Abstract
In this study, siderophore production by various bacteria amongst the plant-growth-promoting rhizobacteria was quantified by a rapid and efficient method. In total, 23 siderophore-producing bacterial isolates/strains were taken to estimate their siderophore-producing ability by the standard method (chrome azurol sulphonate assay) as well as 96 well microplate method. Production of siderophore was estimated in percent siderophore unit by both the methods. It was observed that data obtained by both methods correlated positively with each other proving the correctness of microplate method. By the modified microplate method, siderophore production by several bacterial strains can be estimated both qualitatively and quantitatively at one go, saving time, chemicals, making it very less tedious, and also being cheaper in comparison with the method currently in use. The modified microtiter plate method as proposed here makes it far easier to screen the plant-growth-promoting character of plant-associated bacteria.
Keywords: Siderophore, PGPR, CAS assay, Microplate, Plate reader
Introduction
Siderophores are low-molecular weight secondary metabolites with iron-chelating potential. These are compounds with small peptidic molecules having side chains and functional groups which have high-affinity ligand to bind ferric ions and transport them through the cell membrane (Raymond et al. 2015; Niehus et al. 2017). Siderophores are produced by various microorganisms and are classified into four main classes (carboxylate, hydroxamates, catecholates, and mixed type) on the basis of their structural features, functional groups, and types of ligands (Table 1) (Ali and Vidhale 2013; Kumar et al. 2017; Miethke and Marahiel 2007; Aznar et al. 2015). Diverse bacterial and fungal genera ranging from human pathogens to environmental microbes such as plant-growth-promoting rhizobacteria (PGPR) are reported to produce siderophores.
Table 1.
Type of siderophores produced by plant-growth-promoting bacteria.
Modified from Saha et al. (2015)
| S. no. | Siderophore type | Characteristic functional group | Example with microbial source | References |
|---|---|---|---|---|
| 1 | Hydroxamate | Esters or acid chlorides or carboxylic acids | Ferrioxamine B–Pseudomonas fluorescence | (Maurer et al. 1968); Radhakrishnan et al. (2014) |
| 2 | Catecholates | Phenolate or 2,3-dihydroxy benzoate (DHB) binding groups | Enterobactin–Escherichia coli | Dave et al. (2006); Grobelak and Hiller (2017) |
| 3 | Carboxylates | Hydroxyl carboxylate and carboxylates | Rhizobactin–Rhizobium meliloti | Smith and Neilands (1984); Ghavami et al. (2017) |
| 4 | Mixed type | Mixture of above mentioned functional groups | Pyoverdine–Pseudomonas aeruginosa | Leong and Neilands (1982); Behnsen and Raffatellu (2016) |
One of the key mechanisms of PGPR in promoting plant growth involves the production of secondary metabolites such as siderophores (Verma et al. 2011; Ghavami et al. 2017). Although iron is abundantly available in soil, most of it is unavailable to the plant or other organisms, because it forms insoluble complexes. Hence, iron deficiency is a major global issue. Siderophores produced by PGPR help in fulfilment of the iron requirement of plants by causing its solubilisation and chelation from organic or inorganic complexes present in soil (Wandersman and Delepelaire 2004; Arora et al. 2013; Singh et al. 2017). Microbial siderophores strongly chelate iron and enhance iron uptake by forming a ferric–siderophore complex even at very low concentrations (Dimkpa et al. 2009; Fernández-Scavino and Pedraza 2013; Boiteau et al. 2016). Siderophores thus not only help in enhancing plant growth, but also play a very important role in providing iron to other organisms including humans. Siderophores produced by PGPR also help in protection of plant from phytopathogens (Arora 2015; Saha et al. 2016). Phytopathogens are inhibited in rhizosphere by siderophore-producing PGPR because of iron starvation or due to competitive exclusion in iron-deficient conditions (Beneduzi et al. 2012; Parmar and Chakraborty 2016; Dalvi and Rakh 2017). Besides plant growth promotion, siderophores also play an important role in bioremediation of heavy metals from contaminated sites by binding to the toxic metals such as Cr3+, Al3+, Pb2+, Cd2+, Hg2+, etc. (Saha et al. 2015). Siderophore-producing microorganisms can thus be used to detoxify heavy metal contamination by mobilization of insoluble heavy metals (Dimkpa et al. 2008; Rajkumar et al. 2010; Hao et al. 2014; Mishra et al. 2017). Siderophore-producing microbes can thus be used in a variety of ways including bioremediation, sustainable agriculture as biosensors, and even in medicine.
Siderophore production ability of microorganisms is commonly detected by the chrome azurol sulphonate (CAS) assay as given by Schwyn and Neilands (1987). For quantitative estimation of siderophore production, supernatants of microbial cultures are used. Solid CAS agar media are also used for detection of siderophore production qualitatively (Raaska et al. 1993). In CAS assay, competition is for iron uptake between the siderophore and ferric complex of the CAS dye (CAS–iron–detergent complex). Siderophore strongly chelate the iron from iron–dye complex and dye becomes free in the media which causes change in colour from blue to orange (Louden et al. 2011).
Quantity of siderophore produced by microorganisms is measured by spectrophotometric estimation. In this traditional method, CAS reagent is mixed with microbial supernatant and amount of siderophore is estimated by taking optical density of each sample individually. However, this method requires large amount of chemical, time, labour, and space. Keeping this in mind, a modified method of siderophore estimation was developed which is far cheaper, time saving, and less laborious. This method of quantitative estimation of siderophore production was developed principally from the classical method of Schwyn and Neilands (1987) using 96 well microtiter plate and plate reader thus enabling the screening of several PGP strains at a time.
Materials and methods
Bacteria and growth conditions
In the present study, 23 siderophore-producing bacterial strains were taken from Culture Collection of Rhizosphere Microbiology Laboratory, Department of Environmental Microbiology, BBA University, Lucknow (Uttar Pradesh, India). Bacterial strains were grown on Luria–Bertani (LB) agar media (Himedia, Mumbai) at 28 °C for 48 h. All strains were preserved in LB slants at 4 °C and in 25% glycerol stock solution at − 80 °C.
Siderophore estimation assay
Bacterial strains were checked for siderophore-producing ability by universal CAS assay (Schwyn and Neilands 1987). Before starting the experiment, glassware was rinsed with 3 mol/l hydrochloric acid (HCl) to remove iron and subsequently washed in deionized water (Cabaj and Kosakowska 2007). Both qualitative and quantitative methods were used to estimate the siderophore production by bacterial strains. For both the methods, CAS reagent was prepared as per Schwyn and Neilands (1987). Briefly, 121 mg CAS was dissolved in 100 ml distilled water and 20 ml of 1 mM ferric chloride (FeCl3·6H2O) solution prepared in 10 mM HCl. This solution was added to 20 ml hexadecyl trimethyl ammonium bromide (HDTMA) solution under stirring. HDTMA solution was prepared by mixing 729 mg HDTMA in 400 ml distilled water. The CAS-HDTMA solution was sterilized before further use.
Qualitative method
This assay was performed according to modified method given by Hu and Xu (2011). CAS agar plates were prepared by mixing 100 ml CAS reagent in 900 ml sterilized LB agar medium. Four bacterial strains were spot inoculated on each plate. An un-inoculated plate was taken as control. After inoculation, plates were incubated at 28 °C for 5–7 days and observed for the formation of orange zone around the bacterial colonies (Louden et al. 2011).
Quantitative method
Quantitative estimation of siderophore production by bacterial strain was done by (i) traditional method and (ii) modified microplate method.
Traditional method
Quantitative estimation of siderophore was done by taking supernatant of bacterial cultures grown in LB broth medium (Hu and Xu 2011). For this, 1 ml broth was taken in 1.5 ml centrifuge tube (Thomas Scientific, US) (one for each bacterial culture) and after sterilization inoculated with 10 µl of freshly grown bacterial culture (108 colony forming units (cfu) per ml). Four replicates (tubes) were taken for each strain. Apart from this, control tube (un-inoculated broth) was also maintained. After incubation at 28 °C for 48 h, bacterial cultures were centrifuged at 10,000 rpm for 10 min, cell pellets were discarded, and supernatant was used to estimate siderophore. Supernatant (0.5 ml) of each bacterial culture was mixed with 0.5 ml CAS reagent and after 20 min optical density was taken at 630 nm (Spectrophotometer: Thermo Scientific, Evolution 201). Siderophore produced by strains was measured in percent siderophore unit (psu) which was calculated according to the following formula (Payne 1993):
where A r = absorbance of reference (CAS solution and un-inoculated broth), and A s = absorbance of sample (CAS solution and cell-free supernatant of sample).
Modified microplate method
The modified method for estimating siderophore production was carried out using microtiter plate. Supernatant was obtained from 0.5 ml inoculated (5 μl inoculum containing 108 cfu/ml) broth in microcentrifuge tube (Thomas Scientific, US). Supernatant (100 µl) of each bacterial culture was added in separate wells of microplate (CLS3474 Sigma) followed by the addition of 100 µl CAS reagent. After incubation, optical density of each sample (placed in wells of microplate) was recorded at 630 nm using microplate reader (Spectra Max M5e). Four replicates were taken for each strain in 96 well plate and siderophore estimated by the same formula as mentioned above.
Correlation analysis between traditional method and modified microplate method
Correlation between the data obtained from both the methods (traditional method and microplate method) was calculated to observe the similarity. Correlation coefficient was checked by software statistical package for the social science (SPSS) (2016) for windows.
Results and discussion
Bacterial strains
The 23 siderophore-producing bacterial strains taken in the study belong to species amongst diverse genera including Pseudomonas, Rhizobium, Enterobacter, Chronobacter, Kosakonia, Beijerinckia, and Pantoea. Details of the strains taken in the study with accession number and reference are mentioned in Table 2. All of these bacterial genera and species are well-known PGPR (Farina et al. 2012; Ahemad and Kibret 2014; Majeed et al. 2015; Naqqash et al. 2016) and common inhabitants of rhizosphere. The study also included endophytic strains from family Enterobacteriaceae, namely, E. cloacae, P. agglomerans, and C. sakazakii which are earlier reported to be siderophore producers (Mokracka et al. 2004; Grim et al. 2012; Walpola and Yoon 2013; Pandey et al. 2016). A novel PGPR strain of Kosakonia pseudosacchari, which has not been reported to produce siderophore earlier, was also included in this study. The study thus included very diverse PGPR from different locations of plants including rhizosphere, root nodules, and plant tissues (endophyte) (Table 2).
Table 2.
Detail of bacterial strains used for siderophore production assay
| S. no. | Bacterial strains | Host plant | Collection site | Accession number (Genbank/ MTCC/MCC) |
References |
|---|---|---|---|---|---|
| 1 | P. aeruginosa (KA19) | Brassica campestris | Rhizospheric soil | – | Mishra and Arora (2012a) |
| 2 | P. aeruginosa (TO3) | – | Rhizospheric soil | FJ685995 | Khare and Arora (2010) |
| 3 | Kosakonia pseudosacchari (LN) | Leucaena leucocephala | Root nodule | KY392997 | – |
| 4 | P. fluorescence (JM1) | – | Rhizospheric soil | KT734728 | – |
| 5 | Enterobacter cloacae (CV5) | Crotalaria juncea | Root nodule | MF416432 | – |
| 6 | Pseudomonas sp. (NDN1) | Lycopersicum esclantum | Rhizospheric region | – | Arora et al. (2008) |
| 7 | P. aeruginosa (RB1) | Withania somnifera | Plant tissue Endophyte | KT761191 | – |
| 8 | Kosakonia sp. (ClU1) | Clitoria ternatea | Root nodule | KY392994 | – |
| 9 | E. cloacae (ClU2) | C. ternatea | Root nodule | KY178303 | – |
| 10 | R. meliloti (RMP3) | Mucuna pruriens | Root nodule | – | Arora et al. (2001) |
| 11 | R. meliloti (RMP5) | M. pruriens | Root nodule | – | Arora et al. (2001) |
| 12 | Rhizobium sp. (RASH6) | Leguminous plant | Root nodule | – | Singh et al. (2014) |
| 13 | P. fluorescence (TO7) | Brassica sp. | Rhizospheric soil | HQ457044 | Mishra and Arora (2012b) |
| 14 | Pantoea agglomerans (CV2) | Crotalaria juncea | Root nodule | KY178304 | – |
| 15 | Rhizobium pusense (LM) | L. leucocephala | Root nodule | KY392995 | – |
| 16 | R. pusense (AB3) | Abrus precatorius | Root nodule |
KY392993 MCC 3409 |
– |
| 17 | P. tropicalis (EKi) | L. esclantum | Rhizospheric soil |
FJ816019 MTCC 9737 |
Khare et al. (2011) |
| 18 | Cronobacter sakazakii (CGJ) | C. juncea | Root nodule | MF416433 | – |
| 19 | P. aeruginosa (PF07) | Helianthus annus | Rhizospheric | – | Tewari and Arora (2014a) |
| 20 | P. aeruginosa (PF23) | – | Rhizospheric soil | KF598858 | Tewari and Arora (2014b) |
| 21 | Beijerinckia fluminensis (AB1) | Abrus precatorius | Root nodule | MF400858 | – |
| 22 | Rhizobium radiobacter (LB2) | L. leucocephala | Root nodule | KY392996 | – |
| 23 | P. fluorescence (PF17) | H. annus | Rhizospheric soil | KU201600 | Tewari and Arora (2016) |
Siderophore estimation
Formation of orange-coloured zone around the bacterial colonies was observed which indicated siderophore production by bacterial strains. It was observed that all the bacterial strains taken in the study were positive for siderophore production. KA19 (P. aeruginosa) showed maximum siderophore production ability on CAS agar (Table 3). The production of siderophore was roughly estimated on the basis of size of halo formation on CAS agar. CAS agar method can only give rough idea and is not a perfect method for quantification of siderophore production. Hence, quantitative estimation of siderophore is done using liquid culture media and CAS reagent.
Table 3.
Results of siderophore production from bacterial strains and their estimation by qualitative analysis and quantitative analysis (traditional method and modified microplate method)
| Bacterial strains | Qualitative analysis | Quantitative analysis | % increase in absorbance by microplate method | |
|---|---|---|---|---|
| Traditional method (psu) | Microplate method (psu) | |||
| Control | 1.12 ± 0.01 | 1.12 ± 0.01 | 0.00 | |
| KA19 | +++ | 69.16 ± 0.71 | 69.81 ± 0.16 | 0.93 |
| TO3 | ++ | 41.45 ± 0.44 | 43.26 ± 0.06 | 4.18 |
| LN | ++ | 40.44 ± 0.59 | 41.44 ± 0.09 | 2.41 |
| JM1 | + | 24.99 ± 0.60 | 25.50 ± 0.14 | 2.00 |
| CV5 | + | 35.51 ± 0.53 | 35.77 ± 0.04 | 0.72 |
| NDN1 | + | 36.64 ± 0.73 | 37.35 ± 0.12 | 1.90 |
| RB1 | ++ | 44.43 ± 0.33 | 44.44 ± 0.16 | 0.02 |
| ClU1 | ++ | 30.39 ± 0.18 | 30.64 ± 0.10 | 0.81 |
| ClU2 | + | 21.46 ± 0.52 | 21.90 ± 0.15 | 2.00 |
| RMP3 | + | 24.12 ± 0.62 | 24.49 ± 0.07 | 1.51 |
| RMP5 | ++ | 41.12 ± 0.42 | 41.35 ± 0.05 | 0.55 |
| RASH6 | + | 14.13 ± 0.24 | 14.67 ± 0.04 | 3.68 |
| TO7 | + | 22.58 ± 0.60 | 23.50 ± 0.15 | 3.91 |
| CV2 | + | 27.82 ± 0.52 | 27.90 ± 0.14 | 0.06 |
| LM | + | 12.63 ± 0.15 | 13.01 ± 0.01 | 2.92 |
| AB3 | + | 33.34 ± 0.03 | 33.61 ± 0.03 | 0.80 |
| EKi | ++ | 47.19 ± 0.72 | 47.43 ± 0.18 | 0.50 |
| CGJ | + | 32.55 ± 0.47 | 33.06 ± 0.15 | 1.54 |
| PF07 | + | 27.62 ± 0.37 | 27.71 ± 0.07 | 0.32 |
| PF23 | ++ | 45.99 ± 0.59 | 46.33 ± 0.09 | 0.73 |
| AB1 | + | 07.97 ± 0.58 | 08.33 ± 0.08 | 4.32 |
| LB2 | ++ | 45.12 ± 0.05 | 45.64 ± 0.04 | 1.13 |
| PF17 | ++ | 48.42 ± 0.26 | 49.54 ± 0.13 | 2.26 |
Data are represented by the mean of four replicates ± standard deviation, (+++), high production; (++), medium production; (+), low production
In the traditional method, after growth, cell-free supernatant (0.5 ml) is taken for spectrophotometric estimation in cuvette. However, in the proposed method, supernatant (only 100 μl) was poured in wells of microtiter plate. While 0.5 ml CAS reagent was used per tube in the traditional method, only 100 μl was employed in case of microplate method. Thus, there was drastic reduction in the amount of reagents and broth being used. In fact, there is 80% reduction in the requirement for CAS reagent and 50% decrease in the amount of broth used. Apart from this, the proposed method required far less time. As per our calculation in terms of total time required to quantify the siderophore produced (after incubation and centrifugation to get cell-free supernatant), there was 91.7% reduction. Hence, the proposed method is not only more economical, but is also time saving (Table 4). By the 96 well microplate method, siderophore quantification can be done for several strains at one go (Fig. 1). Although other workers have also reported that microplate method is time saving and efficient (Lapinski et al. 1978; Frac et al. 2016), but this is the first report demonstrating the efficiency of 96 well microplate method for siderophore estimation in terms of time, money and being even more efficient than the traditional spectrophotometric method.
Table 4.
Comparative analysis between traditional and microplate methods of siderophore estimation
| Comparative analysis | Methods | |
|---|---|---|
| Traditional method | Microplate method | |
| Labour | Requires high labour input | Requires less labour input |
| Media | 96 ml | 48 ml |
| Reagent (for 23 strains plus control in quadruplicates) |
48 ml | 9.6 ml |
| Accuracy | Less accurate because several samples are handled individually which may cause high handling error | More accurate because several samples (96) can be handled collectively in only one plate which reduces the handling error |
| Time | 180 min (3 h) | 15 min |
Fig. 1.
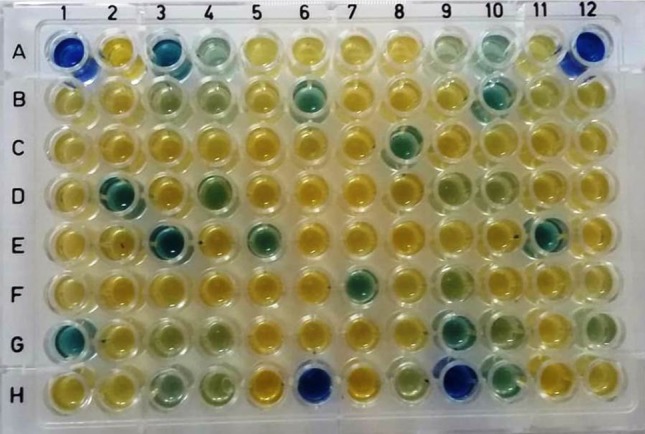
Estimation of siderophore production by microplate method. Diverse bacterial strains producing different amounts of siderophore
Amount of siderophore produced by all the 23 strains was checked and compared for both the traditional and proposed methods so as to determine the efficiency. The absorbance of reference (A r) or control (un-inoculated broth and CAS reagent) was significantly similar both in case of traditional and modified microplate method. Concentration of siderophore produced by bacterial strains varied from 7.97 ± 0.58 to 69.16 ± 0.71 psu when measured by the traditional method, while when quantified through the proposed microplate method, it was from 8.33 ± 0.08 to 69.81 ± 0.16 psu (Table 3). Quantitatively also P. aeruginosa (KA19) produced maximum amount of siderophore and the readings were significantly similar whether taken by spectrophotometer (traditional method) or by the microplate reader (proposed method). In fact, for all the 23 strains, results were significantly similar whether checked spectrophotometrically or by microplate reader (Table 3). This proves the similarity of both the methods.
Results by both the methods indicated that different strains showed variable siderophore-producing abilities. The present study also proved that fluorescent pseudomonads were the most proficient siderophore producers in comparison with other strains. Many researchers have reported fluorescent pseudomonads to be prolific producers of siderophores (Pandey et al. 2005; Subramanian and Satyan 2014; Pattan et al. 2017; Kotasthane et al. 2017). In fact, P. aeruginosa are amongst the most efficient producers of siderophores reported from rhizosphere or other habitats as shown by this work also (de Villegas et al. 2002; Unni et al. 2014; Sasirekha and Srividya 2016). Strains of P. aeruginosa (including TO3, RB1, and PF23) were found to be most efficient siderophore producers in comparison with others. Members of the family Rhizobiaceae (except R. radiobacter LB2 and R. meliloti RMP5) were not very efficient producers of siderophore, and in general, also researchers have not found rhizobia to be prolific producers of siderophores (Joseph et al. 2007; de Souza et al. 2015). Very few studies report that rhizobia are good producers of siderophores (Berraho et al. 1997; Arora et al. 2001; Duhan 2013; Wdowiak-Wróbel et al. 2017). Endophytic strain K. pseudosacchari (LN) is being reported for the first time as an efficient siderophore producer. Although production of siderophore is a common phenomenon among PGPR present in rhizosphere (Tewari and Arora 2013; de Souza et al. 2015), recent researches have also shown their production by endophytes residing in the plant tissues and role in plant growth promotion (Zhao et al. 2015; Santoyo et al. 2016; Perez-Rosales et al. 2017).
Correlation analysis between traditional and modified microplate method
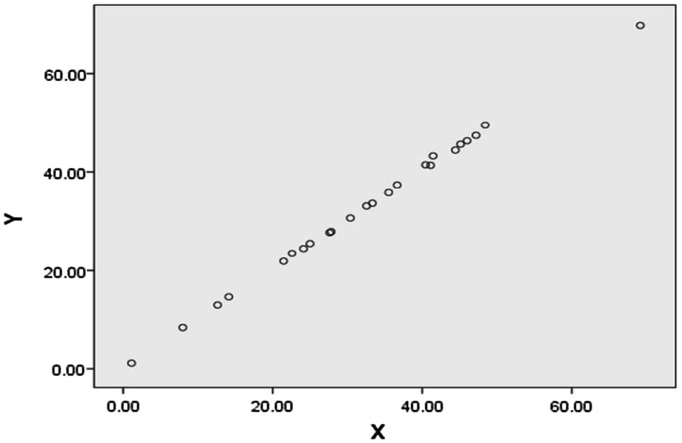
Correlation between both the methods was checked so as to measure the similarity between them. It was found that data from both the methods were highly correlated with each other with R value of 0.999 (Fig. 2). This is a very strong positive correlation. The value of coefficient of determination (R 2) was 0.999 which indicates that both methods were almost similar in efficiency; however, microplate method is far more rapid and economical. In addition, if the results of quantification by both the methods are observed, it can be seen that microplate method shows slightly higher readings (ranging from 0.02 to 4.32%) in case of all the strains. This proves that microplate method is more accurate in comparison with traditional spectrophotometric method. This may be because absorbance of all the samples was taken at one go using 96 well microplate which reduced the handling error when compared to the traditional method, where absorbance of all the samples is taken individually. The accuracy factor was further confirmed by small value of standard deviation (SD) in case of microplate method (SD was within 0.18) compared to the traditional method (SD up to 0.73).
Fig. 2.
Correlation between results of siderophore production by two methods: (i) Traditional method = X value and (ii) Microplate method = Y value
Since the classical assay given by Schwyn and Neilands (1987) to check the siderophore activity by bacteria and fungi, some modifications have appeared from time to time (Ames-Gottfred et al. 1989; Milagres et al. 1999; Machuca and Milagres 2003; Pérez-Miranda et al. 2007; Hu and Xu 2011). However, all these modifications were mainly for qualitative analysis only and were not for quantitative estimation. However, here we report a far more economical, time saving, and accurate method for quantitative estimation of siderophore by microbes.
Conclusion
Siderophore production is considered a very important trait of PGPR involved both in growth promotion and in biocontrol of phytopathogens. Siderophore production is also known by other groups of microbes including other soil bacteria and human pathogens. Traditionally, siderophore production and quantification is done by colorimetric/spectrophotometric method. However, in the present study, 96 well microplate method using microplate reader is proposed for estimation of siderophore production by bacteria. The proposed method is far cheaper, consumes less time, and is even more accurate. The suggested method can be used for quantification of siderophore by any bacteria as a better alternative of the routine colorimetric method. Saving chemicals (particularly CAS dye), the proposed method will also be far less harmful to the environment.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
References
- Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci. 2014;26:1–20. doi: 10.1016/j.jksus.2013.05.001. [DOI] [Google Scholar]
- Ali SS, Vidhale NN. Bacterial siderophore and their application: a review. Int J Curr Microbiol Appl Sci. 2013;2:303–312. [Google Scholar]
- Ames-Gottfred NP, Christie BR, Jordan DC. Use of the chrome azurol S agar plate technique to differentiate strains and field isolates of Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol. 1989;55:707–710. doi: 10.1128/aem.55.3.707-710.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora NK, editor. Plant Microbes Symbiosis: applied facets. India: Springer; 2015. p. 381. [Google Scholar]
- Arora NK, Kang SC, Maheshwari DK. Isolation of siderophore inducing strains of Rhizobium meliloti and their biocontrol potential against Macrophomina phaseolina that causes charcoal rot of ground nut. Curr Sci. 2001;81:673–677. [Google Scholar]
- Arora NK, Khare E, Verma A, et al. In vivo control of Macrophomina phaseolina by a chitinase and β-1,3-glucanase-producing pseudomonad NDN1. Symbiosis. 2008;46:129–135. [Google Scholar]
- Arora NK, Tewari S, Singh R. Multifaceted plant-associated microbes and their mechanisms diminish the concept of direct and indirect PGPRs. In: Arora NK, editor. Plant microbe symbiosis—fundamentals and advances. India: Springer; 2013. pp. 411–449. [Google Scholar]
- Aznar A, Chen NW, Thomine S, Dellagi A. Immunity to plant pathogens and iron homeostasis. Plant Sci. 2015;240:90–97. doi: 10.1016/j.plantsci.2015.08.022. [DOI] [PubMed] [Google Scholar]
- Behnsen J, Raffatellu M. Siderophores: more than stealing iron. M Bio. 2016;7(6):e01906–e01916. doi: 10.1128/mBio.01906-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneduzi A, Ambrosini A, Passaglia LM. Plant growth promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet Mol Biol. 2012;35:1044–1051. doi: 10.1590/S1415-47572012000600020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berraho EL, Lesueur D, Diem HG, et al. Iron requirement and siderophore production in Rhizobium ciceri during growth on an iron-deficient medium. World J Microbiol Biotechnol. 1997;13(5):501–510. doi: 10.1023/A:1018553022960. [DOI] [Google Scholar]
- Boiteau RM, Mende DR, Hawco NJ, et al. Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proc Natl Acad Sci. 2016;113(50):14237–14242. doi: 10.1073/pnas.1608594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabaj A, Kosakowska A. Iron-dependent growth of and siderophore production by two heterotrophic bacteria isolated from brackish water of the southern Baltic Sea. Microbiol Res. 2007;164(5):570–577. doi: 10.1016/j.micres.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Dalvi SM, Rakh RR. Siderophore producing Pseudomonas cf. monteilii 9 for assured biological control of Sclerotium rolfsii causing stem rot of groundnut. Biosci Discov. 2017;8(3):546–555. [Google Scholar]
- Dave BP, Anshuman K, Hajela P. Siderophores of halophilic archaea and their chemical characterization. Ind J Exp Biol. 2006;44:340–344. [PubMed] [Google Scholar]
- de Souza R, Ambrosini A, Passaglia LM. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Mol Biol. 2015;38(4):401–419. doi: 10.1590/S1415-475738420150053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villegas MD, Villa P, Frias A. Evaluation of the siderophores production by Pseudomonas aeruginosa PSS. Rev Latinoam Microbiol. 2002;44(3/4):112–117. [PubMed] [Google Scholar]
- Dimkpa CO, Svatoš A, Dabrowska P, et al. Involvement of siderophores in the reduction of metal-induced inhibition of auxin synthesis in Streptomyces spp. Chemosphere. 2008;74:19–25. doi: 10.1016/j.chemosphere.2008.09.079. [DOI] [PubMed] [Google Scholar]
- Dimkpa CO, Merten D, Svatoš A, et al. Siderophores mediate reduced and increased uptake of cadmium by Streptomyces tendae F4 and sunflower (Helianthus annuus), respectively. J Appl Microbiol. 2009;107:1687–1696. doi: 10.1111/j.1365-2672.2009.04355.x. [DOI] [PubMed] [Google Scholar]
- Duhan JS. Tn5 siderophore producing mutants of Rhizobium and its role in nitrogen fixation and iron uptake in pigeonpea. Afric J Microbiol Res. 2013;7(16):1459–1464. doi: 10.5897/AJMR12.223. [DOI] [Google Scholar]
- Farina R, Beneduzi A, Ambrosini A, et al. Diversity of plant growth-promoting rhizobacteria communities associated with the stages of canola growth. Appl Soil Ecol. 2012;55:44–52. doi: 10.1016/j.apsoil.2011.12.011. [DOI] [Google Scholar]
- Fernández-Scavino A, Pedraza RO. The role of siderophores in plant growth-promoting bacteria. In: Maheshwari DK, Saraf M, Aeron A, editors. Bacteria in agrobiology: crop productivity. Heidelberg: Springer; 2013. pp. 265–285. [Google Scholar]
- Frąc M, Gryta A, Oszust K, et al. Fast and accurate microplate method (Biolog MT2) for detection of Fusarium fungicides resistance/sensitivity. Front Microbiol. 2016;7:489. doi: 10.3389/fmicb.2016.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavami N, Alikhani HA, Pourbabaei AA, et al. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J Plant Nut. 2017;40:736–746. doi: 10.1080/01904167.2016.1262409. [DOI] [Google Scholar]
- Grim CJ, Kothary MH, Gopinath G, et al. Identification and characterization of Cronobacter iron acquisition systems. Appl Environ Microbiol. 2012;78(17):6035–6050. doi: 10.1128/AEM.01457-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobelak A, Hiller J. Bacterial siderophores promote plant growth: screening of catechol and hydroxamate siderophores. Int J Phytoremediation. 2017;19(9):825–833. doi: 10.1080/15226514.2017.1290581. [DOI] [PubMed] [Google Scholar]
- Hao X, Taghavi S, Xie P, et al. Phytoremediation of heavy and transition metals aided by legume-rhizobia symbiosis. Int J Phytoremediation. 2014;16:179–202. doi: 10.1080/15226514.2013.773273. [DOI] [PubMed] [Google Scholar]
- Hu QP, Xu JG. A simple double-layered chrome azurol S agar (SDCASA) plate assay to optimize the production of siderophores by a potential biocontrol agent Bacillus. Afr J Microbiol Res. 2011;5:4321–4327. [Google Scholar]
- Joseph B, Ranjan PR, Lawrence R. Characterization of plant growth promoting rhizobacteria associated with chickpea (Cicer arietinum L.) Int J Plant Prod. 2007;2:141–152. [Google Scholar]
- Khare E, Arora NK. Effect of indole-3-acetic acid (IAA) produced by Pseudomonas aeruginosa in suppression of charcoal rot disease of chickpea. Curr Microbiol. 2010;61(1):64–68. doi: 10.1007/s00284-009-9577-6. [DOI] [PubMed] [Google Scholar]
- Khare E, Singh S, Maheshwari DK, et al. Suppression of charcoal rot of chickpea by fluorescent pseudomonas under saline stress condition. Curr Microbiol. 2011;62:1548–1553. doi: 10.1007/s00284-011-9895-3. [DOI] [PubMed] [Google Scholar]
- Kotasthane AS, Agrawal T, Zaidi NW, et al. Identification of siderophore producing and cynogenic fluorescent Pseudomonas and a simple confrontation assay to identify potential bio-control agent for collar rot of chickpea. 3 Biotech. 2017;7:137. doi: 10.1007/s13205-017-0761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Menon S, Agarwal H, et al. Characterization and optimization of bacterium isolated from soil samples for the production of siderophores. Resour Eff Technol. 2017 [Google Scholar]
- Lapinski FJ, Crowley KM, Merrit CA, et al. Use of microplate methods in paternity testing. Am J Clin Pathol. 1978;70(5):766–769. doi: 10.1093/ajcp/70.5.766. [DOI] [PubMed] [Google Scholar]
- Leong SA, Neilands JB. Siderophore production by phytopathogenic microbial species. Arch Biochem Biophys. 1982;281:351–359. doi: 10.1016/0003-9861(82)90356-3. [DOI] [PubMed] [Google Scholar]
- Louden BC, Haarmann D, Lynne AM. Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ. 2011;12(1):51–53. doi: 10.1128/jmbe.v12i1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca A, Milagres AM. Use of CAS-agar plate modified to study the effect of different variables on the siderophore production by Aspergillus. Lett Appl Microbiol. 2003;36:177–181. doi: 10.1046/j.1472-765X.2003.01290.x. [DOI] [PubMed] [Google Scholar]
- Majeed A, Abbasi MK, Hameed S et al (2015) Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front Microbiol 6:198. doi:10.3389/fmicb.2015.00198 [DOI] [PMC free article] [PubMed]
- Maurer B, Muller A, Keller-Schierlein W, et al. Ferribactin, ein siderochrom aus Pseudomonas fluorescens Migula. Arch Microbiol. 1968;60:326–339. [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milagres AM, Machuca A, Napoleao D. Detection of siderophores production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J Microbiol Methods. 1999;37:1–6. doi: 10.1016/S0167-7012(99)00028-7. [DOI] [PubMed] [Google Scholar]
- Mishra S, Arora NK. Evaluation of rhizospheric Pseudomonas and Bacillus as biocontrol tool for Xanthomonas campestris pv campestris. World J Microbiol Biotechnol. 2012;28:693–702. doi: 10.1007/s11274-011-0865-5. [DOI] [PubMed] [Google Scholar]
- Mishra S, Arora NK. Management of black rot in cabbage by rhizospheric Pseudomonas species and analysis of 2,4-diacetylphloroglucinol by qRT-PCR. Biol Control. 2012;61:32–39. doi: 10.1016/j.biocontrol.2011.12.011. [DOI] [Google Scholar]
- Mishra J, Singh R, Arora NK. Alleviation of heavy metal stress in plants and remediation of soil by rhizosphere microorganisms. Front Microbiol. 2017;8:1706. doi: 10.3389/fmicb.2017.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokracka J, Koczura R, Kaznowski A. Yersiniabactin and other siderophores produced by clinical isolates of Enterobacter spp. and Citrobacter spp. FEMS Immunol Med Microbiol. 2004;40(1):51–55. doi: 10.1016/S0928-8244(03)00276-1. [DOI] [PubMed] [Google Scholar]
- Naqqash T, Hameed S, Imran A et al (2016) Differential response of potato toward inoculation with taxonomically diverse plant growth promoting rhizobacteria. Front Plant Sci 7:144. doi:10.3389/fpls.2016.00144 [DOI] [PMC free article] [PubMed]
- Niehus R, Picot A, Oliveira NM, et al. The evolution of siderophore production as a competitive trait. Evolution. 2017;71(6):1443–1455. doi: 10.1111/evo.13230. [DOI] [PubMed] [Google Scholar]
- Pandey P, Kang SC, Gupta CP, et al. Rhizosphere competent Pseudomonas aeruginosa GRC1 produces characteristic siderophore and enhances growth of Indian mustard (Brassica campestris) Curr Microbiol. 2005;51(5):303–309. doi: 10.1007/s00284-005-0014-1. [DOI] [PubMed] [Google Scholar]
- Pandey PK, Singh MC, Singh S, et al. Inside the plants: endophytic bacteria and their functional attributes for plant growth promotion. Int J Curr Microbiol Appl Sci. 2016;6(2):11–21. doi: 10.20546/ijcmas.2017.602.002. [DOI] [Google Scholar]
- Parmar HY, Chakraborty H. Effect of siderophore on plant growth promotion. Int J Appl Pure Sci Agric. 2016;2(3):60–68. [Google Scholar]
- Pattan J, Kajale S, Pattan S. Isolation, production and optimization of siderophores (iron chilators) from Pseudomonas fluorescence NCIM 5096 and Pseudomonas from soil rhizosphere and marine water. Int J Curr Microbiol App Sci. 2017;6(3):919–928. doi: 10.20546/ijcmas.2017.603.109. [DOI] [Google Scholar]
- Payne SM. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993;1:66–69. doi: 10.1016/0966-842X(93)90036-Q. [DOI] [PubMed] [Google Scholar]
- Pérez-Miranda S, Cabirol N, George-Téllez R, et al. O-CAS, a fast and universal method for siderophore detection. J Microbiol Methods. 2007;70(1):127–131. doi: 10.1016/j.mimet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Perez-Rosales E, Alcaraz-Meléndez L, Puente ME, et al. Isolation and characterization of endophytic bacteria associated with roots of jojoba (Simmondsia chinensis (Link) Schneid) Curr Sci. 2017;112(2):396–401. doi: 10.18520/cs/v112/i02/396-401. [DOI] [Google Scholar]
- Raaska L, Viikari L, Mattila-Sandholm T. Detection of siderophores in growing cultures of Pseudomonas spp. J Ind Microbiol. 1993;11:181–186. doi: 10.1007/BF01583720. [DOI] [Google Scholar]
- Radhakrishnan M, Samshath KJ, Balagurunathan R. Hydroxamate siderophore from Bacillus sp SD12 isolated from iron factory soil. Curr World Environ. 2014;9(3):990. doi: 10.12944/CWE.9.3.53. [DOI] [Google Scholar]
- Rajkumar M, Ae N, Prasad MNV, Freitas H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010;28:142–149. doi: 10.1016/j.tibtech.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Raymond KN, Allred BE, Sia AK. Coordination chemistry of microbial iron transport. Acc Chem Res. 2015;48:2496–2505. doi: 10.1021/acs.accounts.5b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res. 2015;23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- Saha M, Sarkar S, Sarkar B, et al. Microbial siderophores and their potential applications: a review. Environ Sci Poll Res. 2016;23(5):3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- Santoyo G, Moreno-Hagelsieb G, del Carmen OMM, et al. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Sasirekha B, Srividya S. Siderophore production by Pseudomonas aeruginosa FP6, a biocontrol strain for Rhizoctonia solani and Colletotrichum gloeosporioides causing diseases in chilli. Agric Nat Resour. 2016;50:250–256. [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Singh S, Gupta G, Khare E, et al. Phosphate solubilizing rhizobia promote the growth of chickpea under buffering conditions. Int J Pure App Biosci. 2014;2(5):97–106. [Google Scholar]
- Singh R, Pandey DK, Kumar A, et al. PGPR isolates from the rhizosphere of vegetable crop Momordica charantia: characterization and application as biofertilizer. Int J Curr Microbiol App Sci. 2017;6(3):1789–1802. doi: 10.20546/ijcmas.2017.603.205. [DOI] [Google Scholar]
- Smith MJ, Neilands JB. Rhizobactin, a siderophore from Rhizobium meliloti. J Plant Nutr. 1984;7:449–458. doi: 10.1080/01904168409363211. [DOI] [Google Scholar]
- Subramanian J, Satyan K. Isolation and selection of fluorescent pseudomonads based on multiple plant growth promotion traits and siderotyping. Chil J Agric Res. 2014;74(3):319–325. doi: 10.4067/S0718-58392014000300010. [DOI] [Google Scholar]
- Tewari S, Arora NK. Transactions amongst microorganisms and plant in the composite rhizosphere habitat. In: Arora NK, editor. Plant microbe symbiosis—fundamentals and advances. India: Springer; 2013. pp. 143–149. [Google Scholar]
- Tewari S, Arora NK. Talc based exopolysaccharides formulation enhancing growth and production of Hellianthus annuus under saline conditions. Cell Mol Biol. 2014;60(5):73–81. [PubMed] [Google Scholar]
- Tewari S, Arora NK. Multifunctional exopolysaccharides from Pseudomonas aeruginosa PF23 involved in plant growth stimulation, biocontrol and stress amelioration in sunflower under saline conditions. Curr Microbiol. 2014;69:484–494. doi: 10.1007/s00284-014-0612-x. [DOI] [PubMed] [Google Scholar]
- Tewari S, Arora NK. Fluorescent Pseudomonas sp. PF17 as an efficient plant growth regulator and biocontrol agent for sunflower crop under saline conditions. Symbiosis. 2016;68(1–3):99–108. doi: 10.1007/s13199-016-0389-8. [DOI] [Google Scholar]
- Unni KN, Priji P, Sajith S, et al. Pseudomonas aeruginosa BUP2—a novel strain isolated from Malabari goat produces type 2 pyoverdine. Adv Biosci Biotechnol. 2014;5:874–885. doi: 10.4236/abb.2014.511102. [DOI] [Google Scholar]
- Verma VC, Singh SK, Prakash S. Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica. A Juss J Basic Microbiol. 2011;51:550–556. doi: 10.1002/jobm.201000155. [DOI] [PubMed] [Google Scholar]
- Walpola BC, Yoon MH. Isolation and characterization of phosphate solubilizing bacteria and their co-inoculation efficiency on tomato plant growth and phosphorous uptake. Afric J Microbiol Res. 2013;7(3):266–275. [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Ann Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Wdowiak-Wróbel S, Marek-Kozaczuk M, Kalita M, et al. Diversity and plant growth promoting properties of rhizobia isolated from root nodules of Ononis arvensis. Antonie Van Leeuwenhoek. 2017;110(8):1087–1103. doi: 10.1007/s10482-017-0883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xu Y, Lai XH, et al. Screening and characterization of endophytic Bacillus and Paenibacillus strains from medicinal plant Lonicera japonica for use as potential plant growth promoters. Braz J Microbiol. 2015;46(4):977–989. doi: 10.1590/S1517-838246420140024. [DOI] [PMC free article] [PubMed] [Google Scholar]