Abstract
It is recently appreciated that many bacterial chemoreceptors have ligand-binding domains (LBD) of the dCACHE family, a structure with two PAS-like subdomains, one membrane-proximal and the other membrane-distal. Previous studies had implicated only the membrane-distal subdomain in ligand recognition. Here, we report the 2.2 Å resolution crystal structure of dCACHE LBD of the Helicobacter pylori chemoreceptor TlpC. H. pylori tlpC mutants are outcompeted by wild type during stomach colonisation, but no ligands had been mapped to this receptor. The TlpC dCACHE LBD has two PAS-like subdomains, as predicted. The membrane-distal one possesses a long groove instead of a small, well-defined pocket. The membrane-proximal subdomain, in contrast, had a well-delineated pocket with a small molecule that we identified as lactate. We confirmed that amino acid residues making contact with the ligand in the crystal structure—N213, I218 and Y285 and Y249—were required for lactate binding. We determined that lactate is an H. pylori chemoattractant that is sensed via TlpC with a K D = 155 µM. Lactate is utilised by H. pylori, and our work suggests that this pathogen seeks out lactate using chemotaxis. Furthermore, our work suggests that dCACHE domain proteins can utilise both subdomains for ligand recognition.
Introduction
Helicobacter pylori is a motile, gram-negative bacterium that infects over 50% of the world’s population1. H. pylori selectively colonises the gastric epithelium and is able to survive in the host stomach for years. Although the majority of the infected people remain asymptomatic, H. pylori infection can be associated with a range of gastroduodenal diseases, including gastritis, gastric and duodenal ulcers, and different types of cancer including mucosa-associated lymphoid tissue (MALT) lymphoma and gastric adenocarcinoma2–4. Directed motility, or chemotaxis, is important for the ability of H. pylori to swim through the highly acidic lumen towards the epithelium and to survive in the host environment under the conditions of constant turnover of the gastric mucosa. Non-motile or non-chemotactic mutants have been shown to be less effective in colonising the gastric mucosa and do not attain full infection compared to the wild type in animal models5–8.
Chemotaxis allows motile bacteria to sense chemical cues and find optimal environments for growth by, for example, swimming towards favourable chemicals (chemoattractants) and away from harmful ones (repellents). Extracellular chemicals are sensed by chemoreceptors, also termed transducer-like proteins (Tlps). Most of the characterised Tlps are dimeric membrane proteins that comprise an extracytoplasmic ligand-binding domain (LBD), the transmembrane region, the HAMP (histidine kinases, adenylyl cyclases, methyl-accepting protein, and phosphatases) domain and the methyl-accepting (MA) domain (Fig. 1a), the latter transmitting information to a signalling cascade. The signal is relayed through the coupling protein CheW to the histidine protein kinase, CheA, which phosphorylates the response regulator protein, CheY, altering its affinity to the flagellar motors and, as a consequence, the direction (clockwise or counter-clockwise) in which they rotate9.
Figure 1.
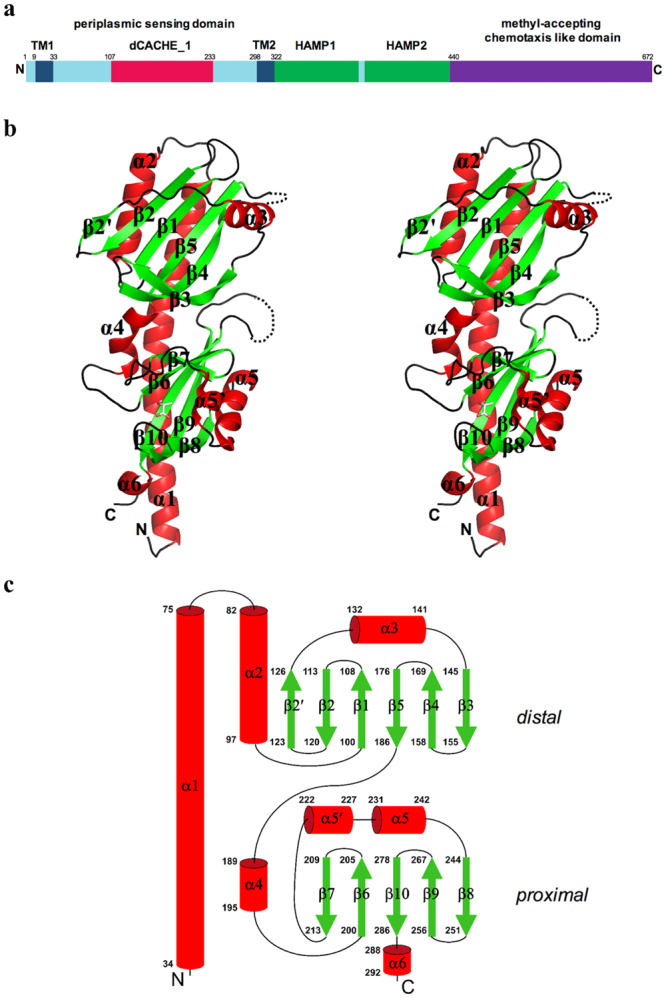
Overall fold of TlpC dCACHE LBD. (a) Domain organisation of TlpC, showing LBD location with respect to other structural elements. Transmembrane region (TM, dark blue); dCACHE_1 domain (red), HAMP domains (green); methyl-accepting chemotaxis-like domain (purple). (b) Stereo representation of structure of TlpC LBD monomer. (c) Topology of secondary structure elements of TlpC LBD. The α-helices are represented by rods and β-strands by arrows. The membrane-distal and membrane-proximal subdomains are labelled.
The chemotaxis pathway has been extensively studied in Escherichia coli 10–12. Recognition of cue molecules in this bacterium is mediated by five different chemoreceptors9,12,13. Four of them contain a periplasmic LBD with a 4-helix bundle (4HB) fold. The fifth receptor, Aer, has a cytosolic Per–Arnt–Sim (PAS) LBD and is involved in aerotaxis14.
Chemoreceptors have been classified according to the size of their LBD into cluster I (~150 amino acids) or cluster II (~250 amino acids)15. Much of what is known about bacterial chemoreceptors comes from studies on cluster I chemoreceptors with a 4HB LBD. However, more recent studies have shown that extra-cytoplasmic LBDs of chemoreceptors from different bacteria vary largely in their amino acid sequence and three-dimensional structure16 and, to date, additional structural families have been identified, including single CACHE (sCACHE)17–19, helical biomodular (HBM)20,21 and double CACHE (dCACHE) domains19,22–25.
In H. pylori, four chemoreceptors have been identified based on full genome sequence analysis: TlpA, TlpB, TlpC, and TlpD. TlpD is a soluble, cytoplasmic chemoreceptor that is involved in energy taxis26 and the repulsion response to reactive oxygen species27 and acid28. TlpA, TlpB, and TlpC are integral membrane proteins29,30. TlpA has been linked to recognition of bicarbonate and arginine as attractants31 and acid as a repellent28, whilst TlpB has been reported to detect acidic pH17,30 and the quorum-sensing molecule autoinducer-2 (AI-2)32 as repellents and direct the chemoattraction response to urea33. No signals have been associated with TlpC.
H. pylori additionally exhibits chemotactic responses to various other signals, including amino acids (aspartate, glutamate, asparagine, glutamine, histidine, proline, tyrosine, valine, leucine, serine and glycine)34, cholesterol35, bile acids (such as glycocholic, taurocholic, glycodeoxycholic, taurodeoxycholic, glycochenodeoxycholic and taurochenodeoxycholic acid)34, ZnCl2 36 and NiCl2 31. However, the recognition of these molecules has not been attributed to a specific chemoreceptor, and the mechanism by which these signals are sensed is currently unknown.
Amongst the four H. pylori chemoreceptors, only the periplasmic LBD of TlpB has been structurally characterised17. It is a homodimer of sCACHE modules17,19 – a feature that contrasts to the helical-bundle (4HB) modules of the extensively characterised aspartate receptor Tar from Salmonella typhimurium 37, the serine receptor Tsr from E. coli 38, and the McpS chemoreceptor from Pseudomonas putida 20. It is now recognised that the CACHE domain, either in its single sCACHE or double dCACHE form, is the most abundant extracellular sensing domain in prokaryotes, and is commonly found in two-component histidine kinases and chemoreceptors16,19,22,23,39–41.
TlpC is the least characterised chemoreceptor in H. pylori, and its natural ligand was unknown. H. pylori tlpC mutants are outcompeted by wild type during stomach colonisation, and TlpC modulates the chemotactic response to acid29,36. A BLAST search with the sequence of the sensing domain of TlpC against the structures deposited in the Protein Data Bank (PDB) identified no structural homologues of this domain. However, a pairwise comparison of profile Hidden Markov Models using the HHpred server42 predicted homology at the level of secondary structure to the sensing domains of family 1 histidine kinases (PDB entries 3lia, 3lib, 3lic, 3lid, 3lif)43 and chemoreceptors Tlp1 and Tlp3 from Campylobacter jejuni (PDB entries 4wy9 and 4xmr)23,24. These sensing modules belong to the recently redefined dCACHE_1 structural family19.
dCACHE domains consist of two structurally similar subdomains that each adopt a PAS-domain-like fold and are arranged in tandem, with one membrane- proximal and the other membrane-distal. dCACHE domain proteins can recognise their signal molecules directly or indirectly. Directly recognised ligands include amino acids22,23,44–46, pyrimidines47 and purines48. In all previously characterised dCACHE domains, direct sensing involves binding of the signal molecule to the membrane-distal, rather than membrane-proximal, subdomain, and no role for the membrane-proximal subdomain has been determined22,23,43,44,49.
In this paper, we report the crystal structure of LBD of H. pylori TlpC in complex with a small-molecule ligand that co-purified with the protein. The ligand was bound to the membrane-proximal subdomain. Based on the analysis of the electron density maps and the chemical nature of the ligand-binding pocket, we hypothesised and confirmed the ligand to be lactate. The location of the binding site has been validated by mutagenesis. We furthermore verified that lactate acts as an attractant for H. pylori, and that TlpC mediates the chemoattractant response. To the best of our knowledge, this is the first example of the dCACHE domain that directly recognises its ligand via the membrane-proximal module.
Results
Overall structure of TlpC LBD
The three-dimensional structure of recombinant H. pylori TlpC LBD (residues 34–297 plus six additional residues (GIDPFT) at the N-terminus, introduced as an artifact of the cloning procedure), was determined by X-ray crystallography using a single-wavelength anomalous dispersion (SAD) technique to a resolution of 2.2 Å. The TlpC LBD crystals (hereafter referred to as form A) belonged to the space group C2, with three molecules in the asymmetric unit related to each other by a three-fold pseudo-symmetry. The coordinates of these molecules were refined independently, and in the final model, they showed very similar backbone conformations that could be superimposed in a pairwise fashion with an overall root mean square deviation (r.m.s.d.) for the Cα atoms of 0.5–0.7 Å. Disordered regions 170–175, 271–274 and 295–297 were not seen in the electron density maps and could not be modelled.
In common with family 1 histidine kinases, the TlpC LBD has a dCACHE fold, and is composed of a membrane-proximal and membrane-distal PAS-like modules folding against the N-terminal and C-terminal halves of a long stalk helix, respectively (Fig. 1b). The TlpC LBD structure comprises six α-helices and 11 β-strands (Fig. 1c). The membrane-distal subdomain (residues 63–186) contains a six-stranded antiparallel β-sheet with the strand order 2′ 2 1 5 4 3, which is flanked on one side by an antiparallel two-helix bundle formed by helix α2 and the C-terminal half of helix α1, and on the other side by helix α3. The membrane-proximal subdomain (residues 34–62, 189–292) contains a five-stranded antiparallel β-sheet with the strand order 7 6 10 9 8. This β-sheet is flanked by an antiparallel two-helix bundle formed by helix α4 and the N-terminal half of helix α1 on one side, and by helices α5 and α5′ on the other side. Finally, an additional helix α6 forms an extension of strand β10 at the C-terminal end of TlpC LBD. The membrane-distal and membrane-proximal subdomains are intimately associated with each other, with a total buried surface area of 1169 Å2, which is equivalent to 16% of the total buried surface area (BSA) of an individual subdomain.
Analysis of the packing of monomers in the crystal lattice revealed head-to-tail arrangement of molecules, where the membrane-proximal subdomain of one subunit packs against the membrane-distal subdomain of the other, which is not likely to represent a physiologically relevant assembly. To determine the oligomeric state of TlpC LBD in solution, size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) experiments were performed. TlpC LBD eluted as a single monodispersed peak in all conditions tested. The derived molecular weight of 28.8 kDa was close to the value calculated from the amino acid sequence of a monomer (30 kDa). Furthermore, the apparent hydrodynamic radius Rh of the particles in this peak (25 Å) was close the Rh value calculated from the crystal structure of a single TlpC LBD subunit (26 Å). In line with these results, analysis of the crystal packing using the PDBe PISA server (http://www.ebi.ac.uk/pdbe/pisa/) identified no quaternary structure and suggested that TlpC LBD is a monomer in the crystal.
Comparison of TlpC LBD structure to other extracytoplasmic sensing domains
In comparison of TlpC LBD atomic coordinates against structures deposited in the Protein Data Bank using PDBeFold50, the closest structural similarities were found with the dCACHE_1 sensory modules of chemoreceptors Tlp1 and Tlp3 from C. jejuni 23,24, and bacterial family 1 histidine kinases (HK) HK1s-Z8 (Vibrio parahaemolyticus), HK1s-Z3 and HK1s-Z2 (Methanosarcina mazei)43. TlpC LBD structure can be superimposed well over those of Tlp1 (Fig. 2a), HK1s-Z8 (Fig. 2b), HK1s-Z3, HK1s-Z2 and Tlp3 [root-mean-square deviation (r.m.s.d.) of 2.1, 2.2, 2.3, 2.5 and 2.9 Å for 285, 266, 271, 262 and 254 aligned Cα atoms from Tlp1, Z8, Z3, Z2 and Tlp3, respectively], despite the low overall sequence identity (<17%). The dCACHE fold adopted by the TlpC LBD has also been previously observed in sensing domains of chemoreceptor Mlp3751 and C4-dicarboxylate transport sensory HK DctB from V. cholerae 52 (Fig. 2c). Furthermore, this fold is remotely similar to the tandem-PAS fold of LBD of luminescence (lux) system HK LuxQ from V. harveyi 53 (Fig. 2c).
Figure 2.

Comparison of dCACHE LBD of TlpC with structures of other dCACHE domains. (a,b) Structural superposition of TlpC LBD (Cα trace, in red) with dCACHE domains of (a) Tlp1 (Cα, in green, PDB entry 4wy924) and (b) HK1s-Z8 (Cα, in blue, PDB entry 3lie43). (c) LBDs of TlpC, Tlp1 (PDB entry 4wy924), HK1s-Z8 (PDB entry 3lie43), Tlp3 (PDB entry 4xmq23), HK1s-Z3 (PDB entry 3lib43), Mlp37 (PDB entry 5ave51), DctB (PDB entry 3by952) and LuxQ (PDB entry 1zhh53).
Analysis of putative ligand-binding sites
We next examined the putative ligand binding sites, starting with the membrane-distal module. Inspection of the structure of the membrane-distal subdomain around the region implicated in binding of small-molecule ligands in other dCACHE-containing proteins revealed a well-defined groove that runs along the full length of the β-sheet and is flanked on one side by helix α3 and the stretch of amino acid residues connecting β2′ and α3, and on the other side, by the β3-β4-tongue (Fig. 3a and b). This cleft is lined by mostly aliphatic and small hydrophilic residues and has the following approximate dimensions: 30 Å in length, 11 Å in width, and 9 Å in depth. Structural comparisons show that the groove in the membrane-distal domain of TlpC is significantly larger than the small pocket present in Tlp3 and other dCACHE sensing domains that recognise their small-molecule ligands directly23,24. Superimposition of the membrane-distal subdomains of TlpC and Tlp3 over 121 Cα atoms (r.m.s.d. of 2.9 Å) shows that helix α3 and the β3-β4-tongue in TlpC are positioned significantly further apart than the equivalent α-helix and β-tongue in Tlp3 (Fig. 3b). The cleft in TlpC appears too large to be a small-molecule-ligand binding site and could hypothetically fit a molecule of the size of a peptide, such as, for example, a loop or a terminal peptide of an-as-yet unidentified PBP.
Figure 3.
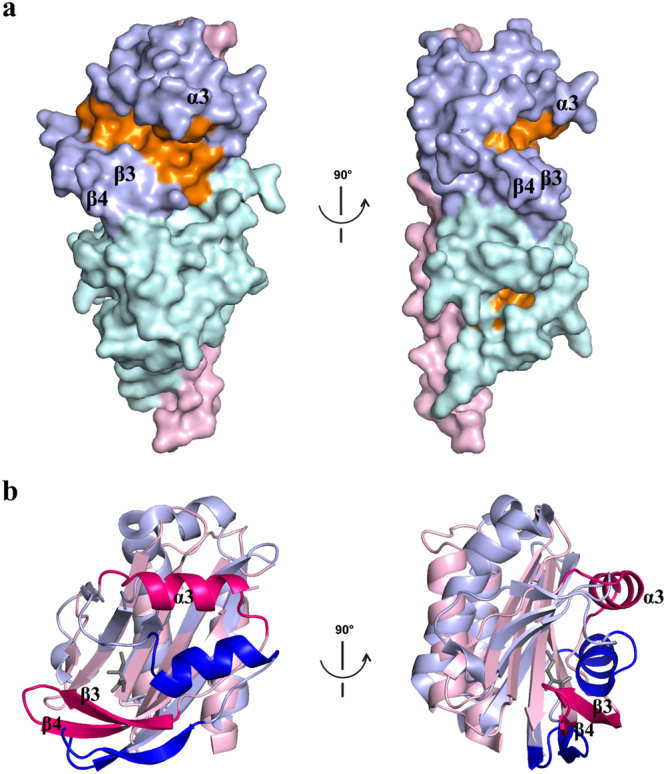
Putative ligand-binding sites of TlpC LBD. (a) Molecular surface of TlpC LBD with cavities and pockets coloured orange. The stalk helix is coloured pink, the membrane-distal module – light blue and the membrane-proximal module – cyan. (b) Structure superposition of membrane-distal modules of TlpC (pink) and Tlp3 (light blue) highlighting differences in position of helix α3 and β3-β4-tongue (coloured hot pink and blue in TlpC and Tlp3, respectively). Isoleucine bound to the membrane-distal module of Tlp3 is shown as grey sticks.
We then analysed the molecular surface of the membrane-proximal subdomain of TlpC LBD using CASTp54 with a probe radius of 1.4 Å. We detected a putative ligand-binding pocket with the surface area and solvent-accessible volume of 203 Å and 196 Å3, respectively (Fig. 3a). There was a clear electron density for a non-protein molecule bound in this pocket (Fig. 4). However, its shape did not match any of the components of the purification or crystallisation buffers, which suggested that the ligand trapped in the crystal could be a molecule that was present in the refolding mix or a product of proteolytic degradation of the sample.
Figure 4.

Architecture of ligand-binding site in membrane-proximal module of TlpC dCACHE domain. The (mFo - DFc) σA-weighted electron density for lactate is shown in green. The map was calculated at 2.2 Å resolution and contoured at the 3.0 σ level. The lactate molecule is shown in all-atom ball-and-stick representation with C atoms coloured orange. The protein side chains that form direct contacts with lactate are shown in stick representation.
Identification of lactate as a ligand for TlpC
To identify the ligand captured by TlpC LBD, the protein was denatured to release the small molecules, and these were analysed by liquid chromatography-electrospray ionisation mass spectrometry (LC-ESI-MS). The negative ionisation mode MS data showed a small peak at m/z = 89.022 that was absent in the buffer control (Supplementary Fig. 1). Within the experimental error, this peak matched the chemical formula C3H5O3 (m/z 89.024). A search in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/search/) identified 47 different compounds matching this formula. The shape of four of these (lactate, 1,1-dihydroxypropan-2-one, hydron-2-hydroxypropanoate and prop-2-ene-1,1,2-triol) matched the shape of the electron density in the membrane-proximal pocket. As lactate is the only one of these four compounds that is a natural metabolite produced during E. coli growth, we hypothesised that during refolding, the lactate was captured by the protein from the cell lysate.
Isothermal titration calorimetry (ITC) measurements confirmed that L-lactate binds exothermically to TlpC LBD with an apparent K D of 155 μM ± 5 μM (Fig. 5). The binding is driven by a favourable enthalpy change (∆H = −20 kcal mol−1) and is associated with a minor unfavourable entropy change (T∆S = −1.29 kcal mol−1). This binding appears specific to lactate because no significant heat release or absorption was observed with pyruvate, malate or oxaloacetate, that are chemically similar and metabolically exchangeable with lactate (Supplementary Fig. 2).
Figure 5.

ITC titrations of TlpC LBD and its Y249F variant with lactate. The upper panel shows raw titration data, where each peak corresponds to the injection of 10 μl of 5 mM sodium L-lactate into a 1.45-ml reaction cell containing 10 μM protein. The lower panel shows the integrated and dilution-corrected peak areas of the titration plot.
Validation of lactate binding site in membrane-proximal module of TlpC dCACHE domain
To establish whether lactate binds to the membrane-distal or membrane-proximal module, we determined the crystal structure of TlpC dCACHE domain co-crystallised with 10 mM L-lactate. The co-crystals with lactate were isomorphous to the form A crystals grown with no lactate in the crystallisation mix. Superposition of the protein contents of the two asymmetric units based on the overlap of 767 Cα atoms with an r.m.s.d. of 0.32 Å showed that, within the limit of the experimental error in the coordinates (0.33 Å for the co-crystal with lactate), their structures were essentially identical. Analysis of the electron density maps revealed no lactate binding sites other than the one in the membrane-proximal subdomain. This subdomain contained a lactate molecule bound in a very similar mode to that observed in the form A crystals grown with no added lactate (Fig. 4).
The lactate binding site is located in a pocket formed by residues F202, L210, N213, I218, L223, Y249, L252, S253, and Y285. Calculation of the accessible surface area (ASA) showed that lactate becomes almost completely shielded from the solvent upon binding to TlpC LBD, with 99.5% of its ASA buried by the protein. The carboxyl and hydroxyl groups of lactate form hydrogen bonds with the side chains of N213, Y249 and Y285, and with the main-chain amides of L252 and S253. The TlpC LBD/lactate complex is further stabilised by hydrophobic interactions between the methyl group of lactate and the side chains of F202, L210, I218 and L223 (Fig. 4).
To evaluate the contribution of individual amino acids to the lactate binding, N213, I218 and Y285 were individually replaced with alanine and Y249 with phenylalanine, and the effect of the single-amino acid substitutions was assessed by isotitration calorimetry. Comparison of the circular dichroism spectra of the variants with that of native TlpC LBD showed no significant differences, indicating that the amino acid substitutions did not alter the secondary structure (Fig. 6). ITC measurements demonstrated that each of the N213A, I218A, Y285A and Y249F substitutions abolished the binding of lactate to TlpC LBD (Fig. 4 and Table 1). To further confirm that the membrane-distal subdomain does not bind lactate, the TlpC residues S104, Y151 and K153 – occupying the positions structurally equivalent to the ligand-binding residues Tyr118, Tyr167 and Thr170 in the membrane-distal subdomain of Tlp3 – were individually substituted with alanine. In contrast to the effect on the binding to the membrane-proximal domain, these substitutions only resulted in 2–3 fold reduction in the affinity to lactate, likely due to partial fold destabilisation rather than loss of interactions with the ligand.
Figure 6.
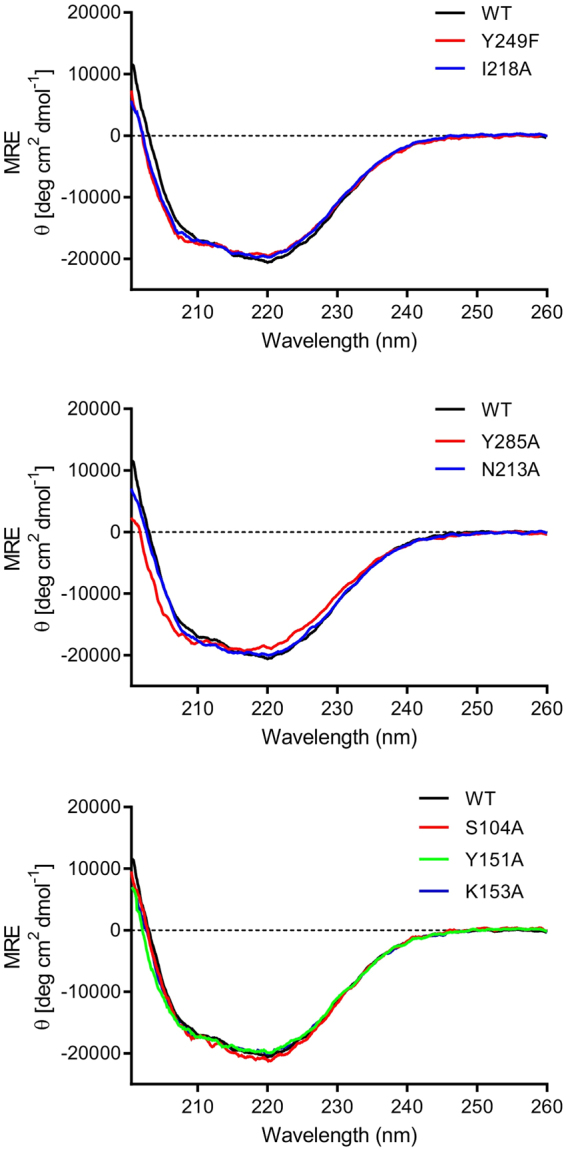
CD spectra of native TlpC LBD (WT) and its S104A, Y151A, K153A, N213A, I218A, Y285A and Y249F variants.
Table 1.
Thermodynamic parameters of lactate binding to TlpC LBD and its variants derived from ITC measurements.
| Protein | K D | Enthalpy, ∆H (cal/mol) | Entropy, ∆S (cal/mol/degree) |
|---|---|---|---|
| TlpC LBD native | 155.0 ± 5.0 µM | −21,323.3 ± 713.0 | −54.1 ± 2.0 |
| TlpC LBD N213A | >3,000 | — | — |
| TlpC LBD I218A | 3.1 ± 0.6 mM | −18,145.0 ± 49.0 | −49.4 ± 0.2 |
| TlpC LBD Y249A | >3,000 | — | — |
| TlpC LBD Y285F | >3,000 | — | — |
| TlpC LBD F202A | >3,000 | — | — |
| TlpC LBD K223A | >3,000 | — | — |
| TlpC LBD S104A | 359.0 ± 3.0 µM | −13,105 ± 318.0 | −28.2 ± 1.0 |
| TlpC LBD Y151A | 278.5 ± 2.0 µM | −12,040 ± 250.2 | −24.1 ± 1.0 |
| TlpC LBD K153A | 467.2 ± 5.0 µM | −12,730 ± 345.0 | −27.5 ± 1.0 |
H. pylori TlpC mediates positive chemotactic response to lactate
To test the physiological relevance of the observed specific interaction between TlpC and lactate, we assessed the lactate chemotactic response of H. pylori. Although there are isolates of the laboratory H. pylori strain 26695 that are motile55 this strain is prone to motility loss and difficult to use in motility evaluation. We therefore used the human isolate pre-mouse SS1 (PMSS1)56, which displays a high level of reliable motility, and has been studied for chemotaxis responses in recent publications28. The TlpC ligand-binding domain from 26695 and PMSS1 are identical (Supplementary Fig. 3), so we reasoned both proteins would respond similarly to lactate. We assessed whether lactate is an H. pylori attractant or repellent using a swimming assay that enumerates flagellar-based bacterial reversals, a common read out for a chemotactic response57. In this assay, attractants cause decreased and repellents cause increased direction changes26,27,32. Wild-type H. pylori showed a significant response to 0.1 mM lactate in this assay, but lost the response at higher concentrations (Fig. 7).
Figure 7.

Lactate triggers a TlpC-dependent attractant response in H. pylori. Cultures of isogenic wild-type (WT) and ΔtlpC (∆TlpC) H. pylori PMSS1 were grown in BB10 overnight, treated with various concentrations of sodium L-lactate or control compounds as indicated, and then immediately filmed. Direction changes were enumerated over a 3 second period in at least 200 cells per treatment in two biological replicates. 10 mM HCl or 50 μM dipyridyl serve as known repellent and attractant response controls, respectively. Error bars represent the standard error of the mean. *p < 0.05; **p < 0.01, comparisons performed using a two-way ANOVA, followed by Tukey’s pairwise comparisons (α = 0.05). There were no significant differences in the basal behaviour between wild type and its tlpC mutant.
To account for possible chemotactic effects due to pH change upon sodium L-lactate treatment of BB10, the pH of the media with and without treatment was assessed. While treatment with 10 mM HCl decreased the pH by more than 1.5 pH units, treatment with any concentration of sodium L-lactate only decreased the BB10 pH by less than 0.05 pH units. Furthermore, the pH difference between the highest and lowest amount of sodium L-lactate (10 mM and 0.1 mM) was only ~0.01. This analysis suggests that sodium L-lactate did not substantially change the medium pH, and thus any effect due to pH change upon sodium L-lactate treatment were likely negligible compared to chemotactic effects due to sodium L-lactate itself.
We next examined whether TlpC was required for this lactate chemotaxis response. We generated an isogenic null mutant strain lacking tlpC (∆tlpC). tlpC is part of a single gene operon58,59, and thus mutations are unlikely to have polar effects. Consistent with this idea of specificity, the ∆tlpC mutant retained chemotaxis responses to signals that act through other chemoreceptors: the known repellent HCl that acts through TlpB, TlpA, and TlpD, as well as the attractant dipyridyl that acts through TlpD27,28,30 (Fig. 7). These responses confirmed that the ∆tlpC mutant was not generally chemotaxis defective. Deletion of tlpC, however, abolished the chemotactic response to lactate at all tested concentrations (Fig. 7). Additionally, while ∆tlpC PMSS1 H. pylori displayed fewer reversals on average compared to WT PMSS1, this difference in basal reversal frequency was not significant (Fig. 7). This data thus supports that TlpC is the chemoreceptor for lactate in H. pylori.
Discussion
dCACHE domains of bacterial chemoreceptors and histidine kinases consist of two subdomains, membrane-distal and membrane-proximal, each of which could, in principle, harbour a binding site for a small signal or regulatory molecule. In all previously characterised dCACHE domains, including LBDs of Tlp3 from C. jejuni 23, Mlp37 from V. paraehemoliticus 51, McpB and McpC from B. subtilis 22,45, and Mlp24 from V. cholerae 60, direct sensing involved binding of the signal molecule to the membrane-distal subdomain. Our analysis of the structural basis of lactate recognition by H. pylori chemoreceptor TlpC changes this paradigm regarding the mechanism of sensing by dCACHE domain by providing the first example where direct sensing of the signal molecule is mediated by the ligand binding to the membrane-proximal, rather than membrane-distal, subdomain.
This result has important implications for the conceptual framework of dCACHE-mediated sensing and signal transmission across the membrane. As this and previous studies showed, the membrane-distal and membrane-proximal modules of dCACHE are intimately associated with each other, and are therefore structurally and dynamically coupled23,24. For example, we previously demonstrated using X-ray crystallography that, upon binding to an attractant, the dCACHE membrane-distal subdomain of C. jejuni Tlp3 closes around the ligand and loses its tight association with the membrane-proximal domain, which, as a result, adopts a more open conformation23. The structural coupling of the membrane-distal and membrane-proximal subdomains is consistent with the finding that signalling across the membrane presumably can be triggered by direct ligand binding to either subdomain – the membrane-proximal subdomain, as in TlpC, or membrane-distal subdomain as in Tlp3, Mlp37, McpB or McpC. Furthermore, one cannot eliminate the possibility that different ligands may signal through the same receptor, with some binding to the membrane-distal and others to the membrane-proximal subdomain. Indeed, all membrane-proximal subdomains of dCACHE sensing domains characterised to date contain a putative small ligand-binding pocket, including those that sense ligands with the membrane-distal domain23,24,43,51. For example, the structural study of LBD of C. jejuni chemoreceptor Tlp1 revealed an acetate ion bound to the dCACHE membrane-proximal module24. Acetate has not yet been found to trigger a chemotaxis response in C. jejuni 61. However, it has been shown to induce either a positive or negative chemotactic response in other species18,62–64.
Apart from implicating the membrane-proximal, rather than membrane-distal, subdomain in direct sensing of a small molecule ligand, our crystallographic analysis revealed one more difference between the dCACHE domain of TlpC and that of other structurally characterised chemoreceptors of this type – the presence of a long groove in the membrane-distal subdomain, instead of a small well-defined pocket23,43,51. This groove might represent a putative binding site for a larger molecule, such as an intermediate ligand-binding protein from the periplasmic binding protein (PBP) family. PBP-mediated sensing is used for chemotaxis in other bacteria, where some PBPs have a function of a primary chemoreceptor that recognises and binds a small molecule in the periplasm, and, in the ligand-bound form, associates with its cognate, membrane-bound transducer-like protein, initiating the signal65. There are at least six putative PBPs encoded in the H. pylori genome which have been shown to bind autoinducer 266 and nickel67, and proposed to bind other compounds including peptides68, molybdenum, amino acids, and iron69. Whilst the membrane-proximal domain of TlpC mediates direct sensing of lactate, its membrane-distal domain – the shape of which does not imply small-molecule binding – may partner with a PBP to sense some other ligand.
Our analysis of the chemotactic behaviour of wild-type H. pylori showed that lactate induced an attractant response in a concentration-dependent manner, and that this response was drastically reduced in a chemotactically competent isogenic ΔtlpC mutant, demonstrating that TlpC is the primary chemoreceptor for lactate in H. pylori. Within the tested range of concentrations of lactate, the response was strongest at 0.1 mM, detectable at 1.0 mM, and not detectable at 10 mM. Putting the observed TlpC-dependent chemotactic behaviour towards L-lactate in the context of the receptor-ligand interactions, we note that the order of the optimal concentration at which lactate is sensed by H. pylori as attractant (0.1 mM) is the same as the order of the dissociation constant K D (0.155 mM) for its binding to the TlpC dCACHE domain. L-lactate, secreted by gastric mucous cells, reaches the concentration of 0.3–1 mM in gastric juice70,71. Presumably lactate forms a gradient with its highest amount at the source, the cells, but the stomach distribution of lactate is not known. Lactate is known to promote H. pylori growth in the stomach72 and in media that is lacking dextrose, suggesting it can serve as either a carbon or energy source, or both73. Metabolically, lactate can be generated by lactate dehydrogenase (LDH) from pyruvic acid as the end product of glycolysis. However, LDH can also catalyse the reverse reaction, converting lactate into pyruvate74,75. Thus, if exogenous lactate was imported into H. pylori, it could be oxidised into pyruvate, which would then enter the tricarboxylic acid cycle. Alternatively, lactate can donate electrons to NADH and, in turn, to the electron transport chain to enhance proton motive force and bacterial energy levels76. In H. pylori, the proteins necessary for the import and utilisation of lactate have been identified69,73, including two lactate permeases and two LDHs73. At least one of the lactate utilisation genes has been shown to be required for stomach colonisation, supporting the importance of this process in vivo 77.
TlpC mutants have mouse stomach colonisation defects but only when competing with wild type29. This phenotype is consistent with the idea that either lactate is limiting and wild type utilises it more efficiently, or that wild type follows a lactate gradient and occupies key niches before the tlpC mutant can get there. Thus, the in vivo fitness defect observed with isogenic TlpC mutants is consistent with an inability to efficiently access and catabolise lactate in vivo 29.
Interestingly, several H. pylori lab strains appear to lack TlpC protein expression27. One of these strains, G27, has a single base indel that creates a frameshift and results in loss of TlpC expression27. Another, B128, also has a tlpC frameshift but its gerbil-selected daughter strain, 7.13, has regained TlpC expression27. These findings suggest that TlpC is not required for lab growth. The stomach may provide different selective pressures such that TlpC expression is an advantage. In support of this idea, four clinical H. pylori isolates analysed all expressed TlpC27.
The dissociation constant for lactate binding to the TlpC dCACHE domain falls within the middle of the range of values reported for ligand binding by other CACHE domains (e.g. 23–356 µM for Pseudomonas syringae pv. actinidiae PscD LBD78, 0.6–373 µM for P. putida KT2440R McpA LBD79, 1–1000 µM for B. subtilis McpC LBD22). Lactate is also a chemoattractant for Pseudomonas aeruginosa 18. P. aeruginosa senses lactate via an sCACHE domain receptor named McpP. While this receptor has not yet been crystallised, it is known to bind lactate with similar affinity to TlpC, with a K D of 107 µM18. McpP additionally binds acetate, pyruvate, and propionate. McpP is similar to several other sCACHE chemoreceptors, suggesting chemotaxis toward lactate and related C2 and C3 carboxylic acids may be widespread.
Our observation that a higher concentration of lactate (10 mM) did not elicit a positive chemotactic response in H. pylori is in agreement with the reports that, at a concentration of 10 mM or above, lactate has an inhibitory effect on H. pylori growth72,80. The anti-H. pylori activity of high levels of exogenous lactate was first observed in co-cultures with lactic acid bacteria (LAB)81–83. Lin et al.84 demonstrated that short chain fatty acids (SCFA) (acetic, formic, propionic, butyric and lactic acid) secreted by LAB, and the associated low pH values reduce H. pylori viability, with lactic acid exhibiting the strongest inhibitory effect out of all tested SCFA84,85. Although the exact mechanism by which high levels of lactic acid exert anti-H. pylori activity remains unclear, it is likely a combination of its inhibitory effect on H. pylori urease activity and the reduced ability of H. pylori to survive at low pH in the absence of urea80,84–87.
Full-length chemoreceptors function as trimers of dimers88. Although the degree and mechanism of the contribution dCACHE LBDs make to oligomerisation in vivo remains to be established, previous crystallographic studies on the dCACHE domains of C. jejuni Tlp323, M. mazei HK1s-Z343 and V. cholerae DctB52 suggested that they likely dimerise through their stalk helix, with the twofold axis approximately perpendicular to the membrane plane. The dimerisation forces between isolated dCACHE domains are weak, as all domains of this type characterised so far, including that of H. pylori TlpC (this study), Tlp1 and Tlp3 from C. jejuni 23,24, CtaA and CtaB from Pseudomonas fluorescens 89,90, VfcA from Vibrio fischeri 91, and PctA from P. putida 92, are monomeric in solution. Our analysis showed that the isolated recombinant dCACHE LBD of TlpC is monomeric in the crystal as well. However, the observed structural similarity between LBDs of TlpC, Tlp3, HK1s-Z3 and DctB allows for the possibility that, in the context of the membrane-embedded full-length receptor, TlpC LBD may also dimerise through its stalk helix.
In conclusion, this study reports the first example of the dCACHE type chemoreceptor that directly senses its ligand via its membrane-proximal subdomain, and that H. pylori seeks out lactate using chemotaxis. It adds to the mounting evidence that dCACHE sensing domains have evolved to recognise their ligands via several different direct and indirect mechanisms that may utilise either the membrane-distal, or the membrane-proximal, subdomain, or both. This raises an intriguing question about whether, despite this diversity, different dCACHE sensing domains share a common mechanism of signal transduction across the membrane.
Methods
Site-directed mutagenesis, protein expression, and purification
The expression vectors for single-point variants of TlpC LBD in which S104, Y151, K153, N213, I218 or Y285 were replaced by alanine, and Y249 by phenylalanine, were prepared from a TlpC-expressing plasmid described previously93. This plasmid expresses codon-optimised H. pylori 26695 TlpC LBD consisting of amino acid residues 34–297 (Fig. 1a). Mutants were created using the QuikChange Mutagenesis Kit (Stratagene). TlpC LBD and its variants were expressed and purified following the previously published procedure93.
Crystallisation, data collection and structure determination
Form A crystals of TlpC LBD were obtained as described93. The crystals grew in space group C2 (Table 2) and contained three monomers in the asymmetric unit. Co-crystallisation with 10 mM sodium L-lactate under similar conditions produced the crystals of the TlpC LBD/lactate complex that were isomorphous with form A crystals (Table 2). Two platinum derivatives were obtained by soaking the TlpC LBD crystals in either potassium tetrachloroplatinate (1 mM) or potassium hexachloroplatinate (1 mM). The derivative crystals belonged to space group P321 (form B) with a monomer in the asymmetric unit (Table 2). Native X-ray diffraction data (λ = 0.95 Å) and SAD data for the derivatives (λ = 1.07 Å) were collected on the MX1 and MX2 beamlines of the Australian Synchrotron (AS)94. All data were processed with iMOSFLM 95 and scaled with AIMLESS96 from the CCP4 software suite97 (Table 2).
Table 2.
X-ray Data collection and processing statistics. Values in parentheses are for the highest resolution shell.
| Dataset | Native | K2PtCl4 | K2PtCl6 | Co-crystal with 10 mM lactate |
|---|---|---|---|---|
| Space group | C2 | P321 | P321 | C2 |
| a, b, c (Å) | 189.3, 103.2, 61.8 | 102.7, 102.7, 62.4 | 102.5, 102.5, 63.0 | 188.5, 102.6, 61.2 |
| β (°) | 98.3 | 98.5 | ||
| Resolution range (Å) | 30.6–2.2 | 62.4–3.3 | 51.4–3.3 | 30.0–2.5 |
| (2.31–2.20) | (3.48–3.30) | (3.48–3.30) | (2.60–2.50) | |
| Rmerge | 0.062 (0.280) | 0.095 (0.364) | 0.096 (0.302) | 0.069 (0.341) |
| Average I/σ(I) | 13.3 | 18.2 | 18.1 | 58.5 |
| Completeness (%) | 98 (96) | 99.9 (99.9) | 99.9 (99.9) | 99.8 (99.9) |
| Redundancy | 3.6 | 10.5 | 10.5 | 3.6 |
| Anomalous redundancy | 5.5 | 5.4 | ||
| Observed reflections | 215,101 | 62,341 | 62,541 | 505,592 |
| Unique reflections | 59,028 | 5,941 | 5,975 | 40,012 |
The two isomorphous SAD data sets were used to locate the platinum sites and calculate the phases for the form B crystals with Autosol98 from the PHENIX software suite99. The resulting phase set (overall figure of merit of 0.30 for data between 51.4 and 3.3 Å) was used to generate an initial partial model for a TlpC LBD monomer with AutoBuild (PHENIX). This model was used for phasing the form A data for the native crystal and the co-crystal with lactate by molecular replacement. Refinement statistics and stereochemistry are given in Table 3. For both models, all the non-glycine residues lie in permitted regions of the Ramachandran plot, with 97% of these in the most favoured regions.
Table 3.
Refinement statistics.
| Data set | TlpC native | Co-crystal with 10 mM lactate |
|---|---|---|
| Resolution range (Å) | 30.6–2.2 | 30.0–2.5 |
| No. reflections | 59,028 | 40,012 |
| Rwork/Rfree a | 0.182/0.218 | 0.192/0.251 |
| No. atoms | ||
| Protein | 6310 | 6212 |
| Water | 452 | 66 |
| Lactate | 18 | 18 |
| B-factors | ||
| Average B (protein atoms) (Å2) | 46 | 42 |
| Average B (water molecules) (Å2) | 52 | 37 |
| Average B (lactate) (Å2) | 41 | 41 |
| R.m.s. deviations from ideality | ||
| Bond lengths (Å) | 0.017 | 0.008 |
| Bond angles (°) | 1.4 | 1.1 |
aThe Rfree was calculated on 5% of the data omitted at random.
SEC-MALS analysis
The hydrated molecular mass and hydrodynamic radius of TlpC LBD in solution were determined by SEC-MALS. Protein was dialysed against buffer A containing 100 mM Tris-HCl pH 8.0 and 150 mM NaCl, and concentrated to 3 mg ml−1. A 100 µl sample was loaded onto a WTC-030S5 SEC column (Wyatt Technology Corporation) pre-equilibrated with the same buffer flowing at 0.4 ml min−1. The eluate was passed through an inline DAWN HELEOS light scattering detector, an Optilab T-rEX differential refractive index detector and a quasi-elastic light scattering detector (WyattQELS, Wyatt Technology Corporation). The experiment was repeated in the presence of 10 mM sodium L-lactate. Calculations of the molecular mass and hydrodynamic radius from the intensity of the scattered light and refractive index were performed using ASTRA 6.0 (Wyatt) (Table 4). Theoretical calculations of the hydrodynamic radius from the crystal structure were carried out using HYDROPRO version 10100.
Table 4.
Dynamic light-scattering results.
| Sample | Polydispersity | Molecular weight (kDa) | R h (nm) |
|---|---|---|---|
| TlpC LBD | 1.0 | 28.8 | 2.5 |
| TlpC LBD + sodium L-lactate | 1.0 | 27 | 2.6 |
| BSA | 1.0 | 63.9 | 3.6 |
LC-ESI-MS analysis
Identification of the ligand captured by TlpC LBD was achieved by extracting small molecules from the purified protein and measuring their masses by LC-ESI-MS. TlpC LBD (30 µM in buffer A) was unfolded by boiling at 100 °C for 15 min and then pelleted by centrifugation. Buffer A subjected to the same procedure was used as a negative control. 200 µl of the supernatant was directly infused into MicrOTOF-Q quadrupole time-of-flight (TOF) mass spectrometer (Bruker Daltonics), and then nebulised and ionised using the Bruker electrospray source. Data were acquired in both positive and negative ion ESI modes over the mass range of 70 to 200 Daltons. The spectra were processed using the Data Analysis software version 3.4 (Bruker Daltonics).
CD analysis
CD spectroscopy was used to compare secondary structure composition of TlpC LBD and its single-point variants. Protein samples (0.1 mg ml−1) were dialysed against buffer B (10 mM sodium phosphate pH 7.4, 150 mM NaCl). Far-UV CD spectra were recorded over the wavelength range 200–260 nm at 25 °C with the scan rate of 20 nm min−1 using a JASCO J-815 spectropolarimeter. The spectra were measured in triplicate, averaged and smoothed using the Savitzky-Golay algorithm with a radius of 25101. Raw data were converted to mean residue ellipticity θ (in deg cm2 dmol−1)102.
ITC experiments
TlpC LBD and its variants were dialysed against buffer A. 5 mM solutions of sodium lactate, sodium pyruvate, sodium malate and sodium oxaloacetate were prepared by dissolving them in the dialysis buffer. Measurements were performed on a MicroCal VP-ITC instrument microcalorimeter (MicroCal) at 25 °C. Protein (10 µM) in a 1.45-ml sample cell was injected with 25 successive 10-μl aliquots of the lactate solution. Binding isotherms were generated by plotting the heat change evolved per injection versus molar ratio of lactate to protein. The data was fitted to a single-site binding model using non-linear least-squares regression (Origin 7, OriginLab, USA), yielding the binding enthalpy ∆H, dissociation constant K D, and binding entropy ∆H. Each experiment was repeated three times.
Construction of isogenic ΔtlpC mutant in H. pylori strain PMSS1
The PMSS1 ΔtlpC mutant was created by natural transformation of wild-type PMSS1 with 5 μg of ∆tlpC::cat SS1 genomic DNA29,56. Chloramphenicol-resistant mutants were selected using 10 μg/ml chloramphenicol on Columbia Horse Blood Agar as previously described29. Mutation of tlpC was confirmed by PCR and western blot.
Chemotaxis assay
Swimming behaviour assays were done with H. pylori PMSS1 strains described above grown in Brucella broth (BD BBL/Fisher) with 10% FBS (Life Technologies) (BB10), with shaking, at 37 °C, under microaerobic conditions of 5% O2, 10% CO2, balance N2. Overnight cultures (~OD600 0.25–0.5) were diluted to an OD600 of 0.1 in fresh BB10, and then incubated as above until an OD600 of 0.12–0.15 was reached. Motile, OD600 0.12–0.15, cultures were treated with sodium L-lactate (0.1 mM, 1 mM, 10 mM) or an equal volume of H2O as an untreated control. As a repellent control, 10 mM HCL was used as done previously30,32,36. As an attractant control, 50 μM dipyridyl was used as done previously27. Dipyridyl results in fewer directions changes, an attractant response, dependent on chemotaxis in general and TlpD specifically. Dipyridyl induces and attractant response as it counters reactive oxygen species via chelation of iron27. The pH of BB10 upon treatment was independently assessed using a Denver Instruments pH meter. Cultures were filmed immediately after ligand addition at 400x magnification using a Hamamatsu Digital Camera C4742-95 with the μManager software (Version 1.4.22), mounted on a Nikon Eclipse E600 phase contrast microscope103 (Supplementary videos 1–12). Videos were relabeled to blind the observer to the strain identity. For each sample, >100 3-s-long bacterial tracks from two independent cultures were analysed manually to identify stops followed by direction changes and to calculate the average number of direction changes in 3 s. Statistical analysis of the data for treated versus untreated samples was performed using a Student’s t-test.
PDB submission codes
The atomic coordinates and structure factors of the TlpC LBD/lactate complex obtained at 2.2 Å resolution have been deposited in the Protein Data Bank (http://www.rcsb.org) under accession code 5wbf.
Electronic supplementary material
Acknowledgements
We thank Dr. Danuta Maksel and Dr. Robyn Gray at the Monash Macromolecular Crystallisation Facility for their assistance in setting up automated crystallization screens. We additionally thank Dr. Susan Williams (UC Santa Cruz) for early work on lactate chemotaxis. Part of this research was undertaken on the MX1 and MX2 beamlines of the AS, Victoria, Australia. We thank the AS staff for their assistance with data collection. The described project was supported by a National Institutes of Health National Institute of Allergy and Infectious Disease (NIAID) grant number RO1AI116946 (to K.M.O.). Mayra A. Machuca is indebted to the Departamento Admistrativo de Ciencia, Tecnología e Innovación COLCIENCIAS for a doctoral scholarship. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author Contributions
A.R., Y.C.L. and K.M.O. conceived the study. M.A.M., K.S.J., Y.C.L., D.L.S. performed the experiments. All authors analyzed the data. M.A.M., K.S.J., Y.C.L. and A.R. prepared the Figures. M.A.M., Y.C.L. and A.R. drafted the manuscript. K.S.J. and K.M.O. edited the manuscript. All authors reviewed and approved the final version.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14372-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Karen M. Ottemann, Email: ottemann@ucsc.edu
Anna Roujeinikova, Email: anna.roujeinikova@monash.edu.
References
- 1.Hunt RH, et al. Helicobacter pylori in developing countries. World Gastroenterology Organisation Global Guideline. J. Gastrointestin. Liver Dis. 2011;20:299–304. [PubMed] [Google Scholar]
- 2.Uemura N, et al. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 3.Peek RM, Jr., Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 4.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 5.Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terry K, Williams SM, Connolly L, Ottemann KM. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 2005;73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keilberg D, Ottemann KM. How Helicobacter pylori senses, targets and interacts with the gastric epithelium. Environ. Microbiol. 2016;18:791–806. doi: 10.1111/1462-2920.13222. [DOI] [PubMed] [Google Scholar]
- 8.Foynes S, et al. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 2000;68:2016–2023. doi: 10.1128/IAI.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller LD, Russell MH, Alexandre G. Diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 2009;66:53–75. doi: 10.1016/S0065-2164(08)00803-4. [DOI] [PubMed] [Google Scholar]
- 12.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 13.Yeh JI, et al. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 14.Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in. Escherichia coli. J. Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacal J, Garcia-Fontana C, Munoz-Martinez F, Ramos JL, Krell T. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010;12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 16.Bardy, S. L., Briegel, A., Rainville, S. & Krell, T. Recent advances and future prospects in bacterial and archaeal locomotion and signal transduction. J. Bacteriol, 10.1128/JB.00203-17 (2017). [DOI] [PMC free article] [PubMed]
- 17.Goers Sweeney E, et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia V, et al. Identification of a Chemoreceptor for C2 and C3 Carboxylic Acids. Appl. Environ. Microbiol. 2015;81:5449–5457. doi: 10.1128/AEM.01529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upadhyay, A. A., Fleetwood, A. D., Adebali, O., Finn, R. D. & Zhulin, I. B. Cache Domains That are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 12, e1004862; 1004810.1001371/journal.pcbi.1004862, (2016). [DOI] [PMC free article] [PubMed]
- 20.Pineda-Molina E, et al. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc. Natl. Acad. Sci. USA. 2012;109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Mora D, et al. McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ. Microbiol. 2016;18:3284–3295. doi: 10.1111/1462-2920.13030. [DOI] [PubMed] [Google Scholar]
- 22.Glekas GD, et al. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J. Biol. Chem. 2012;287:39412–39418. doi: 10.1074/jbc.M112.413518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, Roujeinikova A. Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr. D Biol. Crystallogr. 2015;71:2127–2136. doi: 10.1107/S139900471501384X. [DOI] [PubMed] [Google Scholar]
- 24.Machuca MA, Liu YC, Beckham SA, Gunzburg MJ, Roujeinikova A. The Crystal Structure of the Tandem-PAS Sensing Domain of Campylobacter jejuni Chemoreceptor Tlp1 Suggests Indirect Mechanism of Ligand Recognition. J. Struct. Biol. 2016;194:205–213. doi: 10.1016/j.jsb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Matilla MA, Krell T. Chemoreceptor-based signal sensing. Curr. Opin. Biotechnol. 2017;45:8–14. doi: 10.1016/j.copbio.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Schweinitzer T, et al. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol. 2008;190:3244–3255. doi: 10.1128/JB.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins KD, et al. The Helicobacter pylori CZB Cytoplasmic Chemoreceptor TlpD Forms an Autonomous Polar Chemotaxis Signaling Complex That Mediates a Tactic Response to Oxidative Stress. J. Bacteriol. 2016;198:1563–1575. doi: 10.1128/JB.00071-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang JY, Goers Sweeney E, Guillemin K, Amieva MR. Multiple Acid Sensors Control Helicobacter pylori Colonization of the Stomach. PLoS Pathog. 2017;13:e1006118. doi: 10.1371/journal.ppat.1006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andermann TM, Chen YT, Ottemann KM. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 2002;70:5877–5881. doi: 10.1128/IAI.70.10.5877-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 2006;188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerda O, Rivas A, Toledo H. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 2003;224:175–181. doi: 10.1016/S0378-1097(03)00423-3. [DOI] [PubMed] [Google Scholar]
- 32.Rader BA, et al. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology. 2011;157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JY, et al. Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium. Cell Host Microbe. 2015;18:147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worku ML, Karim QN, Spencer J, Sidebotham RL. Chemotactic response of Helicobacter pylori to human plasma and bile. J. Med. Microbiol. 2004;53:807–811. doi: 10.1099/jmm.0.45636-0. [DOI] [PubMed] [Google Scholar]
- 35.Wunder C, et al. Cholesterol glucosylation promotes immune evasion by Helicobacter pylori. Nat. Med. 2006;12:1030–1038. doi: 10.1038/nm1480. [DOI] [PubMed] [Google Scholar]
- 36.Sanders L, Andermann TM, Ottemann KM. A supplemented soft agar chemotaxis assay demonstrates the Helicobacter pylori chemotactic response to zinc and nickel. Microbiology. 2013;159:46–57. doi: 10.1099/mic.0.062877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milburn MV, et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 38.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 39.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 40.Pappalardo L, et al. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 2003;278:39185–39188. doi: 10.1074/jbc.C300344200. [DOI] [PubMed] [Google Scholar]
- 41.Neiditch MB, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKellar JL, Minnell JJ, Gerth ML. A high-throughput screen for ligand binding reveals the specificities of three amino acid chemoreceptors from Pseudomonas syringae pv. actinidiae. Mol. Microbiol. 2015;96:694–707. doi: 10.1111/mmi.12964. [DOI] [PubMed] [Google Scholar]
- 45.Glekas GD, et al. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J. Biol. Chem. 2010;285:1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb BA, Hildreth S, Helm RF, Scharf BE. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl. Environ. Microbiol. 2014;80:3404–3415. doi: 10.1128/AEM.00115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Wood PL, Parales JV, Parales RE. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J. Bacteriol. 2009;191:2909–2916. doi: 10.1128/JB.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez M, Morel B, Corral-Lugo A, Krell T. Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol. Microbiol. 2016;99:34–42. doi: 10.1111/mmi.13215. [DOI] [PubMed] [Google Scholar]
- 49.Zhou YF, et al. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 51.Nishiyama, S. et al. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci. Rep. 6, 20866, 10.21038/srep20866 (2016). [DOI] [PMC free article] [PubMed]
- 52.Cheung J, Hendrickson WA. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 54.Dundas J, et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Josenhans C, Eaton KA, Thevenot T, Suerbaum S. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 2000;68:4598–4603. doi: 10.1128/IAI.68.8.4598-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnold IC, et al. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology. 2011;140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terry K, Go AC, Ottemann KM. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol. Microbiol. 2006;61:871–882. doi: 10.1111/j.1365-2958.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- 58.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 59.Lertsethtakarn, P., Draper, J. & Ottemann, K. M. Chemotactic Signal Transduction in Helicobacter pylori in Two-Component Systems in Bacteria (eds R. Gross & D. Beier) Ch. 17, 355–370 (Caister Academic Press, 2012).
- 60.Nishiyama S, et al. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immun. 2012;80:3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hugdahl MB, Beery JT, Doyle MP. Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 1988;56:1560–1566. doi: 10.1128/iai.56.6.1560-1566.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Repaske DR, Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J. Bacteriol. 1981;145:1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kihara M, Macnab RM. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J. Bacteriol. 1981;145:1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamblin PA, Maguire BA, Grishanin RN, Armitage JP. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol. Microbiol. 1997;26:1083–1096. doi: 10.1046/j.1365-2958.1997.6502022.x. [DOI] [PubMed] [Google Scholar]
- 65.Bi S, Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol. Life Sci. 2015;72:691–708. doi: 10.1007/s00018-014-1770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson, J. K. et al. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. MBio6, e00379; 10.01128/mBio.00379-00315 (2015). [DOI] [PMC free article] [PubMed]
- 67.Shaik MM, Cendron L, Salamina M, Ruzzene M, Zanotti G. Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores. Mol. Microbiol. 2014;91:724–735. doi: 10.1111/mmi.12487. [DOI] [PubMed] [Google Scholar]
- 68.Weinberg MV, Maier RJ. Peptide transport in Helicobacter pylori: roles of dpp and opp systems and evidence for additional peptide transporters. J. Bacteriol. 2007;189:3392–3402. doi: 10.1128/JB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 70.Piper DW, Fenton BH, Goodman LR. Lactic, pyruvic, citric, and uric acid and urea content of human gastric juice. Gastroenterology. 1967;53:42–48. [PubMed] [Google Scholar]
- 71.Armstrong CP, Dent DM, Berman P, Aitken RJ. The relationship between gastric carcinoma and gastric juice lactate (L + D) and lactate dehydrogenase. Am. J. Gastroenterol. 1984;79:675–678. [PubMed] [Google Scholar]
- 72.Takahashi T, et al. L-lactic acid secreted from gastric mucosal cells enhances growth of Helicobacter pylori. Helicobacter. 2007;12:532–540. doi: 10.1111/j.1523-5378.2007.00524.x. [DOI] [PubMed] [Google Scholar]
- 73.Iwatani, S. et al. Identification of the genes that contribute to lactate utilization in Helicobacter pylori. PLoS One9, e103506, 10.101371/journal.pone.0103506 (2014). [DOI] [PMC free article] [PubMed]
- 74.Garvie EI. Bacterial lactate dehydrogenases. Microbiol. Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Everse J, Kaplan NO. Lactate dehydrogenases: structure and function. Adv. Enzymol. Relat. Areas Mol. Biol. 1973;37:61–133. doi: 10.1002/9780470122822.ch2. [DOI] [PubMed] [Google Scholar]
- 76.Smith H, Yates EA, Cole JA, Parsons NJ. Lactate stimulation of gonococcal metabolism in media containing glucose: mechanism, impact on pathogenicity, and wider implications for other pathogens. Infect. Immun. 2001;69:6565–6572. doi: 10.1128/IAI.69.11.6565-6572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baldwin DN, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect. Immun. 2007;75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brewster JL, et al. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci. Rep. 2016;6:35198. doi: 10.1038/srep35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corral-Lugo, A. et al. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ. Microbiol. 10.1111/1462-2920.13170 (2015). [DOI] [PubMed]
- 80.Aiba Y, Suzuki N, Kabir AM, Takagi A, Koga Y. Lactic acid-mediated suppression of Helicobacter pylori by the oral administration of Lactobacillus salivarius as a probiotic in a gnotobiotic murine model. Am. J. Gastroenterol. 1998;93:2097–2101. doi: 10.1111/j.1572-0241.1998.00600.x. [DOI] [PubMed] [Google Scholar]
- 81.Bhatia SJ, Kochar N, Abraham P, Nair NG, Mehta AP. Lactobacillus acidophilus inhibits growth of Campylobacter pylori in vitro. J. Clin. Microbiol. 1989;27:2328–2330. doi: 10.1128/jcm.27.10.2328-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Canducci F, et al. Probiotics and Helicobacter pylori eradication. Dig. Liver Dis. 2002;34(Suppl 2):S81–83. doi: 10.1016/S1590-8658(02)80172-4. [DOI] [PubMed] [Google Scholar]
- 83.Gotteland M, Brunser O, Cruchet S. Systematic review: are probiotics useful in controlling gastric colonization by Helicobacter pylori? Aliment. Pharmacol. Ther. 2006;23:1077–1086. doi: 10.1111/j.1365-2036.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- 84.Lin WH, et al. Anti-Helicobacter pylori activity of fermented milk with lactic acid bacteria. J. Sci. Food Agric. 2011;91:1424–1431. doi: 10.1002/jsfa.4327. [DOI] [PubMed] [Google Scholar]
- 85.Midolo PD, Lambert JR, Hull R, Luo F, Grayson ML. In vitro inhibition of Helicobacter pylori NCTC 11637 by organic acids and lactic acid bacteria. J. Appl. Bacteriol. 1995;79:475–479. doi: 10.1111/j.1365-2672.1995.tb03164.x. [DOI] [PubMed] [Google Scholar]
- 86.Sgouras D, et al. In vitro and in vivo inhibition of Helicobacter pylori by Lactobacillus casei strain Shirota. Appl. Environ. Microbiol. 2004;70:518–526. doi: 10.1128/AEM.70.1.518-526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin WH, et al. Antagonistic activity of spent culture supernatants of lactic acid bacteria against Helicobacter pylori growth and infection in human gastric epithelial AGS cells. J. Food Sci. 2009;74:M225–230. doi: 10.1111/j.1750-3841.2009.01194.x. [DOI] [PubMed] [Google Scholar]
- 88.Briegel A, et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry. 2014;53:1575–1585. doi: 10.1021/bi5000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ud-Din AI, Roujeinikova A. Cloning, purification, crystallization and X-ray crystallographic analysis of the periplasmic sensing domain of Pseudomonas fluorescens chemotactic transducer of amino acids type A (CtaA) Biosci. Trends. 2016;10:320–324. doi: 10.5582/bst.2016.01059. [DOI] [PubMed] [Google Scholar]
- 90.Salah Ud-Din AIM, Roujeinikova A. The periplasmic sensing domain of Pseudomonas fluorescens chemotactic transducer of amino acids type B (CtaB): Cloning, refolding, purification, crystallization, and X-ray crystallographic analysis. Biosci. Trends. 2017;11:229–234. doi: 10.5582/bst.2016.01218. [DOI] [PubMed] [Google Scholar]
- 91.Salah Ud-Din AI, Roujeinikova A. The periplasmic sensing domain of Vibrio fischeri chemoreceptor protein A (VfcA): cloning, purification and crystallographic analysis. Acta Crystallogr. F Struct. Biol. Commun. 2016;72:382–385. doi: 10.1107/S2053230X16005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rico-Jimenez M, et al. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA) Mol. Microbiol. 2013;88:1230–1243. doi: 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 93.Liu YC, Roujeinikova A. Expression, refolding, purification and crystallization of the sensory domain of the TlpC chemoreceptor from Helicobacter pylori for structural studies. Protein Expr. Purif. 2015;107:29–34. doi: 10.1016/j.pep.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 94.McPhillips TM, et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/S0909049502015170. [DOI] [PubMed] [Google Scholar]
- 95.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ortega A, Amoros D, Garcia de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Savitzky A, Golay MJE. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964;36:1627–1639. doi: 10.1021/ac60214a047. [DOI] [Google Scholar]
- 102.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Edelstein, A. D. et al. Advanced methods of microscope control using μManager software. J. Biol. Methods1, e10, 10.14440/jbm.12014.14436 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


