Recent work on Model Informed Drug Discovery and Development (MID3) has noted the need for clarity in model description used in quantitative disciplines such as pharmacology and statistics.1, 2, 3 Currently, models are encoded in a variety of computer languages and are shared through publications that rarely include original code and generally lack reproducibility. The DDMoRe Model Description Language (MDL) has been developed primarily as a language standard to facilitate sharing knowledge and understanding of models.
MOTIVATION
Models are now used not just for data analysis, but for knowledge representation integrating across a wide range of data sources and model types.4 One fundamental problem is a lack of standards in expressing the model across different quantitative disciplines and across this breadth of scope. While the mathematical and statistical aspects of the model may be commonly understood, implementation typically varies across the various software tools employed by different users of a model. Sharing knowledge through sharing computer code becomes difficult because of this, hampering knowledge transfer and impacting reproducibility.
MDL provides the means to describe models in a clear and consistent manner for modelers and those using the models, and, together with the other DDMoRe exchange standards—Pharmacometrics Markup Language (PharmML),5 which defines the XML‐based software interchange standard; probability distribution ontology and knowledge‐base (ProbOnto),6 which provides a consistent basis for definition of probability distributions across MDL and PharmML and how these distributions are encoded in various target software tools; and the Standard Output (SO) definition,7 which defines a consistent XML representation of output from target software tools—provides standards for model definition, software input, output and interoperability, and knowledge management through metadata annotation using suitable ontologies. With a growing number of open‐source tools for pharmacometric analysis and inference, there is also a benefit to providing model exchange standards that are similarly open and extensible.
STRUCTURE AND USE OF MDL
MDL has been designed as a declarative language, based on the model hierarchical structure to aid clarity of model definition and to facilitate reuse of the model definition for a variety of modeling and simulation tasks. Information regarding the model, data, design, parameters, priors, and tasks have been split into independent entities which we call “objects.” MDL Objects are organized in blocks and sub‐blocks of code which group model information and make it easier for the reader to understand what is being defined. Figure 1 illustrates the different MDL Objects and the blocks and sub‐blocks within each.
Figure 1.
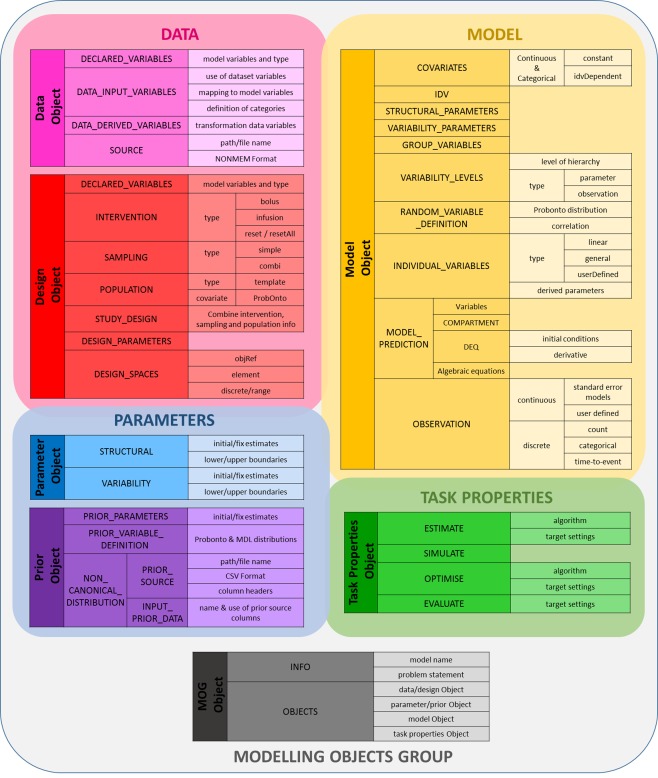
Schematic representation of MDL structure and objects.
The Model Object is the core element of the MDL. It describes the mathematical and statistical properties of the model by defining the structural model prediction, covariate, hierarchical model random variability, and observation components of the model.
The Data Object describes the source of the data and the attributes of each of the data variables. It allows the user to define the content of the different variables and indicate how they are to be used within the model definition. A Design Object can be specified to replace the Data Object when performing simulation or optimal design tasks.
The Parameter Object provides values for both structural and variability parameters defined within the Model Object. The values can be used as initial values with associated constraints for parameter estimation or can be fixed, for example, in simulation and optimal design tasks. Having a separate Parameter Object allows the user to easily change or update values in the model, depending on the task being performed, without having to change the Model Object definition. The Prior Object provides prior distributions of model parameters and replaces the Parameter Object for use in Bayesian tasks.
The Task Properties Object contains settings specific to the task—both general settings and target software specific settings—which will be passed on to the target software tool; e.g., when estimating parameters it will define the estimation algorithm and the associated settings.
The Modeling Object Group (MOG) defines a collection of the MDL objects required for executing a modeling and simulation task. Modularity is a key feature of MDL and this makes it possible to craft reproducible workflows where elements change across tasks, while the core Model Object is retained unchanged. A key attribute of the Model Object is that it should be agnostic to the target software to be used for the modeling and simulation task. Table 1 provides an example of how the MOG may change across a typical pharmacometric modeling and simulation workflow. Task Properties Objects describe relevant settings for the specific target tool to be used in a given task. They can be set up to provide consistent settings for one software tool, for example when estimating parameters across models, or tailored to ensure the reproducibility of results across software for a single model. Currently, conversion tools exist for translating MDL to NONMEM (v. 7.3), Monolix (v. 4.3.2), WinBUGS (v. 1.4), PFIM (v. 4.0), and PopED (v. 0.3).
Table 1.
MDL objects used in the Model Object Group (MOG) definition for various modeling and simulation tasks.
|
Pharmacometric activity |
MDL objects used in the MOG definition | |||||
|---|---|---|---|---|---|---|
| Data object | Design object | Parameter object | Prior object | Model object | Task properties obj.a | |
| Estimation | X | Initial Values | X | MLX task properties | ||
| Bayesian estimation | X | X | X | BUGS task properties | ||
| Visual Predictive Check | X | Estimated Parameters | X | NONMEM task properties | ||
| Prediction/simulation | (X)b | X | Estimated Parameters | X | simulx task properties | |
| Optimal design/evaluation | X | Estimated Parameters | X | PFIM or PopED task properties | ||
The Task Properties Object contains settings for the specific modeling and simulation task relevant to the target software tool for that task.
For prediction or simulation, a Data Object can be used as an alternative to the Design Object.
Example models have been encoded in MDL and are provided with the DDMoRe Interoperability Framework software illustrating how to encode a variety of model features (https://sourceforge.net/p/ddmore/use.cases/ci/master/tree/MDL/Product5.1/). An MDL user guide is also available: https://modeldefinitionlanguage.github.io/MDLUserGuide/.
Supplementary Material for this article shows one of the MDL example models, describing a Poisson count model. This model shows MDL's organization into named blocks of code defining each model component. The MDL shows how the Model Object clearly defines the model hierarchy, relationship of fixed and random effects, and the distributional properties of random effects and outcome. Equivalent models in NMTRAN, MLXTRAN, are provided for comparison. These have been automatically generated from the PharmML obtained from the MDL representation of the model via the DDMoRe interoperability framework. The Data, Parameter, Task Properties, and MOG are not shown for sake of brevity.
MDL AS A COMMUNICATION TOOL
Without clear communication of all aspects of the model, including structural form, model hierarchy, distributional properties of random variables, covariate relationships, mathematical and statistical aspects there is little hope of accurately conveying knowledge imbued within the model.
The Model Object in particular has been designed such that the population, individual, structural, and observation models are easily identified and understood.
For example, as illustrated in the Poisson example in the Supplementary Material:
The distribution of random effects in MDL is explicitly described in the Model Object using ProbOnto definitions rather than being inferred based on the parameter name or its use.
Random variables are generated according to their level in the random effect hierarchy, allowing easy extension beyond the common two‐level hierarchy of parameters and observation.
The linear relationship (after transformation) between fixed and random effects in specifying the individual variables in the model is identified explicitly, rather than inferred based on equation structure.
The distribution of the observed outcome is explicit using the ProbOnto definition and is consistent regardless of whether estimating or simulating the outcome. The user does not need to write likelihood functions for distributions that are described via ProbOnto definitions.
As a whole, the intention of MDL is for anybody reading the model to identify what the model does without having to understand tool‐specific implementation tricks. This is a key feature of knowledge representation—the modeler does not have to know the target software program syntax in order to use it. MDL has been developed taking into account features that provide clarity in model description while retaining the flexibility to describe complex models.
MDL AND ITS IMPLEMENTATION IN THE DDMORE PLATFORM
The DDMoRe Interoperability Framework (https://sourceforge.net/projects/ddmore/files/) has provided a proof of concept implementation and demonstrated the utility of interoperability without manual intervention. An MDL Editor8 is provided to assist the user in writing valid MDL and software tools are provided to convert from MDL to target software and return results from modeling and simulation tasks as R objects using the DDMoRe software exchange standards PharmML and SO. An R package, “ddmore,” distributed with the DDMoRe Interoperability Framework, has been written to allow the user to read, write, and work with the MDL Objects within a pharmacometrics workflow. Using an R script to define all tasks with a given model facilitates an unbroken workflow and dataset ensures reproducibility of modeling steps. DDMoRe has achieved this aim, by demonstrating the following tasks within a single R script: exploration of data using R; estimation of parameters using NONMEM, Monolix, and BUGS; model qualification using PsN and Xpose; simulation of new outcomes using simulx (R package “mlxR”); optimal design using PFIM and PopED. The Supplementary Material shows an R script illustrating this workflow and associated output.
FUTURE PERSPECTIVE OF MDL
The definition and evolution of the Systems Biology Markup Language (SBML) has revolutionized systems biology and quantitative systems pharmacology.10 The DDMoRe exchange standards, including MDL, and DDMoRe model repository have the potential to be similarly transformative for pharmacometrics and MID3.
The current implementation of MDL is only a first step, with the initial scope covering the majority of population pharmacokinetic (PK) pharmacokinetic/pharmacodynamic (PK/PD), and disease progression models. Since MDL, the DDMoRe exchange languages and associated tools are open‐source standards, it is possible and desirable for the modeling and simulation community to suggest enhancements and contribute code for implementation of these enhancements. Source code for MDL, and the user guide, are available via a Github project (https://github.com/ModelDefinitionLanguage/).
MDL ensures accurate knowledge representation and facilitates model sharing across disciplines without the constraints or requirements of knowing software‐specific implementations or tricks. The DDMoRe project has shown with the model exchange standards and the interoperability framework that there should be no major hurdles to technical implementation of the concept of interoperability. With sufficient interest and uptake of MDL by the community, it is hoped that software developers will adopt the model exchange standards to facilitate interoperability across an increasing number of software tools.
There is considerable activation energy required to engage the modeling and simulation community to adopt these standards. However, the use of DDMoRe exchange standards in the model repository (http://repository.ddmore.eu) and the growing number of models published in the repository covering disease progression and therapeutic intervention provides an incentive to use MDL. MDL could also provide a consistent model description standard for many of the open‐source tools for estimation, simulation, and optimal design, leaving these tool developers to concentrate on the implementation of algorithms and functionality of their tools, requiring only conversion from and to the DDMoRe standards to integrate their tool into a pharmacometrics workflow.
CONCLUSION
By creating a clear, modular, flexible, explicit, and unambiguous language for model description, MDL presents a step forward for improved accurate knowledge representation through models. This step represents a paradigm shift in pharmacometrics as a discipline, enabling knowledge‐based decision making. It will also improve productivity of pharmacometricians and other modelers. Together with the other DDMoRe standards, MDL is anticipated to increase quality, efficiency, and cost‐effectiveness of modeling in drug development and therapeutic applications such as those described by MID3.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Acknowledgments
The authors thank the many colleagues across the DDMoRe project who have contributed to developing, refining, implementing, and providing training for MDL. We also thank the constructive comments and contributions of an unknown reviewer whose suggestions have improved the article. This study received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115156, resources of which are composed of financial contributions from the European Union's Seventh Framework Programme (FP7/2007–2013) and EFPIA companies' in kind contribution. The DDMoRe project is also financially supported by contributions from Academic and SME partners.
Conflict of Interest
The authors declared no conflicts of interest.
References
- 1. Marshall, S.F. et al Good practices in model‐informed drug discovery and development: practice, application, and documentation. CPT Pharmacometrics Syst. Pharmacol. 5; 93–122. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Kelly, M. , Anisimov, V. , Campbell, C. & Hamilton, S. Proposed best practice for projects that involve modelling and simulation. Pharm. Stat. 16, 107–113 (2016). [DOI] [PubMed] [Google Scholar]
- 3. Mentré, F. Lewis Sheiner ISoP/UCSF Lecturer Award: From drug use to statistical models and vice versa. CPT Pharmacometrics Syst. Pharmacol. 3, 1–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Milligan, P. et al Model‐based drug development: a rational approach to efficiently accelerate drug development. Clin. Pharmacol. Ther. 93 6, 502–514 (2013). [DOI] [PubMed] [Google Scholar]
- 5. Swat, M.J. et al Pharmacometrics Markup Language (PharmML): opening new perspectives for model exchange in drug development. CPT Pharmacometrics Syst. Pharmacol. 4, 316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swat, M.J. , Grenon, P. & Wimalaratne, S. ProbOnto‐Ontology and knowledge base of probability distributions. Bioinformatics 32, 2719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Terranova, N. et al Standardized output: flexible and tool‐independent storage format of typical M&S results. Abstr 3599. <www.page-meeting.org/?abstract=3599> (2015).
- 8. Kokash, N. , Moodie, S.L. , Smith, M.K. & Holford, N. Implementing a domain‐specific language for model‐based drug development. Proc. Comput. Sci. 63, 308–316 (2015). [Google Scholar]
- 9. Dunlavey, M.R. , Leary, R.H. Rationale for PML Design. ACOP 5. <https://isop.memberclicks.net/assets/Legacy_ACOPs/ACOP5/Poster_Abstracts/w-034.pdf> (2014).
- 10. Hucka, M. et al Evolving a lingua franca and associated software infrastructure for computational systems biology: the Systems Biology Markup Language (SBML) project. Syst. Biol. 1, 41 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


