore, the ability to utilize FFPE tissue with this platform will facilitate enhanced integration of molecular and histological features.
The inclusion of sequential protocol biopsies afforded the opportunity to analyze the natural course of gene expression over time. All of the NHP recipients that developed AMR exhibited at least transiently elevated endothelial gene expression. In 28% of these recipients, molecular evidence of microcirculation injury preceded histological features of AMR. This suggests that gene expression testing is more sensitive than histology for the detection of early AMR. The 11 non-AMR samples that exhibited transient AMR 3-gene set expression elevation without subsequent evidence of AMR may represent examples of either self-resolving antibody-mediated injury not captured by Banff criteria or alternative causes of transient microcirculation injury.
The PCA results validate recent updates to the 2013 Banff classification, specifically the incorporation of gene expression testing as a potential diagnostic criterion for AMR (26). Using current Banff criteria, two samples in the suspicious for AMR group would be upgraded to a full diagnosis of AMR with incorporation of the AMR 3-gene set. PCA also highlighted the ambiguity of intimal arteritis (v-lesions), peritubular capillaritis (ptc-lesions), vasculopathy (cv-lesions), and IFTA (ci- and ct-lesions) between AMR and TCMR. This confirms recent observations in human patients (28, 29) and provides further evidence for the relative non-specificity of these lesions as well as the complex interplay between antibody and cell-mediated pathways of injury. However, a relatively closer relationship between chronic histologic lesions and AMR is consistent with antibody-mediated injury being associated with a greater degree of cumulative damage later in the post-transplant period in this NHP model. The phenomenon of mixed rejection, with AMR often following TCMR, was identified in more than half of the AMR NHP recipients, suggesting that this represents the true natural pattern of rejection. However, PCA demonstrated closer proximity of the mixed rejection cases to AMR. Borderline and TCMR, on the other hand, appear to represent a molecular-histological continuum as previously described in humans (30).
The AMR 3-gene set derived in this study demonstrated excellent discrimination of AMR and non-AMR pathologies (AUC=0.92). These three genes (VWF, DARC, CAV1) are all endothelium-related transcripts with long-standing associations with AMR in humans (31–33). Their biological functions include endothelial injury, repair, activation, and angiogenesis. Elevated endothelial-associated transcript expression has been reported in non-AMR conditions, including acute tubular injury, TCMR, and viral and bacterial infection, although, in the absence of DSA, this has not been associated with inferior allograft survival (27). The discrepancy in diagnostic performance between NHP and human samples, in which the complete 34-gene set performed better, may be related to differences in gene expression platforms, recipient treatment, comorbidities, and sample selection (i.e. retrospective protocol biopsies versus prospective indication biopsies). In particular, the conditioning protocols used in this NHP model, which included BMT, irradiation, and immunosuppression weaning, differ significantly from standard clinical practice and may have contributed to gene expression discrepancies. However, most biopsies were taken more than 6 months post-transplantation, suggesting that the rejection seen in these animals represents failure of the conditioning protocol and, with that, the natural course of rejection given that they did not receive any specific anti-rejection treatment. Due to the additional confounders in humans, a larger gene set might be more applicable in the clinical setting but should likely include the three top genes identified here, as they appear to have the strongest association with a pure AMR phenotype.
To successfully translate molecular transplant diagnostics into routine clinical practice, consensus must be generated on specific molecules to be assessed, methods and settings in which to assess them, and diagnostic thresholds to use. These elements can be validated in retrospective and prospective multicenter studies including standardized molecular assessment in comparable clinical contexts. The analytic approach utilized in this study represents a potential model for such validation studies utilizing large archives of well-annotated human FFPE samples with long-term follow-up. This will enable wider application and eventual adoption of molecular transplant diagnostics as standard of care.
Supplementary Material
Table S1: Probe sequences for 38 genes analyzed with NanoString® nCounter®.
Table S2: Diagnostic performance of individual antibody-mediated rejection (AMR)-related genes in discriminating AMR from non-AMR in 197 nonhuman primate samples.
Table S3: Diagnostic performance of antibody-mediated rejection (AMR) gene sets in discriminating AMR from non-AMR in 197 nonhuman primate samples.
Figure S1: Temporal patterns of antibody-mediated rejection (AMR) 3-gene set expression in nonhuman primate recipients with at least four sequential samples. Recipients with AMR demonstrate higher gene set expression than those with T-cell mediated rejection (TCMR) only or no evidence of rejection. This generally corresponds with the development of AMR-related immunopathologic features. The ROC curve-derived diagnostic threshold for AMR (−0.092) is indicated by the dashed line. Asterisks indicate euthanasia samples. Histological categories: 'histo+' = pure AMR and mixed rejection; 'histo−' = suspicious for AMR, TCMR, borderline, no rejection, other, normal.
Acknowledgments
This work was supported by funding from Astellas Pharma Canada, Inc., Canadian Foundation for Innovation, and National Institutes of Health (NIH PO1 HL18646).
M.M. received consultancy honoraria from Astellas Pharma Canada, Inc.
ABBREVIATIONS
- AMR
antibody-mediated rejection
- AUC
area under the curve
- BMT
bone marrow transplant
- cg
transplant glomerulopathy
- ci
interstitial fibrosis
- ct
tubular atrophy
- cv
arterial fibrous intimal thickening
- DSA
donor specific antibody
- FFPE
formalin-fixed paraffin-embedded
- g
glomerulitis
- i
interstitial inflammation
- IFTA
interstitial fibrosis and tubular atrophy
- mm
mesangial matrix increase
- NHP
nonhuman primate
- PC
principal component
- PCA
principal component analysis
- ptc
peritubular capillary margination
- ROC
receiver operating characteristic
- t
tubulitis
- TCMR
T-cell mediated rejection
- ti
total interstitial inflammation
- v
intimal arteritis
Footnotes
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. The other authors have no conflicts of interest to disclose.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Adam B, Mengel M. Molecular nephropathology: ready for prime time? Am J Physiol Renal Physiol. 2015;309(3):F185–8. doi: 10.1152/ajprenal.00153.2015. [DOI] [PubMed] [Google Scholar]
- 2.Akalin E, Hendrix RC, Polavarapu RG, Pearson TC, Neylan JF, Larsen CP, et al. Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation. 2001;72(5):948–53. doi: 10.1097/00007890-200109150-00034. [DOI] [PubMed] [Google Scholar]
- 3.Stegall MD, Park WD, Kim D, Kremers W. Gene expression during acute allograft rejection: novel statistical analysis of microarray data. Am J Transplant. 2002;2(10):913–25. doi: 10.1034/j.1600-6143.2002.21007.x. [DOI] [PubMed] [Google Scholar]
- 4.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–38. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 5.Scherer A, Krause A, Walker JR, Korn A, Niese D, Raulf F. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation. 2003;75(8):1323–30. doi: 10.1097/01.TP.0000068481.98801.10. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann SC, Hale DA, Kleiner DE, Mannon RB, Kampen RL, Jacobson LM, et al. Functionally significant renal allograft rejection is defined by transcriptional criteria. Am J Transplant. 2005;5(3):573–81. doi: 10.1111/j.1600-6143.2005.00719.x. [DOI] [PubMed] [Google Scholar]
- 7.Halloran PF, Einecke G. Microarrays and transcriptome analysis in renal transplantation. Nat Clin Pract Nephrol. 2006;2(1):2–3. doi: 10.1038/ncpneph0066. [DOI] [PubMed] [Google Scholar]
- 8.Halloran PF, de Freitas DG, Einecke G, Famulski KS, Hidalgo LG, Mengel M, et al. The molecular phenotype of kidney transplants. Am J Transplant. 2010;10(10):2215–22. doi: 10.1111/j.1600-6143.2010.03267.x. [DOI] [PubMed] [Google Scholar]
- 9.Allanach K, Mengel M, Einecke G, Sis B, Hidalgo LG, Mueller T, et al. Comparing microarray versus RT-PCR assessment of renal allograft biopsies: similar performance despite different dynamic ranges. Am J Transplant. 2008;8(5):1006–15. doi: 10.1111/j.1600-6143.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- 10.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 11.Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adam B, Afzali B, Dominy K, Chapman E, Gill R, Hidalgo L, et al. Multiplexed color-coded probe-based gene expression assessment for clinical molecular diagnostics in formalin-fixed paraffin-embedded human renal allograft tissue. Clin Transplant. 2016;30(3):295–305. doi: 10.1111/ctr.12689. [DOI] [PubMed] [Google Scholar]
- 13.Afzali B, Chapman E, Racapé M, Adam B, Bruneval P, Gil F, et al. Molecular assessment of microcirculation injury in formalin-fixed human cardiac allograft biopsies with antibody-mediated rejection. Am J Transplant. 2017;17(2):496–505. doi: 10.1111/ajt.13956. [DOI] [PubMed] [Google Scholar]
- 14.Cosimi AB, Burton RC, Kung PC, Colvin RB, Goldstein G, Lifter J, et al. Evaluation in primate renal allograft recipients of monoclonal antibody to human T-cell subclasses. Transplant Proc. 1981;13(1 Pt 1):499–503. [PubMed] [Google Scholar]
- 15.Kawai T, Sogawa H, Boskovic S, Abrahamian G, Smith RN, Wee SL, et al. CD154 blockade for induction of mixed chimerism and prolonged renal allograft survival in nonhuman primates. Am J Transplant. 2004;4(9):1391–8. doi: 10.1111/j.1600-6143.2004.00523.x. [DOI] [PubMed] [Google Scholar]
- 16.Smith RN, Kawai T, Boskovic S, Nadazdin O, Sachs DH, Cosimi AB, et al. Chronic antibody mediated rejection of renal allografts: pathological, serological and immunologic features in nonhuman primates. Am J Transplant. 2006;6(8):1790–8. doi: 10.1111/j.1600-6143.2006.01351.x. [DOI] [PubMed] [Google Scholar]
- 17.Smith RN, Kawai T, Boskovic S, Nadazdin O, Sachs DH, Cosimi AB, et al. Four stages and lack of stable accommodation in chronic alloantibody-mediated renal allograft rejection in Cynomolgus monkeys. Am J Transplant. 2008;8(8):1662–7. doi: 10.1111/j.1600-6143.2008.02303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada Y, Boskovic S, Aoyama A, Murakami T, Putheti P, Smith RN, et al. Overcoming memory T-cell responses for induction of delayed tolerance in nonhuman primates. Am J Transplant. 2012;12(2):330–40. doi: 10.1111/j.1600-6143.2011.03795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oura T, Hotta K, Lei J, Markmann J, Rosales I, Dehnadi A, et al. Immunosuppression with CD40 costimulatory blockade plus rapamycin for simultaneous islet-kidney transplantation in nonhuman primates. Am J Transplant. 2017;17(3):646–56. doi: 10.1111/ajt.13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiebe C, Gibson I, Blydt-Hansen T, Karpinski M, Ho J, Storsley L, et al. Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant. 2012;12(5):1157–67. doi: 10.1111/j.1600-6143.2012.04013.x. [DOI] [PubMed] [Google Scholar]
- 21.Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, et al. The Banff 2015 Kidney meeting report: Current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017;17(1):28–41. doi: 10.1111/ajt.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeve J, Sellarés J, Mengel M, Sis B, Skene A, Hidalgo LG, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013;13(3):645–55. doi: 10.1111/ajt.12079. [DOI] [PubMed] [Google Scholar]
- 23.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 24.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–5. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Loupy A, Vernerey D, Tinel C, Aubert O, Duong van Huyen JP, Rabant M, et al. Subclinical rejection phenotypes at 1 year post-transplant and outcome of kidney allografts. J Am Soc Nephrol. 2015;26(7):1721–31. doi: 10.1681/ASN.2014040399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, et al. Banff 2013 meeting report: inclusion of C4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant. 2014;14(2):272–83. doi: 10.1111/ajt.12590. [DOI] [PubMed] [Google Scholar]
- 27.Sis B, Jhangri GS, Bunnag S, Allanach K, Kaplan B, Halloran PF. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9(10):2312–23. doi: 10.1111/j.1600-6143.2009.02761.x. [DOI] [PubMed] [Google Scholar]
- 28.Lefaucheur C, Loupy A, Vernerey D, Duong Van Huyen JP, Suberbielle C, Anglicheau D, et al. Antibody-mediated vascular rejection of kidney allografts: a population-based study. Lancet. 2013;381(9863):313–9. doi: 10.1016/S0140-6736(12)61265-3. [DOI] [PubMed] [Google Scholar]
- 29.Hill GS, Nochy D, Bruneval P, Duong van Huyen JP, Glotz D, Suberbielle C, et al. Donor-specific antibodies accelerate arteriosclerosis after kidney transplantation. J Am Soc Nephrol. 2011;22(5):975–83. doi: 10.1681/ASN.2010070777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Freitas DG, Sellarés J, Mengel M, Chang J, Hidalgo LG, Famulski KS, et al. The nature of biopsies with "borderline rejection" and prospects for eliminating this category. Am J Transplant. 2012;12(1):191–201. doi: 10.1111/j.1600-6143.2011.03784.x. [DOI] [PubMed] [Google Scholar]
- 31.Lagoo AS, Buckley PJ, Burchell LJ, Peters D, Fechner JH, Tsuchida M, et al. Increased glomerular deposits of von Willebrand factor in chronic, but not acute, rejection of primate renal allografts. Transplantation. 2000;70(6):877–86. doi: 10.1097/00007890-200009270-00005. [DOI] [PubMed] [Google Scholar]
- 32.Segerer S, Regele H, Mack M, Kain R, Cartron JP, Colin Y, et al. The Duffy antigen receptor for chemokines is up-regulated during acute renal transplant rejection and crescentic glomerulonephritis. Kidney Int. 2000;58(4):1546–56. doi: 10.1046/j.1523-1755.2000.00316.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto I, Horita S, Takahashi T, Kobayashi A, Toki D, Tanabe K, et al. Caveolin-1 expression is a distinct feature of chronic rejection-induced transplant capillaropathy. Am J Transplant. 2008;8(12):2627–35. doi: 10.1111/j.1600-6143.2008.02421.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Probe sequences for 38 genes analyzed with NanoString® nCounter®.
Table S2: Diagnostic performance of individual antibody-mediated rejection (AMR)-related genes in discriminating AMR from non-AMR in 197 nonhuman primate samples.
Table S3: Diagnostic performance of antibody-mediated rejection (AMR) gene sets in discriminating AMR from non-AMR in 197 nonhuman primate samples.
Figure S1: Temporal patterns of antibody-mediated rejection (AMR) 3-gene set expression in nonhuman primate recipients with at least four sequential samples. Recipients with AMR demonstrate higher gene set expression than those with T-cell mediated rejection (TCMR) only or no evidence of rejection. This generally corresponds with the development of AMR-related immunopathologic features. The ROC curve-derived diagnostic threshold for AMR (−0.092) is indicated by the dashed line. Asterisks indicate euthanasia samples. Histological categories: 'histo+' = pure AMR and mixed rejection; 'histo−' = suspicious for AMR, TCMR, borderline, no rejection, other, normal.