Abstract
To investigate the genetic variation in Haemophilus parasuis (HPS) in Sichuan, China, 11 isolates were analyzed based on the outer membrane protein P2 (OMPP2) sequence. Sequence analysis showed that the 11 isolates shared 93.0 to 100% nucleotide homology with 15 reference strains, and the consistency between the 26 strains was 89.0%. The isolates of HPS-1, 2, 4, 5, 6, 7, 8, 10 and 11 had a 69-base deletion from 770 base pairs (bp) to 850 bp, which was infrequent in China. The phylogenetic tree showed that HPS-3 and HPS-8 had closer relationships with European and Japanese strains, but shared 98.7% nucleotide homology with the SW114 Japanese strain.
Keywords: haemophilus parasuis, outer membrane protein, sequence analysis
Haemophilus parasuis (HPS) is a commensal colonizer of the porcine upper respiratory tract, and it causes high morbidity and mortality in all age groups of pigs [1, 4]. HPS has at least 15 confirmed serotypes and others that are non-typeable. The main serotypes in China are 4, 5, 13, 12 and 14 [2]. The outer membrane is an important structure of gram-negative bacteria, and OMPP2, which was confirmed to be a virulence factor of HPS, is a member of the porin family [5, 7]. OMPP2 is diverse and easy to mutate and the OMPP2 genes could be divided into 2 genotypes, which were named as genetic type-I and type-II [3]. Recently, OMPP2 was found to induce proinflammatory cytokine mRNA expression in porcine alveolar macrophages, suggesting that OMPP2 may play an important role in the pathogenesis of disease caused by HPS [6]. In summary, the sequence analysis of HPS OMPP2 may provide new information regarding gene mutations that could be useful for vaccine research, pathogenesis, and heritable variation of HPS.
The 11 HPS isolates were provided by the Animal Quarantine Laboratory, Sichuan Agricultural University (Table 1). A pair of primers was designed based on the published GenBank sequence (FJ685756.1). The sense primer was 5ʹ-CGGGGTACCATGAAAAAAACACTAG-3ʹ and the antisense primer was 5ʹ-CGCGGATCCTTACCATAATACAC-3ʹ. PCR was performed in a 25-µl mixture containing 12.5 µl of 2× Master Mix, 2 µl of 10 µM sense and antisense primers, 2 µl template, and 6.5 µl ddH2O. The PCR was carried out as follows: 94°C for 5 min, followed by 30 cycles of denaturing at 94°C for 30 sec, annealing at 56°C for 30 sec, and extension at 72°C for 1 min, and a final elongation was performed at 72°C for 10 min. The OMPP2 recombinant plasmid was confirmed by DNA sequencing (Life Technology Inc., Shanghai, China). Sequences of the 11 isolates were analyzed, by comparing them with those of 15 reference strains using DNASTAR software and the Clustal W algorithm of MegAlign.
Table 1. HPS strain summary.
| Name | Location | Year | Isolation site | Serovar | GenBank |
|---|---|---|---|---|---|
| HPS-1 | Suining | 2013 | Lung | 4 | KU375454 |
| HPS-2 | Pujiang | 2013 | Lung | 4 | KU508597 |
| HPS-3 | Suining | 2013 | Lung | 5 | KU508598 |
| HPS-4 | Nanchong | 2014 | Lung | 10 | KU508599 |
| HPS-5 | Mianyang | 2014 | Lung | Unknown | KU508600 |
| HPS-6 | Guangan | 2014 | Lung | Unknown | KU508601 |
| HPS-7 | Meishan | 2014 | Lung | 4 | KU508602 |
| HPS-8 | Suining | 2014 | Lung | Unknown | KU508603 |
| HPS-9 | Pujiang | 2014 | Joint | 12 | KU508604 |
| HPS-10 | Xichang | 2015 | Joint | 13 | KU508605 |
| HPS-11 | Pixian | 2015 | Joint | Unknown | KU508606 |
The HPS OMPP2 was amplified and a 1,071-bp fragment was separated via agarose gel electrophoresis. The results of the analysis showed that the 26 sequences, including 11 isolates and serotypes 1–15 of the reference strains, shared 93.0 to 100.0%, 86.3 to 100.0% and 89.00% nucleotide sequence homology, amino acid homology, and nucleotide sequence consistency, respectively. HPS-2, 3, 4, 5 and 8 had the highest homology with Japanese strains SW140 and SW114, with nucleotide and amino acid sequence homology at 98.7 to 99.2% and 96.5 to 97.8%, respectively. HPS-1, 7, 9 and 10 had the highest homology with the 84-22113 U.S.A. strain, with greater than 99.0% nucleotide and amino acid sequence homology. Sequence analysis showed two main differences between isolates and reference strains. In the 430 to 490-bp region of HPS OMPP2, HPS-1, 6, 7, 9, 10 and 11 had a 31-bp deletion, but HPS-2, 4 and 5 were only missing 25 bases. Furthermore, in the 770 to 850-bp region of HPS OMPP2, the other 9 strains had a 69-bp deletion, with the exception of HPS-3 and 8, which had deletions similar to those in strains 174, 84-15995, 84-17975, 84-22113, F042, H425, NO.4, SW124 and SW140 OMPP2 (Table 2).
Table 2. OMPP2 nucleotide and amino acid sequence homology across strains.
| HPS-1 | HPS-2 | HPS-3 | HPS-4 | HPS-5 | HPS-6 | HPS-7 | HPS-8 | HPS-9 | HPS-10 | HPS-11 | No.4 | SH0165 | SW114 | SW124 | SW140 | 84-15995 | 84-17957 | 84-22113 | 131 | 174 | C5 | D74 | F042 | H425 | H465 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPS-1 | 96.2 | 93.8 | 96 | 96 | 97.6 | 99.9 | 93.8 | 99.6 | 99.7 | 97.9 | 98.1 | 99 | 93.6 | 97.3 | 96.8 | 98.1 | 97.5 | 97.9 | 94.1 | 99 | 94 | 93.3 | 97.2 | 99.7 | 95.9 | |
| HPS-2 | 91.1 | 93.5 | 99.3 | 99.3 | 97.8 | 96.3 | 93.5 | 96 | 96.1 | 97.9 | 95.9 | 96.2 | 93.2 | 97.7 | 99.2 | 97 | 97.6 | 97.9 | 93.4 | 96.4 | 93.3 | 93 | 97.3 | 96.5 | 94.1 | |
| HPS-3 | 89.7 | 88.3 | 93.3 | 94 | 94 | 93.9 | 99.8 | 93.7 | 93.8 | 93.9 | 95.2 | 93.1 | 98.7 | 93.4 | 94 | 94 | 93.8 | 93.9 | 97.6 | 94.3 | 98.2 | 97.8 | 93.5 | 94.1 | 97.2 | |
| HPS-4 | 90.8 | 98.6 | 88 | 99.1 | 97.6 | 96.1 | 93.3 | 95.8 | 95.9 | 97.7 | 95.7 | 96 | 93 | 97.5 | 99 | 96.9 | 97.4 | 97.7 | 93.2 | 96.2 | 93.1 | 92.8 | 97.1 | 96.3 | 93.9 | |
| HPS-5 | 90.3 | 98.1 | 89.4 | 97.8 | 98 | 96.1 | 94 | 95.8 | 95.9 | 97.7 | 95.9 | 96 | 93.7 | 97.5 | 99.2 | 96.9 | 97.4 | 97.7 | 93.7 | 96.2 | 93.6 | 93.4 | 97.1 | 96.3 | 94.4 | |
| HPS-6 | 94.4 | 94.4 | 89.4 | 94.2 | 94.7 | 97.7 | 94 | 97.4 | 97.5 | 99.7 | 97.6 | 97.4 | 93.8 | 98.7 | 97.9 | 98.6 | 99.4 | 99.7 | 94.1 | 97.4 | 94 | 93.5 | 99.1 | 97.9 | 95 | |
| HPS-7 | 99.7 | 91.4 | 89.9 | 91.1 | 90.5 | 94.7 | 93.9 | 99.7 | 99.8 | 98 | 98.1 | 99.1 | 93.7 | 97.4 | 96.9 | 98.1 | 97.6 | 98 | 94.2 | 99.1 | 94.1 | 93.4 | 97.3 | 99.8 | 96 | |
| HPS-8 | 89.7 | 88.3 | 99.5 | 88 | 89.4 | 89.4 | 89.9 | 93.7 | 93.8 | 93.9 | 95.2 | 93.1 | 98.7 | 93.4 | 94 | 94 | 93.8 | 93.9 | 97.6 | 94.3 | 98.2 | 97.8 | 93.5 | 94.1 | 97.2 | |
| HPS-9 | 99.4 | 91.1 | 89.7 | 90.8 | 90.3 | 94.4 | 99.7 | 89.7 | 99.5 | 97.7 | 97.9 | 98.8 | 93.4 | 97.1 | 96.7 | 97.9 | 97.3 | 97.7 | 93.9 | 98.8 | 93.9 | 93.1 | 97 | 99.5 | 95.7 | |
| HPS-10 | 99.7 | 91.4 | 89.9 | 91.1 | 90.5 | 94.7 | 100 | 89.9 | 99.7 | 97.8 | 98 | 98.9 | 93.5 | 97.2 | 96.8 | 98 | 97.4 | 97.8 | 94 | 98.9 | 93.9 | 93.2 | 97.1 | 99.6 | 95.8 | |
| HPS-11 | 95.3 | 94.7 | 89.1 | 94.4 | 93.9 | 99.2 | 95.5 | 89.1 | 95.3 | 95.5 | 97.7 | 97.7 | 93.7 | 99 | 97.8 | 98.9 | 99.6 | 100 | 94.2 | 97.7 | 94.1 | 93.4 | 99.4 | 98.1 | 95.1 | |
| No.4 | 96.7 | 91.1 | 91.6 | 90.8 | 90.8 | 95.3 | 96.9 | 91.6 | 96.7 | 96.9 | 95.5 | 97.6 | 94.9 | 97.8 | 96.4 | 97.9 | 97.3 | 97.7 | 95.4 | 98.3 | 95.3 | 94.8 | 97 | 98.3 | 95.6 | |
| SH0165 | 97.8 | 91.6 | 89.3 | 91.4 | 90.8 | 94.2 | 98.1 | 89.3 | 97.8 | 98.1 | 95 | 95.5 | 92.7 | 97.4 | 96.7 | 98.1 | 97.5 | 97.7 | 93.4 | 98.3 | 93.2 | 92.5 | 97.2 | 99.1 | 95.1 | |
| SW114 | 89.1 | 87.2 | 96.5 | 86.9 | 88.3 | 88.8 | 89.4 | 96.5 | 89.1 | 89.4 | 88.5 | 91.1 | 88.7 | 93.1 | 93.7 | 93.8 | 93.5 | 93.7 | 97.8 | 94 | 98.8 | 98.9 | 93.2 | 93.8 | 97.1 | |
| SW124 | 94.4 | 94.7 | 89.4 | 94.4 | 93.9 | 97.2 | 94.7 | 89.4 | 94.4 | 94.7 | 98.1 | 95.5 | 95 | 88.8 | 97.4 | 98.4 | 99 | 99 | 93.7 | 97.3 | 93.6 | 93 | 98.7 | 97.6 | 94.7 | |
| SW140 | 92.5 | 98.1 | 89.7 | 97.8 | 97.8 | 94.7 | 92.8 | 89.7 | 92.5 | 92.8 | 94.4 | 92.5 | 92.5 | 88.5 | 93.9 | 97.5 | 97.5 | 97.8 | 93.7 | 97 | 93.6 | 93.4 | 97.2 | 97.1 | 94.4 | |
| 84-15995 | 95.5 | 92.8 | 89.9 | 92.5 | 91.9 | 96.9 | 95.8 | 89.9 | 95.5 | 95.8 | 97.8 | 95.8 | 95.5 | 89.4 | 96.9 | 94.2 | 99.1 | 98.9 | 94.3 | 97.9 | 94.2 | 93.5 | 98.8 | 98.3 | 95.2 | |
| 84-17957 | 94.2 | 93.9 | 88.8 | 93.6 | 93.1 | 98.1 | 94.4 | 88.8 | 94.2 | 94.4 | 98.9 | 94.4 | 94.4 | 88.3 | 98.1 | 93.6 | 98.3 | 99.6 | 94 | 97.3 | 93.9 | 93.2 | 99.5 | 97.8 | 94.9 | |
| 84-22113 | 95.3 | 94.7 | 89.1 | 94.4 | 93.9 | 99.2 | 95.5 | 89.1 | 95.3 | 95.5 | 100 | 95.5 | 95 | 88.5 | 98.1 | 94.4 | 97.8 | 98.9 | 94.2 | 97.7 | 94.1 | 93.4 | 99.4 | 98.1 | 95.1 | |
| 131 | 90.2 | 88 | 94.9 | 87.7 | 88.5 | 89.4 | 90.5 | 94.9 | 90.2 | 90.5 | 89.6 | 92.2 | 89.8 | 95.5 | 89.9 | 88.8 | 90.5 | 89.4 | 89.6 | 94.4 | 98.2 | 97.9 | 93.9 | 94.4 | 96.6 | |
| 174 | 98.1 | 91.6 | 90.2 | 91.4 | 90.8 | 94.4 | 98.3 | 90.2 | 98.1 | 98.3 | 95.3 | 96.9 | 96.7 | 89.7 | 94.7 | 93 | 95.5 | 94.2 | 95.3 | 90.8 | 94.3 | 93.8 | 97 | 99.3 | 95.5 | |
| C5 | 90.2 | 88 | 95.7 | 87.7 | 88.5 | 89.4 | 90.5 | 95.7 | 90.2 | 90.5 | 89.6 | 92.2 | 89.8 | 97.7 | 89.9 | 88.8 | 90.5 | 89.4 | 89.6 | 95.9 | 90.8 | 98.6 | 93.9 | 94.3 | 97.3 | |
| D74 | 88.8 | 86.6 | 94.7 | 86.3 | 87.7 | 88.3 | 89.1 | 94.7 | 88.8 | 89.1 | 88 | 90.8 | 88.4 | 97.7 | 88.5 | 88 | 88.8 | 87.7 | 88 | 95.7 | 89.4 | 97.2 | 92.9 | 93.6 | 96.6 | |
| F042 | 94.2 | 93.9 | 88.8 | 93.6 | 93.1 | 98.1 | 94.4 | 88.8 | 94.2 | 94.4 | 98.9 | 94.4 | 94.4 | 88.3 | 98.1 | 93.6 | 98.3 | 99.4 | 98.9 | 89.4 | 94.2 | 89.4 | 87.7 | 97.5 | 94.8 | |
| H425 | 99.2 | 91.9 | 90.2 | 91.6 | 91.1 | 95.3 | 99.4 | 90.2 | 99.2 | 99.4 | 96.1 | 97.2 | 98.1 | 89.7 | 95 | 93.3 | 96.4 | 95 | 96.1 | 90.8 | 98.6 | 90.8 | 89.4 | 95 | 96.1 | |
| H465 | 93.8 | 88.7 | 94.6 | 88.4 | 89.2 | 91.2 | 94.1 | 94.6 | 93.8 | 94.1 | 91.5 | 93.2 | 93.6 | 94.9 | 91.5 | 89.5 | 92.1 | 90.9 | 91.5 | 93.6 | 93.2 | 94.4 | 93.9 | 90.9 | 94.3 |
Homologous comparison of nucleotide sequences on the right upper and homologous comparison of amino acid sequences on the bottom left.
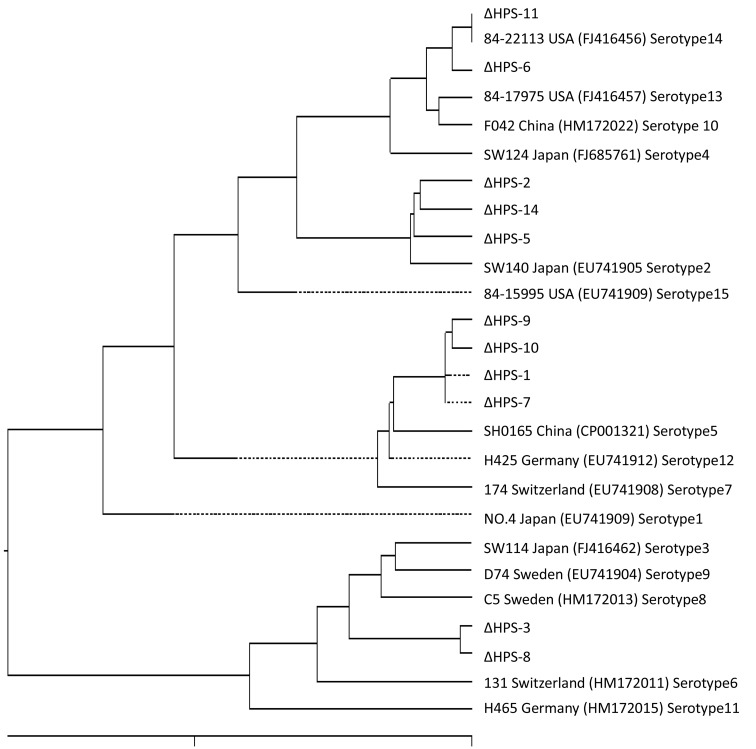
Phylogenetic tree (Fig. 1) analysis revealed clustering between HPS-6 and HPS-11 and they were closest to a cluster containing U.S.A. strain 84-22113, HPS-2, HPS-4, HPS-5, and Japanese strain SW140. HPS-1, 7, 9 and 10 were clustered with the classic Chinese strain SH0165. HPS-3 and HPS-8 were in a single cluster, in a branch with C5, Swedish strain D74, and Japanese strain SW114.
Fig. 1.
Phylogenetic tree analysis of HPS based on OMPP2 nucleotide sequences.
OMPP2 nucleotide sequence homology showed highest homology (98.7%) between the 2013 and 2014 Suining Sichuan province strains HPS-3 and HPS-8 with SW114. The nucleotide sequence consistency reached to 99.83% between these strains, with only two single base differences found in OMPP2. Therefore, we speculated that HPS-3 and HPS-8 were most likely introduced from Japan and the Suining Sichuan province may have persistent infection of these strains.
In general, 11 isolates showed significant differences in the OMPP2 sequence, especially in the 770 to 850-bp region, where 9 isolates had a 69-bp deletion. This deletion was important for the virulence of HPS. Therefore, since strong HPS virulence may be prevalent in the Sichuan area, pig farms should work toward the prevention of HPS infections and be aware of the potential crisis associated with HPS.
Acknowledgments
We gratefully acknowledge financial support for this work from the planning subjects of “the Twelfth Five-Year Plan” in the national science and technology for rural development in China (2013BAD12B04).
REFERENCES
- 1.Bello-Ortí B., Deslandes V., Tremblay Y. D., Labrie J., Howell K. J., Tucker A. W., Maskell D. J., Aragon V., Jacques M.2014. Biofilm formation by virulent and non-virulent strains of Haemophilus parasuis. Vet. Res. (Faisalabad) 45: 104. doi: 10.1186/s13567-014-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai X., Chen H., Blackall P. J., Yin Z., Wang L., Liu Z., Jin M.2005. Serological characterization of Haemophilus parasuis isolates from China. Vet. Microbiol. 111: 231–236. doi: 10.1016/j.vetmic.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 3.Kim J., Chung H. K., Jung T., Cho W. S., Choi C., Chae C.2002. Postweaning multisystemic wasting syndrome of pigs in Korea: prevalence, microscopic lesions and coexisting microorganisms. J. Vet. Med. Sci. 64: 57–62. doi: 10.1292/jvms.64.57 [DOI] [PubMed] [Google Scholar]
- 4.Oliveira S., Pijoan C.2004. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet. Microbiol. 99: 1–12. doi: 10.1016/j.vetmic.2003.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Olvera A., Pina S., Pérez-Simó M., Oliveira S., Bensaid A.2010. Virulence-associated trimeric autotransporters of Haemophilus parasuis are antigenic proteins expressed in vivo. Vet. Res. 41: 26. doi: 10.1051/vetres/2009074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou S., He X., Xu C., Zhang B., Feng S., Zou Y., Li J., Liao M.2014. The outer membrane protein P2 (OmpP2) of Haemophilus parasuis induces proinflammatory cytokine mRNA expression in porcine alveolar macrophages. Vet. J. 199: 461–464. doi: 10.1016/j.tvjl.2013.12.010 [DOI] [PubMed] [Google Scholar]
- 7.Zhou S. M., Xu C. G., Zhang B., Feng S. X., Zhang L. Y., Zou Y., Liao M.2013. Natural IgG antibodies in normal rabbit serum are involved in killing of the ompP2 mutant of Haemophilus parasuis SC096 strain via the classical complement pathway. Vet. J. 196: 111–113. doi: 10.1016/j.tvjl.2012.09.007 [DOI] [PubMed] [Google Scholar]